Abstract
Background and aims
Genetic testing presents a unique opportunity for diagnosis and management of genetic kidney diseases (GKD). Here, we describe the clinical utility and valuable impact of a specialized GKD clinic, which uses a variety of genomic sequencing strategies.
Methods
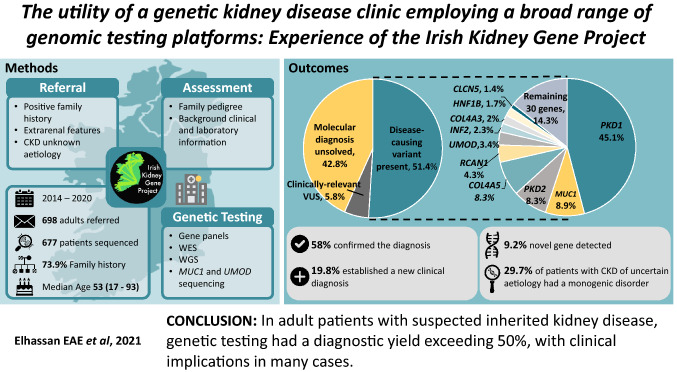
In this prospective cohort study, we undertook genetic testing in adults with suspected GKD according to prespecified criteria. Over 7 years, patients were referred from tertiary centres across Ireland to an academic medical centre as part of the Irish Kidney Gene Project.
Results
Among 677 patients, the mean age was of 37.2 ± 13 years, and 73.9% of the patients had family history of chronic kidney disease (CKD). We achieved a molecular diagnostic rate of 50.9%. Four genes accounted for more than 70% of identified pathogenic variants: PKD1 and PKD2 (n = 186, 53.4%), MUC1 (8.9%), and COL4A5 (8.3%). In 162 patients with a genetic diagnosis, excluding PKD1/PKD2, the a priori diagnosis was confirmed in 58% and in 13% the diagnosis was reclassified. A genetic diagnosis was established in 22 (29.7%) patients with CKD of uncertain aetiology. Based on genetic testing, a diagnostic kidney biopsy was unnecessary in 13 (8%) patients. Presence of family history of CKD and the underlying a priori diagnosis were independent predictors (P < 0.001) of a positive genetic diagnosis.
Conclusions
A dedicated GKD clinic is a valuable resource, and its implementation of various genomic strategies has resulted in a direct, demonstrable clinical and therapeutic benefits to affected patients.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s40620-021-01236-2.
Keywords: Chronic kidney disease, Inherited kidney diseases, Next-generation sequencing, Polycystic kidney genetics, Genetic kidney disease
Introduction
Testing for genetic kidney diseases (GKD), encompasses an array of more than 150 rare monogenic disorders, uncovers new horizons for diagnosis and management of patients and their families [1, 2]. Up to 35% of adults with chronic kidney disease (CKD) report a positive family history, suggesting a hereditary element [3]. While a strong genetic component of certain forms of GKD such as autosomal dominant polycystic kidney disease (ADPKD) is well recognised, other forms of adult GKD have historically been overlooked, and equally can be complex and multifaceted [4, 5].
The establishment of a specialised genetics service, utilising a multidisciplinary team (MDT) of clinical nephrologists, clinical geneticists, genetics counsellors, nurses, pathologists, and research geneticists/bioinformaticians is warranted to diagnose, manage, and treat patients with GKD [6]. Therefore, to determine the efficacy of genomic sequencing technologies, including next-generation sequencing (NGS), and the underlying genetic cause of disease in patients with suspected GKD, we established a research program known as the Irish Kidney Gene Project (IKGP) and an associated clinical service called the Genetic Kidney Disease Clinic (GKDC), at Beaumont Hospital, Dublin, Ireland [7, 8].
To diagnose GKD, several approaches can be adopted. Recent studies have focused on the utility of whole-exome sequencing (WES) in diagnosing GKD, with a diagnostic yield of 9–37% for monogenic disease depending on the patient population [9–12]. While WES is undoubtedly useful, it may not be practical or cost-efficient for all GKD clinics. Indeed, WES has been described as ineffective for diagnosis of ADPKD, the most common form of GKD [13]. NGS-based targeted gene panels may be considered as an alternative, with several studies reporting diagnostic rates ranging from 20 to 78% [14–16]. Other techniques may be required for specific genetic diseases. For example, the MUC1 gene contains a highly repetitive region with a high guanosine/cytosine content, resulting in the inability of WES and NGS panels to identify MUC1 variants, one of the most common causes of autosomal dominant tubulointerstitial kidney disease (ADTKD) [17]. Specialized testing is required for this condition [18]. Finally, the use of WES or indeed whole genome sequencing (WGS) allows future-proofing of diagnostic tests, allowing for reanalysis of data as novel genes are discovered. Through integration with research centres, WES and WGS can be utilised to assist in the identification of novel GKD genes. Thus, a multi-faceted approach provides the best opportunity to supply genetic diagnoses.
In this prospective study, we describe our overall experience of the IKGP over the 7-year period from 2014 to 2020 and the clinical impact of GKDC on patients, including some patient subsets that have been previously described [11, 19–24].
Methods
Patient data
Adult patients attending a university-based academic Department of Nephrology, the Irish National Kidney Transplant Centre, at Beaumont Hospital, Dublin were recruited into this prospective cohort study. Ethical approval was sought and granted by the Ethics Review Board of Beaumont Hospital (REC 19/28). All patients gave explicit informed consent to participate.
