Abstract
Agricultural soils are typically fumigated to provide effective control of nematodes, soilborne pathogens, and weeds in preparation for planting of high-value cash crops. The ability of soil microbial communities to recover after treatment with fumigants was examined using culture-dependent (Biolog) and culture-independent (phospholipid fatty acid [PLFA] analysis and denaturing gradient gel electrophoresis [DGGE] of 16S ribosomal DNA [rDNA] fragments amplified directly from soil DNA) approaches. Changes in soil microbial community structure were examined in a microcosm experiment following the application of methyl bromide (MeBr), methyl isothiocyanate, 1,3-dichloropropene (1,3-D), and chloropicrin. Variations among Biolog fingerprints showed that the effect of MeBr on heterotrophic microbial activities was most severe in the first week and that thereafter the effects of MeBr and the other fumigants were expressed at much lower levels. The results of PLFA analysis demonstrated a community shift in all treatments to a community dominated by gram-positive bacterial biomass. Different 16S rDNA profiles from fumigated soils were quantified by analyzing the DGGE band patterns. The Shannon-Weaver index of diversity, H, was calculated for each fumigated soil sample. High diversity indices were maintained between the control soil and the fumigant-treated soils, except for MeBr (H decreased from 1.14 to 0.13). After 12 weeks of incubation, H increased to 0.73 in the MeBr-treated samples. Sequence analysis of clones generated from unique bands showed the presence of taxonomically unique clones that had emerged from the MeBr-treated samples and were dominated by clones closely related to Bacillus spp. and Heliothrix oregonensis. Variations in the data were much higher in the Biolog assay than in the PLFA and DGGE assays, suggesting a high sensitivity of PLFA analysis and DGGE in monitoring the effects of fumigants on soil community composition and structure. Our results indicate that MeBr has the greatest impact on soil microbial communities and that 1,3-D has the least impact.
Application of methyl bromide (MeBr) to agricultural soils before planting of high-value cash crops has been the mainstay for the control of nematodes, soilborne pathogens, and weeds for many years in warm regions of the United States. The chemistry and air pollution potential of this fumigant have been documented (2, 7, 8). It has also been shown that MeBr may have the ability to destroy stratospheric ozone (25, 43), and a ban on its production in and importation into the United States is scheduled to be fully implemented by 2005 (35). 1,3-Dichloropropene (1,3-D), methyl isothiocyanate (MITC), and chloropicrin (CP) have been proposed as most likely to be chemical alternatives to MeBr. While most of these fumigants are known to have broad biocidal activity (1), their effects on soil microbial communities are largely unknown. Recently, the effect of MITC (the toxic degradation product of metam sodium) on soil microbial community structure and function was studied by the use of traditional heterotrophic activity measures, catabolic potential, and biochemical assay (18). Those authors showed that abundances of indicator fatty acids for bacteria after 5 weeks of incubation were correlated to MITC doses but that after 18 weeks very few were related to MITC dose. MITC was also observed to reduce populations of culturable organisms dramatically in the Biolog assay.
Garland and Mills (11) adapted the Biolog redox technology based on community-level carbon source utilization patterns to characterize and classify microbial communities from environmental (soil, aquatic, and rhizosphere) samples. In a comparative study of rhizosphere bacterial communities and hydrocarbon-polluted environments, Garland and Mills (12) and Wünsche et al. (41) showed that substrate utilization patterns can be used as an indicator of community structure and function. Although the Biolog assay has been proposed as a measure of functional diversity (44), assay responses are attributed mainly to a small subset of heterotrophic bacteria in the soil.
White and Findlay (37) developed a community-level approach to characterize microbial community structure by evaluating shifts in phospholipid fatty acids (PFLA) from environmental samples. Different groups of bacteria are characterized by specific PLFA profiles; therefore, a change in the phospholipid pattern in soil would indicate a change in the bacterial composition of that soil. This concept has resulted in the identification and quantification of viable biomass and community structure in sediments (3, 27, 29) and in agricultural soils (45, 46).
Analysis of cloned ribosomal gene sequences retrieved from the environment can provide detailed information about the community composition and the structural diversity of the environment without bias. To monitor the structural diversity of microbial communities, denaturing gradient gel electrophoresis (DGGE) was introduced by Muyzer et al. (21). This technique is based on the separation of ribosomal gene sequences directly amplified from community DNA by using conserved primers on a denaturing gel according to their melting properties. This allows direct comparison of many samples with different treatments (19). Our objectives were to monitor the biocidal effects of these compounds and compare their ecotoxicological effects on a soil microbial community in response to the application of fumigants.
MATERIALS AND METHODS
Soil type and experimental design.
Soil samples (Arlington sandy loam) were taken from the top 15 cm in a fallow field at the University of California Riverside Agricultural Experiment Station. Soil samples were collected with a 10-cm-diameter stainless-steel auger that was washed with diluted methanol between samplings to avoid lipid contamination. There has been no history of fumigant treatment on this plot. The soil had a pH of 7.2 and an organic carbon content of 0.92%. Moist field soil was passed through a 4-mm sieve, and the water content of the original soil was determined and readjusted to 12%. Soil samples were stored at room temperature for 48 h before being used in the experiment.
Soil samples were placed in microcosms containing about 1.5 kg (dry weight) of soil. The experimental design consisted of four fumigants at three different concentrations and a control in three replicate microcosms (Table 1). Fumigants were added as freshly prepared aqueous solutions. Table 1 shows the test ranges used in the different microcosms. Microcosms were sealed for 24 h after fumigant application and were vented continuously through a small opening on the cover. Samples for community analysis were taken at 1, 8, and 12 weeks after fumigant treatment.
TABLE 1.
Concentrations of fumigants used in the microcosm experiments
| Fumigant | Concn (mg/kg [dry wt] of soil) useda |
|---|---|
| MeBr | 160 (1) |
| 320 (2) | |
| 3,200 (3) | |
| MITC | 160 (1) |
| 320 (2) | |
| 3,200 (3) | |
| 1,3-D | 80 (1) |
| 160 (2) | |
| 1,600 (3) | |
| CP | 40 (1) |
| 80 (2) | |
| 800 (3) |
Numbers in parentheses indicate concentrations of fumigants at 50% (1), 100% (2), and 1,000% (3) of the recommended agricultural dose (see figures).
Substrate utilization patterns of microbial populations determined using the Biolog system.
