Abstract
Two abundant, low-redox-potential cytochromes c were purified from the facultative anaerobe Shewanella oneidensis strain MR1 grown anaerobically with fumarate. The small cytochrome was completely sequenced, and the genes coding for both proteins were cloned and sequenced. The small cytochrome c contains 91 residues and four heme binding sites. It is most similar to the cytochromes c from Shewanella frigidimarina (formerly Shewanella putrefaciens) NCIMB400 and the unclassified bacterial strain H1R (64 and 55% identity, respectively). The amount of the small tetraheme cytochrome is regulated by anaerobiosis, but not by fumarate. The larger of the two low-potential cytochromes contains tetraheme and flavin domains and is regulated by anaerobiosis and by fumarate and thus most nearly corresponds to the flavocytochrome c-fumarate reductase previously characterized from S. frigidimarina to which it is 59% identical. However, the genetic context of the cytochrome genes is not the same for the two Shewanella species, and they are not located in multicistronic operons. The small cytochrome c and the cytochrome domain of the flavocytochrome c are also homologous, showing 34% identity. Structural comparison shows that the Shewanella tetraheme cytochromes are not related to the Desulfovibrio cytochromes c3 but define a new folding motif for small multiheme cytochromes c.
Shewanella is a versatile genus of facultatively anaerobic, gram-negative bacteria that is capable of growth under a variety of conditions. The most remarkable characteristic of Shewanella species is their ability to reduce and dissolve insoluble metal oxides, including those of iron and manganese (31). Shewanella oneidensis strain MR1 (formerly called Shewanella putrefaciens strain MR-1 [42]) was specifically isolated as a metal oxide-reducing organism that could couple the reduction of metal oxides to the oxidation of organic carbon (28). While the mechanism of reduction of metal oxides by strain MR1 (or any other metal oxide reducer) remains unknown, the process has received much attention because of its potential importance. Some of the impacts of the process relate to the cycling of organic carbon, particularly in freshwater environments, to the potential use of dissimilatory metal oxide-reducing bacteria as agents of bioremediation and potential roles in processes, such as metal leaching and corrosion (19, 31). Given these potential impacts, it is not surprising that MR1 was chosen for genome sequencing, a project that is nearly completed (www.TIGR.org).
Although the details of the mechanism(s) of metal oxide reduction remain elusive, there is agreement that electron transport processes are involved, carrying reducing equivalents from the cell to the metal oxides at or near the cell surface. For this reason, many studies have focused on the characterization of the electron transport components of various Shewanella species. Cytochromes are abundant in S. oneidensis MR1 and have been implicated in the reduction of metal oxides (31). In fact, the preliminary genome sequence shows that it has more c-type cytochromes than any other species examined to date (approximately 38 at last count in contrast to the 7 found in Escherichia coli). We report here a continuation of our studies of the structure and function of cytochromes of strain MR1. Through this approach, we hope to begin to understand at many levels (global regulation, protein synthesis, and enzyme activity) the processes involved in anaerobic metabolism by this versatile organism.
Myers and Myers (24) showed that 80% of the membrane-bound cytochrome (actually heme) is localized to the surface of the outer membrane, presumably where insoluble iron and manganese oxides are reduced. Myers and Myers (25) also showed that there are at least four distinct outer membrane cytochromes with masses of 150, 83, 65, and 53 kDa, and they isolated and purified the 83-kDa cytochrome. The 83-kDa cytochrome is induced by anaerobiosis and is more abundant in cells grown on fumarate than in those grown on soluble iron citrate as the terminal electron acceptor. The cytochrome gene (omcA) was cloned by Myers and Myers (29) from S. oneidensis strain MR1 and found to encode a lipoprotein with an N-terminal diacylglyceride binding site (providing the membrane anchor) and to contain 10 hemes. Other species and strains of Shewanella contain high-molecular-weight membrane-bound cytochromes, and at least two strains have homologs of OmcA (29). A much larger, 14-kb DNA fragment of strain MR1 in the region of omcA was cloned by Beliaev and Saffarini (6; GenBank accession no. AF083240) and found to contain five 10-heme cytochrome genes including omcA. There are two 73- to 76-kDa lipoprotein homologs of OmcA, called MtrC and MtrF, and two 40-kDa, apparently periplasmic, 10-heme cytochromes related through a recent gene duplication, called MtrA and MtrD. It is unknown how the most recently discovered 10-heme cytochromes are related to the cytochromes observed on gels by Myers and Myers (25).
In previous studies of Shewanella, using low-temperature electron spin resonance spectroscopy, both a succinate dehydrogenase and two different types of fumarate reductase, soluble and membrane bound, were thought to be present (38). The succinate dehydrogenase operon from Shewanella frigidimarina (also known as S. putrefaciens) NCIMB400 has been cloned and shown to have the usual flavoprotein and iron-sulfur protein subunits as well as a cytochrome b membrane anchor (EMBL accession no. Y13760). The structural organization of membrane-bound fumarate reductase would presumably be similar to that of succinate dehydrogenase (for reviews, see references 10 and 41). However, studies with knockout mutants, in which the genes for the soluble fumarate reductase were inactivated, suggest that it is the only functional fumarate reductase present in Shewanella (9, 27).
Morris et al. (22) thoroughly characterized the soluble fumarate reductase from S. frigidimarina, and the gene was cloned by Pealing et al. (33), who showed that it results from the fusion of a small tetraheme cytochrome c gene to the 5′ end of a flavoprotein gene that is related to membrane-bound succinate dehydrogenase and fumarate reductase genes. The gene for a second flavocytochrome c (ifcA) from S. frigidimarina, which is homologous to the soluble fumarate reductase and which is induced by growth on soluble iron citrate, was cloned and characterized by Dobbin et al. (7). Although IfcA has fumarate reductase activity in vitro, it appears that it is nonfunctional in fumarate reduction in vivo because of the way it is regulated. ifcA mutants in which the gene was inactivated show no impairment in their ability to grow on iron citrate (or fumarate) as the electron acceptor, which suggests alternative pathways. Moreover, at least two other cytochromes are induced by growth on iron citrate, a soluble 35-kDa protein, further enhanced in the knockout mutants, and a 45-kDa membrane cytochrome. A soluble 52-kDa cytochrome is also apparent in the iron citrate-induced cultures.