Letters were sent out to nephrologists nationwide to inform them of the service and invite them to refer any adult patient (age ≥ 18 years) with CKD who had either a positive family history, extrarenal features, or had CKD of ‘’uncertain aetiology’’ (uCKD). Depending on the clinical and histological findings of the nephrologists’ referrals, patients were grouped into seven categories of a priori clinical diagnoses. A detailed description of these categories and the diagnostic genomic methods used are listed in the Supplementary Material.
All patients referred to the GKDC, were reviewed and counselled by two among the following trained nephrologists—PC, CK, DC, SM, EE—with an interest in GKD and underwent research genetic testing guided by the a priori diagnosis.
Genetic diagnosis
The choice of genetic testing was guided by the patient’s a priori diagnosis, the likely success of sequencing strategies and cost considerations. Sequencing and bioinformatics analyses for gene panels [19, 23], WES [11, 22, 25] and WGS [24] were performed by DMC, PCH, FH, GLC, KAB as described previously(see Supplementary Material). MUC1 genotyping [18] was undertaken at the Broad Institute, while immunostaining for MUC1fs in urinary cell smears or kidney biopsy [20] followed by entire MUC1 sequencing using either Illumina [20] or PacBio Single Molecule, Real-Time (SMRT) Sequencing [26] were provided by Charles University in Prague by KK, AJB, MZ, SK. In each case these results of genomic testing were assessed by the MDT of clinical nephrologists with specific experience and training in GKD together with experts in clinical genetics and bioinformatics. Variants were prioritised and classified as per the American College of Medical Genetics (ACMG) guidelines [27].
Where a genetic diagnosis was made, patients were invited by the clinic to undergo a confirmatory genetic testing at an accredited clinical lab using a second sample, along with counselling on the implications of the results on their management and on other family members. This was required to ensure correct governance when including the test result in the clinical record and ensured complete accuracy of both the variant identification and interpretation. Only ACMG likely pathogenic/pathogenic results replicated at the clinical lab were returned to patients.
Statistics
Patient characteristics and genetic diagnosis were collected, and descriptive statistics were expressed as mean ± SD, percentages or median [interquartile range, IQR]. We evaluated clinical predictors favouring identification of a genetic diagnosis using logistic regression analyses. Data were analysed using STATA SE (version 16 StataCorp, College Station, TX, USA). Probability of a type 1 error less than 0.05 was statistically significant.
Results
Cohort description
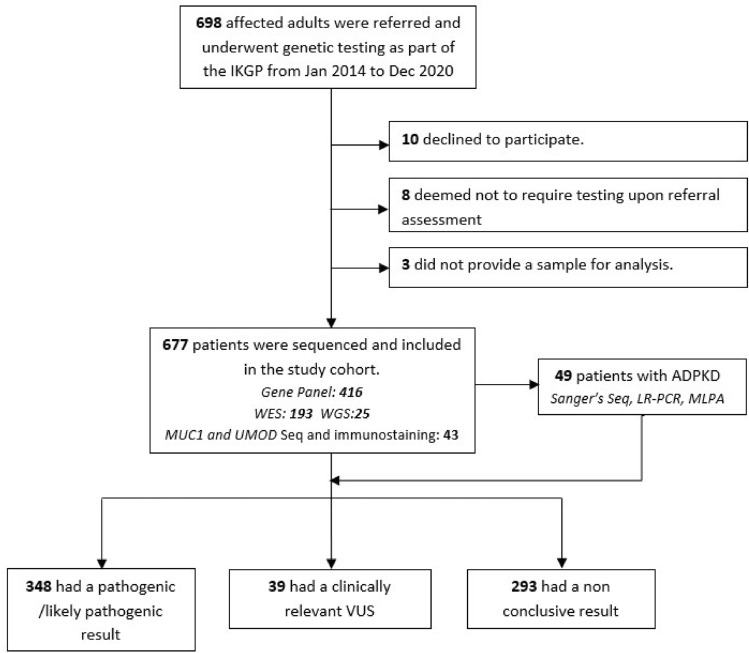
A total of 698 affected adult individuals (n = 522 families) were referred to the GKD service, none of whom had undergone prior genomic testing. Twenty-one patients were excluded from analysis as they declined participation, were deemed not to require testing upon referral assessment, or did not provide a sample for analysis (Fig. 1). The remaining 677 adults (n = 501 families) underwent genetic sequencing and formed the study cohort. Table 1 presents the baseline characteristics of the patients stratified by their genomic sequencing status.
Fig. 1.
Flowchart of genetic kidney disease clinic recruitment, sequencing technologies, and outcome. IKGP Irish Kidney Gene Project, MUC1 mucin 1 gene, MLPA multiplex ligation-dependent probe amplification, LR-PCR long-range polymerase chain reaction, UMOD Uromodulin, VUS variant of uncertain significance, WES whole–exome sequencing, WGS whole–genome sequencing
Table 1.