Biolog GN (gram-negative) microtiter plates were used to analyze functional diversity through the substrate utilization patterns shown by soil microorganisms. The Biolog GN plates contained 95 separate carbon sources and a blank well with no substrate. In order to obtain substrate utilization patterns of whole soil communities, the cell suspensions were prepared by extracting the soils with phosphate buffer solution, serially diluting, and adding a 150-μl suspension of a 10−3 dilution to the Biolog plates with an eight-channel repeating pipette. Plates were inoculated with three replicates of soil extract and incubated at 25°C, and absorbance data were recorded at 595 nm at 0, 24, 48, 72, and 96 h with a Bio-Rad (Richmond, Calif.) Plate Reader. The patterns of sole carbon source utilization in Biolog plates were expressed as an index of color development in each of the wells as reported by Wünsche et al. (41) and modified by Ibekwe and Kennedy (16). The index was obtained by subtracting the color response reading in the spectrophotometer for each of the 95 wells from that for the control well and then dividing by the reading for the control well according to the formula WE = (WA − W0/W0)100, where WA is the absorbance in each well from well A2 to well H12 and W0 is the absorbance in the blank well (no color development). A WE value of greater than 100 was regarded as indicating a positive reaction (evidence of substrate utilization), and a WE value of less than 100 was regarded as indicating a negative reaction. The variables were coded as binary values (1 for a positive reaction and 0 for a negative reaction). This approach was used to eliminate the problem of inoculum cell density and produced a weighted data set that could be used in principal-component analysis (PCA). PCA was used to reduce the number of variables (95 variables) to the number of principal components (PCs) that explain 80% or more of the variance (six PCs in this study). Since we used the correlation matrix to compute variables (substrates), all substrates with eigenvalues of greater than 1 were used in our analysis. We computed the correlation between the PCs and the treatments to examine the effects of substrates.
Phospholipid extraction and separation.
Triplicate soil samples (5 g from each microcosm) were extracted by using the modified method of Bligh and Dyer (38) as described by Petersen and Klug (24). The total lipid extract was fractionated into glycolipids, neutral lipids, and polar lipids (13, 19). The polar lipid fraction was transesterified with mild alkali to recover the PLFA as methyl esters in hexane. The PLFA were separated, quantified, and identified by gas chromatography-flame ionization detection (19). Samples were run for 38 min, which is long enough for fatty acids with up to 28 carbons to elute from the column. The system consisted of a gas chromatograph (HP6980; Hewlett-Packard, Wilmington, Del.) with a flame ionization detector and HP3365 ChemStation software.
Fatty acid nomenclature.
The suffixes c for cis and t for trans refer to geometric isomers. The prefixes i, a, and me refer to isomethyl, anteisomethyl, and mid-chain methyl branching, respectively, with cyclopropyl rings indicated by cy (17).
DNA extraction, PCR primers, and DGGE analysis.
Total bacterial community DNA was extracted to assess the effects of fumigants on bacterial community diversity. DNA was extracted by placing 500 mg of soil in FastPrep tubes (Bio 101, Vista, Calif.) containing lysing matrix and shaken for 30 s. Isolation of total DNA was accomplished with a FastPrep DNA isolation kit according to the protocols of the manufacturer (Bio 101).
PCR was performed using 20 to 80 ng of template DNA with primers PRBA338f and PRUN518r, located at the V3 region of the 16S rRNA genes of bacterioplankton (23). PRBA338f consists of a region that is conserved among the domain Bacteria, and PRUN518r is located at a universal conserved region. PCR mixtures contained Ready-To-Go PCR beads from Amersham-Pharmacia Biotech (Piscataway, N.J.), 10 pmol of each primer, 4 μg of bovine serum albumin, template DNA, and sterile distilled water in a final volume of 25 μl. PCR conditions were 92°C for 2 min, followed by 30 cycles of 92°C for 1 min, 55°C for 30 s, and 72°C for 1 min and a single final extension at 72°C for 6 min.
DGGE was performed with 8% (wt/vol) acrylamide gels containing a linear chemical gradient ranging from 30 to 70% denaturant, with 100% defined as 7 M urea and 40% formamide. Gels were run for 3 h at 200 V with a Dcode Universal Mutation System (Bio-Rad). DNA was visualized after ethidium bromide staining by UV transillumination and photographed with a Polaroid camera. Major bands were excised for identification of bacterial species. Bands were placed into sterilized vials with 20 μl of sterilized distilled water and stored overnight at 4°C to allow the DNA to passively diffuse out of the gel strips. Ten microliters of eluted ribosomal DNA (rDNA) was used as the DNA template with eubacterial primers. The sizes of the PCR products were checked on a 1.5% agarose gel, and the DNA was cloned into a pGEM-T Easy vector (Promega, Madison, Wis.) and transformed into Escherichia coli JM109. Isolation of plasmids from E. coli was performed using standard protocols from the Qiagen (Valencia, Calif.) plasmid minikit. The purified plasmids were sequenced with the ABI PRISM Dye Terminator Cycle Sequencing Kit with AmpliTaq DNA polymerase, FS (Perkin-Elmer).
Statistical analysis of PLFA profiles and DGGE bands.
Data analysis of PLFA profiles was performed using SAS (30). Analyses of variances, means, and standard deviations for the individual fatty acids in triplicate-sample PLFA profiles were determined to compare the moles percent of each PLFA in each sample. Correspondence analysis was used to compare PLFA profiles among sampling times. The mean moles percent data were presented as cluster analysis or a two-dimensional plot for better understanding of relationships. Minitab statistical software (version 13) was used to cluster observations into groups.
DNA fingerprints obtained from the 16S rDNA banding patterns on the DGGE gels were photographed and digitized using ImageMaster Labscan (Amersham-Pharmacia Biotech, Uppsala, Sweden). The lanes were normalized to contain the same amount of total signal after background subtraction. The gel images were straightened and aligned using ImageMaster 1D Elite 3.01 (Amersham-Pharmacia Biotech, Uppsala, Sweden) and analyzed to give a densitometric curve for each gel. Band positions were converted to Rf values between 0 and 1, and profile similarity was calculated by determining Dice's coefficient for the total number of lane patterns. Dendrograms were constructed by using the unweighted pair group method with mathematical averages (UPGMA). Dice's similarity coefficients were generated, converted into x-y line plots, and transferred to Excel files. Community similarities based on peak areas from the Excel files for the different bacterial groups (16S rDNA bands) were analyzed by correspondence analysis (CANACO 4.0; Microcomputer Power, Ithaca, N.Y.) to generate an ordination diagram as described by Yang and Crowley (42). Dice's similarity coefficients generated from the three sampling points were integrated and analyzed using ImageMaster 1D database 2.01 (Amersham-Pharmacia Biotech, Uppsala, Sweden). The data obtained were used for the construction of a library to determine the best-fit profile and to integrate the area under each peak for every gel and for the construction of a dendrogram between treatments. For analysis of diversity, each band was presumed to represent the ability of that bacterial species to be amplified. The Shannon-Weaver index of diversity (H) was used to compare changes in diversity of microbial communities within the four treatments at each time (31) by using the function H = −(Pi log Pi), where Pi = ni/N, ni is the peak height, and N is the sum of all peak heights in the curve.
Phylogenetic analysis.