Wolinella succinogenes, which was isolated as a fumarate-respiring organism, shows several interesting similarities to Shewanella. Wolinella has the usual three-subunit membrane-bound fumarate reductase (FrdABC) (16, 17). In addition, Wolinella has a soluble fumarate reductase like that of Shewanella, but in this case, separate genes for the flavoprotein (fccA) and cytochrome c (fccB) were found to be associated with another tetraheme cytochrome c gene (fccC) to form an operon (fccABC) (36). A Shewanella 21-kDa membrane-bound tetraheme cytochrome c gene (cymA) was cloned by Myers and Myers (26, 30) that is required for reduction of fumarate, nitrate, and iron oxide, but it is not the terminal enzyme. This gene is closely related to Wolinella fccC.
MATERIALS AND METHODS
Growth conditions, cytochrome quantification, and protein preparation.
S. oneidensis strain MR1 was grown at a temperature of 30°C usually for 24 to 48 h on lactate (20 mM) plus oxygen or anaerobically on lactate (20 mM) plus fumarate (15 mM) Luria-Bertani (LB) medium (37). Cytochromes were quantified once they were separated from the other cytochromes by column chromatography but before they were completely purified. The extinction coefficients at the alpha peak for pure protein were used to determine the amount of heme present; this was divided by the heme content per protein to determine the amount of protein present. The cytochromes c were purified as described previously (39). Briefly, an extract from fumarate-grown cells was adsorbed to DEAE-cellulose from 10 mM Tris-HCl, pH 8, and the column was developed with a stepwise salt gradient. Flavocytochrome c eluted at about 60 to 100 mM NaCl, and the small cytochrome c eluted at 200 to 300 mM NaCl. The cytochromes c were chromatographed on Sephadex G-75. The small cytochrome c was fractionated by 60 to 80% ammonium sulfate precipitation and chromatographed on DEAE-Sepharose using a linear gradient from 200 to 400 mM NaCl from which it eluted at about 270 mM NaCl. There was no noticeable 280-nm peak, and the 280-nm absorbance/408-nm absorbance ratio for pure protein was 0.066. The extinction coefficient was 23 mM−1 cm−1 heme−1 at 552 nm, assuming a value of 30 mM−1 cm−1 for the pyridine hemochromogen. Flavocytochrome c precipitated at 50 to 70% ammonium sulfate and eluted from DEAE-Sepharose at 160 mM NaCl. The 280-nm absorbance/408-nm absorbance ratio for pure protein was 0.24. The extinction coefficient was 33 mM−1 cm−1 heme−1 at 552 nm. Both proteins showed single bands on sodium dodecyl sulfate (SDS)-polyacrylamide gels using 15% T and 3% C gels run by the method of Laemmli (15).
Protein modification.
Heme was removed from the small cytochrome c by overnight treatment with HgCl2 in acidified urea by the method of Ambler and Wynn (3). Desalting was performed by gel filtration through a Sephadex G-25 column (0.32 by 10 cm; Pharmacia, Uppsala, Sweden), equilibrated, and eluted with 5% formic acid. Cysteines in the apoprotein were alkylated with 3-bromopropylamine by the method of Jue and Hale (14).
Enzymatic digestions.
Five nanomoles of the alkylated apoprotein of the small cytochrome c was digested with LysC endoprotease (Wako, Osaka, Japan) for 3.5 h at 37°C in 20 mM Tris-HCl buffer, pH 8.03, at an enzyme/substrate ratio (wt/wt) of 1/20. The same amount of modified apoprotein was used for digestion with GluC endoprotease (Boehringer, Mannheim, Germany) at an enzyme/substrate ratio of 1/40. The protein was incubated overnight at room temperature in 25 mM ammonium bicarbonate buffer, pH 7.85. Finally, an AspN (Boehringer) digestion was performed on 3.2 nmol of apoprotein. This digestion was incubated for 2 h at 37°C in 20 mM Tris-HCl buffer, pH 8.0, at an enzyme/substrate ratio (wt/wt) of 1/40.
Peptide purification.
Peptides from the enzymatic digestions were separated on a C2C18 3.2/3 column on a SMART chromatographic system (Pharmacia) with gradient elution in which solvent A was 0.1% trifluoroacetic acid–H2O and solvent B was 0.08% trifluoroacetic acid–70% acetonitrile–H2O.
Amino acid sequence analysis.
N-terminal and peptide sequence analyses were performed on a 477A or 476A pulsed liquid sequenator equipped with an on-line phenylthiohydantoin amino acid analyzer (all from Perkin-Elmer Biosystems, Foster City, Calif.). The C-terminal sequence analysis was performed on a Procise Sequencer (Perkin-Elmer Biosystems).
Mass analysis and NMR measurement.
Electrospray mass spectrometry was performed on a Bio-Q quadrupole mass spectrometer equipped with an electrospray ionization source (Micromass, Altrincham, United Kingdom). Ten microliters of sample solution in 50% acetonitrile–0.5% formic acid was injected manually in the 10-ml loop of the Rheodyne injector and pumped to the source at a flow rate of 5 ml/min. The solvent was delivered by a solvent delivery system (model 140A; Perkin-Elmer Biosystems). Scans of 12 s over the mass range of 400 to 1,600 atomic mass units were collected for 2 min. The instrument was calibrated with 50 pmol of horse myoglobin (Sigma). Matrix-assisted laser desorption mass spectrometry was performed on a TofSpec SE Time-of-Flight instrument using a nitrogen laser (337-nm wavelength) (Micromass, Wythenshawe, United Kingdom). Scans were accumulated over 20 to 70 laser shots, using alpha-cyanohydroxycinnamic acid as the matrix. External calibration was performed using both angiotensin II and bovine insulin (Sigma). Nuclear magnetic resonance (NMR) spectra were obtained with a Bruker DRX-400 NMR spectrometer. All NMR measurements were done at a temperature of 303 K and at a pD of 9.0.