Characteristics of the 677 affected individuals (501 families) sequenced by the Irish Kidney Gene Project (IKGP)
| Characteristics | Total sequenced (n = 677) | Total variants identified (n = 387) | Unsolved (n = 290) | P value |
|---|---|---|---|---|
| A priori clinical diagnosis, n (%) | ||||
| PKD | 241 (35.6) | 205 (53) | 36 (12.4) | < 0.001 |
| CAKUT | 85 (12.6) | 12 (3.1) | 73 (25.2) | |
| Chronic GN | 112 (16.5) | 24 (6.2) | 88 (30.3) | |
| TIKD | 75 (11.1) | 49 (12.7) | 26 (9) | |
| AS/FSGS | 72 (10.6) | 57 (14.7) | 15 (5.2) | |
| Others | 18 (2.7) | 13 (3.3) | 5 (1.7) | |
| uCKD | 74 (10.9) | 27 (7) | 47 (16.2) | |
| Recruited from, n (%)1 | ||||
| Monogenic kidney disease study | 138 (20.4) | 56 (14.5) | 82 (28.3) | < 0.001 |
| PKD study | 208 (30.7) | 177 (45.7) | 31 (10.7) | |
| GKD clinic | 331 (7.8) | 154 (39.8) | 177 (61) | |
| Median age, yrs (range) | 53 (18–93) | 54 (18–88) | 51 (18–93) | 0.018 |
| Age in years at onset of disease, n (%) | ||||
| < 18 (childhood onset) | 118 (17.4) | 66 (17.1) | 52 (17.9) | 0.142 |
| ≥ 18 (adult onset) | 450 (66.5) | 274 (70.8) | 176 (60.7) | |
| Unavailable | 109 (16.1) | 47 (12.1) | 62 (21.4) | |
| ESKD, n (%) | ||||
| Yes | 440 (65) | 228 (58.9) | 212 (73.1) | 0.001 |
| No | 215 (31.8) | 142 (36.7) | 73 (25.2) | |
| Missing | 22 (3.2) | 17 (4.4) | 5 (1.7) | |
| Median age at onset of ESKD, [IQR] | 30 [20–43] | 30 [21–44] | 30 [18–43] | 0.425 |
| Sex | ||||
| Male | 358 (52.9) | 196 (50.6) | 162 (55.8) | 0.163 |
| Female | 319 (47.1) | 191 (49.4) | 128 (44.2) | |
| FHx of CKD, n (%) 2 | ||||
| Yes | 500 (73.9) | 336 (86.8) | 164 (56.6) | < 0.001 |
| No | 140 (20.7) | 43 (11.1) | 97 (33.4) | |
| Unavailable | 37 (5.4) | 8 (2.1) | 29 (10) | |
| Self-reported ethnicity | ||||
| Irish | 646 (95.4) | 373 (96.4) | 273 (94.2) | 0.551 |
| Other Europeans | 18 (2.7) | 8 (2.1) | 10 (3.4) | |
| Black | 8 (1.2) | 4 (1) | 4 (1.4) | |
| Asian | 5 (0.7) | 2 (0.5) | 3 (1) | |
AS Alport syndrome, CAKUT congenital anomalies of the kidney and urinary tract, CKD Chronic kidney disease, ESKD end stage kidney disease, GKD genetic kidney disease, IQR Interquartile range, FHx family history, FSGS focal segmental glomerulosclerosis, GN glomerulonephritis, PKD polycystic kidney disease, uCKD CKD of uncertain aetiology, TIKD tubulointerstitial kidney disease, Yrs years
1In total, 677 patients were reviewed in the genetic kidney disease (GKD) clinic, recruited and bio-banked for the evaluation and management of nephropathy. Included in this large cohort, two groups of patients were previously published in the Monogenic Kidney Disease Study [11] and PKD study [23]; these are grouped separately for clarity
2A positive family history of kidney disease that was reported by the patient (either a 1st-degree relative (parent, child, or sibling) or a 2nd -degree relative (grandparent, aunt, uncle, niece, nephew, or cousin)
The study population had a slight male preponderance, with 358 (52.9%) males. One-hundred and eighteen (17.4%) patients had disease onset at < 18 years of age with a mean age of 10.2 ± 5.7 years, and 450 (66.5%) patients presented as adults, with a mean age of 37.2 ± 13 years. Five hundred (73.9%) participants reported a family history of renal disease (P = < 0.001). Sixty-five% of patients reached end stage kidney disease (ESKD) at the last review, with a median age at ESKD of 30 (interquartile range (IQR) 20–43) years. Two hundred fifteen (31.8%) patients had CKD, defined as decreased estimated glomerular filtration rate lower than 60 ml/min/1.73 m2 for 3 months or longer, by 47 (IQR 37–59) years of age. A total of 664 (98%) were self-reported as Caucasian, and more than 95% of the cohort were Irish, which is representative of the Irish population [28]. None of the patients reported consanguinity.
