Abstract
Purpose
Ovarian cancer is the seventh most frequent form of malignant diseases in women worldwide and over 150,000 women die from it every year. More than 70 percent of all ovarian cancer patients are diagnosed at a late-stage disease with poor prognosis necessitating the development of sufficient screening biomarkers. MicroRNAs displayed promising potential as early diagnostics in various malignant diseases including ovarian cancer. The presented study aimed at identifying single microRNAs and microRNA combinations detecting ovarian cancer in vitro and in vivo.
Methods
Intracellular, extracellular and urinary microRNA expression levels of twelve microRNAs (let-7a, let-7d, miR-10a, miR-15a, miR-15b, miR-19b, miR-20a, miR-21, miR-100, miR-125b, miR-155, miR-222) were quantified performing quantitative real-time-PCR. Therefore, the three ovarian cancer cell lines SK-OV-3, OAW-42, EFO-27 as well as urine samples of ovarian cancer patients and healthy controls were analyzed.
Results
MiR-15a, miR-20a and miR-222 showed expression level alterations extracellularly, whereas miR-125b did intracellularly across the analyzed cell lines. MicroRNA expression alterations in single cell lines suggest subtype specificity in both compartments. Hypoxia and acidosis showed scarce effects on single miRNA expression levels only. Furthermore, we were able to demonstrate the feasibility to clearly detect the 12 miRNAs in urine samples. In urine, miR-15a was upregulated whereas let-7a was down-regulated in ovarian cancer patients.
Conclusion
Intracellular, extracellular and urinary microRNA expression alterations emphasize their great potential as biomarkers in liquid biopsies. Especially, miR-15a and let-7a qualify for possible circulating biomarkers in liquid biopsies of ovarian cancer patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00404-021-06287-1.
Keywords: microRNAs, Ovarian cancer, Liquid biopsies, Urine, Disease biomarker, Urinary microRNAs, Hypoxia, Acidosis
Background
Ovarian cancer (OC) is the seventh most frequent form of malignant diseases in women worldwide with an incidence of nearly 300,000 in 2018 [1]. Due to difficulties in detecting it at an early stage, over 150,000 women die from it every year, placing it as the most lethal malignant gynecologic disease in terms of mortality [2]. Over 70 percent of OC patients are diagnosed at an advanced, incurable stage (stages III and IV) with a five-year survival rate of 29% and only 15% are diagnosed when the disease is still localized [3, 4]. Established diagnostic tools such as TVS or CA-125 biomarker have had limited success in the early detection of OC (sensitivity in stage I/II OC < 60% and overall < 88.6%) [5–7].
Where the latter diagnostic methods could not provide satisfying results, miRNAs have shown promising potential as biomarkers in various malignant diseases such as breast cancer (BC) [8–10]. Urinary miRNAs, in particular, have shown great potential in BC [11], as well as in other cancer entities like urothelial and pancreatic cancer [12, 13]. MiRNAs are small, single-stranded and non-coding RNA molecules counting approximately 20–22 nucleotides and could be detected in body fluids of healthy and diseased patients [14–16]. Incorporated into the RNA-induced silencing complex (RISC), miRNAs play a key role in specifically regulating messenger RNAs (mRNAs) posttranscriptionally [15, 17, 18]. They are involved in apoptosis, carcinogenesis, metastasis, invasion, proliferation and chemoresistance in general and in OC specifically, play an important role as oncogenes and tumor suppressor genes and are up-/down-regulated during carcinogenesis since they are binding to fragile regions on several chromosomes [15, 19–22]. Furthermore, multiple analyses have proven exosomal trafficking of miRNAs which confirms their role in cell–cell communication and therefore as possible circulating biomarkers [23, 24]. MiRNAs in contrast to proteins as biomarkers display different advantages: expression level alterations occur simultaneously to the underlying biological mechanisms, stability in most endogenous and exogenous fluids and under extreme conditions like high temperatures, long-term storage and extreme pH values like in urine [14, 23, 25]. The existing body of literature on miRNAs in OC includes OC-specific miRNAS in vitro, in tissue and circulating miRNAs. An overview of miRNA studies on OC is provided in Table 1.
Table 1.
Overview of all included studies on miRNA expression levels in OC
| Reference | Histology | Analyzed matrix | Findings |
|---|---|---|---|
| Zhang et al. [48] |
EOC FIGO: all |
In vitro |
Upregulated: miR-26b, miR-182, miR-103, miR-26a Downregulated: miR-127, miR-134, miR-154*, miR-410, miR-377, miR-100, miR-432, miR-368, miR-154, miR-495, miR-376a, miR-323, miR-376b, miR-370, miR-299, let-7d, miR-155, miR-140, miR-222, miR-337, miR-124a, miR-99a, miR-331, miR-104, miR-150, miR-184, miR-152, miR-145, miR-424, miR-224, miR-302c |
| Dahiya et al. [58] |
All subtypes FIGO: all |
Tissue In vitro |
Upregulated: miR-221, miR-146b, miR-508 Downregulated: let-7f, miR-106b, miR-134, miR-155, miR-21, miR-346, miR-422a, miR-424, miR-519a, miR-648, miR-662 |
| Iorio et al. [38] |
EOC FIGO: all |
Tissue In vitro |
Upregulated: miR-200a, miR-200b, miR-200c, miR-141 Downregulated: miR-140, miR-199a, miR-199b, miR-145, miR-143, miR-125a, miR-125b, miR-101, miR-212, miR-222 |
| Yang et al. [41] |
EOC FIGO: all |
Tissue In vitro |
Upregulated: miR-199a, miR-424, miR-302d, miR-320, miR-214, miR-200a, miR-29a Downregulated: miR-493, miR-494, miR-125b, miR-100, let-7a, let-7b, let-7c |
| Nam et al. [39] |
SEOC FIGO: all |
Tissue |
Upregulated: miR-200b, miR-21, miR-200c, miR-141, miR-20a, miR-27a, miR-16, miR-93 Downregulated: miR-145, miR-125b, miR-100, miR-99a, miR-26a, miR-10b, miR-143, miR-214, let-7b, miR-29a, miR-125a |