Gene cloning.
The general strategy for obtaining the nucleotide sequence of the small cytochrome c gene was as follows. Two oligonucleotide primers were designed for PCR based upon the amino acid sequence as follows: primer SAS1, GCI GAY GGY GCI TTY GAR TT, and primer SAS2, TCR CAI GTI GGY TTY TGR CC. PCR with chromosomal DNA isolated from S. oneidensis strain MR1 as a template gave four bands, including a band of ca. 90 bp which was expected from the known amino acid sequence. This band was sequenced and found to code for the small tetraheme cytochrome c. The PCR product was then used to probe digests of chromosomal DNA. After hybridization and washing the filter for 30 min at 62°C in 0.2× SSC (NaCl and sodium citrate buffer [pH 7]) and 0.5% SDS (35), we obtained a number of single bands using 18 different endonucleases. We chose the 2.7-kb PstI fragment for subsequent DNA analysis. The PstI fragment was ligated with pUC18, which had also been digested with PstI. The ligation product was used for PCR as a set of templates using oligonucleotide SAS1 or SAS2 as one primer. For a second primer, we used standard forward or reverse primers from M13. These PCR products were then ligated into pGEM-T for sequencing.
To clone the flavocytochrome c gene, we applied a strategy similar to the one described above. For initial PCR, we used primers which were designed based upon the N-terminal sequence of the protein and that of a peptide as follows: primer SAS7, GCD CCW GAR GTI YTD GCD GAY TT, and primer SAS8, TGR CAI SWR TCR CAY TC. We isolated and purified a PCR product of ca. 500 bp and found it to have the correct sequence. We performed Southern analysis with this fragment as a probe and chose a 2.2-kb SphI fragment for subsequent DNA analysis.
RESULTS AND DISCUSSION
Induction of cytochrome synthesis.
When grown on LB medium with lactate, Shewanella produces a large quantity of cytochromes as well as a variety of cytochromes. There are three major soluble cytochromes: a small, high-potential monoheme cytochrome (HP cyt); a small, low-potential tetraheme cytochrome (ST cyt); and a large, low-potential tetraheme flavocytochrome (FL cyt) (21, 39). We completely purified the two low-potential cytochromes as shown by homogeneity by column chromatography, by SDS-polyacrylamide gel electrophoresis, and by a low absorbance at 280 nm. The absorption spectra of the two proteins are similar to one another (data not shown), but there were two prominent differences. There is no 280-nm peak, and the alpha peak of the small tetraheme cytochrome is broad and has a lower extinction coefficient than that of the large cytochrome. Cells grown to log phase with high levels of oxygen in batch culture contained the smallest quantity of soluble cytochrome c, and it had a high redox potential (1.3 μmol of HP cyt, 0.1 μmol of FL cyt, and 0.15 μmol of ST cyt [all values per 100 g of cells]). As the cells became limited for oxygen when grown to stationary phase with less vigorous aeration, cytochrome synthesis was dramatically increased. The quantity of HP cyt did not change very much with reduced oxygenation levels, but the synthesis of the two low-potential soluble cytochromes c, large and small, was increased by a factor of 10 (1.1 μmol of FL cyt and 1.4 μmol of ST cyt [both per 100 g of cells]). When the strain was grown anaerobically with fumarate, there was a further increase in the synthesis of the two low-potential cytochromes, with the large cytochrome half again as abundant as the smaller cytochrome (2.5 μmol of FL cyt and 1.7 μmol of ST cyt [both per 100 g of cells]). Thus, it appears that both low-potential cytochromes are induced by anaerobiosis, although the large cytochrome is specifically induced by growth on fumarate.
Amino acid sequence of the ST cyt c.
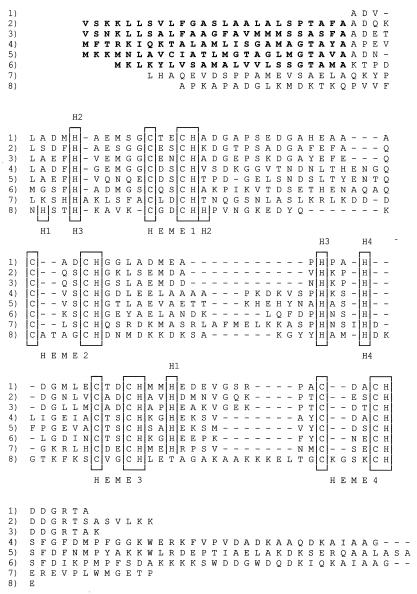
The complete amino acid sequence of the S. oneidensis strain MR1 ST cyt shows that the protein contains a total of 91 amino acid residues and four heme binding sites. The 12,210-Da mass deduced from the mass of the mature protein sequence plus the mass of the four hemes is in excellent agreement with that measured by mass spectroscopy, 12,210.5 Da (the 12,120-Da mass reported previously [39] was a typographic error). The S. oneidensis ST cyt is 64% identical to that from S. frigidimarina (EMBL accession no. AJ000006), and they are 55% identical to that from bacterial strain H1R (2). The ST cyt's are 34% identical to the N-terminal tetraheme domain of S. frigidimarina fumarate reductase (33). The amino acid sequences of these cytochromes are compared in Fig. 1.
FIG. 1.
Comparison of the sequences of ST cyt's and the tetraheme domains or subunits of FL cyt's from different species. Bacterial strain H1R ST cyt (2) (rows 1), S. oneidensis MR1 ST cyt (rows 2), S. frigidimarina NCIMB400 ST cyt (EMBL accession no. AJ000006) (rows 3), S. oneidensis MR1 FL cyt (rows 4), S. frigidimarina NCIMB400 FL cyt (33) (rows 5), S. frigidimarina NCIMB400 IfcA cytochrome (7) (rows 6), W. succinogenes (36) (rows 7), and D. vulgaris Miyazaki cytochrome c3 (12) (rows 8) sequences are shown. The heme binding sites and sixth heme ligand histidines are boxed. The sixth ligands to Shewanella hemes 1 to 4 are labeled H1 to H4 above the sequence and those for the Desulfovibrio hemes are labeled H1 to H4 below the sequence. To conserve space, no attempt to precisely align the N and C termini was made. Gaps introduced to optimize alignment are indicated by the dashes.