Sequence identification was performed by using the BLAST database (National Center for Biotechnology Information [www.ncbi.nlm.nih.gov]) and the Sequence Match Facility of the Ribosomal Database Project (www.cme.msu.edu/RDP).
RESULTS AND DISCUSSION
Analysis of heterotrophic microbial communities by using Biolog GN microplates.
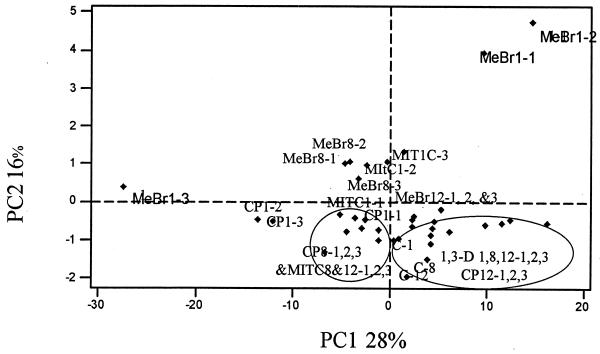
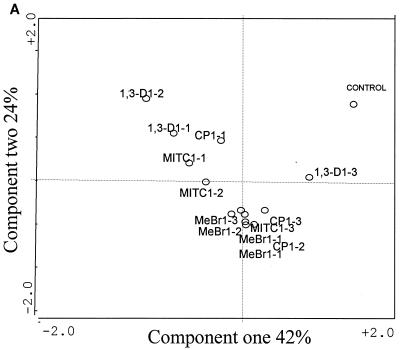
The results of the PCA performed on the mean values of the Biolog GN fingerprints of the different soil microbial communities obtained after 72 h of incubation are shown in Fig. 1. The functional abilities of the heterotrophic soil microbial communities were altered by the application of the fumigants, especially MeBr, during the first week of the experiment. The PCA plot (Fig. 1) for the four microbial communities and the control showed that the first component accounted for 28% of the variance, while the second component accounted for 16% and the third component accounted for 14%, with six PCs accounting for over 80% of the variation. The control soil and the soil treated with 1,3-D were separated along PC1, and their coefficients were positively correlated to the right of PC1. Analysis of MeBr-treated communities did not show any pattern of groupings except that communities from the first week of treatments were positively correlated along PC2 and grouped with MITC after weeks 8 and 12. Pairwise comparison showed that the MeBr-treated communities were significantly different (P < 0.05) from the control and 1,3-D-treated communities.
FIG. 1.
PCA performed on Biolog GN fingerprints of soil extracts treated with MeBr, MITC, 1,3-D, and CP and of nonfumigated soil (C). Numbers after the abbreviations indicate week 1, 8, or 12, and those after the hyphen indicate concentrations of fumigants used in the experiment as listed in Table 1. Since most of the 1,3-D samples clustered together along PC1, they are not differentiated by time and concentrations.
The Biolog assay showed that functional characteristics of the control and 1,3-D treatments were similar when compared to those for the other three fumigants. The problem with the Biolog system is that color development in the Biolog plates is due first to the respiratory activities of fast-growing heterotrophic bacteria, resulting in the stimulation or reduction of the catabolism of 95 carbon substrates (6). The shifts in microbial communities observed in the Biolog assays are due to organisms that grow rapidly because of their high population in a sample. For example, Pseudomonas species, which are found in most of the samples in this study, respond well in Biolog assays (9, 10, 14). Analysis of the microbial communities of the Biolog GN microplates by DGGE has been shown to confirm that carbon source utilization profiles obtained with Biolog GN plates do not necessarily discriminate the numerically dominant members of the microbial community used as the inoculum (6, 32).
PLFA analysis.
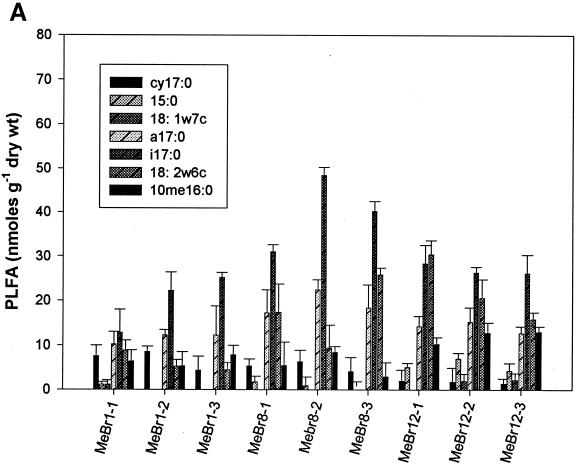
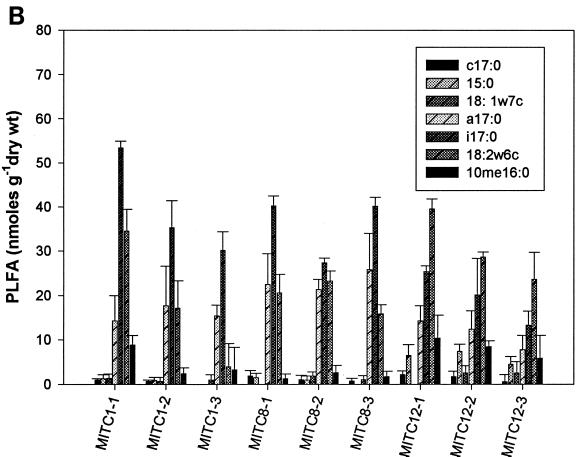
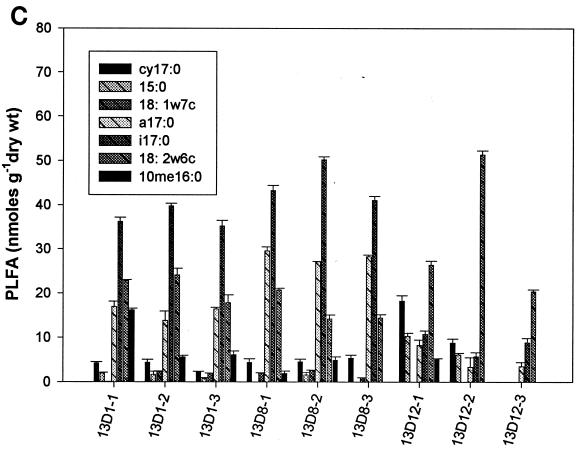
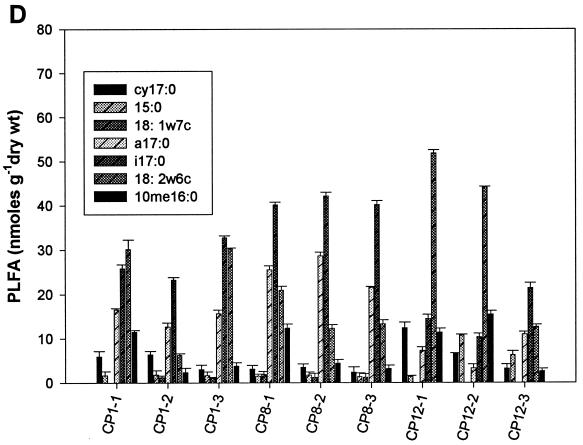
Analysis of PLFA profiles for all four treatments from weeks 1, 8, and 12 was carried out in triplicate. The content of individual biomarker peaks ranged from a minimum of 1.3 nmol per g (dry weight) for the four fumigants in week 1 to a maximum of 55 nmol per g (dry weight) for the 1,3-D- and CP-treated samples in week 12 (Fig. 2). Biomass contents as indicated by total PLFA were significantly different at different time points for some treatments (P < 0.05). At week 1, biomass contents in MeBr-amended microcosms were significantly lower than those at weeks 8 and 12 (Fig. 2A) and were also lower than those for microcosms amended with the three other fumigants (Fig. 2B, C, and D). There was also a decrease in biomass of some gram-negative biomarkers (cy17:0, 15:0, and 18:1ω7c) and fungal biomarkers (18:2ω6c) with the increase in MeBr concentration during the first week of the experiment (Fig. 2A). However, there was a significant increase in biomass for gram-positive bacteria (a17:0 and i17:0), fungi (18:2ω6c), and actinomycetes (10me16:0) in weeks 8 and 12. The effects of MITC followed the same trend as those of MeBr, except that the recovery of gram-negative bacterial biomass did not occur during week 8 (Fig. 2B). 1,3-D and CP had the strongest effects on actinomycetes, resulting in a significant decrease in biomass for most treatments (Fig. 2C and D).