| Wyman et al. [40] |
All subtypes FIGO: III + IV |
Tissue |
Upregulated: miR-182, miR-200c, miR-142-3p, miR-200b, miR-135b, miR-200a, miR-195, miR-126*, miR-26b, miR-10b, miR-126, miR-199b-5p, miR-107, miR-30b, miR-192, miR-335, miR-32, miR-20a, miR-30c, miR-143, miR-92a, miR-199b-3p, miR-99a, miR-26a, miR-18a, miR-16, miR-15a, miR-30e, miR-194, miR-29c, miR-30d, miR-106b, Downregulated: miR-127-3p, miR-377*, miR-382, miR-493, miR-409-3p, miR-193a-5p, miR-210, miR-935, miR-100, miR-31, miR-22, miR-152, miR-379, miR-185, miR-221, miR-744, miR-21*, let-7a*, miR-574-5p, miR-31*, miR-130b, miR-149, miR-423-5p, miR-1308, miR-629, miR-320a |
| Calura et al. [47] |
EOC FIGO: I |
Tissue (histotype specificity examined) |
Upregulated: miR-30a, miR-30a*, miR-192/194 cluster Downregulated: none |
| Taylor et al. [26] |
SEOC FIGO: all |
Serum |
Upregulated: miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, miR-214 Downregulated: none |
| Chung et al. [27] |
SEOC FIGO: all |
Serum |
Upregulated: none Downregulated: miR-132, miR-26a, let-7b, miR-145 |
| Surayawanshi et al. [28] |
SEOC + others FIGO: all |
Plasma |
Upregulated: miR-16, miR-21, miR-191, miR-16, miR-191, miR-4284 Downregulated: none |
| Resnick et al. [29] |
SEOC + others FIGO: all |
Serum |
Upregulated: miR-21, miR92, miR-93, miR-126, miR-29a Downregulated: miR-155, miR-127, miR-99b |
| Häusler et al. [30] |
SEOC + others Recurrent disease |
Whole blood |
Upregulated: miR-30c-1 Downregulated: miR-342-3p, miR-181a, miR-450-5p |
| Zheng et al. [55] |
All subtypes FIGO: all |
Plasma |
Upregulated: miR-205 Downregulated: let-7f |
| Meng et al. [53] |
SEOC + others FIGO: all |
Serum |
Upregulated: miR-7, miR-429 Downregulated: miR-25, miR-93 |
| Kapetanakis et al. [52] |
All subtypes FIGO: all |
Upregulated: miR-200b Downregulated: none |
|
| Kan et al. [51] |
SEOC FIGO: III + IV |
Serum |
Upregulated: miR-182, miR-200a, miR-200b, miR-200c Downregulated: none |
| Shapira et al. [54] |
SEOC FIGO: all |
Plasma |
Upregulated: miR-1274a, miR-625-3p, miR-720 Downregulated: miR-106b, miR-126, miR-150, miR-17, miR-20a, miR-92a |
| Zuberi et al. [32] |
SEOC FIGO: all |
Serum |
Upregulated: miR-200a, miR-200b, miR-200c Downregulated: none |
| Gao et al. [31] |
All subtypes FIGO: all |
Serum |
Upregulated: miR-141, miR-200c Downregulated: none |
| Liang et al. [34] |
All subtypes FIGO: all |
Serum |
Upregulated: none Downregulated: miR-145 |
| Hong et al. [33] |
SEOC + others FIGO: all |
Serum |
Upregulated: miR-221 Downregulated: none |
| Zhou et al. [35] |
SEOC FIGO: all |
Urine |
Upregulated: miR-30-5p Downregulated: 37 different miRNAs |
| Zavesky et al. [36] |
All subtypes FIGO: all |
Urine |
Upregulated: miR-92a, miR-200b Downregulated: miR-106b, miR-100 |
EOC epithelial ovarian cancer, SEOC serous epithelial ovarian cancer, FIGO Fédération Internationale de Gynécologie et d'Obstétrique
OC-specific circulating microRNAs
Upcoming approaches target the investigation of disease specific miRNA expression profiles in human body fluids such as blood and urine aiming at an early diagnosis. The comparison of tissue and serum miRNA expression levels revealed a definite relationship between tissue miRNAs and tumor-derived miRNAs in human body fluids [26]. Mir-132, miR-26a, let-7b and mir-145 showed promising potential as novel biomarkers in serous EOC, since they exhibited to be down-regulated in serum specimens [27]. Surayawanshi et al. also compared tissue and plasma miRNA expression profiles using global profiling. They concluded that different expression profiles in both media might account for another origin of plasma miRNAs than the ovarian malignancy [28].
Resnick et al. applied qRT-PCR to compare miRNA expression profiles of 21 miRNAs in 28 EOC patients and 19 HCs [29]. Five miRNAs were upregulated and three miRNAs were down-regulated in the serum of EOC patients (see Table 1) [29]. Another study on whole blood OC samples of 24 OC patients with recurrent disease revealed the deregulation of 147 miRNAs in OC patients compared to HCs, with miR-30c1 upregulated and miR-342-3p, miR-181a and miR-450b-5p down-regulated [30]. Further blood-based studies on miRNA expression profiles in OC comprise the studies by Zheng et al., Meng et. al., Kapetanakis et al., Kan et al. and Shapira et al.. For detailed information, see Table 1.
The miRNA-200-family underwent extensive research as diagnostic biomarker in OC. MiR-200a, miR-200b, miR-200c and miR-141 were found upregulated in the serum of OC patients in two independent studies and showed significant correlation with prognosis, tumor stage and histology [31, 32]. MiR-145 as well as miR-221 showed promising potential in the serum-based discrimination OC patients from HCs [33, 34].
Zhou et al. investigated exosomal miRNA expression in urine samples of 39 serous EOC patients pre- and postsurgically, 26 patients presenting with another gynecological disease and 30 HCs applying qRT-PCR [35]. First, they found miR-30a-5p upregulated as well as 37 miRNAs down-regulated comparing presurgical OC and HC samples. Second, stratifying for stage and metastatic status, they showed a distinct association between miR-30a-5p and early-stage OC as well as lymph node metastasis. Third, in urine samples of gastric and colon cancer patients miR-30a-5p showed to be down-regulated, supporting its OC specificity. And finally, in postsurgical OC samples of the same patients, the expression levels of miR-30a-5p were clearly lower than presurgically suggesting OC strongly as its origin [35]. Interestingly, miR-30a-5p was upregulated in the supernatant of OC cell lines, which displays a possible excretion mechanism. They also performed a knockout of miR-30a-5p resulting in a significant decrease of OC cell proliferation as well as migration [35].
