Summary
Digital health solutions, with apps, virtual care, and electronic medical records, are gaining momentum across all medical disciplines, and their adoption has been accelerated, in part, by the COVID-19 pandemic. Personal wearables, sensors, and mobile technologies are increasingly being used to identify health risks and assist in diagnosis, treatment, and monitoring of health and disease. Genomics is a vanguard of digital healthcare as we witness a convergence of the fields of genomic and digital medicine. Spurred by the acute need to increase health literacy, empower patients’ preference-sensitive decisions, or integrate vast amounts of complex genomic data into the clinical workflow, there has been an emergence of digital support tools in genomics-enabled care. We present three use cases that demonstrate the application of these converging technologies: digital genomics decision support tools, conversational chatbots to scale the genetic counseling process, and the digital delivery of comprehensive genetic services. These digital solutions are important to facilitate patient-centered care delivery, improve patient outcomes, and increase healthcare efficiencies in genomic medicine. Yet the development of these innovative digital genomic technologies also reveals strategic challenges that need to be addressed before genomic digital health can be broadly adopted. Alongside key evidentiary gaps in clinical and cost-effectiveness, there is a paucity of clinical guidelines, policy, and regulatory frameworks that incorporate digital health. We propose a research agenda, guided by learning healthcare systems, to realize the vision of digital health-enabled genomics to ensure its sustainable and equitable deployment in clinical care.
The current landscape for digital health-enabled genomics
Digital health is gaining momentum across all medical disciplines and has been accelerated, in part, by the COVID-19 pandemic. Digital health apps, virtual care, personal wearables, sensors, and mobile technologies are increasingly being used to identify health risks and assist with diagnosis, treatment, and monitoring of health and disease across cardiology, internal medicine, and infectious disease.
In many respects, genomics is a vanguard of digital healthcare, as we witness a convergence of the fields of genomic and digital medicine. Spurred by the acute need to increase health literacy, empower patients’ preference-sensitive decisions, fill critical genetics workforce shortages, or integrate vast amounts of complex genomic data into the clinical workflow, there has been an emergence of digital tools in genomic medicine. We review the current landscape and evidence base as it pertains to our use cases of digital support tools, chatbots, portals, and platforms in genomics-enabled care.1
Evidence of the value of digital health-enabled genomics is beginning to emerge. Current evidence on patient experience suggests a relatively high level of acceptability of pre-test counseling tools from a range of contexts, with majorities of patients recommending and reporting high levels of satisfaction with these tools.2, 3, 4, 5 While patient-centered care is important, it is not often used to support reimbursement by healthcare insurance providers. Instead, clinical outcomes and service efficiency are often considered the gold-standard for coverage decisions. The evidence is mixed: a review of interactive e-counselling tools for genetic pre-test decisions found that across most studies, participants who used the e-counselling tools experienced equivalent or lower decisional conflict, distress, or anxiety as compared to those who did not use a digital tool.2,6, 7, 8, 9 Tool users who were initially undecided about taking a test or receiving cancer treatment were also more likely to reach a decision compared to those who did not use digital tools.2,7 Further, a number of studies reported that knowledge acquisition about genetics was equivalent or greater in users of digital tools as compared to the control cohort.2,6,10, 11, 12, 13, 14, 15, 16 There are limited data about service efficiency, but most studies indicate that participants that used digital tools spend 10–40 min less with clinicians as compared to participants who received standard pre-test counseling.3,9,14
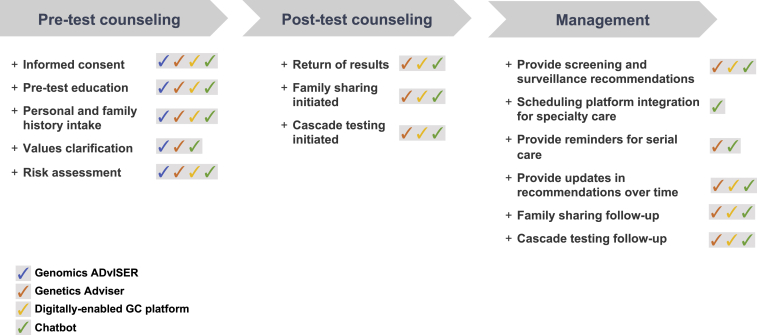
Here, we present three use cases that demonstrate the potential opportunity of these converging technologies: digital genomics decision support tools, conversational chatbots to scale the genetic counseling (GC) process, and the digital delivery of comprehensive genetic services. These use cases highlight leading digital innovations developed to address clinical needs and challenges that arise across the continuum of care in genomics—from pre-test and post-testing counseling to the full spectrum of genomic service delivery (Figure 1)—including digital decision aids, chatbots, and digital portals.
Figure 1.
Digital technologies across the clinical genomics pathway
Digital genomics decision support tools
One of the fundamental functions of genomic counseling is to provide patient-centered decisional support for genetic testing; decision support tools advance this goal.