FIG. 2.
Biomass contents (nanomoles of PLFA per gram [dry weight] of soil) of samples collected after weeks 1, 8, and 12 from MeBr (A)-, MITC (B)-, 1,3-D (C)-, and CP (D)-fumigated soils (n = 3). Numbers after the abbreviations indicate times and concentrations as described in the legend to Fig. 1. Error bars represent standard deviations.
Analysis of microbial community structure by PLFA.
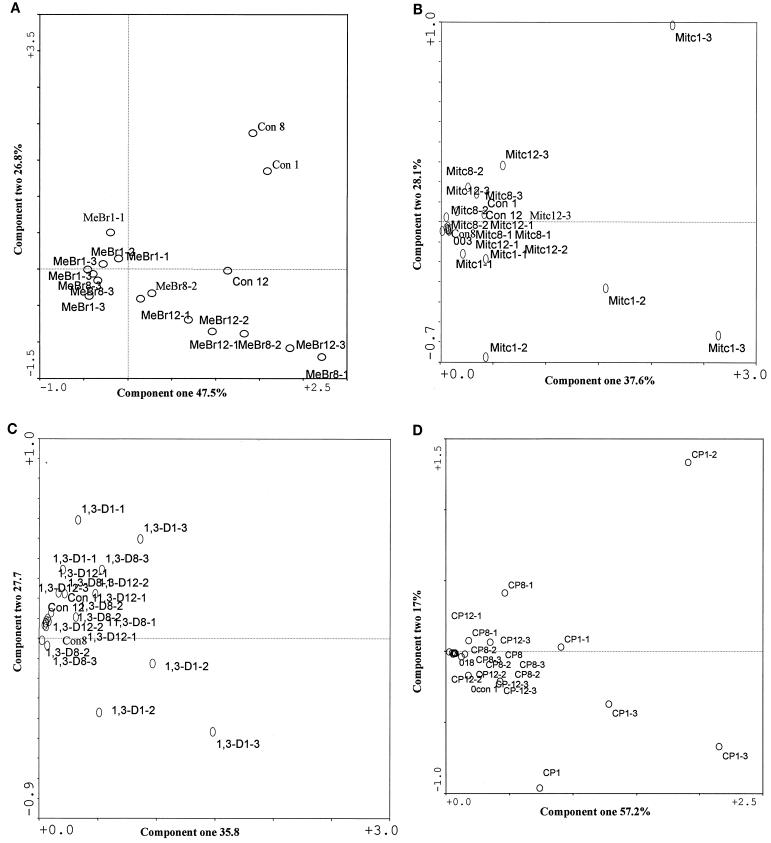
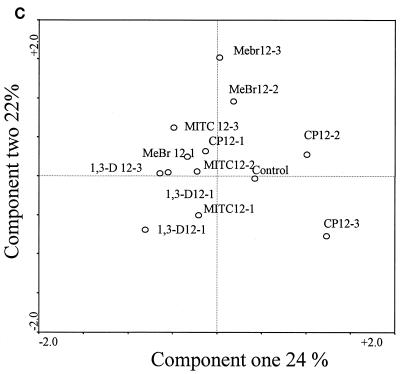
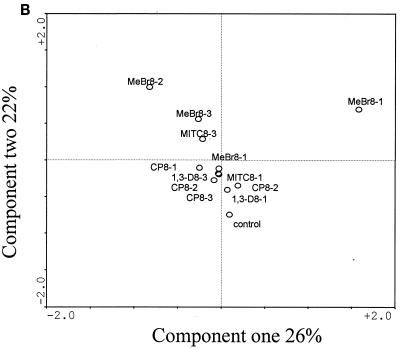
The community structures of fumigant-treated microcosms from weeks 8 and 12, measured by PLFA analysis, shifted away from those during the first week of sampling. The shift was greatest with MeBr, which showed a 47.5% variation in component one and a 26.8% variation in component two (Fig. 3A). The major difference in the PLFA profiles between the MeBr-treated and the control microcosms was that the MeBr-treated microbial communities contained significantly more branched-chain PLFA (specifically, a17:0, i17:0, a15:0, and i15:0 [P < 0.05]), indicative of gram-positive bacteria (15, 34, 37). For all microcosms treated with MeBr, the relative proportion of PLFA indicative of fungal biomass (13), specifically, 18:2ω6c and 18: 3ω6c, increased over time. Correspondence analysis of the PLFA profiles showed similar results, with all of the samples in the week 12 treatment forming a distinct cluster (Fig. 3A). Analyses comparing PLFA profiles of MITC-, 1,3-D-, and CP-treated microcosms to those of the control samples consistently revealed the same patterns. At week 1, the profiles were furthest away from the control; at weeks 8 and 12, PLFA profiles of samples treated with the three fumigants more closely resembled each other and that of the control sample (Fig. 3B, C, and D).
FIG. 3.
Scatter plots of the results from correspondence analysis of the PLFA contents described in Fig. 2. Con, control.
Cluster analysis of the PLFA profiles (complete squared Euclidean distance with mean moles percent data) showed the major trends within the four treatments during 12 weeks of the study (Fig. 4). MeBr-amended microcosm samples from week 1 clustered according to time and concentrations. 1,3-D- and CP-treated samples did not show any pattern of clustering throughout the 12 weeks of the study. In fact, for all of the fumigants except MeBr, bacteria recovered from the initial effects after 12 weeks and clustered closer to the control.