Sequence of the ST cyt gene.
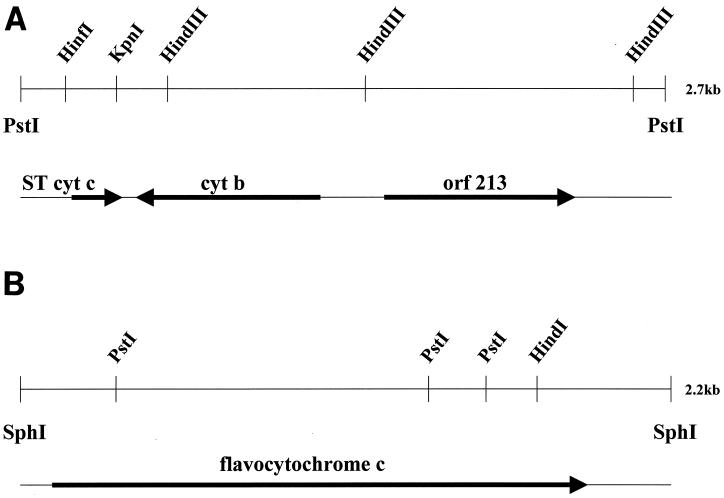
The S. oneidensis MR1 ST cyt gene was cloned by PCR using primers based on the amino acid sequence. A 2.7-kb PstI fragment of chromosomal DNA was isolated as shown in the physical map of Fig. 2 and sequenced. The start codon appears to be GTG (rather than the more usual ATG), and there is a possible ribosome binding site 8 bases upstream as well as a signal peptide of 25 amino acid residues which is cleaved in the mature protein. The translated gene sequence of the ST cyt is in complete agreement with the amino acid sequence obtained by Edman degradation. There is a palindromic region (bases 37 to 60) upstream and downstream of the ST cyt gene (bases 727 to 762).
FIG. 2.
Physical map of the cloned S. oneidensis MR1 ST cyt gene (A) and FL cyt gene (B).
Cytochrome b.
There is a 744-bp open reading frame (ORF) 85 bases downstream of the end of the gene encoding the ST cyt, but the orientation of this ORF is opposite that of the ST cyt gene. The 248-residue protein encoded by this ORF is homologous to a number of membrane-spanning cytochromes b, such as those that are part of hydrogenase operons (43). The Shewanella cytochrome b gene is not preceded by hydrogenase genes, and it appears to be independently transcribed. Based upon sequence homology, it is most similar to the cytochrome b from Chromatium vinosum (27% identity) that is adjacent to a cytochrome c′ gene (8). However, we have no evidence that would link the ST cyt c to cytochrome b in an electron transfer chain. The ST cyt gene has also been cloned from S. frigidimarina NCIMB400 (EMBL accession no. AJ000006). However, the genetic context of the NCIMB400 gene is different from that of the MR1 gene in that it is followed, 288 bases downstream, by the gene for an assimilatory nitrate reductase. This lack of conservation of the genetic context suggests that MR1 ST cyt is not functionally associated with cytochrome b or with nitrate reductase.
Transcriptional regulator homolog.
Another 581 bases downstream of the start of the gene for cytochrome b, there is a gene (ORF213) that apparently encodes a transcriptional regulator in the same orientation as the ST cyt gene. The C-terminal 65 residues of ORF213 are clearly homologous to a large family of DNA binding proteins exemplified by E. coli NarL for which there is a crystal structure (4). NarL is responsible in part for nitrate-dependent induction of nitrate reductase, nitrite export, and formate dehydrogenase genes. Most of the transcriptional regulators homologous to NarL contain an N-terminal chemotactic CheY-like domain, which changes conformation upon phosphorylation, either activating or inactivating the other domain. However, the N terminus of the supposed Shewanella transcriptional regulator is not related to CheY and has only one homolog (with 28% identity), the CsgD protein from E. coli which is involved in regulation of synthesis of Curli polymers (11). The MR1 transcriptional regulator gene is also followed by a palindrome, and there are no other obvious ORFs in the remaining 266-base sequence of the clone.
Gene sequence of flavocytochrome c.
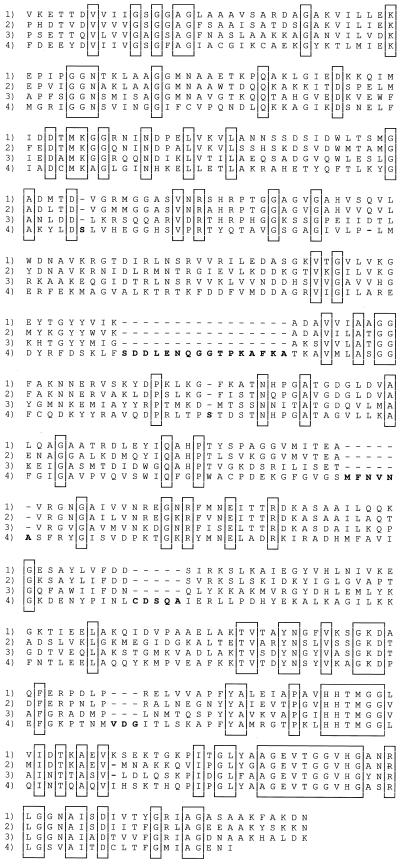
In addition to the ST cyt gene, we cloned and sequenced the FL cyt gene from S. oneidensis strain MR1. In S. frigidimarina, this gene had been shown to be a fusion of genes for a tetraheme cytochrome c at the 5′ end and the flavoprotein moiety of fumarate reductase at the 3′ end (33). This also proved to be the case for the flavocytochrome c of MR1, as shown in the sequence alignments in Fig. 1 and 3. The FL cyt's of the two Shewanella species show 59% identity overall and contain three small insertions and deletions. Most of the identity lies in the flavoprotein region, with only 48% identity in the cytochrome (N-terminal) domain. An isozyme of fumarate reductase, called IfcA, is induced in S. frigidimarina by growth on iron citrate (7). Overall, it has slightly higher identity to the S. oneidensis fumarate reductase than to the S. frigidimarina fumarate reductase (45 versus 41%), and the heme domain shows a consistently lower similarity (34 versus 31%). The sequences of the ST cyt's and the tetraheme domains of the FL cyt's from Shewanella species are also homologous and show a relatively low identity of 34%. There is only 32% overall identity to the Wolinella fumarate reductase, and there are 11 insertions and deletions. This sequence is interesting, because it shows that fumarate reductase can exist as a single gene for a protein of two domains or it can have separate genes for cytochrome and flavoprotein subunits.