Zavesky et al. analyzed cell-free urine of eleven OC as well as endometrial cancer patients [36]. They compared not only pre-and postsurgical specimen of the same patients, but also contrasted them to three HCs. Among the 18 included miRNAs, miR-92a, miR-200b, miR-106b and miR-100 exhibited a significant deregulation comparing OC and HC. Mir-92a and mir-200b were upregulated, while miR-100 and miR-106b were down-regulated [36].
Methods
Cohorts and sampling
After the positive ethical vote (Number 36/12 and 386/16 approved by the Institutional Ethical Review Board of the University of Freiburg) as well as positive informed consent each patient included into the study provided an indefinite volume of urine. In the presented case–control study, thirteen patients and 17 HCs are included for comparative analysis. The urine samples were collected at the Department of Gynecology and Obstetrics of the University Medical Center Freiburg (ten cancer samples, 17 HCs) between January 2015 and May 2016 and of the University Clinic of Bergen (Norway) (three cancer samples). Inclusion criterion for the disease group was newly diagnosed primary OC (FIGO I–IV), whereas for the control group, a detailed gynecological examination without evidence of any gynecologic malignancy prior to probes collection was necessary. Exclusion criteria were previous cancer diseases, simultaneous malignancies, chemotherapy or radio-chemotherapy prior to sample collection, autoimmune diseases, diabetes mellitus type 1 and infections. OC and HC specimen were collected simultaneously and age matched (see Table 2).
Table 2.
Ovarian cancer patients included into the study
| Sample no. | Age | Histologic subtype | Stage | TNM | Grading | Confounding diagnosis |
|---|---|---|---|---|---|---|
| 1 | 47 | Serous Adenocarcinoma | FIGO IIIc | pT3c, pN1, cM0 | G3 | None |
| 2 | 53 | Serous Adenocarcinoma | FIGO IIIc | pT2a, pN1, cM0 | G3 | Zoeliakie |
| 3 | 56 | Serous Adenocarcinoma | FIGO IIIc | pT3c, pN1, cM0 | G3 | None |
| 4 | 76 | Serous Adenocarcinoma | FIGO IV | pT3c, pN1, cM0 | G3 | Gastritis |
| 5 | 48 | Serous Adenocarcinoma | FIGO IIIc | pT3a, pN1, cM0 | G3 | Hypothyreosis |
| 6 | 79 | Serous Adenocarcinoma | FIGO IV | pT3c, pN1, cM0 | G3 | None |
| 7 | 65 | Serous Adenocarcinoma | FIGO IIIc | pT3c, pN1, cM0 | G2 | Esophagitis, bulbitis, gastritis |
| 8 | 24 | Sertoli-Leydig-Cell Tumor | FIGO Ia | pT1a, pN0, cM0 | – | None |
| 9 | 63 | Serous Adenocarcinoma | FIGO IIa2 | pT3a, pN1, cM1 | G3 | None |
| 10 | 71 | Serous Adenocarcinoma | FIGO IIIc | pT3c, pN1, cM0 | G3 | None |
| 11 | – | Serous Adenocarcinoma | – | – | – | – |
| 12 | – | Serous Adenocarcinoma | – | – | – | – |
| 13 | – | Serous Adenocarcinoma | – | – | – | – |
TNM tumorstatus identified by pathologist, G grading, FIGO tumorstatus according to the French organization Fédération Internationale de Gynécologie et d'Obstétrique (FIGO)
All urine samples were collected in 100 ml sterile lockable urine collection cups (Sarstedt, Germany). After the urine acquisition, all samples were stored at − 80 °C until further processing. Prior to the final analysis, extensive centrifugation at 4,000 rpm for five minutes was performed.
Cell culture
The established human tumor cell lines, SK-OV-3 and EFO-27 and OAW-42 were cultured in a humidified incubator (37 °C, 5% CO2, 95% air) and maintained according to the recommended cell line-specific culturing conditions. Cells were transferred into 25 cm2 cell culture flasks (Greiner Bio-One, Frickenhausen, Germany) until they reached 70% confluency. For hypoxia experiments, cells were placed in a hypoxic chamber (3% O2; mentioned as hypoxia). For acidosis experiments, culture media were supplemented with 2-Hydroxypropionic acid (Carl Roth, Karsruhe, Germany 0.2%, pH 6.2). Cells were cultured in parallel experiments under normal conditions (used as control). All treatments lasted for 24 h. Triplicates were generated. Afterwards, cells and cell culture media were processed separately. Cells underwent direct lysis according to the RNA isolation protocol, whereas cell culture media underwent extensive centrifugation (4000 rpm for ten minutes) before further processing.
RNA isolation, reverse transcription, poly-A-tailing and pre-amplification
Two RNA isolation protocols were used: analytic Jena´s innuPREP Micro RNA Kit (Analytic Jena, Jena, Germany) for the cells and Exiqon´s miRCURY RNA Isolation Kit—Biofluids (Exiqon, Qiagen GmbH, Hilden, Germany) for the liquid specimen. Before performing Exiqon´s miRCURY RNA Isolation Kit—Biofluids, extensive centrifugation steps eradicated all cellular material and separated DNA from RNA. The NanoDrop®-ND-1000-Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) served to determine the RNA concentration spectrometrically. The isolated RNA was finally stored at − 20 °C until further processing.
Subsequently, a reverse transcription (RT) protocol was performed (miScript Reverse Transcription Kit, Qiagen GmbH, Hilden, Germany) that generated cDNA of miRNAs only.
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (qPCR) served as detection method for determining miRNA expression levels in cell culture, cell culture media and urine samples. We processed qPCR on the Eppendorf 480 Mastercycler (Eppendorf, Hamburg, Germany). Duplicates of each sample were analyzed. For the qPCR, one µl cDNA and nine µl of an in-house qPCR mastermix (containing TRIS pH 8.1, dATP, dCTP, dGTP, dTTP, magnesium, potassium ammonium, SYBRGreen (Jena Bioscience, Jena, Germany), enhancers, HotStart Taq Polymerase (Jena Bioscience)) were used. A negative control (ten µl mastermix, no cDNA) and a RT (no RNA for reverse transcription, 1 µl unspecific cDNA, 9 µl mastermix) were added, to evaluate if specific or unspecific products were amplified. For primer sequences, see Table 3.
Table 3.