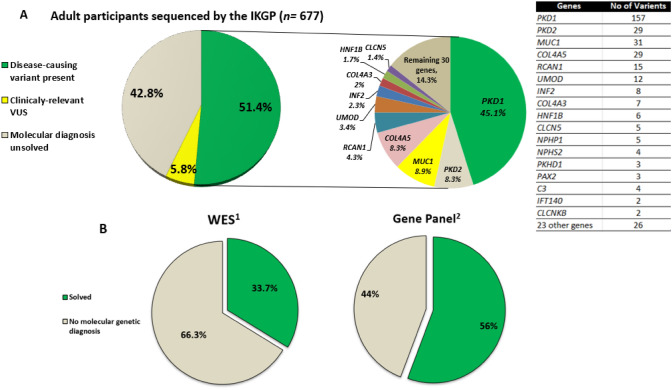
According to the criteria adopted, we achieved a genetic diagnosis in 46.5% of the 501 families, corresponding to 51.4% of the 677 patients (Table 2, Supplementary Table S1). Two-thirds of patients with a reported family history of kidney disease achieved a genetic diagnosis (67.2% (336/500)) versus 31% (43/140) in patients with no family history of kidney disease (P = < 0.001). Segregation analysis was required for 56 (9%) families. Amongst the 40 identified monogenic disorders, ACMG pathogenic or likely-pathogenic variants within four genes accounted for 70.7% of all identified causative variants; PKD1 (n = 157/348; 123 families), PKD2 (n = 29/348; 22 families), MUC1 (n = 31/348; 10 families), and COL4A5 (n = 29/348; 18 families). The remaining 29.3% of patients with a genetic diagnosis contained variants across a further 36 genes (Figs. 2 and 3).
Table 2.
Distribution of diagnostic yield per a priori clinical diagnosis and sequencing technology
| A priori Clinical diagnosis | Patients underwent gene Sequencing, n | Families per a priori clinical diagnosis, n | Clinical characteristics | Sequencing technology | Diagnostic yield | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median age at onset of ESKD, years (range) | CKD only in adulthood, n (%) | ESKD in adulthood, n (%) | ESKD in childhood, n (%) | Missing data of Renal Status | Gene-panel, n (%) | WES/WGS, n (%) | MUC1 and UMOD sequencing, n (%) | Disease-causing variant per patients, n (%) | Disease-causing variant per families, n (%) | |||
| PKD | 241 | 188 | 50 (6–83) | 85 (35.3) | 149 (61.8) | 3 (1.2) | 4 (1.7) | 235 (97.5) | 6 (2.5) | 0 (0) | 191 (79.2) | 148 (78.7) |
| CAKUT | 85 | 73 | 29 (3–66) | 20 (23.5) | 53 (62.4) | 10 (11.8) | 2 (2.3) | 31 (36.5) | 54 (63.5) | 0 (0) | 11 (12.8) | 7 (9.6) |
| TIKD | 75 | 39 | 34 (5–72) | 23 (30.7) | 33 (44) | 7 (9.3) | 12 (16) | 12 (16) | 23 (30.7) | 40 (53.3) | 49 (65.3) | 18 (46.2) |
| Chronic GN | 112 | 74 | 38.5 (9–68) | 35 (31.3) | 69 (61.6) | 5 (4.5) | 3 (2.6) | 52 (46.4) | 59 (52.7) | 1 (0.9) | 15 (13.4) | 6 (8.1) |
| FSGS | 39 | 20 | 40 (8–77) | 15 (38.4) | 18 (46.2) | 5 (12.8) | 1 (2.6) | 13 (38.1) | 26 (61.9) | 0 (0) | 24 (61.5) | 7 (35) |
| AS | 33 | 32 | 28 (15–77) | 13 (39.4) | 18 (54.5) | 2 (6.1) | 0 (0) | 25 (63.4) | 8 (36.6) | 0 (0) | 24 (72.7) | 18 (56.2) |
| Tubular | 4 | 4 | 38 | 3 (80) | 1 (20) | 0 (0) | 0 (0) | 2(50) | 2(50) | 0 (0) | 4 (100) | 4 (100) |
| uCKD | 74 | 61 | 40 (9–71) | 16 (21.6) | 54(73) | 4 (5.4) | 0 (0) | 34 (45.9) | 38 (51.4) | 2 (2.7) | 22 (29.7) | 19 (31.1) |
| Others | 14 | 10 | 32 (6–62) | 5 (35.8) | 8 (57.1) | 1 (7.1) | 0 (0) | 12 (85.7) | 2 (14.3) | 0 (0) | 8 (57.1) | 6 (50) |
| Total | 677 | 501 | 30 (20–43) | 215 (31.8) | 403 (59.5) | 37 (5.5) | 22 (3.2) | 416 (61.4) | 218 (32.2) | 43 (6.4) | 348 (51.4) | 233 (46.5) |
AS Alport syndrome, CAKUT congenital anomalies of the kidney and urinary tract, CKD Chronic kidney disease, ESKD end stage kidney disease, FSGS focal segmental glomerulosclerosis, MUC1 mucin 1 gene, GN glomerulonephritis, PKD polycystic kidney disease, uCKD CKD of uncertain aetiology, TIKD tubulointerstitial kidney disease, WES whole-exome sequencing, WGS whole-genome sequencing
Fig. 2.
a Breakdown of disease-causing genes containing a pathogenic variant detected by the Irish Kidney Gene Project in whole cohort. b Pathogenic detection rate in whole-exome sequencing (WES) and a targeted gene panel. IKGP Irish Kidney Gene Project, VUS variant of unknown significance. 1More than 475 known chronic kidney disease genes; see references [11, 25]. 2Roche SeqCap EZ Choice (227 genes panel) and Roche NimbleGen HeatSeq panel (11 genes panel); see reference [23]
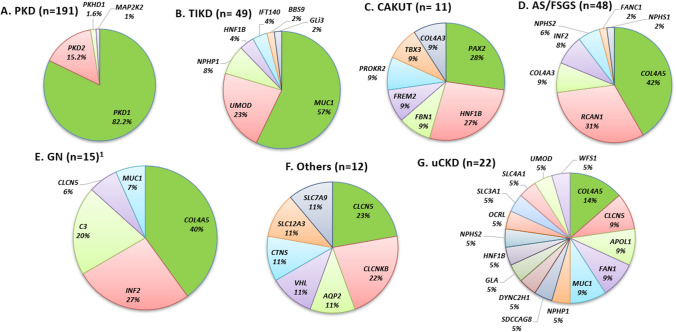
Fig. 3.