Digital decision aids have been increasingly used to support pre-test counseling, education, and decision making in light of the workforce shortages in genomics. Decision support tools are best suited to meet this challenge because they improve knowledge,2,12,13 support shared decision making,5 promote decisions congruent with patients’ values, and reduce decisional conflict.17 Better decision quality17 translates into improved health outcomes18,19 and patient-centered care.20 The Genomics ADvISER provides a leading example of a digital decision aid.
The Genomics ADvISER is a freely available web-based interactive decision aid to guide patients’ selection of secondary findings (SFs).21 Based on the Ottawa Decision Support Framework,22 this tool is intended for adult patients and/or for parents of pediatric patients to promote their involvement in clinical decision making about which of their SFs they want to learn when they have their genomes sequenced. All clinically relevant SFs are included: medically actionable and non-medically actionable conditions, pharmacogenomic variants, polygenic risks/SNPs, and carrier results.23,24 The Genomics ADvISER consists of an educational whiteboard video that reviews key concepts of genomic sequencing and the risks and benefits of learning SFs. The video is followed by a values-clarification exercise to obtain feedback about the preferences of the user and a knowledge quiz to reinforce key concepts (with correct answers provided afterwards). The tool ends by asking participants to select among five categories of SFs, consistent with frameworks that “bin” SFs into categories based on medical actionability and potential distress.25, 26, 27, 28
The Genomics ADvISER was found to be highly acceptable21 and effective9 and enhanced patient-centered care in the delivery of genomic counseling.20 The prototype was evaluated in a usability study with adult cancer patients. It demonstrated strong face validity, acceptability, and high content comprehension.21 Patients found the length and amount of information “just right,” clear, and balanced. All participants liked the educational video, felt that the prototype provided enough information to make a decision, and would recommend the tool to others. The Genomics ADvISER was also found to provide quality patient-centered care through a secondary analysis of pre-test GC sessions that were conducted over the course of a randomized controlled trial evaluating the effectiveness of the Genomics ADvISER with adult cancer patients. In the delivery of genomic counseling, the Genomics ADvISER contributed to enhancing counseling by (1) promoting informed dialogue, (2) facilitating preference-sensitive deliberation, and (3) deepening personalization of decisions, all of which represent fundamental principles of patient-centered care: providing clear, high-quality information; respecting patients' values, preferences, and expressed needs; and providing emotional support.20 In a subsequent clinical trial, the intervention group (using the Genomics ADvISER) had higher knowledge scores (mean difference: 0.39, 95% CI 0.18–0.59; p < 0.001) and spent less time talking to a genetic counselor (mean difference: 24.40 min, 95% CI 27.72–21.07; p < 0.001),9 suggesting that the Genomics ADvISER is an effective educational tool, reducing in-clinic time and, potentially, costs.
As part of the trial, a qualitative study identified patient profiles—hypothetical representations of target users—based on shared characteristics, preferences, and/or attitudes in an effort to tailor information to match the needs of specific clinical populations. Five profiles were identified:29 “Information enthusiasts” self-identified as “planners” and valued learning most or all SFs to enable planning and disease prevention. “Concerned individuals” were reluctant to learn SFs, anticipating negative psychological impacts from SFs. “Contemplators” weighed health benefits with the impacts of not being able to “un-know” information. “Individuals of advanced life stage” primarily considered their implications for family members. “Reassurance seekers” were reassured by previous negative genetic test results, which shaped their expectations for receiving no SFs. These profiles can help target counseling and inform other digital tools aimed at tailoring support by providing a framework to address common values, concerns, and misconceptions.
Overall, the evidence demonstrated that the Genomics ADvISER digital tool enhanced patient-centered care and was an effective and potentially cost-effective tool for genomic counseling. However, some key lessons and potential pitfalls were also revealed through the evaluative studies. For example, despite the enthusiasm and recommendation the Genomics ADvISER received, some patients still wanted to speak with a clinician to check their decisions,21 indicating a need for further decision support structures. Others wanted more robust decisional support features, such as the ability to provide personalized feedback tailored to their preferences and characteristics. Incorporating patient profiles up front can make the user experience more patient centered. Further, building in conversational entities, like chatbots, to answer questions in real time can be used to simulate a more interactive user experience. There was a desire to make the tool more adaptable so that clinicians could turn on or turn off certain SF categories to align the options with their specific lab and clinical capabilities. These lessons learned provided opportunities to iterate and improve the tool. The Genomics ADvISER has been transformed into the "Genetics Adviser," an interactive, patient-centered digital interface that integrates the genetics service pathway from pre-test, waiting period, and post-test return of results, providing continuity of care that is fully adaptable to any testing platform, population, indication, and lab or clinical setting (Figure 1) (www.geneticsadviser.com). Although the original Genomics ADvISER significantly reduced counseling times without compromising patient-centered care, it is unclear whether these benefits translate to downstream healthcare cost savings, reduced wait times, and improved accessibility and uptake of genetics services. Research is underway to examine service efficiencies, accessibility, and equity by evaluating whether the Genetics Adviser increases access and addresses literacy and a diversity of ethnocultural perspectives. Further outcomes research is needed to provide evidence of utility and cost-effectiveness for adoption, as described in the research agenda below.