List of primers used for qPCR
| Universal antisense | 5′-GAA CAG TAT GTG TCA CAG ACG TAC-3′ |
| Let-7a | 5′-GCGG TGAGGTAGTAGGTTGTAT-3′ |
| Let-7d | 5′-GCGG AGAGGTAGTAGGTTGCATA-3′ |
| miR-10a | 5′-GCATG TAC CCT GTA GAT CCG A-3′ |
| miR-15a | 5′-GCGG TAGCAGCACATAATGGTT-3′ |
| miR-15b | 5′-CATG CAT AGC AGC ACA TCA TG -3′ |
| miR-19b | 5′-CATG TGT GCA AAT CCA TGC A -3′ |
| miR-20a | 5′-GCGG TAAAGTGCTTATAGTGCAG-3′ |
| miR-21 | 5′-GCATGCA TAG CTT ATC AGA CTG -3′ |
| miR-25 | 5′ -TCA TTG CAC TTG TCT CGG T -3´ |
| miR-100 | 5′-GCATT AAC CCG TAG ATC CGA-3′ |
| miR-103 | 5′-CGG AGCAGCATTGTACAGG-3′ |
| miR-125b | 5′-GCAT TCC CTG AGA CCC TAA C-3′ |
| miR-155 | 5′-GCATGCA TTA ATG CTA ATC GTG A -3′ |
| miR-191 | 5′-GCGG CGG AAT CCC AAA AGC AG-3′ |
| miR-222 | 5′-GCATG CTCAGTAGCCAGTGTAG-3′ |
The part of capital letters represents the miRNA-specific primer, the uncapitalized part represents the melting temperature overhang
Analysis and statistics
We applied a multivariable linear regression model to the log-transformed expression values with cell line (SK-OV-3, EFO-27, OAW-42) and its two-way and three-way interactions with treatment (control, hypoxia, acidosis) and compartment (intra-/extracellularly) as independent variables. All statistical methods involve ΔCt values normalized against the mean value of miR-25, -103 and -191. To calculate the influence of hypoxia and acidosis ΔCt values of treated and untreated probes were compared statistically which is represented in the relative expression (= 2−ΔCT).
For the interpretation of the multivariable analysis, all miRNA expression levels are compared to the intercept. The intercept represents cell line EFO-27 under control condition in the intracellular compartment. All statistical results with a p value of 0.05 or lower were interpreted as significant.
Results
Summarizing, all analyzed miRNAs except miR-155 could be detected intra- and extracellularly. MiR-155 was detectable extracellularly only, however, to an extremely low degree (ΔCt values between 22.34 and 29.20). Additionally, miR-21 did not show any statistically significant results neither on the intra- nor extracellular level under any analyzed condition in neither of the cell lines. The following paragraph pictures relevant findings only. All expression level regulations of miRNAs must be interpreted in comparison to the expression level of the same miRNA in the arbitrary reference (EFO-27, untreated, intracellularly). For a full list of results as well as the expression levels of each miRNA for the intercept and its confidence intervals, see supplemental data 1–3.
Intracellular expression level alterations
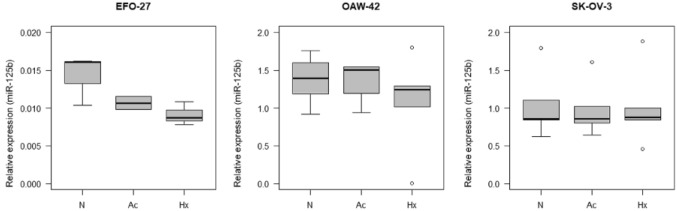
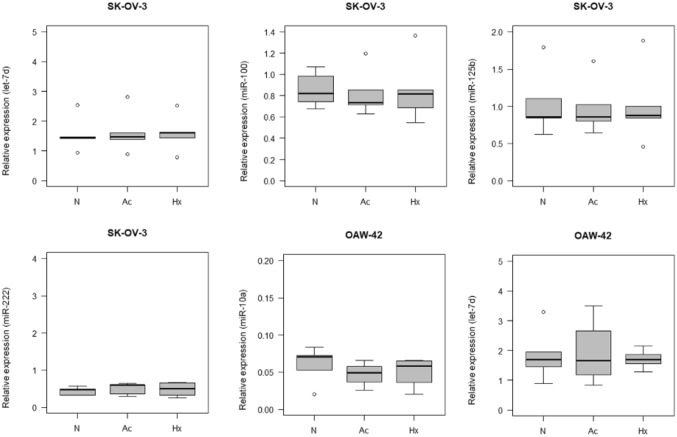
Focusing on intracellular expression levels only, miR-125b expression levels showed significant alterations in all analyzed cell lines. Expression levels were higher in SK-OV-3 (70.14; CI 10.71–459.31; p < 0.001) and OAW-42 (95.99; CI 14.66–628.61; p < 0.001) cells, compared to EFO-27 (see Fig. 1). Furthermore, in SK-OV-3, let-7d (0.35; CI 0.15–0.83; p = 0.02) and miR-222 (0.16; CI 0.05–0.53; p = 0.004) showed lower expression levels, miR-100 (10.29; CI 2.17–48.83; p = 0.005) showed higher expression levels compared to EFO-27. Additionally, miR-10a (0.06; CI 0.01–0.44; p = 0.008) and let-7d (0.40; CI 0.17–0.95; p = 0.041) showed lower intracellular expression levels in OAW-42 cells (see Fig. 2).
Fig. 1.
Intracellular relative expression of miR-125b in EFO-27, OAW-42 and SK-OV-3 cells. N control, Ac acidosis, Hx hypoxia
Fig. 2.