Disease-causing genes detected in the IKGP participants separated according to a priori diagnosis. CAKUT congenital anomalies of the kidney and urinary tract, AS/FSGS Alport syndrome/focal segmental glomerulosclerosis, GN glomerulonephritis, PKD polycystic kidney disease, uCKD chronic kidney disease of uncertain aetiology, TIKD tubulointerstitial kidney disease. 1In family F87, the proband had initial presentation of CKD stage 5, proteinuria, and family history of CKD, and referred with a priori diagnosis of GN. Proteinuria is thought to be a result of chronic changes rather than representing nature of the primary disease tubulointerstitial disease following the establishment of genetic diagnosis by MUC1 sequencing and immunostaining for MUC1fs
In addition, variants of uncertain significance (VUS) considered to be clinically interesting by the MDT were detected in 11.8% (39/329; 32 families) of patients without a disease-causing variant. Segregation analysis is underway in 29 of these VUS families in an effort to reclassify these variants. In the remaining 236 families (47.1%), we were not able to obtain a genetic diagnosis.
A priori diagnosis and identification of causative variants
Polycystic kidney disease (PKD): PKD was the most prevalent a priori diagnosis (241/677, 35.6%). The a priori clinical diagnoses are listed in Table 1, with a detailed description in Supplementary Material. We identified a disease-causing variant in 78.7% (148/188) of families recruited with a priori diagnosis of PKD (Table 2), and targeted gene-panels were used as the primary sequencing technology. In 191 PKD patients with ACMG likely pathogenic/pathogenic variants, 157 carried a disease-causing variant in PKD1, and 29 in PKD2 (Supplementary Table S1). Of the remaining five patients, three patients were found to have PKHD1 variants associated with autosomal recessive PKD (ARPKD), and two carried MAP2K2 variants associated with cardio-facio-cutaneous syndrome. Ten PKD families were identified to harbour clinically relevant VUS, whereas 30 families had no diagnostic results (16%).
Non-Cystic GKD: Among 313 families with non-cystic GKD, the diagnostic yield was 27.1%. The diagnostic yield varied within each diagnostic subgroup (Table 2, Supplementary Fig. S1).
Alport syndrome (AS)/focal segmental glomerulosclerosis (FSGS): Within the AS/FSGS cohort, we identified ACMG pathogenic/likely pathogenic variants within seven genes accounting for 48/72 individuals (66.7%). COL4A-related variants (n = 24) were the most frequent in this patient group and COL4A5 predominated (n = 20, 41.6%). In one large family with autosomal dominant FSGS, we discovered a heterozygous NM_004414:p.Ile162Thr RCAN1 variant, responsible for the patients’ FSGS [24]. Eight patients (3 families) were identified with disease-causing INF2 variants, with a clear positive family history of proteinuric renal disease.
Glomerulonephritis (GN) and IgA Nephropathy: We identified a genetic diagnosis in 6 out of 74 families referred with chronic GN (8.1%). We detected three variants segregating in families with IgA nephropathy as reported by Stapleton et al. [22]. We did not identify a genetic diagnosis in any families with MPGN/C3GN. In family F87, the proband presented with advanced CKD stage 5, proteinuria and family history of CKD. Following re-examination of the kidney biopsy specimen, immunostaining for MUC1fs in urinary cell smears was positive confirming the diagnosis of MUC1-ADTKD, which was not suspected on clinical grounds before this study, hence correcting the clinical diagnosis from GN to ADTKD. Proteinuria was thought to be related to chronic changes.
Tubulointerstitial kidney disease (TIKD): The well-established MUC1 cytosine duplication variant was detected in 6 families (25 patients) with ADTKD. In four ADTKD families (7 patients) where a MUC1 variant was not identified, we used non-invasive immunohistochemical urinary smear or kidney biopsies to confirm the presence of the frameshifted MUC1 protein (MUC1fs). Alternative genetic diagnoses were made in seven ADTKD families (18 patients); five with ADTKD-UMOD (12 patients), and two with ADTKD-HNF1B (6 patients). Amongst ADTKD families, TIKD predominated as an a priori diagnosis, but eight patients were initially referred with uCKD (n = 4), congenital anomalies of the kidney and urinary tract (CAKUT) (n = 3), or GN (n = 1). WES identified disease-causing variants in further eight patients with a priori diagnosis of TIKD and inconclusive biopsy findings.
uCKD: Amongst 61 families (74 patients) referred with a priori diagnosis of uCKD, 31.1% (19/61; 22 patients) were found to have a known monogenic disorder, bringing their diagnostic odyssey to an end, and emphasising the difficulties in making a clinical diagnosis in these very rare conditions without genetics support (Supplementary Table S1). In patients with a priori diagnosis of CAKUT, we detected 11 pathogenic/likely pathogenic variants in 11 individuals, corresponding to 7 of 73 families (9.6%). WES on three families with a child who had prune-belly syndrome did not reveal any underlying genetic cause.
