Abstract
Background
Acute myeloid leukemia (AML) is of heterogeneous pathogenesis and caused by alterations of multiple genes. CircRNAs act as oncogenes or tumor suppressors in numerous tumors and could be novel diagnostic and prognostic biomarkers. Few studies had incorporated circRNAs in AML.
Aim of the Work
Assessment of circANXA2, circ0075001, and circFBXW7 gene expressions in AML patients. Evaluation of their relations with clinical, cytogenetic, and overall survival outcome to emphasize their diagnostic role and prognostic impact.
Methods
This study was carried out on 120 subjects (66 AML patients and 54 controls). All subjects were subjected to gene expressions assay for circANXA2, circ0075001, circFBXW7 by quantitative real-time polymerase chain reaction.
Results
Prominent overexpression of circANAX2 and circ0075001 in patients than control (P < 0.001), whereas circFBXW7 was markedly downregulated in patients than in control (P < 0.001). Moreover, circANXA2 with AUC 0.824, P <0.001, had a sensitivity of 74.24%, specificity 88.89% whereas circ0075001 with AUC 0.855, P < 0.001, had the highest sensitivity of 83.33% and specificity 79.63%, and circFBXW7 with AUC 0.826, P < 0.001, had a sensitivity of 75.76% and specificity 74.07% in the distinction of AML patients from controls. Additionally, we find out that high expression of circANXA2 and circ0075001 correlated significantly with splenomegaly, hepatomegaly, less differentiated FAB subtypes (M5, M7), short overall survival, and had an adverse cytogenetic pattern.
Conclusion
CircANXA2, circ0075001, and circFBXW7 gene expressions could serve as potential diagnostic biomarkers for AML disease. Moreover, CircANXA2 and circ0075001 exert poor prognostic effects on AML patients.
Keywords: acute myeloid leukemia, CircRNAs, CircANXA2, circ0075001, circFBXW7
Introduction
Acute myeloid leukemia (AML) is a malignant neoplasm that compromises hematopoietic stem cell (HSC) differentiation. Thus, infiltration of the bone marrow, peripheral blood, and extramedullary tissues (eg, spleen, the lymph nodes, and central nervous system) by immature myeloid precursors occurs.1 AML displays various genetic features and different clinical outcomes. It was evident that many patients still relapse and later die within 2 years despite the effectiveness of intensive chemotherapy and innovative drugs.2
AML has diverged from being an acute leukemia entity to a heterogeneous pattern of AML subgroups distinguished by distinct clinical, pathophysiologic, cytogenetic, and molecular profiles that benefit from customized selective therapies and have enormously diverse outcomes.3
Molecular markers are attractive to risk stratify patients into subgroups1,4,5 including cytogenetic risk subgroups, molecular genetic alterations such as FMS-like tyrosine kinase 3 internal tandem repeat (FLT3-ITD), CCAAT/enhancer-binding protein (CEBPA) mutations, and nucleophosmin-1 (NPM1).5 And besides they outline prognosis and therapeutic goals. Furthermore, in patients with a diploid karyotype, single mutations and mutation combinations act together differently.3 Next-generation sequencing recognized numerous recurrent somatic mutations in >90% of AML patients. Commonly mutated genes (frequency >5%) are IDH1, NPM1, DNMT3A, FLT3, IDH2, RUNX1, TET2, p53, NRAS, CEBPA, WT1.6,7
Circular RNAs (CircRNAs) are non-coding RNAs endogenous (ncRNAs), abundant in eukaryotic cells, and have extended interest recently. They are produced by exon or intron cyclization by ligating the 5′ terminal cap and 3′ terminal poly (A) tail to compose a circular structure. They are present in the cytoplasm or stored in exosomes and are stably expressed as they resist damage by RNA exonucleases.8,9
The diversity of circRNA and their distinct biological functions are a consequence of various formation mechanisms. Such as, circRNAs which can act as miRNA sponge,10 be translated into proteins,11 bind functional proteins,12 control RNA splicing13 and regulate transcription.14
Past reports recognized an important task of circRNAs in cancer progression by modifying cancer features,15 such as being involved in proliferative signaling,16 evasion of growth suppressors,17 loss of differentiation signals, influence on tumor metastasis and invasion,18 and provocation of angiogenesis.19
CircRNAs are novel molecules that have been implicated in the basis of numerous diseases and malignancies. However, their role in AML had not been fully understood yet.20 In this study, the light was shaded on the gene expression of circANXA2 (Annexin A2), circ0075001, and circFBXW7 in newly diagnosed Egyptian AML patients. We aimed to investigate their possible relations with clinical, cytogenetic, survival outcomes and validate their risk stratification in those patients. Thus, elucidating their diagnostic role and prognostic impact.
Materials and Methods
Subjects
This study was performed from January 2019 to June 2021 in the department of Clinical Pathology in collaboration with the departments of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Menoufia University. Newly diagnosed AML patients were recruited from the Hematology Unit of Internal Medicine Department, Clinical Oncology and Nuclear Medicine, Menoufia University hospitals, Nasser, and National cancer institutes.
This study included 120 studied subjects that were divided into the following: Group I composed of 66 newly diagnosed AML patients without beginning care. Exclusion criteria: pediatric age group, patients who started treatment or were diagnosed with other hematologic malignancies or disorders, and patients who discontinued the study. AML patients were further classified based on morphology and cytochemistry of The French American British (FAB) system.21
In addition, they were risk-stratified according to ELN 2017 into favorable, intermediate, and adverse cytogenetic criteria.22 We had considered the following items from all patients: History, clinical examination, laboratory, and radiological investigations.
The protocol of treatment of AML patients started with induction chemotherapy with a combination of cytosine arabinoside and daunorubicin (3 and 7 protocol)23 for eligible patients. Patients who achieved complete remission and had favorable cytogenetics received consolidation chemotherapy with high dose Ara-C.
Patients with adverse or intermediate-risk cytogenetics were transferred for allogeneic bone marrow transplantation. While those who relapsed after conventional chemotherapy or failed to achieve CR despite optimal induction were treated with second induction and then were transferred for allogeneic bone marrow transplantation.
Patients with acute promyelocytic leukemia (APL) received induction treatment with ATRA and maintenance with ATRA, 6 mercaptopurine, and methotrexate.24 Older age and frail patients received low dose Ara-C.25 All patients were followed up for 12 months after induction chemotherapy. Overall survival of the patients was calculated from the date of diagnosis to the date of death or the end of the follow-up period.
Group II constituted 54 age and gender-matched control subjects with the AML group. Each participant in the study provided informed written consent and written formal consent was obtained from the parent of child participants. The study approval was obtained from the Research Ethics Committee at Menoufia Faculty of Medicine with accord to Helsinki Declaration and regulations.