Intracellular relative expression of let-7d, miR-100, miR-125b and miR-222 in SK-OV-3 cells and of let-7d and miR-10a in OAW-42 cells. N control, Ac acidosis, Hx hypoxia
Extracellular expression level alterations
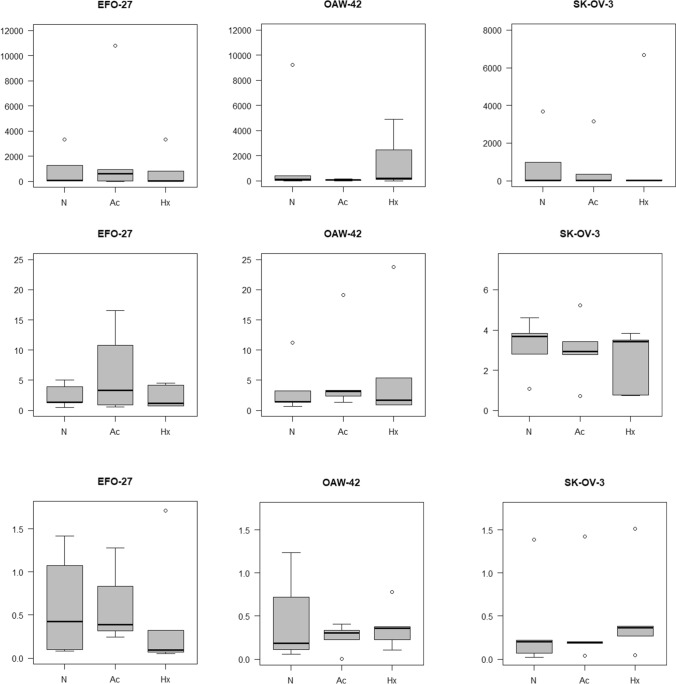
Extracellularly, compared to the intercept, miR-15a and miR-20a showed altered expression levels in the analyzed OC cell lines, whereas they were stable in the intracellular compartment. MiR-15a (SK-OV-3: 536.59; CI: 29.26–9838.90; p < 0.001; EFO-27: 428.64; CI 23.38–7859.65; p < 0.001; OAW-42: 438.58; CI 23.92–8041.82; p < 0.001) and miR-20a (SK-OV-3: 5.08; CI 1.84–14.00; p = 0.002; EFO-27: 3.73; CI 1.35–10.28; p = 0.013; OAW-42: 5.74; CI 2.08–15.83; p = 0.001) showed higher expression levels. In EFO-27 and OAW-42, miR-222 showed lower expression levels (EFO: 0.12; CI 0.03–0.42; p = 0.002; OAW-42: 0.14; CI 0.04–0.48; p = 0.003) (see Fig. 3 and supplemental data for box plots, expression levels and confidence intervals). Additionally, there were expression level alterations in the extracellular compartment of the EFO-27 cell line compared to the reference: let-7a (0.11; CI 0.03–0.40; p = 0.001), let-7d (0.18; CI 0.07–0.42; p < 0.001), miR-15b (0.48; 0.23–1.02; p = 0.060) and miR-125b (6.05; CI 0.92–39.61; p = 0.065) and expressed lower, whereas miR-15a, miR-20a (see above) and miR-19b (3.15; CI 0.95–10.44; p = 0.065) expressed higher. In OAW-42 cells supernatant, let-7a (0.26; CI 0.07–0.97; p = 0.048) expressed lower compared to the intercept.
Fig. 3.
Extracellular expression alterations: miR-15a, miR-20a and miR-222 in EFO-24, OAW-42 and SK-OV-3 cells. N control, Ac acidosis, Hx hypoxia
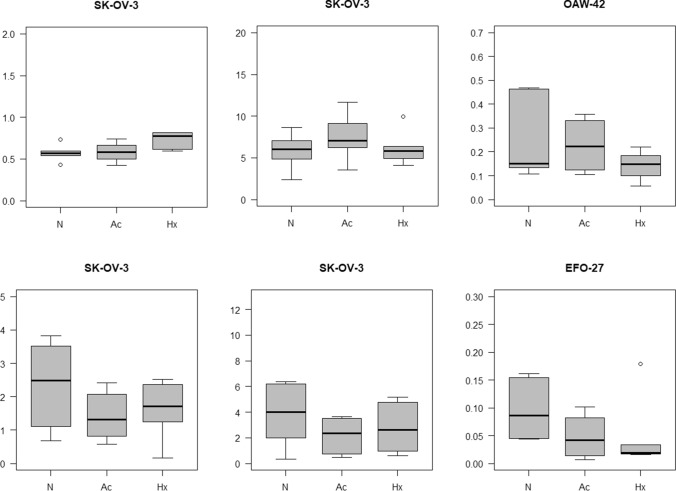
Expression level alterations under hypoxia and acidosis
Regarding hypoxia and acidosis, no significant miRNA alterations were visible after the application of the multivariable linear regression model. Still, the raw data and the boxplots revealed some promising miRs: in the intracellular compartment of SK-OV-3 cells, hypoxia led to an upregulation of miR-20a while acidosis led to an upregulation of miR-21. Inside OAW-42 cells, acidosis led to an upregulation of miR-19b (see Fig. 5). In EFO-27 cells, no alterations occurred. In the extracellular compartment of the SK-OV-3 cell line, hypoxia led to a downregulation of let-7d. Simultaneously, acidosis led to an even stronger downregulation of miRNAs let-7a and let-7d. In the OAW-42 cell line, neither hypoxia nor acidosis led to a significant alteration of the same miRNAs. In the extracellular compartment of the EFO-27 cell line, acidosis triggered a downregulation of miR-125b (see Fig. 4; for all changes caused by hypoxia and acidosis see supplemental data 1–3).
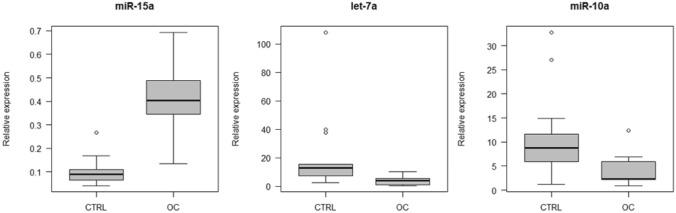
Fig. 5.
Deregulated miRNAs in the urine of OC patients compared to HC. Relative expression is shown. CTRL healthy controls, OC ovarian cancer patients
Fig. 4.
Regulated miRs under hypoxia and acidosis. N control, Ac acidosis, Hx hypoxia
Urinary results
Finally, the expression level of miR-15a was higher in the urine of OC patients than in the urine of HCs (p = 0.0319) whereas the expression level of let-7a was lower in OC patients (p = 0.0199). MiR-10a tended to be slightly down-regulated in the urine of OC patients (p = 0.0571) (see Fig. 5). MiRNAs let-7d, miR-15b, miR-19b, miR-20a, miR-21, miR-100, miR-125b and miR-222 did not show significant expression level alterations and miR-155 was not detectable.
Discussion
The major source of existing discrepancies across studies lies in differing methodological approaches, a lack of suitable housekeeper miRNAs, disease and tumor heterogeneity as well as in the nature of miRNAs themselves.
The methodological approach and single steps of miRNA analysis have a significant impact on the final results [23, 37]. For example, northern blotting, microarray-based detection, next-generation sequencing and real-time RT-PCR differ in sensitivity and specificity and their random use has led to inconsistent results.
Dahiya et al. describe this issue in their study and literature research [17]. MiRs-21, -155 and let-7d were down-regulated in tissue and cell lines, whereas miR-100 showed to be upregulated in that microarray-based approach. In addition, they found that only 16 of 192 analyzed miRNAs showed consistent expression patterns across studies [17]. The given study demonstrated a downregulation of let-7d in cell culture analyses intra- and extracellularly and found higher expression levels of miR-100 in the intracellular compartment of SK-OV-3 cells only.