FIG. 4.
Cluster analysis of the PLFA contents (complete squared Euclidean distance with moles percent data) described in Fig. 3. Numbers after the abbreviations indicate times and concentrations as described in the legend to Fig. 1. Cont, control.
Advantages of PLFA analysis, in comparison to other techniques, are that PLFA analysis has been regarded as an indicator of total microbial biomass (37, 45) and that certain PLFA can be used as biomarkers for specific groups of microorganisms (34, 39). The presence of large proportions of branched-chain fatty acids (a15:0, i15:0, a17:0, and i17:0), which are markers for gram-positive bacteria (28), suggests that gram-positive bacteria were less affected by the impact of these fumigants than gram-negative bacteria. This is in agreement with the work of Zelles et al. (47), who found that gram-positive bacteria were less injured by chloroform fumigation; they attributed this to protection by the cell wall structure of the bacteria, formation of spores, and ability to adapt to fumigant vapor more quickly.
Analysis of soil microbial community structure by PCR-DGGE.
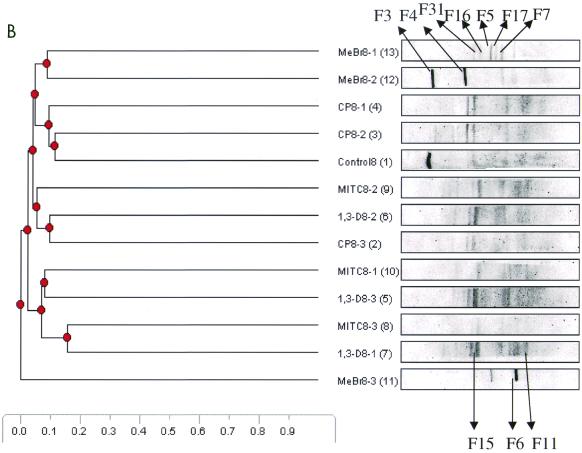
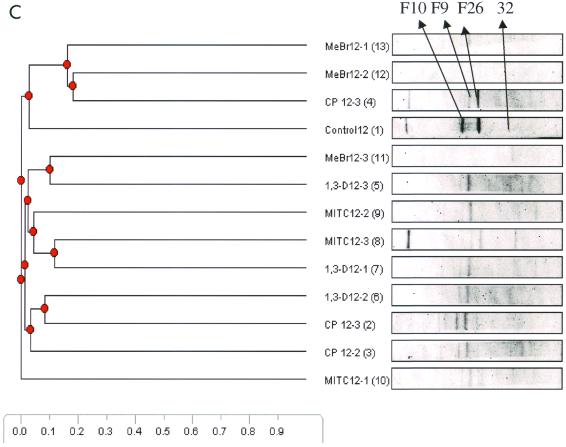
DGGE analysis of 16S rDNA fragments was used to examine the effects of the four fumigants and the control on soil microbial communities. Figure 5 shows cluster analysis of DGGE patterns of the 16S rDNA fragments (primers P338f and P518r) amplified from the four fumigated soils and control soil at 1, 8, and 12 weeks after fumigation. The most drastic effect occurred in the first week of the experiment. During this period, the MeBr treatments clustered away (97% similarity index) from the treatments with the other three fumigants and the control (Fig. 5A). This observation was confirmed by the DGGE banding pattern in Fig. 5A, which shows no dominant bands for MeBr-treated samples during the first week of the experiment. At week 8 there was a significant shift in microbial community structure. The microcosm that was treated with the highest concentration of MeBr clustered away from the other two treatments, the three fumigants, and the control. As shown in Fig. 5B, more bands, which did not occur during the first week after fumigation, began to appear in the MeBr treatments. There was also a decrease in the number of bands as the concentration of MeBr increased (MeBr8-1 to -8-3). At 12 weeks, the microbial communities for all concentrations of MITC, 1,3-D, and CP and the lowest concentrations of MeBr were similar to those for the control (Fig. 5C).
FIG. 5.
DGGE analysis of 16S rDNA fragments of pooled soil samples, at different times after fumigation, collected from triple microcosms treated with different fumigants. Amplified products were separated on a gradient gel of 30 to 70% denaturant. All labeled bands were excised from the gel, reamplified, and subjected to sequence analysis. These reamplification products were cloned and screened as described in the text. (A) Community structures at week 1, 7 days after the initiation of the experiment. Significant differences between the community structures of microcosms treated with MeBr, the highest concentration of MITC, and control treatment had been induced at this time, because all of the major bands in the MeBr samples were undetected, while the samples treated with the other fumigants and the control treatment continued to maintain complex banding patterns. (B) Community structures after 8 weeks of treatment. Most of the samples treated with the other fumigants continued to maintain complex banding patterns, while new community structures emerged with the MeBr-treated samples. (C) Community structures after 12 weeks of treatment. The banding patterns in the MeBr-treated samples continued to emerge, and the communities reverted almost completely to that seen with the control or other fumigants. Numbers 1 through 13 in parentheses refer to the lane numbers in the DGGE gels.
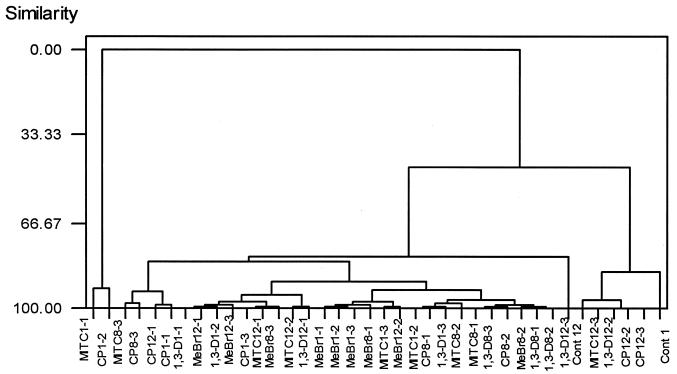
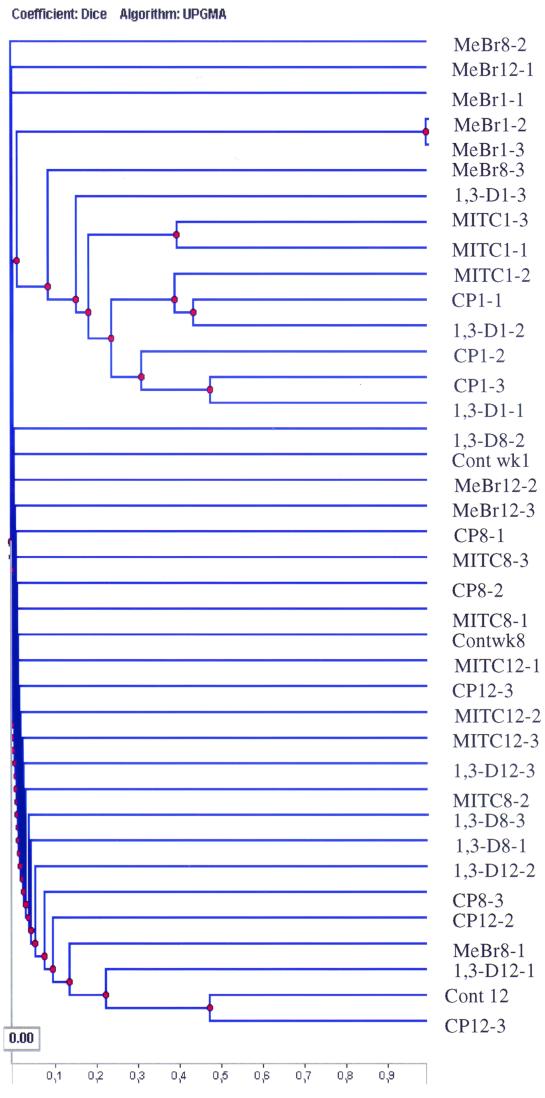
To compare DGGE patterns for the three sampling points, Dice's indices were determined for comparisons of all profiles, and UPGMA was used to create a dendrogram describing pattern similarities (Fig. 6). This analysis clearly showed the impact of the four fumigants during the first week of the experiment and distinguished the effects from those at weeks 8 and 12. All of the week 1 samples were grouped together as most similar, separating them from week 8 and 12 samples.