Blood Sampling and Laboratory Investigations
Venous blood samples (6mL) on EDTA tubes have been obtained from each subject. The following investigations were done; 3mL were withdrawn for complete blood count (CBC) and Leishman-stained film (Sysmex XN-1000, B.M Egypt Company, Japan (19,723)). The remaining 3 mL of blood were utilized for assaying circRNAs gene expressions of CircANXA2, Circ0075001, CircFBXW7, and GAPDH as reference genes by quantitative real-time polymerase chain reaction (qRT- PCR).
Measurement of CircANXA2, Circ0075001, CircFBXW7 Genes Expression
Full RNA isolation from whole blood using Kits supplied by (PureLink® RNA Mini Kit, Life Technologies, Carlsbad, California, USA), followed by RNA quality and purity testing with the aid of nanodrop (ND-1000 spectrophotometer, Thermo Scientific, USA) and extracted RNA was deposited at −80°C till the time of use. At first, cDNA synthesis (reverse transcription step) was done using (RevertAid First Strand cDNA Synthesis kits, Thermofisher Scientific, USA). Initially, cDNA synthesis composed of 1 ul of Nuclease-free- H2O, 1 ul of RT Random Hexamer primer and 10 uL of extracted RNA that were incubated at 65°C for 5min and chilled on ice then followed by adding 4ul of 5x RT Buffer, 2 ul of 10Mm dNTP Mix, 1 ul of RevertAid M-MuLV RT Reverse Transcriptase and 1 ul of RiboLock RNase Inhibitor, to reach 20 ul total reaction. The mixture was incubated for 5 min at 25°C utilizing Applied Biosystems 2720 thermal cycler (Singapore) and cycling conditions were optimized for cDNA reverse transcription at 42°C for 60 min, then at 70°C for 5 min. Finally, cDNA amplification was performed using SYBR green-based quantitative real-time PCR by (SensiFAST TM SYBR Lo-ROX Kit, Thermofisher Scientific, Applied Biosystem), using the following designed primers (Thermofisher Scientific, Invitrogen, USA). CircANXA2 gene primers: forward 5′ CGTTTCCGACACATCTGGC 3′ reverse 5′ CCCTGGCATCCTGGTCAAT 3′. Circ0075001: forward 5’ TTGTTCTCTGGAGCAGCGTT 3′ reverse 5′ GGTCAGTCATCCGGAAGCAA 3’ CircFBXW7: forward 5′ GTCTCCCAAACCTGACTGTC 3′ reverse 5′ GTTGGTTCCCTTCCTCCTTC 3′. Circular RNA gene expression was normalized by using glyceraldehyde 3– p dehydrogenase (GAPDH) as a reference gene forward 5` CTCTGCTCCTCCTGTTCGAC 3` reverse 5` TTAAAAGCAGCCCTGGTGAC 3`.
For (qRT-PCR) amplification, we performed in a total 20ul reaction (10ul of Syber green master mix with low ROX dye, 1 ul of forwarding primer of both target and reference gene, 1ul of reverse primer of both target and reference gene, 3 ul of cDNA product, 5ul of nuclease-free H2O) using Applied BioSystems 7500 software version 2.0.1 for data analysis. The cycling program was as follows: initial denaturation for 10 minutes at 95°C followed by 45 cycles of denaturation for 15 sec at 95°C and annealing/extension for 1 minute at 60°c.
Relative quantification (RQ) of gene expression of CircANXA2 (Figure 1A), Circ0075001 (Figure 1B), CircFBXW7 (Figure 1C) was accomplished using the comparative ∆∆Ct method. The target circular RNA is normalized to an endogenous reference gene (GAPDH gene) and relative to a control. Each run was accomplished using melting curve analysis for CircANXA2 (Figure 1D), Circ0075001 (Figure 1E), CircFBXW7 (Figure 1F) to confirm amplification specificity and primer dimer absence.
Figure 1.
(A) Relative quantification of gene expression of CircANXA2 (B) relative quantification of gene expression of Circ0075001. (C) Relative quantification of gene expression of CircFBXW7. (D) Melting curve analysis for CircANXA2 expression. (E) Melting curve analysis for Circ0075001 expression. (F) Melting curve analysis for CircFBXW7 expression.
Statistical Analysis
The data collected have been tabulated and analyzed on an IBM personal computer by Statistical Package of Social Science (SPSS, version 20; SPSS Inc., Chicago, Illinois, USA).
Results
This study was conducted on 66 AML patients 37 (56.1%) males and 29 (43.9%) females aged from 19.0 to 75.0 (mean ± SD; 51.05±15.30) years and 54 age and gender-matched healthy control 29 (53.7%) males and 25 (46.3%) females, aged from 17.0 to 69.0 (mean ± SD; 47.80 ±12.88) years. Regarding CBC, Hb was (mean ± SD; 8.35 ± 1.85) and PLT median was 55.0 (30.0–72.0). Both were lower in AML patients compared with control group whom their Hb was (mean ± SD;13.38 ± 1.04) (P < 0.001) and PLT median was 266.0 (205.0–300.0) (P < 0.001). However, TLC was not significantly different between both studied groups (P = 0.706).
Concerning circRNAs expressions, we observed meaningful upregulation of circANAX2 with median and interquartile range (IQR) 12.97 (1.80–89.88) (P < 0.001) and circ0075001 with median (IQR) 3.01 (1.70–12.54) (P < 0.001) in patients than in control group circANAX2 1.0 (0.39–1.71) and circ0075001 0.99 (0.57–1.0), respectively. On the other hand, circFBXW7 was considerably downregulated in patients with median (IQR) 0.60 (0.08–1.20) than in control 1.80 (1.0–4.20) (P < 0.001) (Table 1).
Table 1.