FIG. 3.
Comparison of the flavocytochrome c flavoprotein domains from S. oneidensis MR1 (rows 1), S. frigidimarina NCIMB400 (33) (rows 2), S. frigidimarina NCIMB400 IfcA (7) (rows 3), and W. succinogenes (36) (rows 4). Insertions and deletions are boldfaced, and conserved residues are boxed. Gaps introduced to optimize alignment are indicated by the dashes.
The S. frigidimarina fumarate reductase clone has ORFs both upstream and downstream of the FL cyt gene (33). We did not sequence a large enough DNA fragment to determine whether the genetic context was similar. However, the genome sequence of S. oneidensis shows that the upstream gene is not present and the downstream gene is more than 74 kb downstream of the FL cyt gene. The MR1 FL cyt gene is followed 247 bases downstream by a d-lactate dehydrogenase gene. Thus, the FL cyt gene is not part of a multigene cluster, and it is not possible to tell if the same FL cyt is induced by fumarate in the two species, since there are five copies in the MR1 genome.
Structural comparison of tetraheme cytochromes.
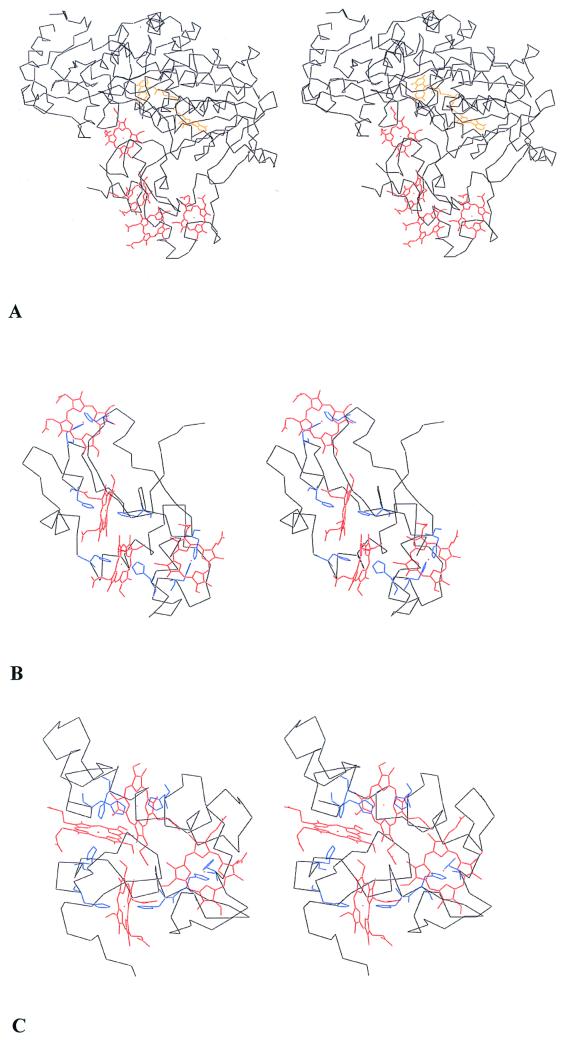
The three-dimensional structures of three different Shewanella soluble fumarate reductases have been determined. We determined the structure of the S. oneidensis FL cyt (18). The S. frigidimarina FccA structure was established by Taylor et al. (37). Bamford et al. (5) deduced the folding pattern of S. frigidimarina IfcA. These structures indicate similar folding patterns for all three but with appropriate insertions and deletions. The folding pattern of the flavoprotein domain is also similar to those of the flavoprotein subunits of the membrane-bound fumarate reductases from E. coli (13) and W. succinogenes (16). The structure of the Shewanella cytochrome domain of the FL cyt is shown in Fig. 4, where it contrasts with Desulfovibrio vulgaris Miyazaki F cytochrome c3 (12), another small tetraheme cytochrome with a low redox potential to which it had been equated. It is apparent that the Shewanella and Desulfovibrio cytochromes c fold quite differently. The cytochromes c3 fold in a compact sphere with the four hemes more or less perpendicular to one another, whereas the FL cyt domain of the Shewanella fumarate reductase is elongated with the hemes arranged in a more or less linear motif. Note that the histidine ligands are positioned differently in the FL cyt sequence for three of the four hemes, as contrasted with cytochrome c3, consistent with the different folding patterns (Fig. 1). Note also that the ST cyt and FL cyt sequences have equivalent numbers of histidine residues, so presumably they are structurally homologous to each other and not to cytochromes c3.
FIG. 4.
Stereo view of the three-dimensional backbone structures of the complete S. oneidensis MR1 flavocytochrome c (18) (A), the heme domain of the FL cyt including residues 1 to 101 (B), and D. vulgaris cytochrome c3 (12) (C). The histidine heme ligands are the only side chains shown. The N terminus is at the bottom in all three representations. Shewanella hemes 1 to 4 are in order from right to left. The Desulfovibrio hemes, starting from the upper right, are in the order 3, 1, 2, and 4.