Four more studies on OC tissue showed diverging results as well [38–41]. Iorio et al. applied microarray analyses and found different miRNAs to be deregulated [38]. MiR-125b1 was down-regulated. MiR-100 showed lower expression levels in OC tissues compared to HCs [38]. Nam et al. showed an upregulation of miR-20a and miR-21 in ovarian tumor tissue [39]. In the presented study, miR-125b showed a similar lower expression in EFO-27 intracellularly, extracellularly and tendentially under acidosis. In SK-OV-3 and OAW-42 cell lines, it showed higher expression levels intracellularly only. This is why it must be hypothesized that miR-125b is subtype specific. Moreover, miR-100 exhibited higher expression levels intracellularly in SK-OV-3 and miR-20a showed higher expression levels in the supernatant of all three analyzed cell lines in our study as well.
Methodologically, Wyman et al. used next-generation sequencing and subsequent qRT-PCR [40]. They found miR-15a and miR-20a to be upregulated like proven extracellularly in the given study. However, they also showed a downregulation of miR-21 and miR-100 [40].
Regarding the impact of hypoxia and acidosis on miRNA expression levels in OC, results are scarce. We found sporadic alterations of miRNA expression levels as well (see results). Giannakakis et al. demonstrated the involvement of miR-210 in the HIF pathway [42]. Compared to our study, hypoxia was methodologically induced using a lower amount of oxygen (1.5% vs. 3%) and a different panel of miRNAs was analyzed [42]. Another study on endometrial cancer cell lines also showed sporadic alterations only: miR-15a, miR-20a, miR-20b, miR-21 under hypoxia and let-7a, miR-22 and miR-125b under acidosis [43]. These results were inconsistent with comparable studies [44].
We hypothesize that OC patients can be distinguished from HC comparing their urinary miRNA expression levels. However, urine as well as cell culture supernatant are both challenging for cancer detection because of their biochemical conditions. The feasibility of urinary miRNA-based detection of cancer has already been proven in other tumor entities like BC [11]. Several studies verified the stability of miRNAs under harsh conditions like extreme pH values [23]. Compared to proteins, miRNAs undergo less degradation through ribonucleases due to their packaging into exosomes and the RISC and also because of their small size [14, 30]. However, the amount of total miRNA in urine and cell culture supernatant is small, which emphasizes that miRNA quantity and quality are crucial for the final detection [25, 45].
Targeting the correlation of tumor cell-derived and urinary miRNAs, one study on OC tissue and serum of the same patients conducted by Taylor et al. found matching expression patterns of eight miRNAs and, therefore, hypothesized that miRNAs in the serum derive directly from the tumor itself [26]. Nakamura et al. also suggest that miRNAs in biofluids reflect tissue miRNA expression levels accurately [23]. Microarray analyses of tissue, ascites and serum of EOC patients also showed uniform alterations [27]. However, in serum, additional miRNAs evolved to be regulated [27]. In our study, we detected differing miRNA signatures in cell culture, in cell culture supernatant and in urine.
Furthermore, OC detection based on urinary miRNAs is hindered by disease heterogeneity. OC is a highly individual disease that differs in histology, stage, metastatic status and molecular tumor characteristics [46]. The studies conducted by Iorio et al. and Calura et al. picture this as well [38, 47]. Both showed histotype-specific miRNA expression signatures. MiR-222 showed specific alteration in endometrioid and clear cell subtypes, whereas miR-21 did in endometrioid subtypes only [38, 47]. In our in vitro study, we were also able to detect subtype specific miRNA alterations. Regarding the patient samples collected in this study, it is important to emphasize the inclusion of one sample of Sertoli–Leydig cell tumor. This also reflects tumor and disease heterogeneity but the statistical significance achieved despite the inclusion indicates the robustness of the found miRNAs and suggests, that the detected miRNA alterations are ovarian cancer specific and not only subtype specific for serous adenocarcinomas.
Not only disease but also tumor heterogeneity complicate miRNA-based OC detection. Additionally, the tumor microenvironment consists of different cells and acellular components connected to different miRNA alterations. According to this, the same tumor might be able to exhibit a varying miRNA expression pattern at different states of its existence. This could explain the inconsistency of miRNA alterations across the published studies.
For example, miR-15a as well as let-7d showed significant downregulation in a cell culture study conducted by Zhang et al. [48]. Interestingly, the degree of downregulation rose with stage and was not detectable in 23.9% of the examined samples. They finally suggested these two miRNAs to be tumor suppressors [48]. MiR-15a was neither down- nor upregulated in the intracellular compartment of the given study but was significantly elevated in the extracellular compartment of all three analyzed cell lines. This suggests that the intracellular downregulation or absence of miR-15a is a possible result of an upregulated trafficking into the tumor microenvironment.
Moreover, the significant downregulation of let-7d in OC cell lines could be proven intracellularly in SK-OV-3 and OAW-42, extracellularly in EFO-27 and OAW-42 and under hypoxia and acidosis in SK-OV-3. Let-7d emerged its role as tumor suppressor by negatively regulating the RAS-pathway in lung cancer [49]. In conjunction with tumor heterogeneity, tumor immunology revealed a crucial role in the development and formation of cancer specific miRNA patterns [50].
Resnick et al. identified miR-21 and miR-155 as possible biomarkers [29]. While miR-21 was upregulated, miR-155 was down-regulated [29]. We observed this in our in vitro study as well, but not in the urine of OC patients. This suggests that urinary miRNAs express differently because of the activity of urine-specific enzymes and several cellular mechanisms. Some other blood-based studies emphasize this hypothesis showing aberrantly altered miRNAs compared to the given urine-based study [30, 51–55]. However, it must be considered that the analyzed OC histologic subtypes and samples as well as the applied methods and the investigated miRNA panels varied tremendously across these six studies.
Finally, patient heterogeneity modifies the results of the given study to an unknown extent, given that each individual presents with many exogeneous as well as endogenous confounders. In detail, cardiovascular, rheumatologic, dermatologic, neurologic, renal and many other diseases lead to specific miRNA expression alterations [25, 56, 57].
Zhou et al. performed qRT-PCR to determine exosomal miRNA expression levels [35]. Not only OC samples but also benign ovarian tumors and gastric as well as colon carcinomas were analyzed and prove OC specificity of miR-30a-5p upregulation and the downregulation of 37 more miRNAs in OC exclusively [35].