Comparison Between the Two Studied Groups According to Different Parameters
| AML (n = 66) | Control (n = 54) | Test of Sig. | p | |||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Sex | ||||||
| Male | 37 | 56.1 | 29 | 53.7 | χ2=0.067 | 0.796 |
| Female | 29 | 43.9 | 25 | 46.3 | ||
| Age (years) | ||||||
| Min. – Max. | 19.0–75.0 | 17.0–69.0 | t=1.242 | 0.217 | ||
| Mean ± SD. | 51.05 ± 15.30 | 47.80 ± 12.88 | ||||
| Median (IQR) | 51.50 (40.0–65.0) | 49.0(38.0–60.0) | ||||
| TLC x 103 | ||||||
| Min. – Max. | 0.80–23.0 | 4.20–11.0 | U=1710.50 | 0.706 | ||
| Mean ± SD. | 8.92 ± 6.57 | 7.06 ± 2.18 | ||||
| Median (IQR) | 7.25(2.70–15.0) | 6.25 (5.30–9.50) | ||||
| Hb g/dl | ||||||
| Min. – Max. | 3.0–11.80 | 11.0–15.10 | t=18.782 | <0.001* | ||
| Mean ± SD. | 8.35 ± 1.85 | 13.38 ± 1.04 | ||||
| Median (IQR) | 8.55 (7.0–9.90) | 13.50 (12.50–14.0) | ||||
| PLT x 103 | ||||||
| Min. – Max. | 10.0–150.0 | 154.0–354.0 | U=0.0 | <0.001* | ||
| Mean ± SD. | 55.17 ± 29.32 | 253.22 ± 50.19 | ||||
| Median (IQR) | 55.0 (30.0–72.0) | 266.0 (205.0–300.0) | ||||
| CircANXA2 | ||||||
| Min. – Max. | 0.02–596.66 | 0.11–4.37 | U=628.0 | <0.001* | ||
| Mean ± SD. | 87.17 ± 148.47 | 1.37 ± 1.13 | ||||
| Median (IQR) | 12.97 (1.80–89.88) | 1.0 (0.39–1.71) | ||||
| Circ0075001 | ||||||
| Min. – Max. | 0.02–825.85 | 0.08–2.50 | U=517.50 | <0.001* | ||
| Mean ± SD. | 72.20 ± 174.93 | 0.84 ± 0.53 | ||||
| Median (IQR) | 3.01 (1.70–12.54) | 0.99 (0.57–1.0) | ||||
| CircFBXW7 | ||||||
| Min. – Max. | 0.01–2.50 | 0.17–4.90 | U= 619.50 | <0.001* | ||
| Mean ± SD. | 0.81 ± 0.76 | 2.41 ± 1.51 | ||||
| Median (IQR) | 0.60 (0.08–1.20) | 1.80 (1.0–4.20) | ||||
Notes: p, p-value for comparing between the studied groups; *statistically significant at p ≤ 0.05.
Abbreviations: IQR, interquartile range; SD, standard deviation; χ2, Chi-square test; t, Student’s t-test; U, Mann Whitney test.
Clinical features of AML patients were described in detail in (Table 2), where numerous patients suffer from splenomegaly 26 (39.4%), and hepatomegaly 25 (37.9%) while just 4 (6.1%) patients had lymphadenopathy and only 2 (3.0%) patients complained of CNS affection. Bone marrow aspiration samples showed a mean blast cell count (mean ± SD;42.71 ± 23.40). AML patients were further subclassified based on FAB classification into M1 8 (12.1%), M2 13 (19.7%), M3 11 (16.7%), M4 11 (16.7%), M5 11 (16.7%), M65 (7.6%) and M7 7 (10.6%).
Table 2.
Distribution of the Studied Cases According to Clinical Data in AML Group (n = 66)
| No. | % | |
|---|---|---|
| Splenomegaly | 26 | 39.4 |
| Hepatomegaly | 25 | 37.9 |
| LN | 4 | 6.1 |
| CNS involvement | 2 | 3.0 |
| B.M BLASTS | ||
| Min. – Max. | 20.0–90.0 | |
| Mean ± SD. | 42.71 ± 23.40 | |
| Median (IQR) | 30.0 (30.0–60.0) | |
| FAB | ||
| M1 | 8 | 12.1 |
| M2 | 13 | 19.7 |
| M3 | 11 | 16.7 |
| M4 | 11 | 16.7 |
| M5 | 11 | 16.7 |
| M6 | 5 | 7.6 |
| M7 | 7 | 10.6 |
| Overall survival (months) | ||
| Min. – Max. | 1.0–12.0 | |
| Mean ± SD. | 7.95 ± 4.32 | |
| Median (IQR) | 9.50 (3.0–12.0) | |
| Outcome | ||
| Died | 29 | 43.9 |
| Alive | 31 | 47.0 |
| Lost during follow up | 6 | 9.1 |
| Cytogenetic criteria | ||
| Favorable | 33 | 50.0 |
| Intermediate | 16 | 24.2 |
| Adverse | 17 | 25.8 |
| CR achievement | 31 | 47.0 |
| ELN CLASS | ||
| Favorable cytogenetic | ||
| t(8:21)+ve | 7 | 10.6 |
| inv(16)+ve | 6 | 9.1 |
| PML-RARA+VE | 11 | 16.7 |
| Mutated NPM1 FLT3-ITD-/low | 9 | 13.6 |
| Intermediate cytogenetic | ||
| t (9:11) | 6 | 9.1 |
| Mutated NPM1 FLT3-ITD high | 10 | 15.2 |
| Adverse cytogenetic | ||
| Inv (3) +ve | 3 | 4.5 |
| Wild NPM1 FLT3-ITD high | 14 | 21.2 |
Abbreviations: IQR, interquartile range; SD, standard deviation.
Follow-up of AML patients was accomplished for 12 months, and we calculated the median (IQR) of overall survival which was 9.50 (3.0–12.0). Regarding outcome, 29 (43.9%) patients died while 31 (47.0%) were alive and occasionally 6 (9.1%) had lost follow-up. Cytogenetic pattern based on ELN classification revealed that 33 of AML patients (50%) had favorable cytogenetics; including 7 patients with t(8:21) (10.6%), 6 patients with inv(16) (9.1%), 11 patients with PML RARA (16.7%), and 9 of them with mutated NPM1 FLT3-ITD/low (13.6%). Intermediate cytogenetics was revealed in 16 patients (24.3%) including mutated NPM1FLT3 –ITD/high 10 (15.2%), and 6 patients with t (9:11) (9.1%). Seventeen patients had adverse cytogenetics pattern (25.7%) including 14 of them with wild NPM1 FLT3-ITD/high (21.2%) and 3 with positive inv (3) (4.5%). Moreover, complete remission (CR) was achieved in 31 (47%) of patients (Table 2).
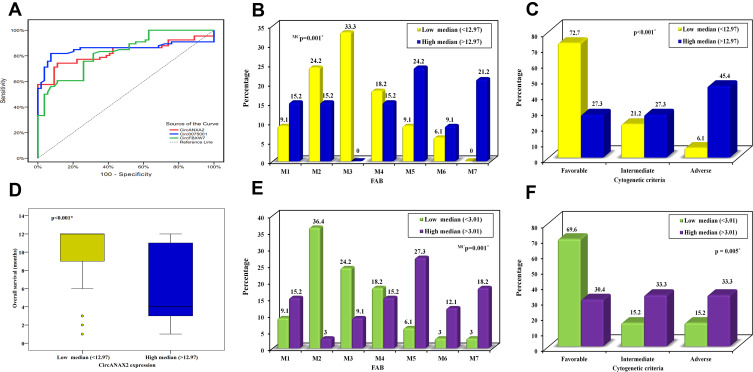
ROC curve was established for the three circRNAs to distinguish the diagnosis of AML from a healthy control. CircANXA2 with AUC 0.824, P < 0.001, cutoff point >1.87 had a sensitivity of 74.24% and specificity 88.89% whereas circ0075001 with AUC 0.855, P < 0.001, cutoff point >1.16 had a sensitivity of 83.33% and specificity 79.63%. Meanwhile, circFBXW7 with AUC 0.826, P < 0.001, cutoff point ≤1.2 had a sensitivity of 75.76% and specificity of 74.07% (Table 3) (Figure 2A).