Kidney donors: Three potential live kidney donors attended the clinic for screening due to a strong family history of CKD. In one case the potential donor was the sister of a patient with documented ADTKD-MUC1, while the other two patients had siblings with AS. In each case, we were able to confirm that the potential donors did not carry the disease-causing variants and were able to progress with living donation assessment.
Diagnostic yield per platform and their clinical utility
WES resulted in a diagnostic rate of approximately 34%, whereas gene panel sequencing resulted in 56% (Fig. 2B). In ADTKD families, MUC1 genotyping and targeted gene panel testing for UMOD, REN and HNF1B at the Broad resulted in a diagnostic rate of 70%. The further addition of urine smear analysis and tissue immunostaining for MUC1fs followed by sequencing of the entire MUC1 gene increased this to 86% in ADTKD patients.
Excluding ADPKD cases, genetic testing confirmed the a priori diagnosis in 58% of individuals. Patients’ diagnoses were refined or a new diagnosis was made in 13% and 20% of non-ADPKD patients, respectively. In 15 of the 162 patients (9%), a new gene was identified as had been reported by Lane et al. [24] (Table 3). A genetic diagnosis facilitated a change in treatment plan in 28/162 (17.3%) patients, while a diagnostic kidney biopsy was deemed unnecessary in 13/162 (8%) patients as a direct result of a genetic diagnosis. In non-ADPKD patients, genetic results prompted reverse phenotyping such as targeted work-up for other associated extra-renal conditions in 51/162 (31.5%) and a further 70/162 (43.2%) patients had appropriate familial cascade testing.
Table 3.
Summary of clinical outcomes of 162 patients (excluding cystic kidney disease) with a confirmed pathogenic diagnosis and the impact of genomic diagnosis on subsequent treatment
| Diagnostic utility, n (%) | |
| Confirmed the a priori diagnosis | 94 (58) |
| Refined the a priori diagnosis* | 21 (13) |
| Established a new diagnosis | 32 (19.8) |
| Novel candidate gene identified | 15 (9.2) |
| Clinical utility, n (%) | |
| Negate Biopsy based on genomic diagnosis | 13 (8) |
| Cascade tests/family counselling | 70 (43.2) |
| Change pharmacological treatment | 28 (17.3) |
| Additional assessment ordered to clarify extra-renal features—reverse phenotyping | 51 (31.5) |
*Genetic testing corrected/reclassified the a prior clinical diagnosis
Clinical lab validation of a genetic diagnosis was undertaken in 89/131 eligible patients, excluding those with ADPKD. Five did not return for clinical validation and 37 patients are awaiting return of results at the time of submission. At the beginning of the project, the median time from being evaluated in GKDC to return of validated results was 570.5 (IQR 317–1385) days, though the turnaround time decreased to 131 (IQR 100–205) days over the last 12 months of the study.
Factors favouring diagnostic outcome
In multivariate logistic regression analysis, patients with causative variants were more than three-fold more likely to report family history of CKD (Odds ratio (OR) 3.69; 95% confidence interval (CI) = 2.1–6.48; P = < 0.001). Patients with the underlying a priori diagnosis of PKD (OR: 14.9; 95% CI = 8.04–27.6; P = < 0.001), TIKD (OR 4.7; 95% CI 2.15–10.3; P = < 0.001), AS (OR 24.6; 95% CI 6.4–93.1; P = < 0.001), FSGS (OR 6.3; 95% CI 2.3–17.4; P = < 0.001), and uCKD (OR 7.6; 95% CI 1.7–32.4; P = < 0.001) had a significantly higher frequency of diagnostic outcome relative to patients with GN. No statistical difference was observed regarding patients’ age (P = 0.246), age at disease onset < 18 years (P = 0.376), or sex (P = 0.471) between cases with or without diagnostic variants (Supplementary Tables S2 and S3).
Discussion
Our study outlines the complexity of monogenic disorders, and the advantages of using genomic testing from diagnostic and clinical perspectives. We identified disease-causing variants in 46.5% (233/501) of GKD families (51.4% (348/677) of patients). The diagnostic yield in our cohort was consistent with several earlier studies [5, 14, 15], but higher than other studies [9, 10]. There are several factors that would explain our relatively high yield. First, we used a variety of genetic techniques to obtain a diagnosis. Earlier studies did not perform specialised genetic analysis for MUC1 variants, which contributed to a significant number (8.9%) of diagnoses in our population. We also used specific clinical criteria to limit our population to a group of patients with a high risk of familial kidney disease. Similar to other studies [3, 9–11, 15], the presence of family history of CKD and the underlying a priori diagnosis were the two most significant predictors of a genetic diagnosis. In addition, a large proportion of the studied patients had clinically suspected PKD, which is known to have a high rate of genetic diagnosis. A targeted gene panel [23], designed to achieve high coverage of PKD1 and PKD2 was utilised in patients with an a priori diagnosis of PKD, which had considerable diagnostic utility (78.7%) in this population. However, studies have reported higher diagnostic rates at 86–94% [29–31], yet our relatively low diagnostic rate can be justified by the broad a priori definition which we adopted, in whom one-fifth of our patients reported no family history.