Zavesky et al. performed qRT-PCR on cell-free urine samples as well and found miR-100 to be down-regulated [36]. The strength of this work was prior assessment of RNA quality and quantity, while the inclusion of carcinomas of the fallopian tube into the study group of only 11 patients is questionable [36].
Conclusion
The landscape of miRNA studies in OC emphasizes their great potential for the detection of OC. With the given study we were able to demonstrate the feasibility of distinct miRNA-based discrimination of OC and HC in urine thanks to a specific miRNA signature. We were also able to widen the panel of miRs that potentially serve as diagnostic urinary biomarker in the detection of OC. However, our study also shows that urinary miRNA expression levels are massively dependent on methodological procedures. Comparing the few previous urine-based studies and the given study, there are several differences in the reported miRNA alterations in OC patients. This mirrors the great necessity of standardized miRNA extraction and detection protocols. Furthermore, to the best of our knowledge this study is the second one to demonstrate the feasibility to detect OC-specific miRNAs in cell culture supernatant and the first to prove the traceability of single miRNAs from the intracellular to the extracellular compartment and finally to urine. MiR-15a was upregulated in OC cell culture supernatant as well as in urine of OC patients which strengthens its OC-specific diagnostic potential observed in various studies before. As this study examines a small number of samples only, future studies are crucial to verify these observations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1 (Additional file 1.docx): Box plots. Shows box plots of relative expression levels of all analyzed miRNAs in the cell lines EFO-27, OAW-42 and SK-OV-3, in both compartments (intra (A)- and extracellular (B)) and under all analyzed treatments (untreated (N), hypoxia (Hx) and acidosis (Ac)) as well as box plots of all analyzed miRNAs in OC patients (OC) compared to healthy controls (CTRL) (DOCX 37098 KB)
Supplementary file 2 (Additional file 2.csv): Raw data. Shows ΔCt values of five cell culture sets (1–5) intra- and extracellularly of the three cell lines EFO-27, OAW-42 and SK-OV-3 under all analyzed treatments (untreated (N), hypoxia (Hx) and acidosis (Ac)) (CSV 11 KB)
Supplementary Additional file 3 (Additional file 3.txt): Results generated by R. Shows relative expression levels, confidence intervals and p values of each cell line after the statistical analysis generated by R (TXT 15 KB)
Acknowledgements
The authors are particularly grateful for continuous organizational support and technical assistance given by Mrs. Claudia Nöthling.
Abbreviations
- BC
Breast cancer
- cDNA
Complementary desoxyribonucleic acid
- EOC
Epithelial ovarian cancer
- FIGO
Fédération international de gynecology et obstetrique
- HC
Healthy controls
- HIF
Hypoxia-inducible factor
- miRNA, miR
MicroRNA
- mRNA
Messenger RNA
- nt
Nano tons
- nm
Nano meters
- OC
Ovarian cancer
- PCR
Polymerase chain reaction
- pre-miRNA
Premature-microRNA
- pri-miRNA
Primary-microRNA
- RISC
RNA-induced silencing complex
- RNA
Ribonucleic acid
- RT
Reverse transcription
- SEOC
Serous epithelial ovarian cancer
- tRNA
Transfer ribonucleic acid
- UTR
Untranslated ending
- WHO
World health organization
Author contributions
TE, MJ and MH devised the project outline and experimental study concept. MJ substantially worked out technical details of analytical assays. KB, MJ, DW and AR performed practical realization of sample handling, processing and data acquisition. KB, DW and AR were responsible for study-relevant patient data administration. KB, TE, MJ, MH and DW conducted a collective literature review. GR conceptually designed and overall guided statistical data assessment. GR performed statistical testing to extract decisive informative value. KB, JA, MH, TE and GR wrote the manuscript in consultation with and under critical revision of AR, DW, IJB and JA. Final data evaluation and determination of conclusive study output were performed by MH, KB, TE, GR and MJ.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. Additional information is available from the corresponding author on reasonable request.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no competing interest in regard to this publication.
Ethical approval and consent to participate
The According Investigation Protocols (36/12 and 386/16) were Approved by the Institutional Ethical Review Board of the University of Freiburg. All patients and healthy controls involved provided written informed consent.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cancer Today 2 (2019) IARC Estimated number of new cancer cases in 2018, worldwide, both sexes, all ages [Online]. Available from: https://gco.iarc.fr/today/online-analysis-table?v=2018&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&nb_items=5&group_cancer=1&include_nmsc=1&include_nmsc_other=1
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds)(2018) SEER cancer statistics review, 1975–2014, National Cancer Institute. Bethesda, MD, [Online]: (based on 2016 SEER data submission, posted to the SEER web site, 2017). Available at https://seer.cancer.gov/csr/1975_2014/
- 4.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA. 2011;305(22):2295–2303. doi: 10.1001/jama.2011.766. [DOI] [PubMed] [Google Scholar]
- 6.Moss EL, Hollingworth J, Reynolds TM. The role of CA125 in clinical practice. J Clin Pathol. 2005;58(3):308–312. doi: 10.1136/jcp.2004.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fung MFK, Bryson P, Johnston M, Chambers A. Screening postmenopausal women for ovarian cancer: a systematic review. J Obstet Gynaecol Can. 2004;26(8):717–728. doi: 10.1016/S1701-2163(16)30643-0. [DOI] [PubMed] [Google Scholar]
- 8.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251(3):499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 9.Velu VK, Ramesh R, Srinivasan AR. Circulating MicroRNAs as biomarkers in health and disease. J Clin Diagn Res. 2012;6(10):1791–1795. doi: 10.7860/JCDR/2012/4901.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wittmann J, Jäck H-M. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806(2):200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Erbes T, Hirschfeld M, Rücker G, Jaeger M, Boas J, Iborra S, et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. 2015;15:193. doi: 10.1186/s12885-015-1190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debernardi S, Massat NJ, Radon TP, Sangaralingam A, Banissi A, Ennis DP, et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am J Cancer Res. 2015;5(11):3455–3466. [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102(3):522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z-H, Xu C-J. Research progress of MicroRNA in early detection of ovarian cancer. Chin Med J. 2015;128(24):3363–3370. doi: 10.4103/0366-6999.171459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer. 2010;17(1):F77–89. doi: 10.1677/ERC-09-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 20.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vecchione A, Belletti B, Lovat F, Volinia S, Chiappetta G, Giglio S, et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci U S A. 2013;110(24):9845–9850. doi: 10.1073/pnas.1305472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406. doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016;15(1):48. doi: 10.1186/s12943-016-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beach A, Zhang H-G, Ratajczak MZ, Kakar SS. Exosomes: an overview of biogenesis, composition and role in ovarian cancer. J Ovarian Res. 2014;7:14. doi: 10.1186/1757-2215-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasparri ML, Casorelli A, Bardhi E, Besharat AR, Savone D, Ruscito I, et al. Beyond circulating microRNA biomarkers: Urinary microRNAs in ovarian and breast cancer. Tumour Biol. 2017;39(5):1010428317695525. doi: 10.1177/1010428317695525. [DOI] [PubMed] [Google Scholar]