Table 3.
Validity (AUC, Sensitivity, Specificity) for CircANXA2, Circ0075001 and CircFBXW7to Discriminate AML (n = 66) from Control (n = 54)
| AUC | p | 95% C. I | Cut off | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|---|---|
| CircANXA2 | 0.824 | <0.001* | 0.746–0.901 | >1.87 | 74.24 | 88.89 | 89.1 | 73.8 |
| Circ0075001 | 0.855 | <0.001* | 0.778–0.932 | >1.16 | 83.33 | 79.63 | 83.3 | 79.6 |
| CircFBXW7 | 0.826 | <0.001* | 0.755–0.898 | ≤1.2 | 75.76 | 74.07 | 78.1 | 71.4 |
Abbreviations: AUC, area under a curve; p value, probability value; CI, confidence intervals; NPV, negative predictive value; PPV, positive predictive value; *, statistically significant at p ≤ 0.05.
Figure 2.
(A) ROC curve for CircANXA2, Circ0075001, and CircFBXW7 to discriminate AML from control. (B) Relation between high and low CircANAX2 expression with FAB classification in AML patients. (C) Relation between high and low CircANAX2 expression with Cytogenetic criteria in AML patients. (D) Relation between high and low CircANAX2 expression with Overall survival in AML patients. (E) Relation between high and low Circ0075001expression with FAB classification in AML. (F) Relation between high and low Circ0075001 expression with Cytogenetic criteria in AML. *Statistically significant at p ≤ 0.05.
AML patients were classified into the high and low expression of circANXA2 circ0075001 and circFBXW7 based on higher or lower than the median value. The results demonstrated that high circANXA2 was significantly expressed in patients with splenomegaly (54.5%) (P = 0.012), hepatomegaly (51.5%) (P = 0.022), less differentiated FAB subtypes M5 (24.2%), M7 (21.2%) (P = 0.001) (Figure 2B) and had adverse cytogenetic pattern (45.4%) (P < 0.001) (Figure 2C), shorter overall survival (P < 0.001) (with median 4 months) (Figure 2D) and most patients died during follow-up period (66.7%), and few of them achieved CR (24.2%) (P < 0.001). In comparison to the low expression group, 24.2% of patients had splenomegaly and hepatomegaly, more differentiated FAB subtypes M3 (33.3%), M2 (24.2%) (Figure 2B), most patients (69.7%) were alive and had a favorable outcome (72.7%)(Figure 2C) and achieved CR (69.7%) and had higher overall survival (with median 12 months) (Figure 2D). The other parameters did not differ between high and low expression groups (Table 4).
Table 4.
Relation Between High and Low Expression of CircANAX2 Expression with Clinical Data in AML Patients (N = 66)
| CircANAX2 Expression | Test of Sig. | p | ||||
|---|---|---|---|---|---|---|
| Low Median (<12.97) (n=33) | High Median (>12.97) (n=33) | |||||
| No. | % | No. | % | |||
| Sex | ||||||
| Male | 18 | 54.5 | 19 | 57.6 | χ2=0.062 | 0.804 |
| Female | 15 | 45.5 | 14 | 42.4 | ||
| Age (years) | ||||||
| Min. – Max. | 19.0–75.0 | 22.0–75.0 | t=1.039 | 0.303 | ||
| Mean ± SD. | 49.09 ± 15.16 | 53.0 ± 15.42 | ||||
| Median (IQR) | 50.0 | 56.0 | ||||
| Splenomegaly | 8 | 24.2 | 18 | 54.5 | χ2=6.346 | 0.012* |
| Hepatomegaly | 8 | 24.2 | 17 | 51.5 | χ2=5.216 | 0.022* |
| Lymphadenopathy | 2 | 6.1 | 2 | 6.1 | χ2=0.0 | FEp=1.000 |
| CNS involvement | 0 | 0.0 | 2 | 6.1 | χ2=2.063 | FEp=0.492 |
| B.M BLASTS | ||||||
| Min. – Max. | 20.0–90.0 | 20.0–90.0 | U=461.0 | 0.278 | ||
| Mean ± SD. | 40.52 ± 23.24 | 44.91 ± 23.70 | ||||
| Median | 30.0 | 33.0 | ||||
| FAB | ||||||
| M1 | 3 | 9.1 | 5 | 15.2 | χ2=23.474 | MCp=0.001* |
| M2 | 8 | 24.2 | 5 | 15.2 | ||
| M3 | 11 | 33.3 | 0 | 0.0 | ||
| M4 | 6 | 18.2 | 5 | 15.2 | ||
| M5 | 3 | 9.1 | 8 | 24.2 | ||
| M6 | 2 | 6.1 | 3 | 9.1 | ||
| M7 | 0 | 0.0 | 7 | 21.2 | ||
| Overall survival (months) | ||||||
| Min. – Max. | 1.0–12.0 | 1.0–12.0 | U=261.50 | <0.001* | ||
| Mean ± SD. | 10.03 ± 3.45 | 5.88 ± 4.13 | ||||
| Median | 12.0 | 4.0 | ||||
| Outcome | ||||||
| Died | 7 | 21.2 | 22 | 66.7 | χ2=15.265 | MCp<0.001* |
| Alive | 23 | 69.7 | 8 | 24.2 | ||
| Lost FU | 3 | 9.1 | 3 | 9.1 | ||
| Cytogenetic criteria | ||||||
| Favorable | 24 | 72.7 | 9 | 27.3 | χ2=17.0 | <0.001* |
| Intermediate | 7 | 21.2 | 9 | 27.3 | ||
| Adverse | 2 | 6.1 | 15 | 45.4 | ||
| CR achievement | 23 | 69.7 | 8 | 24.2 | χ2=13.687 | <0.001* |
| TLC x 103 | ||||||
| Min. – Max. | 0.80–23.0 | 1.10–21.0 | U=538.50 | 0.939 | ||
| Mean ± SD. | 8.88 ± 6.60 | 8.95 ± 6.65 | ||||
| Median | 7.50 | 7.0 | ||||
| Hb gm/dl | ||||||
| Min. – Max. | 3.0–11.80 | 4.90–11.30 | t=0.411 | 0.682 | ||
| Mean ± SD. | 8.45 ± 1.86 | 8.26 ± 1.86 | ||||
| Median | 8.30 | 8.80 | ||||
| PLT x 103 | ||||||
| Min. – Max. | 17.0–120.0 | 10.0–150.0 | U=521.0 | 0.763 | ||
| Mean ± SD. | 55.61 ± 25.84 | 54.73 ± 32.83 | ||||
| Median | 55.0 | 56.0 | ||||
Notes: p, p-value for comparing between different parameters *statistically significant at p ≤ 0.05.