FIG. 6.
UPGMA tree with Dice's coefficient, representing the genetic similarity of the microbial community profiles obtained by PCR-DGGE. Numbers after the abbreviations indicate times and concentrations as described in the legend to Fig. 1. Cont, control.
To quantitatively examine the relative similarities of the communities, the 16S rDNA band profiles were analyzed by peak fitting. Due to artifacts associated with DGGE analysis of images of completely different gels, in which there are subtle differences in the gel gradients, running times, and DNA staining procedures, we decided first to analyze our samples based on time from extraction of DNA from pooled soil samples. This resulted in a direct comparison of the effect of each compound at one time point in one gel. This was done after running gels with triplicate microcosm soil samples that showed banding patterns to be identical (data not shown). The ordination diagrams in which the effects of the four fumigants were compared by correspondence analysis after fumigation (Fig. 7A) showed separation of the treatments from the control. The data explained 42% variability on the first component and 24% on the second component during the first week of the study (Fig. 7A). Variability in the data was smaller at weeks 8 and 12 (Fig. 7B and C).
FIG. 7.
Correspondence analysis of microbial communities generated by the analysis of DGGE 16S rDNA PCR patterns at 1 week (A), 8 weeks (B), and 12 weeks (C) after MeBr, MITC, 1,3-D, and CP treatment.
The second method for determination of the structural diversity was the calculation of the Shannon-Weaver index of diversity (H) from the DGGE banding pattern of the samples. H was calculated on the basis of the number and relative intensities of bands on a gel strip. By avoiding the bias of cultivation and by direct extraction of DNA from fumigated soil samples, H was used as a parameter that reflects the structural diversity of the dominant microbial community. The four fumigant-treated samples showed different levels of diversity (Table 2), ranging between 0.11 and 1.26 at different sampling times. Gel analysis of the four treatments at the three sampling times revealed that MeBr exerted the most significant effects on the structural diversity of the soil. One week after MeBr application, the DGGE banding patterns revealed dramatic changes in the structure of the microbial community (Table 2 and Fig. 5). The number of bands decreased from 16 in the control to almost undetectable numbers in MeBr-treated soil, which established a new cluster far away from the control (Fig. 7A). This indicated the collapse of the microbial community due to the acute toxicity of MeBr. The parameter H decreased from 1.26 in the control to 0.11 in the treatment with the highest concentration of MeBr. The H value increased slightly during week 8 and subsequently, in week 12, to about 0.75 but remained clearly below the average control value (H = 1.26). Communities from samples treated with MITC, 1,3-D, and CP were not as severely affected, although their H values were still below that of the control (Table 2).
TABLE 2.
Effects of fumigants on the Shannon-Weaver index of diversity (H) from DGGE profiles
| Time (wk) |
Ha at the indicated fumigant concn (mg/kg [dry wt] of soil)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP
|
1,3-D
|
MITC
|
MeBr
|
Control (0) | |||||||||
| 40 | 80 | 800 | 80 | 160 | 1,600 | 160 | 320 | 3,200 | 160 | 320 | 3,200 | ||
| 1 | 1.03 | 1.05 | 0.92 | 0.86 | 1.26 | 0.76 | 0.82 | 0.74 | 0.12 | 0.26 | 0.15 | 0.11 | 1.25 |
| 8 | 1.21 | 1.03 | 0.98 | 0.96 | 1.02 | 0.96 | 0.98 | 0.86 | 0.45 | 0.34 | 0.22 | 0.13 | 1.23 |
| 12 | 1.21 | 1.10 | 1.06 | 1.02 | 1.00 | 0.85 | 1.01 | 1.00 | 0.64 | 0.75 | 0.45 | 0.65 | 1.31 |
Diversity indices from DGGE bands calculated as described in Materials and Methods.
In this study we applied the Shannon-Weaver index of diversity to 16S rDNA DGGE from total community DNA separated according to sequence heterogeneity. This approach had been successfully used by Eichner et al. (5) to describe the community structure in activated sludge. Those authors noted that the number and intensities of bands do not equal the number and abundance of species within the microbial community due to features of 16S rDNA-based phylogeny and bias inherent to PCR amplification from complex template mixtures. The reasons for most of the limitations are that DGGE banding patterns are subjected to PCR bias due to DNA extraction methods, potential preferential amplification, and the formation of chimeras (40). Other problems may result from one organism producing more than one DGGE band because of multiple, heterogeneous rRNA operons (4, 22, 26). Also, for some phylogenetic groups of bacteria, 16S rDNA sequences do not allow discrimination between species, so one DGGE band may represent several species with identical rDNA sequences (36).
In a community DNA mixture such as soil, the maximum number of different rDNA fragments separated by DGGE may be vastly underestimated. Torsvik et al. (33) found that there might be as many as 104 different genomes present in soil samples. This shows that DGGE cannot separate all of the 16S rDNA fragments obtained from soil microorganisms, but only the dominant species (20, 21). Therefore, the banding patterns obtained in this study reflect the most abundant rDNA types in the community. In this study the Shannon-Weaver index of diversity was used in combination with the correspondence analysis of the DGGE banding patterns based on the similarity coefficient to monitor a range of community responses after the application of fumigants. First, the changes in community structure within the laboratory microcosms during the first week of the experiment were documented, and the major bands that emerged were identified. Second, the recovery in community structure in the MeBr treatments to a community dominated by gram-positive bacteria may be an indication of how bacteria respond to chemical treatments.