Despite the lack of congruence in three-dimensional structures, sequence comparisons previously suggested a relationship between the Shewanella ST cyt and FL cyt's on the one hand and the class III c-type cytochromes on the other. However, the Shewanella cytochromes have only about 24% sequence identity to the class III cytochromes c3 of Desulfovibrio (12, 20, 23) for which at least seven insertions and deletions are necessary for alignment (Fig. 1). This borderline sequence similarity (which we believe is not significant because of the dominance of heme binding residues) combined with the evidence for gene duplication led Pealing et al. (34), Tsapin et al. (39), Bamford et al. (5), and Turner et al. (40) to equate the Shewanella cytochromes with the Desulfovibrio cytochromes c3. The evidence for gene doubling is stronger for the Desulfovibrio cytochromes c3 than for the Shewanella cytochromes; the sequence evidence is based upon an unusual four-residue spacing between two pairs of heme binding cysteines at hemes 2 and 4 in some of the cytochromes c3 and upon the locations of two pairs of histidines in front of hemes 1 and 3. However, there is no evidence for gene duplication from the three-dimensional structures of either protein in that they fold in single domains rather than in two domains. Thus, despite the apparent sequence homology, the presence of four hemes, and low redox potentials, the ST and FL cyt's cannot be classified as cytochromes c3 or class III cytochromes.
Redox potential and NMR spectra of the ST cyt.
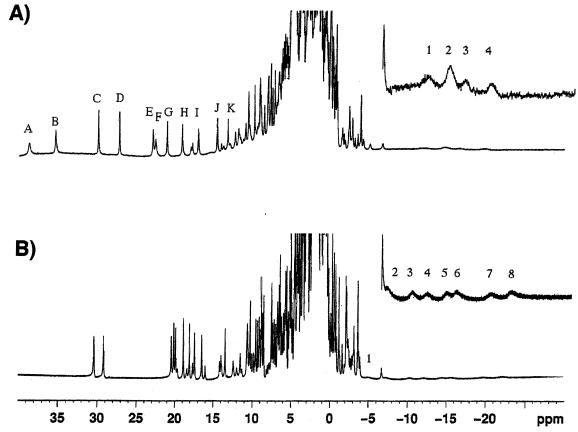
The potentials of the individual hemes of the S. oneidensis ST cyt could not be resolved in a previous redox titration, but the average redox potential was reported to be −233 mV (39). In Desulfovibrio, the close proximity of the four hemes in cytochrome c3 results in heme-heme interactions that modulate the potential as the protein is reduced; thus, there are 32 microscopic redox potentials associated with the four hemes that can be determined by NMR (32). Since a similar behavior in the ST cyt might be expected, we performed a preliminary study to establish whether it is possible to resolve the ST hemes by NMR. 1H NMR spectra of the ST cyt from S. oneidensis and the cytochrome c3 from D. vulgaris Miyazaki F in the oxidized state are presented in Fig. 5 . All four hemes are paramagnetic and low spin. However, there is an interesting difference. The 11 heme methyl proton signals that we have identified in the Shewanella ST cyt (of which there should be 16) are distributed over a wider range in the low-magnetic-field region (up to 40 ppm) than those of D. vulgaris Miyazaki F cytochrome c3 (up to 31 ppm), showing that the spin density distribution is different for the two cytochromes and that there is less spectral overlap.
FIG. 5.
1H NMR spectrum for S. oneidensis MR1 ST cyt (A) and D. vulgaris Miyazaki cytochrome c3 (B) at a pD of 9.0 and 303 K. The heme methyl resonances shown in Table 1 are labeled alphabetically from the low magnetic field. Upfield resonances are presented in an expanded scale to show signals due to heme-coordinated histidine C-2 protons. (A) Resonances 1 to 4 are at −12.23, −14.81, −16.70, and −19.81 ppm. (B) Resonances 1 to 8 are at −5.16, −7.28, −9.98, −11.7, −14.1, −15.1, −19.3, and −21.2 ppm.
By following the movement of the heme methyl resonances toward the high-magnetic-field diamagnetic region as a function of reduction, we were able to classify the 11 heme methyl signals to four groups. Their chemical shifts at a pD of 9.0 are summarized in Table 1. The macroscopic oxidation states S0, S1, S2, S3, and S4 stand for the fully oxidized, one-electron reduced, two-electron reduced, three-electron reduced, and fully reduced states, respectively. The macroscopic redox potentials at pH 9.0 were determined to be −138, −192, −219, and −225 mV by differential pulse polarography (32). The hemes represented by groups 1, 2, and 4 are mainly reduced at the first, second, and fourth macroscopic reduction steps, respectively, judging from the chemical shift changes. The heme represented by group 3, however, does not have a major reduction step, in contrast to D. vulgaris Miyazaki F cytochrome c3 for which each heme has a major macroscopic reduction step. Therefore, it can be said that heme group 3 has unusual characteristics in comparison with others.
TABLE 1.
Chemical shifts of heme methyl signals of S. oneidensis small tetraheme cytochrome c in five macroscopic oxidation states at a pD of 9.0 and 303 K
| Group and signala | Chemical shift (ppm) of signal in oxidation stateb
|
||||
|---|---|---|---|---|---|
| S0 | S1 | S2 | S3 | S4 | |
| Group 1 | |||||
| A | 38.50 | 22.20 | 18.10 | 7.40 | 1.60 |
| H | 19.30 | 11.00 | 10.20 | ||
| I | 17.00 | 10.50 | 8.60 | ||
| Group 2 | |||||
| C | 30.00 | 19.60 | 5.90 | 3.70 | |
| D | 26.90 | 18.50 | 5.85 | 4.15 | |
| Group 3 | |||||
| F | 22.40 | 18.30 | 12.35 | 7.05 | 0.95 |
| G | 21.30 | 19.00 | 18.15 | 9.30 | |
| J | 14.30 | 12.40 | 8.80 | 6.80 | |
| K | 13.25 | 11.40 | 7.95 | 5.90 | 2.70 |
| Group 4 | |||||
| B | 35.30 | 34.90 | 32.80 | 19.60 | 3.25 |
| E | 23.00 | 22.50 | 20.80 | 13.40 | |
The signal labels are given in Fig. 5.
Chemical shifts of heme methyl signals of S. oneidensis small tetraheme cytochrome c in five oxidation states. S0, S1, S2, S3, and S4 stand for the fully oxidized, one-electron reduced, two-electron reduced, three-electron reduced, and fully reduced states, respectively.