Abbreviations: χ2, chi-square test; MC, Monte Carlo; FE, Fisher Exact; t, Student’s t-test, U; Mann Whitney test.
Table 5 shows that patients with high expression of circ0075001 had considerably splenomegaly (63.6%) (P < 0.001), hepatomegaly (60.6%) (P < 0.001), less differentiated FAB subtypes M5 (27.3%), M7 (18.2%) (P = 0.001) (Figure 2E), lower overall survival with median 4 months (P < 0.001), the majority died through follow-up period (57.6%) (P = 0.004) and having adverse cytogenetic pattern (33.3%) (P = 0.005) (Figure 2F). Indeed, few patients achieved CR (24.2%) (P < 0.001) and display lower Hb values (P = 0.032). Whereas circ0075001 was downregulated in few patients with splenomegaly and hepatomegaly (15.2%), with more differentiated FAB subtypes as M2 (36.4%) and M3 (24.2%) (Figure 2E), higher overall survival (66.7% of patients were alive) with median 12 months. Moreover, most of them displayed favorable cytogenetic patterns (69.6%) (Figure 2F), achieved complete remission CR (69.7%), and obtained higher Hb values. The other parameters did not differ between high and low expression groups. As regard circFBXW7, it demonstrated no significant difference between low and high expression AML groups concerning all studied parameters (Table 6). About parameters affecting CR achievement determined by logistic regression analysis, the univariate analysis revealed that age P = 0.006 OR (CI) 0.950 (0.916–0.986), Hb P = 0.011, OR (CI) 1.490 (1.095–2.028), circANXA2 P < 0.001, OR (CI) 7.187 (2.420–21.347) and circ0075001 P < 0.001 OR (CI) 7.187 (2.420–21.347) can predict CR achievement whereas by multivariate analysis circANXA2 P = 0.024, OR (CI) 5.470 (1.250–23.947) can be an independent predictor of CR achievement (Table 7).
Table 5.
Relation Between Low and High Expression of Circ0075001expression with Clinical Data in AML Patients (N = 66)
| Circ0075001 Expression | Test of Sig. | p | ||||
|---|---|---|---|---|---|---|
| Low Median (<3.01) (n=33) | High Median (>3.01) (n=33) | |||||
| No. | % | No. | % | |||
| Sex | ||||||
| Male | 21 | 63.3 | 16 | 48.5 | χ2=1.538 | 0.215 |
| Female | 12 | 36.4 | 17 | 51.5 | ||
| Age (years) | ||||||
| Min. – Max. | 24.0–75.0 | 19.0–75.0 | t=1.236 | 0.221 | ||
| Mean ± SD. | 48.73 ± 13.17 | 53.36 ± 17.06 | ||||
| Median | 49.0 | 58.0 | ||||
| Splenomegaly | 5 | 15.2 | 21 | 63.6 | χ2=16.246 | <0.001* |
| Hepatomegaly | 5 | 15.2 | 20 | 60.6 | χ2=14.488 | <0.001* |
| Lymphadenopathy | 1 | 3.0 | 3 | 9.1 | χ2=1.065 | FEp=0.613 |
| CNS involvement | 0 | 0.0 | 2 | 6.1 | χ2=2.063 | FEp=0.492 |
| B.M BLASTS | ||||||
| Min. – Max. | 20.0–90.0 | 20.0–90.0 | U=522.50 | 0.775 | ||
| Mean ± SD. | 44.88 ± 24.77 | 40.55 ± 22.11 | ||||
| Median (IQR) | 32.0 | 30.0 | ||||
| FAB | ||||||
| M1 | 3 | 9.1 | 5 | 15.2 | χ2=22.144 | MCp=0.001* |
| M2 | 12 | 36.4 | 1 | 3.0 | ||
| M3 | 8 | 24.2 | 3 | 9.1 | ||
| M4 | 6 | 18.2 | 5 | 15.2 | ||
| M5 | 2 | 6.1 | 9 | 27.3 | ||
| M6 | 1 | 3.0 | 4 | 12.1 | ||
| M7 | 1 | 3.0 | 6 | 18.2 | ||
| Overall survival (months) | ||||||
| Min. – Max. | 1.0–12.0 | 1.0–12.0 | U=273.0 | <0.001* | ||
| Mean ± SD. | 10.0 ± 3.44 | 5.91 ± 4.17 | ||||
| Median | 12.0 | 4.0 | ||||
| Outcome | ||||||
| Died | 10 | 30.3 | 19 | 57.6 | χ2=10.693 | MCp=0.004* |
| Alive | 22 | 66.7 | 9 | 27.3 | ||
| Lost FU | 1 | 3.0 | 5 | 15.2 | ||
| Cytogenetic criteria | ||||||
| Favorable | 23 | 69.6 | 10 | 30.4 | χ2=10.25 | P=0.005* |
| Intermediate | 5 | 15.2 | 11 | 33.3 | ||
| Adverse | 5 | 15.2 | 12 | 33.3 | ||
| CR achievement | 23 | 69.7 | 8 | 24.2 | χ2=13.687 | <0.001* |
| TLC x 103 | ||||||
| Min. – Max. | 0.80–23.0 | 1.10–21.0 | U=519.50 | 0.748 | ||
| Mean ± SD. | 9.32 ± 6.93 | 8.51 ± 6.28 | ||||
| Median | 7.50 | 6.20 | ||||
| Hb gm/dl | ||||||
| Min. – Max. | 5.0–11.80 | 3.0–11.30 | t=2.197 | 0.032* | ||
| Mean ± SD. | 8.84 ± 1.54 | 7.87 ± 2.02 | ||||
| Median | 9.0 | 7.50 | ||||
| PLT x 103 | ||||||
| Min. – Max. | 17.0–150.0 | 10.0–100.0 | U=472.0 | 0.352 | ||
| Mean ± SD. | 58.97 ± 30.77 | 51.36 ± 27.74 | ||||
| Median | 55.0 | 50.0 | ||||
Notes: p, p-value for comparing between different parameters *statistically significant at p ≤ 0.05.
Abbreviations: χ2, chi-square test; MC, Monte Carlo; FE, Fisher Exact t: Student’s t-test; U, Mann Whitney test.
Table 6.