- 26.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 27.Chung Y-W, Bae H-S, Song J-Y, Lee JK, Lee NW, Kim T, et al. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patients. Int J Gynecol Cancer. 2013;23(4):673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 28.Suryawanshi S, Vlad AM, Lin H-M, Mantia-Smaldone G, Laskey R, Lee M, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19(5):1213–1224. doi: 10.1158/1078-0432.CCR-12-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 30.Häusler SFM, Keller A, Chandran PA, Ziegler K, Zipp K, Heuer S, et al. Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. 2010;103(5):693–700. doi: 10.1038/sj.bjc.6605833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y-C, Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015;36(6):4843–4850. doi: 10.1007/s13277-015-3138-3. [DOI] [PubMed] [Google Scholar]
- 32.Zuberi M, Mir R, Das J, Ahmad I, Javid J, Yadav P, et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17(10):779–787. doi: 10.1007/s12094-015-1303-1. [DOI] [PubMed] [Google Scholar]
- 33.Hong F, Li Y, Xu Y, Zhu L. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J Int Med Res. 2013;41(1):64–71. doi: 10.1177/0300060513475759. [DOI] [PubMed] [Google Scholar]
- 34.Liang H, Jiang Z, Xie G, Lu Y. Serum microRNA-145 as a novel biomarker in human ovarian cancer. Tumour Biol. 2015;36(7):5305–5313. doi: 10.1007/s13277-015-3191-y. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33(6):2915–2923. doi: 10.3892/or.2015.3937. [DOI] [PubMed] [Google Scholar]
- 36.Záveský L, Jandáková E, Turyna R, Langmeierová L, Weinberger V, Záveská Drábková L, et al. Evaluation of cell-free urine microRNAs expression for the use in diagnosis of ovarian and endometrial cancers. A pilot study. Pathol Oncol Res. 2015;21(4):1027–1035. doi: 10.1007/s12253-015-9914-y. [DOI] [PubMed] [Google Scholar]
- 37.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 39.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 40.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4(4):e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Kong W, He L, Zhao J-J, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 42.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7(2):255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eismann J, Hirschfeld M, Erbes T, Rücker G, Jäger M, Ritter A, et al. Hypoxia- and acidosis-driven aberrations of secreted microRNAs in endometrial cancer in vitro. Oncol Rep. 2017;38(2):993–1004. doi: 10.3892/or.2017.5717. [DOI] [PubMed] [Google Scholar]
- 44.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014;18(3):371–390. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kossaï M, Leary A, Scoazec J-Y, Genestie C. Ovarian cancer: a heterogeneous disease. Pathobiology. 2018;85(1–2):41–49. doi: 10.1159/000479006. [DOI] [PubMed] [Google Scholar]
- 47.Calura E, Fruscio R, Paracchini L, Bignotti E, Ravaggi A, Martini P, et al. MiRNA landscape in stage I epithelial ovarian cancer defines the histotype specificities. Clin Cancer Res. 2013;19(15):4114–4123. doi: 10.1158/1078-0432.CCR-13-0360. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105(19):7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 50.Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. 2016;35:103. doi: 10.1186/s13046-016-0375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kan CWS, Hahn MA, Gard GB, Maidens J, Huh JY, Marsh DJ, et al. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. 2012;12:627. doi: 10.1186/1471-2407-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapetanakis N-I, Uzan C, Jimenez-Pailhes A-S, Gouy S, Bentivegna E, Morice P, et al. Plasma miR-200b in ovarian carcinoma patients: distinct pattern of pre/post-treatment variation compared to CA-125 and potential for prediction of progression-free survival. Oncotarget. 2015;6(34):36815–36824. doi: 10.18632/oncotarget.5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng X, Joosse SA, Müller V, Trillsch F, Milde-Langosch K, Mahner S, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113(9):1358–1366. doi: 10.1038/bjc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapira I, Oswald M, Lovecchio J, Khalili H, Menzin A, Whyte J, et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer. 2014;110(4):976–983. doi: 10.1038/bjc.2013.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng H, Zhang L, Zhao Y, Yang D, Song F, Wen Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS ONE. 2013;8(11):e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Navickas R, Gal D, Laucevičius A, Taparauskaitė A, Zdanytė M, Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang G, Tam L-S, Li EK-M, Kwan BC-H, Chow K-M, Luk CC-W, et al. Serum and urinary cell-free MiR-146a and MiR-155 in patients with systemic lupus erythematosus. J Rheumatol. 2010;37(12):2516–2522. doi: 10.3899/jrheum.100308. [DOI] [PubMed] [Google Scholar]
- 58.Dahiya N, Sherman-Baust CA, Wang T-L, Davidson B, Shih I-M, Zhang Y, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3(6):e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1 (Additional file 1.docx): Box plots. Shows box plots of relative expression levels of all analyzed miRNAs in the cell lines EFO-27, OAW-42 and SK-OV-3, in both compartments (intra (A)- and extracellular (B)) and under all analyzed treatments (untreated (N), hypoxia (Hx) and acidosis (Ac)) as well as box plots of all analyzed miRNAs in OC patients (OC) compared to healthy controls (CTRL) (DOCX 37098 KB)
Supplementary file 2 (Additional file 2.csv): Raw data. Shows ΔCt values of five cell culture sets (1–5) intra- and extracellularly of the three cell lines EFO-27, OAW-42 and SK-OV-3 under all analyzed treatments (untreated (N), hypoxia (Hx) and acidosis (Ac)) (CSV 11 KB)
Supplementary Additional file 3 (Additional file 3.txt): Results generated by R. Shows relative expression levels, confidence intervals and p values of each cell line after the statistical analysis generated by R (TXT 15 KB)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. Additional information is available from the corresponding author on reasonable request.
Not applicable.