The redox potentials of the Shewanella fumarate reductase hemes, obtained by another method, average −170 mV and vary by as much as 136 mV (40). The higher redox potentials of the ST and FL cyt's are consistent with lower solvent exposure than in the cytochromes c3. The lower potential of the ST cyt relative to the FL cyt indicates that heme 4 is likely to be slightly more exposed to solvent, as suggested by the crystal structure of fumarate reductase. The broad signals in the high-magnetic-field region (expanded inserts) can be assigned to C-2 protons of the coordinated imidazole groups (1). While all eight signals due to the histidines are well separated and cover the region between −4 and −22 ppm for D. vulgaris Miyazaki F c3, only four overlapping signals (from the histidines) between −10 and −18 ppm can be seen in the high-magnetic-field region for the Shewanella ST cyt. These signals can be interpreted as being caused by at least five protons. Further work is necessary to completely assign these resonances and to locate the three remaining resonances.
Functional role of Shewanella cytochromes.
The Shewanella genome sequence shows that there are at least six genes for soluble fumarate reductase-like proteins. There are two proteins containing a single subunit and four proteins with separate heme and flavin subunits as in Wolinella. It appears unlikely that they all function as fumarate reductases. The IfcA protein has fumarate reductase activity but is induced by iron citrate (7). It will therefore be interesting to see how the other homologs are regulated.
In summary, Shewanella spp. have the capacity to produce a large number of cytochromes c. To elucidate the functional roles of these proteins, it is first necessary to compare which genes are expressed under different growth conditions and which are regulated. We have cloned the genes for two of the most abundant soluble cytochromes, a small tetraheme cytochrome c, which is induced by anaerobiosis, and a related tetraheme flavocytochrome c, which is induced by anaerobiosis and further induced by growth on fumarate. The genetic contexts of these genes are different for the two species of Shewanella that have been studied, indicating that they are independently transcribed. Nevertheless, they appear to be orthologous, i.e., they have similar structures and appear to be regulated by the same growth conditions.
ACKNOWLEDGMENTS
J.J.V.B. is indebted to the Fund for Scientific Research-Flanders (FWO-Vlaanderen) for grants 3G005497 and 3G006898. The contributions of T.E.M. and M.A.C. to this work were supported by NIH grant GM21277. A.I.T. and K.H.N. were supported by the NASA Astrobiology Program. Preliminary genome sequence data for Shewanella were obtained from The Institute for Genomic Research accomplished with support from the DOE Microbial Genome Program.
REFERENCES
- 1.Akutsu H, Hirasawa M. Nonequivalent nature of the coordinated imidazole rings of cytochrome c3 from D. vulgaris Miyazaki F as studied by 1H NMR. FEBS Lett. 1992;308:264–266. doi: 10.1016/0014-5793(92)81289-x. [DOI] [PubMed] [Google Scholar]
- 2.Ambler R P. Sequence variability in bacterial cytochromes c. Biochim Biophys Acta. 1991;1058:42–47. doi: 10.1016/s0005-2728(05)80266-x. [DOI] [PubMed] [Google Scholar]
- 3.Ambler R P, Wynn M. The amino acid sequences of cytochromes c-551 from three species of Pseudomonas. Biochem J. 1973;131:485–498. doi: 10.1042/bj1310485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baikalov I, Schroeder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 5.Bamford U, Dobbin P S, Richardson D J, Hemmings A M. Open conformation of a flavocytochrome c3 fumarate reductase. Nat Struct Biol. 1999;6:1104–1107. doi: 10.1038/70039. [DOI] [PubMed] [Google Scholar]
- 6.Beliaev A S, Saffarini D A. Shewanella putrefaciens mtrB gene encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J Bacteriol. 1998;180:6292–6297. doi: 10.1128/jb.180.23.6292-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbin P S, Butt J N, Powell A K, Reid G A, Richardson D J. Characterization of a flavocytochrome that is induced during the anaerobic respiration of Fe3+ by Shewanella frigidimarina NCIMB400. Biochem J. 1999;342:439–448. [PMC free article] [PubMed] [Google Scholar]
- 8.Even M T, Kassner R J, Dolata M M, Meyer T E, Cusanovich M A. Molecular cloning and sequencing of cytochrome c′ from the phototrophic purple sulfur bacterium Chromatium vinosum. Biochim Biophys Acta. 1995;1231:220–222. doi: 10.1016/0005-2728(95)00101-n. [DOI] [PubMed] [Google Scholar]
- 9.Gordon E H J, Pealing S L, Chapman S K, Ward F B, Reid G A. Physiological function and regulation of flavocytochrome c3, the soluble fumarate reductase from Shewanella putrefaciens NCIMB400. Microbiology. 1998;144:937–945. doi: 10.1099/00221287-144-4-937. [DOI] [PubMed] [Google Scholar]
- 10.Hagerhall C. Succinate:quinone oxidoreductases. Variations on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 11.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi Y, Kusunoki M, Matsuura Y, Yasuoka N, Kakudo M. Refined structure of cytochrome c3 at 1.8 A resolution. J Mol Biol. 1984;172:109–139. doi: 10.1016/0022-2836(84)90417-0. [DOI] [PubMed] [Google Scholar]
- 13.Iverson T, Luna-Chavez C, Cecchini G, Rees D C. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 14.Jue R A, Hale J E. Identification of cysteine residues alkylated with 3-bromopropylamine by protein sequence analysis. Anal Biochem. 1993;210:39–44. doi: 10.1006/abio.1993.1147. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lancaster C R C, Kroeger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 17.Lauterbach F, Koertner C, Albracht S P J, Unden G, Kroeger A. The fumarate reductase operon of Wolinella succinogenes. Sequence and expression of the frdA and frdB genes. Arch Microbiol. 1990;154:386–393. doi: 10.1007/BF00276536. [DOI] [PubMed] [Google Scholar]