Relation Between Low and High Expression of circFBXW7 Expression with Clinical Data in AML Patients (N = 66)
| circFBXW7 Expression | Test of Sig. | p | ||||
|---|---|---|---|---|---|---|
| Low Median (<0.60) (n=33) | High Median (>0.60) (n=33) | |||||
| No. | % | No. | % | |||
| Sex | ||||||
| Male | 21 | 63.3 | 16 | 48.5 | χ2=1.538 | 0.215 |
| Female | 12 | 36.4 | 17 | 51.5 | ||
| Age (years) | ||||||
| Min. – Max. | 19.0–75.0 | 22.0–72.0 | t=0.973 | 0.334 | ||
| Mean ± SD. | 52.88 ± 15.60 | 49.21 ± 15.0 | ||||
| Median (IQR) | 54.0 | 50.0 | ||||
| Splenomegaly | 14 | 42.4 | 12 | 36.4 | χ2=0.254 | 0.614 |
| Hepatomegaly | 14 | 42.4 | 11 | 33.3 | χ2=0.580 | 0.447 |
| Lymphadenopathy | 4 | 12.1 | 0 | 0.0 | χ2=4.258 | FEp=0.114 |
| CNS involvement | 2 | 6.1 | 0 | 0.0 | χ2=2.063 | FEp=0.492 |
| B.M BLASTS | ||||||
| Min. – Max. | 20.0–90.0 | 20.0–90.0 | U=475.0 | 0.366 | ||
| Mean ± SD. | 39.09 ± 20.74 | 46.33 ± 25.59 | ||||
| Median | 30.0 | 30.0 | ||||
| FAB | ||||||
| M1 | 3 | 9.1 | 5 | 15.2 | χ2=2.844 | MCp=0.856 |
| M2 | 6 | 18.2 | 7 | 21.2 | ||
| M3 | 5 | 15.2 | 6 | 18.2 | ||
| M4 | 6 | 18.2 | 5 | 15.2 | ||
| M5 | 6 | 18.2 | 5 | 15.2 | ||
| M6 | 4 | 12.1 | 1 | 3.0 | ||
| M7 | 3 | 9.1 | 4 | 12.1 | ||
| Overall survival (months) | ||||||
| Min. – Max. | 1.0–12.0 | 1.0–12.0 | U=485.0 | 0.419 | ||
| Mean ± SD. | 8.45 ± 4.28 | 7.45 ± 4.36 | ||||
| Median (IQR) | 12.0 | 7.0 | ||||
| Outcome | ||||||
| Died | 14 | 42.4 | 15 | 45.5 | χ2=0.999 | MCp=0.672 |
| Alive | 17 | 51.5 | 14 | 42.4 | ||
| Lost FU | 2 | 6.1 | 4 | 12.1 | ||
| Cytogenetic criteria | ||||||
| Favorable | 17 | 51.5 | 16 | 48.5 | χ2=1.55 | 0.458 |
| Intermediate | 6 | 18.2 | 10 | 30.3 | ||
| Adverse | 10 | 30.3 | 7 | 21.2 | ||
| CR achievement | 15 | 45.5 | 16 | 48.5 | χ2=0.061 | 0.805 |
| TLC x 103 | ||||||
| Min. – Max. | 0.80–20.10 | 1.20–23.0 | U=539.0 | 0.944 | ||
| Mean ± SD. | 9.18 ± 6.83 | 8.65 ± 6.40 | ||||
| Median | 10.0 | 6.70 | ||||
| Hb gm/dl | ||||||
| Min. – Max. | 4.90–11.80 | 3.0–11.30 | t=0.624 | 0.535 | ||
| Mean ± SD. | 8.49 ± 1.63 | 8.21 ± 2.05 | ||||
| Median | 8.30 | 8.80 | ||||
| PLT x 103 | ||||||
| Min. – Max. | 10.0–120.0 | 10.0–150.0 | U=446.0 | 0.206 | ||
| Mean ± SD. | 50.24 ± 25.42 | 60.09 ± 32.40 | ||||
| Median | 50.0 | 66.0 | ||||
Abbreviations: χ2, chi-square test; MC, Monte Carlo; FE, Fisher Exact; t, Student’s t-test; U, Mann Whitney test p, p-value for comparing between different parameters.
Table 7.
Univariate and Multivariate Logistic Regression Analysis for the Parameters Affecting CR Achievement (31 Vs 35)
| Univariate | #Multivariate | |||
|---|---|---|---|---|
| p | OR (95%C.I) | p | OR (95%C.I) | |
| Female | 0.851 | 1.098(0.415–2.908) | ||
| Age (years) | 0.006* | 0.950(0.916–0.986) | 0.297 | 0.973 (0.924–1.024) |
| TLC | 0.590 | 0.980(0.909–1.055) | ||
| Hb | 0.011* | 1.490(1.095–2.028) | 0.095 | 1.469 (0.936–2.306) |
| PLT | 0.064 | 1.017(0.999–1.036) | ||
| CircANXA2 (Low <12.97) | <0.001* | 7.187 (2.420–21.347) | 0.024* | 5.470 (1.250–23.947) |
| Circ0075001 (Low <3.01) | <0.001* | 7.187 (2.420–21.347) | 0.537 | 1.629 (0.346–7.660) |
| CircFBXW7 (High >0.60) | 0.805 | 1.129 (0.429–2.971) | ||
Notes: #All variables with p<0.05 was included in the multivariate. *Statistically significant at p ≤ 0.05.
Abbreviations: OR, odds ratio; C.I, confidence interval; LL, lower limit; UL, upper limit.