Analysis of predominant bacterial species by PCR-DGGE.
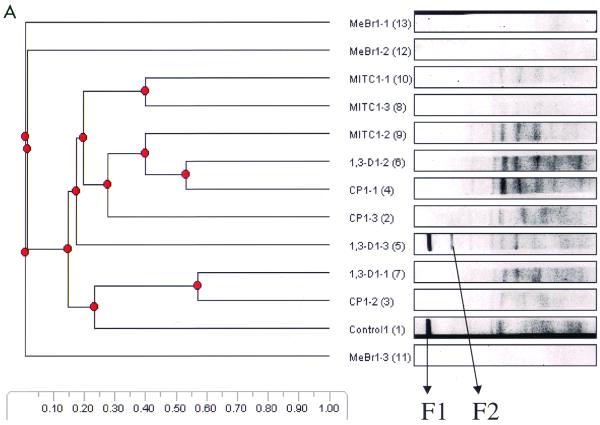
The analysis of predominant bacterial species was carried out with soil samples pooled from triplicate microcosms at weeks 1, 8, and 12. Bands selected for analysis are shown in Fig. 5. Table 3 shows the prominent bands recovered from the DGGE gel. At week 1 (Fig. 5A) all samples generated very complex banding patterns except samples treated with MeBr and the highest concentration of MITC. In addition to the two prominent bands (F1 and F2) observed during the first-week analysis, more than seven visible bands were also present in most of the samples analyzed. The derived sequences from these bands confirmed that F1 was 100% similar to Pseudomonas reactans and that F2 had 99% similarity to Pseudomonas putida. At week 8 new bands appeared in the MeBr treatments. Four dominant bands (F3 to F7) and three other bands (F16, F17, and F31) were also present. Most of the prominent bands visible at week 8 in the other treatments remained strong within each treatment but were not present in the MeBr treatments. The appearance of novel bands (F3 to F7 and F16, F17, and F31) in the MeBr treatment was an indication of the dominance of new microbial species. Bands F3 and F6 showed relationships to the gram-positive species Heliothrix oregonensis and Bacillus subtilis, respectively. The fragment designated F4 was one of the most dominant species in the MeBr treatment community that evolved 8 weeks after fumigation. Its sequence closely matched (97%) that of an unclassified organism (Acidobacterium capsulatum) 16S rRNA gene. Bands F5 and F7 also appeared in the MeBr treatments and matched 100% with uncultured soil bacterium C0111. Three other novel bands (F16, F17, and F31) loosely related to the genus Sphingomonas in the alpha subclass of the Proteobacteria were detected uniquely in the MeBr-treated samples. There were no significant new bands at week 12; therefore, the dominant bands that appeared in all of the samples were analyzed for general information on the dominant species, as shown in Table 3.
TABLE 3.
Sequence analysis of bands excised from DGGE gels derived from bacterial 16S rDNAs extracted from fumigated and nonfumigated soils
| Band | Bacterium with related bacterial sequence | % Similarity | Treatment | Accession no. |
|---|---|---|---|---|
| F1 | Pseudomonas reactans | 100 | Control | AF255337 |
| F2 | Pseudomonas putida | 99 | Control | AJ232794 |
| F3 | Holiothrix oregonensis | 93 | MeBr | LO4675 |
| F4 | Acidobacterium capsulatum | 97 | MeBr | AF214136 |
| F5 | Uncultured bacterium C011 | 100 | MeBr | AF128708 |
| F6 | Bacillus subtilis | 97 | MeBr | AF074970 |
| F7 | Uncultured soil bacterium CO11 | 99 | MeBr | AF128708 |
| F9 | Uncultured bacterium 0319-7F4 | 91 | CP | AF234144 |
| F10 | Uncultured bacterium B10 | 97 | Control | AF257860 |
| F11 | Sphingomonas sp. strain CD | 99 | 1,3-D | AF19022 |
| F17 | Soil bacterium 147 | 100 | MeBr | AF128769 |
| F26 | Legionella sainthelensi | 99 | CP | X73399 |
| F31 | Alpha-subclass proteobacterium RB2256 | 100 | MeBr | AF18422 |
In conclusion, our data have shown that the effects of MeBr and its alternatives on soil microbial composition and function could be evaluated by using a functional assay and biochemical and genetic fingerprints. This study showed that the PLFA technique was the most effective method in evaluating community structure after fumigant treatment, followed by the DGGE approach and the Biolog assay. We have also shown that gram-positive bacteria survived better after fumigant treatment, with 1,3-D exerting the least effect on microbial community structure. The behavior of microbes in the soil after fumigation could be evaluated more accurately by using field samples. However, due to many problems associated with sample collection immediately after fumigation, a laboratory microcosm experiment can be used to give basic results if fumigants are applied at concentrations that reflect field conditions.