Employing WES, the diagnostic yield of around 34% in our adult cohort was comparable with a recent WES study by Jayasinghe et al. [9], which reported a genetic diagnosis in 80/204 (39%) patients across a spectrum of renal phenotype subcategories. In contrast to our study which exclusively involved adult patients, around 40% of the patients reported by Jayasinghe et al. were < 18 years of age.
Up to 10% of total solved cohort had a COL4-related genetic diagnosis, a prominent, yet often unsuspected cause of GKD in adults, which correlated with high diagnostic yield in the AS cohort (56.2%). A diagnostic yield of 35% was obtained using genomic sequencing in the FSGS cohort. Several large studies of adult patients with primary FSGS achieved diagnostic yields ranging from 29 to 37% and 12–30% for familial [32] and sporadic [32, 33] cases, respectively.
In Ireland, ADTKD accounts for 0.5% of ESKD patients [21]. The genetic diagnosis of ADTKD was made in a high percentage in our cohort. The ability to identify MUC1 variants was important for the evaluation of GKD in our cohort, using both MUC1 genetic sequencing and newer techniques to detect the mutant MUC1fs protein.
The ultimate goal of genetic testing is the potential for personalised medicine. We validated and returned most of the research-based testing [34]. In adults, similar to previous studies [9–12], our data demonstrate that GKDC results provide a precise genetic diagnoses with diagnostic and therapeutic implications (Table 3). Genetic testing has numerous other advantages including prognostics and ruling in or out familial kidney donors [35].
A primary weakness of this study was the observational nature of the methodology. A mono-ethnic cohort could limit the generalizability of the results. Also, we cannot exclude potential selection bias by having PKD as the main a priori diagnosis and using specific clinical criteria that limit our cohort to a highly specified group. Lastly, no licensed genetic counsellors or clinical geneticists were involved in reviewing our patients.
Conclusions
In this large prospective cohort, the usage of various genomic testing strategies demonstrates their clinical application value, with a diagnostic yield over 50% supporting the advantageous clinical and therapeutic impact in adult patients with GKD. In our experience, an active renal genetic service requires a variety of genomic strategies and an integrated collaboration between clinical nephrologists and geneticists.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all patients who participated in this study and their physicians. We acknowledge that some of this sequencing was made possible from funding provided by the Punchestown Kidney Research Fund (EPSPD/2019/213) and the contribution of the Irish Kidney association/Health Research Board under the HRCI-HRB Joint funding scheme (Grant code: HRCI-HRB-2020-032).
Author contributions
EE, SLM, DMC, CK, CS, MAL, KK, AJB, MZ, SK, KAB, GLC, PJC conceived and/or designed the work that led to the submission, acquired data and/or played an important role in interpreting the results; drafted or revised the manuscript. EE, SLM, DMC, CK, SC, CC, CS, MAL, KK, AJB, MZ, SK, NKF, BD, AD, MDG, LC, PCH, FH, GLC, KAB, PJC approved the final version. Each author contributed important intellectual content during manuscript drafting or revision, accepts personal accountability for the author’s own contributions and agrees to ensure that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved. The results presented in this article have not been published previously in whole or part, except in abstract form.
Funding
Open Access funding provided by the IReL Consortium.
Declarations
Conflict of interest
E.E reports funds by the Royal College of Surgeons in Ireland Blackrock Clinic StAR MD. S.L.M. reports educational grant from Amgen. D.M.C. was supported by funding from the Health Research Board, Ireland (HPF-2016–1674), and received International Pediatric Research Foundation Early Investigators’ Exchange Program Award and Irish Nephrology Society Amgen Specialist Registrar Research Bursary. F.H. is the William E. Harmon Professor of Paediatrics. This research is supported by a grant from the National Institutes of Health to FH (5R01DK076683-13). MŽ and SK were supported by the Ministry of Health of the Czech Republic (Grant NU21-07-00033), the Ministry of Education of the Czech Republic (Grant LTAUSA19068) and by institutional programs of Charles University in Prague (UNCE/MED/007 and PROGRES-Q26/LF1). The National Center for Medical Genomics (LM2018132) kindly provided MUC1 sequencing. CK, SC, CC, CS, MAL, KK, AJB, MZ, SK, NKF, BD, AD, MDG, LC, PCH, FH, GLC, KAB, and PJC have nothing to disclose.
Ethical approval
Ethical approval was sought and granted by the Ethics Review Board of Beaumont Hospital (REC 19/28).
Informed consent
All patients gave explicit informed consent to participate.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Katherine A. Benson and Peter J. Conlon contributed equally to this work.