Discussion
Acute myeloid leukemia (AML) is a myeloid malignancy in the bone marrow provoked by the oncogenic transformation of hematopoietic progenitors.26 AML is the second prevalent illness amongst hematologic cancers and the widespread disease in adult myeloid neoplasms. Advanced genomics studies stated that AML is recognized by its pathogenic heterogeneity and concurrent modifications in numerous genes.27
CircRNAs include many microRNA response elements (MREs) that absorb and restrain the action of miRNAs and further, disturb the expression of the downstream target mRNAs, and this role of circRNA is termed miRNA sponge.28
Furthermore, CircRNAs behave like oncogenes or tumor suppressors to contribute to the event of manifold tumors and could be innovative diagnostic and prognostic biomarkers.29 The disparity of expression and role of circRNAs in different cancers have been recognized.30 Recently, incipient studies proposed that circRNAs have prominent evidence in the tumorigenesis and progress of hematological neoplasms.31,32 Furthermore, circRNAs are vital in iron metabolism and N6-methyladenosine (m6A) alteration.33,34 There are particular expressions of circRNAs in various types of hematological illnesses. Currently, different circRNAs have been recognized as prognostic models, like circ0003602, circ0005571, circ0001857, circ0007609, circ0012152, circ0074371and circ0001247.35,36 Circ-Foxo3, circ-RPS6KB1, circ-CSMD2, circ-ANXA2, circ-RBM5, circ-PWP2, circ-ZZEF1, circ-GSK3B, and circ-FOXP1 could distinguish AML patients from healthy subjects by roccurve analysis.32,37
This current study was the first one conducted to investigate circRNAs in AML in Egyptian populations. It was carried out on 120 subjects categorized into 66 AML patients and 54 age and sex-matched healthy controls. We investigated circANXA2, circ0075001, and circFBXW7 aiming to explore their deregulated expressions in AML and their relations with prognostic indices including FAB, ELN 2017 classifications, CR (complete remission), and overall survival. We detected noteworthy results as we noticed marked upregulation of circANAX2 in patients than in control. Plotting ROC curve revealed that circANXA2 with AUC 0.824 can significantly discriminate AML patients from healthy controls, P < 0.001. Huang et al emphasized that CircANXA2 augments the transcription of its parental gene ANXA2, which induces fibrinolysis and tumor cell infiltration and restricts the sensitivity of tumor cells to chemotherapy, thus it reduces survival in AML patients.38
As regard relation with prognostic indices, the results demonstrated that high expression of circANXA2 had significant relation with splenomegaly, hepatomegaly, FAB subtypes (M5, M7), shorter overall survival, adverse cytogenetic pattern, and lower CR achievement in comparison to low expression group that related significantly with FAB subtypes (M3, M2), higher overall survival, favorable cytogenetic pattern, and higher CR achievement. Our results coincide with Ding and colleagues, who validated that circANXA2 had a role as a biomarker for predicting the development, progression, and prognosis of AML.37 They added that circANXA2 overexpression was linked to shorter event-free survival (EFS) and overall survival (OS) of AML patients, whereas lower levels of circ-ANAX2 were associated with CR achievement, longer EFS, and OS.37 Ganesan and Seo and their colleagues revealed in their studies that ANXA2 knockdown diminished the proliferation and provoked the apoptosis of the AML cells. Meanwhile, enhanced their chemosensitivity to cytarabine and daunorubicin. CircANXA2 functions as a microRNA sponge that dampens the activity of MiR-23a-5p and miR503-3p, which are known as chemosensitivity enhancers and cancer stem cell proliferation inhibitors.39,40 Notably, Niu and colleagues depicted that ANXA2 exerts a poor prognostic effect on adult AML but a favorable prognostic effect on pediatric AML.41
Regarding circ0075001, we noticed marked up-regulation of patients than control. ROC curve revealed that circ0075001 with AUC 0.855, discriminate AML patients from controls, P < 0.001. Additionally, we found out that circ0075001expression trended to increase in patients with splenomegaly, hepatomegaly FAB subtypes (M5, M7), shorter overall survival, adverse cytogenetic pattern, and lower CR achievement compared with low expression group, which was associated significantly with FAB subtypes (M3, M2), higher overall survival, favorable cytogenetic pattern, and higher CR achievement. Bock et al reported that Circ0075001 was overexpressed in AML subtypes M0 or M1 and show low expression in M2, M4 and M5, thus it can impact AML differentiation pattern.42 Circ0075001 expression was closely related to total NPM1 expression and knockdown of the genes associated with the TLR signaling pathway, which influence the survival of AML cells.42 Furthermore, Brunetti et al elucidated that Circ0075001 was related to the total NPM1 expression in AML. The latter encoded a multifunctional chaperone protein concerned with ribosomal biogenesis, apoptosis, and cell proliferation.43 Concerning circFBXW7, it was extensively downregulated in AML patients. ROC curve with AUC 0.826 shows the considerable difference between patients and controls (P < 0.001).
Papaioannou and colleagues explained that depletion of circFBXW7 affected the phenotype of the AML cell lines and led to an increase in their proliferative capacity, so circFBXW7 is suggested to act as a tumor suppressor gene. Meanwhile, it is related to the positive regulation of signal transduction and leukocyte differentiation. Whereas its lower expression was linked to hematopoiesis and positive regulation of RNA biosynthesis.44
The current study went into the parameters affecting CR achievement. Univariate analysis revealed that age, Hb, circANXA2, and circ0075001 overexpression can predict CR achievement. Meanwhile, multivariate analysis revealed that circANXA2 overexpression can be an independent predictor of CR achievement. It was obvious from previous studies that deregulated circRNAs expressions disturbed the transcription of their parental genes (oncogenes or tumor suppressor genes), which provoked/restricted the aberrant gene expression and the malignant transformation of hematopoietic cells, followed by higher/lower AML risk. Moreover, they could affect carcinogenesis by competitively binding to their target microRNAs. Shang et al added in a study conducted by a high throughput circRNA microarray that the expression of 49 circles differs significantly between naive AML cells and those resistant to doxorubicin cells. This indicates that circRNA was involved in AML drug resistance.30
Finally, CircRNA is crucial in the ncRNA network, which has been recognized as a vital regulator of different cancers. Meanwhile, circRNAs have a fundamental role in anticancer drug resistance. The recent studies on circRNAs ranging from traditional chemotherapeutic drugs to target and immunotherapeutic drugs will develop their clinical potential and provide a research hotspot.45
Conclusion
The current study supports the implication of circRNAs as potential diagnostic and prognostic biomarkers of AML based on prominent relations of overexpression of circANXA2 and circ0075001 with FAB subtypes (M5, M7), shorter overall survival and adverse cytogenetic pattern and lower CR achievement. Moreover CircANXA2 was an independent predictor of CR achievement that could be of value in determining treatment regimen and of novel therapeutic target in AML.
Acknowledgment
We acknowledge all members of central laboratory unit, Faculty of Medicine, Menoufia University for providing all laboratory materials and instruments and moreover their support to accomplish this work.
Funding Statement
There is no funding to report.
Data Sharing Statement
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Ethical Approval and Consent to Participate
The study approval was attained from the Research Ethics Committee at Menoufia Faculty of Medicine with accord to Helsinki Declaration and regulations. Written informed consent was obtained from all participants and from the child participants’ parents.
Consent for Publication
Consent to publish has been received from all co-authors and official institute.
Author Contributions
All authors contributed to the study conception, design, material preparation, the data collection, analysis and interpretation. The first draft of the manuscript was written by all authors and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript and agreed for publication in the journal to which the manuscript has been submitted and agreed to take responsibility and be accountable for the contents of the article.
Disclosure
The authors report no conflicts of interest in relation to this work.