- 18.Leys D, Tsapin A I, Nealson K H, Meyer T E, Cusanovich M A, Van Beeumen J J. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR1. Nat Struct Biol. 1999;6:1113–1117. doi: 10.1038/70051. [DOI] [PubMed] [Google Scholar]
- 19.Little B, Wagner P, Hart K, Ray R, Lavoie D, Nealson K, Aguilar C. The role of biomineralization in microbiologically influenced corrosion. Biodegradation. 1998;9:1–10. doi: 10.1023/a:1008264313065. [DOI] [PubMed] [Google Scholar]
- 20.Matias P M, Morais J, Coelho R, Carrondo M A, Wilson K K, Dauter Z, Sieker L. Cytochrome c3 from Desulfovibrio gigas: crystal structure at 1.8 A resolution and evidence for a specific calcium binding site. Protein Sci. 1996;5:1342–1354. doi: 10.1002/pro.5560050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris C J, Gibson D M, Ward F B. Influence of respiratory substrate on the cytochrome content of Shewanella putrefaciens. FEMS Microbiol Lett. 1990;69:259–262. doi: 10.1016/0378-1097(90)90077-4. [DOI] [PubMed] [Google Scholar]
- 22.Morris C J, Black A C, Pealing S L, Manson F D C, Chapman S K, Reid G A, Gibson D M, Ward F B. Purification and properties of a novel cytochrome: flavocytochrome c from Shewanella putrefaciens. Biochem J. 1994;302:587–593. doi: 10.1042/bj3020587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moura J J G, Costa C, Liu M Y, Moura I, LeGall J. Structural and functional approach toward a classification of the complex cytochrome c system found in sulfate-reducing bacteria. Biochim Biophys Acta. 1991;1058:61–66. doi: 10.1016/s0005-2728(05)80270-1. [DOI] [PubMed] [Google Scholar]
- 24.Myers C R, Myers J M. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J Bacteriol. 1992;174:3429–3438. doi: 10.1128/jb.174.11.3429-3438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers C R, Myers J M. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis and purification of the 83-kDa c-type cytochrome. Biochim Biophys Acta. 1997;1326:307–318. doi: 10.1016/s0005-2736(97)00034-5. [DOI] [PubMed] [Google Scholar]
- 26.Myers C R, Myers J M. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J Bacteriol. 1997;179:1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers C R, Myers J M. Isolation and characterization of a transposon mutant of Shewanella putrefaciens MR-1 deficient in fumarate reductase. Lett Appl Microbiol. 1997;25:162–168. doi: 10.1046/j.1472-765x.1997.00196.x. [DOI] [PubMed] [Google Scholar]
- 28.Myers C R, Nealson K H. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 29.Myers J M, Myers C R. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim Biophys Acta. 1998;1373:237–251. doi: 10.1016/s0005-2736(98)00111-4. [DOI] [PubMed] [Google Scholar]
- 30.Myers J M, Myers C R. Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J Bacteriol. 2000;182:67–75. doi: 10.1128/jb.182.1.67-75.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nealson K H, Saffarini D A. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu Rev Microbiol. 1994;48:311–343. doi: 10.1146/annurev.mi.48.100194.001523. [DOI] [PubMed] [Google Scholar]
- 32.Ohmura T, Nakamura H, Niki K, Cusanovich M A, Akutsu H. Ionic strength-dependent physicochemical factors in cytochrome c3 regulating the electron transfer rate. Biophys J. 1998;75:1483–1490. doi: 10.1016/S0006-3495(98)74067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pealing S L, Black A C, Manson F D C, Ward F B, Chapman S K, Reid G A. Sequence of the gene encoding flavocytochrome c from Shewanella putrefaciens: a tetraheme flavoenzyme that is a soluble fumarate reductase related to the membrane-bound enzymes from other bacteria. Biochemistry. 1992;31:12132–12140. doi: 10.1021/bi00163a023. [DOI] [PubMed] [Google Scholar]
- 34.Pealing S L, Cheesman M R, Reid G A, Thomson A J, Ward F B, Chapman S K. Spectroscopic and kinetic studies of the tetraheme flavocytochrome c from Shewanella putrefaciens NCIMB400. Biochemistry. 1995;34:6153–6158. doi: 10.1021/bi00018a018. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Simon J, Gross R, Klimmek O, Ringel M, Kroeger A. A periplasmic flavoprotein in Wolinella succinogenes that resembles the fumarate reductase of Shewanella putrefaciens. Arch Microbiol. 1998;169:424–433. doi: 10.1007/s002030050593. [DOI] [PubMed] [Google Scholar]
- 37.Taylor P, Pealing S L, Reid G A, Chapman S K, Walkinshaw M D. Structural and mechanistic mapping of a unique fumarate reductase. Nat Struct Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- 38.Tsapin A I, Burbaev D S, Nealson K H, Keppen O I. Investigations of succinate dehydrogenase and fumarate reductase in whole cells of Shewanella putrefaciens (strains MR-1 and MR-7) using electron spin resonance spectroscopy. J Appl Magn Res. 1995;9:509–516. [Google Scholar]
- 39.Tsapin A I, Nealson K H, Meyer T E, Cusanovich M A, Van Beeumen J J, Crosby L D, Feinberg B A, Zhang C. Purification and properties of a low-redox-potential tetraheme cytochrome c3 from Shewanella putrefaciens. J Bacteriol. 1996;178:6386–6388. doi: 10.1128/jb.178.21.6386-6388.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner K L, Doherty M K, Heering H A, Armstrong F A, Reid G A, Chapman S K. Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB 400. Biochemistry. 1999;38:3302–3309. doi: 10.1021/bi9826308. [DOI] [PubMed] [Google Scholar]
- 41.Van Hellemond J J, Tielens A G M. Expression and functional properties of fumarate reductase. Biochem J. 1994;304:321–331. doi: 10.1042/bj3040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venkateswaran K, Moser D P, Dollhopf M E, Lies D P, Saffarini D A, MacGregor B J, Ringelerg D B, White D C, Nishijima M, Sano H, Burghardt J, Stackebrandt E, Nealson K H. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int J Syst Bacteriol. 1999;49:705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- 43.Vignais P M, Toussaint B. Molecular biology of membrane-bound H2 uptake hydrogenases. Arch Microbiol. 1994;161:1–10. doi: 10.1007/BF00248887. [DOI] [PubMed] [Google Scholar]