References
- 1.Devuyst O, et al. Rare inherited kidney diseases: challenges, opportunities, and perspectives. Lancet. 2014;383(9931):1844–1859. doi: 10.1016/S0140-6736(14)60659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soliman NA. Orphan kidney diseases. Nephron Clin Pract. 2012;120(4):c194–c199. doi: 10.1159/000339785. [DOI] [PubMed] [Google Scholar]
- 3.Connaughton DM, et al. The Irish Kidney Gene project-prevalence of family history in patients with kidney disease in Ireland. Nephron. 2015;130(4):293–301. doi: 10.1159/000436983. [DOI] [PubMed] [Google Scholar]
- 4.Cocchi E, Nestor JG, Gharavi AG. Clinical genetic screening in adult patients with kidney disease. Clin J Am Soc Nephrol. 2020;15(10):1497–1510. doi: 10.2215/CJN.15141219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas CP, et al. Initial experience from a renal genetics clinic demonstrates a distinct role in patient management. Genet Med. 2020;22(6):1025–1035. doi: 10.1038/s41436-020-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallett A, et al. A multidisciplinary renal genetics clinic improves patient diagnosis. Med J Aust. 2016;204(2):58–59. doi: 10.5694/mja15.01157. [DOI] [PubMed] [Google Scholar]
- 7.IKGP. The Irish Kidney Gene Porject. 2021; Available from: http://www.beaumont.ie/kidneycentre-aboutus-irishkidneygeneproject. Cited 10 Dec 2021
- 8.HSE. National Service Plan 2020. 2020; Available from: https://www.hse.ie/eng/services/publications/national-service-plan-2020.pdf. Cited 21 May 2021
- 9.Jayasinghe K, et al. Clinical impact of genomic testing in patients with suspected monogenic kidney disease. Genet Med. 2021;23(1):183–191. doi: 10.1038/s41436-020-00963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groopman EE, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380(2):142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connaughton DM, et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95(4):914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lata S, et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med. 2018;168(2):100–109. doi: 10.7326/M17-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali H, et al. PKD1 duplicated regions limit clinical utility of whole exome sequencing for genetic diagnosis of autosomal dominant polycystic kidney disease. Sci Rep. 2019;9(1):4141. doi: 10.1038/s41598-019-40761-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bullich G, et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 2018;94(2):363–371. doi: 10.1016/j.kint.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 15.Domingo-Gallego A et al. (2021) Clinical utility of genetic testing in early-onset kidney disease: seven genes are the main players. Nephrol Dial Transplant [DOI] [PubMed]
- 16.Mansilla MA, et al. Targeted broad-based genetic testing by next-generation sequencing informs diagnosis and facilitates management in patients with kidney diseases. Nephrol Dial Transplant. 2021;36(2):295–305. doi: 10.1093/ndt/gfz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckardt K-U, et al. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management—a KDIGO consensus report. Kidney Int. 2015;88(4):676–683. doi: 10.1038/ki.2015.28. [DOI] [PubMed] [Google Scholar]
- 18.Blumenstiel B, et al. Development and validation of a mass spectrometry-based assay for the molecular diagnosis of Mucin-1 kidney disease. J Mol Diagn. 2016;18(4):566–571. doi: 10.1016/j.jmoldx.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Murray SL, et al. Utility of genomic testing after renal biopsy. Am J Nephrol. 2020;51(1):43–53. doi: 10.1159/000504869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Živná M, et al. Noninvasive immunohistochemical diagnosis and novel MUC1 mutations causing autosomal dominant tubulointerstitial kidney disease. J Am Soc Nephrol. 2018;29(9):2418–2431. doi: 10.1681/ASN.2018020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cormican S, et al. Autosomal dominant tubulointerstitial kidney disease (ADTKD) in Ireland. Ren Fail. 2019;41(1):832–841. doi: 10.1080/0886022X.2019.1655452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapleton CP, et al. An exome sequencing study of 10 families with IgA nephropathy. Nephron. 2020;144(2):72–83. doi: 10.1159/000503564. [DOI] [PubMed] [Google Scholar]
- 23.Benson KA, et al. The genetic landscape of polycystic kidney disease in Ireland. Eur J Hum Genet. 2021;29(5):827–838. doi: 10.1038/s41431-020-00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane BM, et al. A rare autosomal dominant variant in regulator of calcineurin type 1 (RCAN1) gene confers enhanced calcineurin activity and may cause FSGS. J Am Soc Nephrol. 2021;32:1682–1695. doi: 10.1681/ASN.2020081234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CeGAT (2020)
- 26.Vylet'al P, et al. Plasma mucin-1 (CA15–3) levels in autosomal dominant tubulointerstitial kidney disease due to MUC1 mutations. Am J Nephrol. 2021;52:1–10. doi: 10.1159/000513952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Central Statistics Office (2021) Census 2016 results
- 29.Tan YC, et al. Novel method for genomic analysis of PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease. Hum Mutat. 2009;30(2):264–273. doi: 10.1002/humu.20842. [DOI] [PubMed] [Google Scholar]
- 30.Cornec-Le Gall E, et al. Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol. 2013;24(6):1006–1013. doi: 10.1681/ASN.2012070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossetti S, et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2007;18(7):2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 32.Wang M, et al. Contributions of rare gene variants to familial and sporadic FSGS. J Am Soc Nephrol. 2019;30(9):1625–1640. doi: 10.1681/ASN.2019020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gribouval O, et al. Identification of genetic causes for sporadic steroid-resistant nephrotic syndrome in adults. Kidney Int. 2018;94(5):1013–1022. doi: 10.1016/j.kint.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Jarvik GP, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94(6):818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stokman MF, et al. The expanding phenotypic spectra of kidney diseases: insights from genetic studies. Nat Rev Nephrol. 2016;12(8):472–483. doi: 10.1038/nrneph.2016.87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.