References
- 1.Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31(1):63–76. [DOI] [PubMed] [Google Scholar]
- 2.Czerw T, Labopin M, Gorin NC, et al. Long-term follow-up of patients with acute myeloid leukemia surviving and free of disease recurrence for at least 2 years after autologous stem cell transplantation: a report from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation. Cancer. 2016;122(12):1880–1887. [DOI] [PubMed] [Google Scholar]
- 3.Kantarjian H, Kadia T, DiNardo C, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 2021;11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Pan J, Huang S, et al. Development and validation of a novel circular RNA as an independent prognostic factor in acute myeloid leukemia. BMC Med. 2021;19(1):28. doi: 10.1186/s12916-020-01898-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angenendt L, Röllig C, Montesinos P, et al. Chromosomal Abnormalities and Prognosis in NPM1-Mutated Acute Myeloid Leukemia: a Pooled Analysis of Individual Patient Data from Nine International Cohorts. J Clin Oncol. 2019;37(29):2632–2642. doi: 10.1200/JCO.19.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard-Carpentier G, DiNardo CD. Single-agent and combination biologics in acute myeloid leukemia. Hematol Am Soc Hematol Educ Program. 2019;1:548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pradeep C, Nandan D, Das AA, Velayutham D. Comparative transcriptome profiling of disruptive technology, single-molecule direct RNA sequencing. Curr.Bioinf. 2020;15:165–172. [Google Scholar]
- 9.Jiao S, Wu S, Huang S, Liu M, Gao B. Advances in the Identification of Circular RNAs and Research Into circRNAs in Human Diseases. Front Genet. 2021;12:665233. doi: 10.3389/fgene.2021.665233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao T, Hu Y, Cheng L. Deep-DRM: a computational method for identifying disease-related metabolites based on graph deep learning approaches. Brief Bioinform. 2021;22(4):548. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):w1. [DOI] [PubMed] [Google Scholar]
- 13.Qian L, Yu S, Chen Z, et al. The emerging role of circRNAs and their clinical significance in human cancers. Biochimica et Biophysica Acta. 2018;1870(2):247–260. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Yang L, Chen L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. [DOI] [PubMed] [Google Scholar]
- 15.Cheng D, Wang J, Dong Z, et al. Cancer-related circular RNA: diverse biological functions. Cancer Cell Int. 2021;21:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang G, Liu Z, Tan L, et al. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in a hypoxic environment. Anticancer Res. 2017;37(8):4337–4343. [DOI] [PubMed] [Google Scholar]
- 17.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Huang Z, Xu Y, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7(1):5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach D, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S. The characteristics of circRNA as competing endogenous RNA in the pathogenesis of acute myeloid leukemia. BMC Cancer. 2021;21(1):277. doi: 10.1186/s12885-021-08029-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
- 22.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adult patients: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiernik PH, Banks PL. Cytarabine plus idarubicin or daunorubicin as induction and consolidation therapy for previously untreated adult patients with acute myeloid leukemia. Blood. 1992;79(2):313–319. [PubMed] [Google Scholar]
- 24.Tallman MS, Nabhan C, Feusner JH, Rowe JM. Acute promyelocytic leukemia: evolving therapeutic strategies. Blood. 2002;99(3):759. [DOI] [PubMed] [Google Scholar]
- 25.Sekeres MA, Lancet JE, Wood BL, et al. Randomized phase IIb study of low-dose cytarabine and lintuzumab versus low-dose cytarabine and placebo in older adults with untreated acute myeloid leukemia. Haematologica. 2013;98(1):119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, Koh Y, Hong J, Eom HS, Yoon SS. Recent Clinical Update of Acute Myeloid Leukemia: focus on Epigenetic Therapies. J Korean Med Sci. 2021;36(13):e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, Mechanism, and Function in Human Cancers. Int J Mol Sci. 2019;20:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359–365. doi: 10.1136/med [DOI] [PubMed] [Google Scholar]
- 30.Shang J, Chen WM, Liu S, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through the regulation of autophagy. Leuk Res. 2019;85:106198. doi: 10.1016/j.leukres.2019.106198 [DOI] [PubMed] [Google Scholar]
- 31.Che H, Ding H, Jia X. circ_0080145 Enhances imatinib resistance of chronic myeloid leukemia by regulating miR-326/PPFIA1 axis. Cancer Biother Radiopharm. 2020;1:548. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72. doi: 10.1038/s41420-020-00306-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YG, Chen R, Ahmad S, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Su Y, Wang S, et al. Identification of circRNA-lncRNA-miRNA-mRNA Competitive Endogenous RNA Network as Novel Prognostic Markers for Acute Myeloid Leukemia. Genes. 2020;11(8):548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo S, Li B, Chen Y, et al. Hsa_circ_0012152 and Hsa_circ_0001857 Accurately Discriminate Acute Lymphoblastic Leukemia from Acute Myeloid Leukemia. Front Oncol. 2020;10:1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Zhou LY, Tang X, et al. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to a better prognosis in acute myeloid leukemia patients. BMC Cancer. 2019;19(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Y, Dong Y, Lu H, et al. Circular RNA profile of acute myeloid leukemia indicates circular RNA annexin A2 as a potential biomarker and therapeutic target for acute myeloid leukemia. Am J Transl Res. 2020;12(5):1683–1699. [PMC free article] [PubMed] [Google Scholar]
- 38.Huang D, Yang Y, Sun J, et al. Annexin A2- S100A10 heterotetramer is upregulated by PML/RARalpha fusion protein and promotes plasminogen-dependent fibrinolysis and matrix invasion in acute promyelocytic leukemia. Front Med. 2017;11:410–422. [DOI] [PubMed] [Google Scholar]
- 39.Ganesan S, Palani HK, Lakshmanan V, et al. Stromal cells downregulate miR-23a-5p to activate protective autophagy in acute myeloid leukemia. Cell Death Dis. 2019;10:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo M, Kim SM, Woo EY, et al. Stemness-attenuating miR-503-3p as a paracrine factor to regulate the growth of cancer stem cells. Stem Cells Int. 2018;2018:4851949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu Y, Yang X, Chen Y, et al. Distinct prognostic values of Annexin family members expression in acute myeloid leukemia. Clin Transl Oncol. 2019;21(9):1186–1196. [DOI] [PubMed] [Google Scholar]
- 42.Bock S, Murgueitio MS, Wolber G, et al. Acute myeloid leukemia-derived Langerhans-like cells enhance Th1 polarization upon TLR2 engagement. Pharmacol Res. 2016;105:44–53. [DOI] [PubMed] [Google Scholar]
- 43.Brunetti L, Gundry MC, Goodell MA. New insights into the biology of acute myeloid leukemia with mutated NPM1. Int J Hematol. 2019;110(2):150–160. doi: 10.1007/s12185-018-02578-7 [DOI] [PubMed] [Google Scholar]
- 44.Papaioannou D, Volinia S, Nicolet D, et al. Clinical and functional significance of circular RNAs in cytogenetically normal AML. Blood Adv. 2020;4(2):239–251. doi: 10.1182/bloodadvances.2019000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]