REFERENCES
- 1.Anderson J P E. Proceedings of the International Symposium on Environmental Aspects of Pesticide Microbiology. Uppsala, Sweden: Swedish University of Agricultural Science; 1993. Side-effects of pesticides on carbon and nitrogen transformations in soils; pp. 61–67. [Google Scholar]
- 2.Baker L W, Fitzell D L, Seiber J N, Parker T R, Shibamoto T, Poor M W, Longley K E, Tomlin R P, Propper R, Duncan D W. Ambient air concentrations of pesticides in California. Environ Sci Technol. 1996;30:1365–1368. [Google Scholar]
- 3.Balkwill D L, Leach F L, Wilson T J, McNabb J F, White D C. Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate, and direct counts in the subsurface aquifer sediments. Microbial Ecol. 1988;16:73–84. doi: 10.1007/BF02097406. [DOI] [PubMed] [Google Scholar]
- 4.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 5.Eichner C A, Erb R W, Timmis K N, Wagner-Döbler I. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl Environ Microbiol. 1999;65:102–109. doi: 10.1128/aem.65.1.102-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelen B, Meinken K, von Wintzingerode F, Heuer H, Malkomes H-P, Bachaus H. Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl Environ Microbiol. 1998;64:2814–2821. doi: 10.1128/aem.64.8.2814-2821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan J, Yates S R, Crowley D, Becker J O. Acceleration of 1,3-dichloropropene degradation by organic amendments and potential application for emissions reduction. J Environ Qual. 1998;27:408–414. [Google Scholar]
- 8.Gan J, Yates S R, Wang D, Ernst F F. Effects of application methods on 1,3-dichloropropene volatilization from soil under controlled conditions. J Environ Qual. 1998;27:432–438. [Google Scholar]
- 9.Garland J L. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol Ecol. 1997;24:289–300. [Google Scholar]
- 10.Garland J L. Patterns of potential C source utilization by rhizosphere communities. Soil Biol Biochem. 1996;26:223–230. [Google Scholar]
- 11.Garland J L, Mills A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garland J L, Mills A L. A community-level physiological approach for studying microbial communities. In: Ritz K, Dighton J, Giller K E, editors. Beyond the biomass: composition and functional analysis of soil microbial communities. Chichester, United Kingdom: Wiley; 1994. pp. 77–83. [Google Scholar]
- 13.Guckert J B, Antworth C P, Nichols P D, White D C. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol Ecol. 1985;31:147–158. [Google Scholar]
- 14.Haack S K, Garchow H, Odelson D L, Forney L J, Klug M J. Accuracy, reproducibility, and interpretation of fatty acid methyl ester profiles of model bacterial communities. Appl Environ Microbiol. 1994;60:2483–2493. doi: 10.1128/aem.60.7.2483-2493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heipieper H-J, Diefenbach R, Keweloh H. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl Environ Microbiol. 1992;58:1847–1852. doi: 10.1128/aem.58.6.1847-1852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibekwe A M, Kennedy A C. Phospholipid fatty acid profiles and carbon utilization patterns for analysis of microbial community structure under field and greenhouse conditions. FEMS Microbiol Ecol. 1998;26:151–163. [Google Scholar]
- 17.Kates M. Techniques in lipidology: isolation, analysis and identification of lipids. 2nd ed. Amsterdam, The Netherlands: Elsevier Press; 1986. [Google Scholar]
- 18.Macalady J L, Fuller M E, Scow K M. Effects of Metam sodium fumigation on soil microbial activity and community structure. J Environ Qual. 1998;27:54–63. [Google Scholar]
- 19.MacNaughton S J, Stephen J R, Venosa A D, Davis G A, Chang Y-J, White D C. Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol. 1999;65:3566–3574. doi: 10.1128/aem.65.8.3566-3574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two Californian estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Øvreas L, Forney L, Daae F L, Torsvik T. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen S O, Klug M J. Effects of sieving, storage, and incubation temperature on the phospholipid fatty acid profiles of a soil microbial community. Appl Environ Microbiol. 1994;60:2421–2430. doi: 10.1128/aem.60.7.2421-2430.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prather M J, McElroy M R, Wofsy S C. Reduction in ozone at high concentrations of stratospheric halogens. Nature. 1984;31:227–231. doi: 10.1038/312227a0. [DOI] [PubMed] [Google Scholar]
- 26.Rainey F A, Ward-Rainey N L, Janssen P H, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 27.Rajendran N, Matsuda O, Imamura N, Urushigawa Y. Variation in microbial biomass and community in the sediments of eutrophic bays as described by phospholipid ester-linked fatty acids. Appl Environ Microbiol. 1992;58:562–571. doi: 10.1128/aem.58.2.562-571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratledge C, Wilkinson S G. Microbial lipids. Vol. 1. New York, N.Y: Academic Press, Inc.; 1988. [Google Scholar]
- 29.Ringelberg D B, Davis J D, Smith G A, Pfiffner S M, Nichols P D, Nickels J S, Henson J M, Wilson J T, Yates M, Kampbell D H, Reed H W, Stocksdale T T, White D C. Validation of signature phospholipid fatty acids biomarkers for alkaline-utilizing bacteria in soil and subsurface aquifer materials. FEMS Microbiol Ecol. 1988;62:39–50. [Google Scholar]
- 30.SAS Institute. Users guide: statistic version 6. Carey, N.C: Statistical Analytical Institute; 1988. [Google Scholar]
- 31.Shannon C E, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1963. [Google Scholar]
- 32.Smalla K, Wachtendorf U, Heuer H, Liu W, Forney L. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl Environ Microbiol. 1998;64:1220–1225. doi: 10.1128/aem.64.4.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torsvik V, Goksoyr J, Daale F L. Appl. Environ. Microbiol. 56:782–787. 1990. High diversity in DNA of soil bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tunlid A, White D C. Biochemical analysis of biomass, community structure, nutritional status and metabolic activity of the microbial community in soil. In: Bollag J M, Stotzky G, editors. Soil biochemistry. Vol. 7. New York, N.Y: Marcel Dekker, Inc; 1992. pp. 229–262. [Google Scholar]
- 35.U.S. Environmental Protection Agency. Protection of stratospheric ozone. Fed Regist. 1995;58:15014–15049. [Google Scholar]
- 36.Vallaeys T, Topp E, Muyzer G, Macheret V, Laguerre G, Rigaud A, Soulas G. Evaluation of denaturing gradient gel electrophoresis in the detection of 16S rDNA sequence variation in rhizobia and methanotrophs. FEMS Microbiol Ecol. 1997;24:279–285. [Google Scholar]
- 37.White D C, Findlay R H. Biochemical markers for measurement of predation effects on the biomass, community structure, nutritional status, and metabolic activity of microbial biofilms. Hydrobiologia. 1988;159:119–132. [Google Scholar]
- 38.White D C, Davis W M, Nickels J S, King J D, Bobbie R J. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- 39.White D C, Flemming C A, Leung K T, MacNaughton S J. In situ microbial ecology for quantitative assessment, monitoring and risk assessment of pollution remediation in soils, the subsurface, the rhizosphere and in biofilms. J Microbiol Methods. 1998;32:93–105. [Google Scholar]
- 40.Wintzingerode F V, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 41.Wünsche L, Brüggermann L, Wolfgang B. Determination of substrate utilization patterns of soil microbial communities: an approach to assess population changes after hydrocarbon pollution. FEMS Microbiol Ecol. 1995;17:295–306. [Google Scholar]
- 42.Yang C-H, Crowley D E. Rhizosphere microbial community structure in relation to root location and plant iron nutrition status. Appl Environ Microbiol. 2000;66:345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yung Y L, Pinto P, Watson R T, Sander P S. Atmospheric bromine and ozone perturbations in the lower stratosphere. J Atmos Sci. 1980;37:339–353. [Google Scholar]
- 44.Zak J C, Willing M R, Moorehead D L, Wildman H G. Functional diversity of microbial communities: a quantitative approach. Soil Biol Biochem. 1994;26:1101–1108. [Google Scholar]
- 45.Zelles L, Bai Q Y, Beck T, Beese F. Signature fatty acids in phospholipids and lipopolysaccharides as indicators of microbial biomass and community structure in agricultural soils. Soil Biol Biochem. 1992;24:317–323. [Google Scholar]
- 46.Zelles L, Rackwitz R, Bai Q Y, Beck T, Beese F. Discrimination of microbial diversity by fatty acid profiles of phospholipids and lipopolysaccharides in differently cultivated soils. Plant Soil. 1995;170:115–122. [Google Scholar]
- 47.Zelles L, Palojarvi A, Kandeler E, von Lutzow M, Winter K, Bai Q Y. Changes in soil microbial properties and phospholipid fatty acid fractions after chloroform fumigation. Soil Biol Biochem. 1997;29:1325–1336. [Google Scholar]