Conversational chatbots to scale the genetic counseling process
Diverse and innovative service-delivery models have been created and implemented to increase patient access to genetics expertise and to increase the overall efficiency of GC practice. One of the relatively newer innovations that has been developed and tested is that of chatbots.
Chatbots are technology-based, simulated conversational tools used in scaling communications. Some chatbots use artificial intelligence (AI) and natural language processing to answer simple questions, increase and maintain consumer engagement, promote products and services, and provide convenient, easy access between consumers and service providers.30 Multiple industries use chatbots, including banking, insurance, retail, airlines, and hotels, among others.31 Chatbots have also been used in healthcare to promote fertility education and preconception health,32 collect and triage symptoms,33 provide cognitive behavioral therapy,34 promote weight loss and other health behaviors related to diabetes prevention,35 and educate teens on risky health behaviors.36 These conversational digital tools have specific benefits, including deploying by link, with no app download required, and work on multiple device types. They can also be personalized to the end user, with name, pronoun(s), speed of messaging, and specific health and medical information such as genetic risk variant. Back-end analytics can also be accessed to measure utilization, and chatbots can also be integrated into the electronic health record (EHR) for both automation and documentation.
In genetics service delivery, chatbots are being developed and utilized to collect family health history,37 identify individuals at increased risk for hereditary cancer syndromes,38,39 provide education about genetic conditions,40 and support informed decision-making regarding receipt of secondary genomic findings,41 among other use cases. An overarching goal of these chatbots is to free genetic counselors’ and GC assistants’ time for higher-level patient care duties by having the chatbots serve as virtual GC assistants.
In the MyCode Community Health Initiative’s Genomic Screening and Counseling Program (MyCode GSC) at Geisinger, actionable genetic results are returned to patient-participants, with >2,700 results returned to date.42 There are multiple touchpoints with these individuals, which has historically required extensive time and effort on behalf of all of the staff and healthcare providers for the program. To increase efficiency and potentially decrease cost, the GSC program leadership decided to collaborate and co-develop chatbots with Clear Genetics, since acquired by Invitae. Clear Genetics developed “Gia,” the Genetic Information Assistant. The first use cases the team developed were MyCode consent, a family sharing tool (FST) and cascade testing chatbot, and a one-month follow-up chatbot. In addition to developing these first use cases, MyCode genetic counselors also worked with Clear Genetics to build a library of responses for a “SMART FAQ” feature. This AI technology enables chatbots to reply to user-entered questions from a library of programmed responses that learns over time. If the question is new or doesn’t match with a response with high certainty, it is forwarded to the MyCode team. There are also scripted questions and answers throughout the bots in addition to these SMART FAQs.
Diverse stakeholders were engaged in the collaborative development of these chatbots. At Geisinger, the team interfaced with the Information Security Office, legal teams, IT, the IRB, and the in-house digital team to ensure “best practice” standards were followed in the build and deployment of digital health technology. Input was sought from the GC and medical geneticist teams, Ethics Advisory Council, and front-line care providers, as well as two patient advisory boards. In collaboration with Clear Genetics, initial pilot testing was done via usertesting.com. Amazon Mechanical Turk comprehension testing of the consent bot was also conducted. Sixty-two individuals in MyCode participated in six total focus groups to explore acceptability, usability, functionality, and understanding of the initial chatbot use cases.43
MyCode GSC initiated clinical deployment of the chatbots and began obtaining consent from individuals to receive “electronic communications” in August 2018. To date, approximately 60% of patients receiving results from the MyCode GSC Program have opted to utilize the bots. Since Geisinger provides care to individuals located in a largely rural area in central Pennsylvania, and since the median age of individuals receiving results is 63 years, the uptake in this relatively older, rural population is encouraging. Most patients prefer to access the chatbots via the EHR portal (52.1%), followed by e-mail (28.6%), and then text (19.3%). EHR integration includes populating the chatbot with certain patient demographics, genetic variants, and disease risk, as well as automated import of the patient’s chat transcript into their chart (Figure 1).
The engagement with the FST, cascade chatbot, and follow-up bots have also been evaluated. Preliminary data indicate that individuals who consented to be sent the FST generated more completed cascade tests in their families than those who declined this tool. As part of the NHLBI-funded R01 IMPACT-FH (Identification Methods, Patient Activation, and Cascade Testing for Familial Hypercholesterolemia), dyadic interviews and surveys have been conducted with individuals and their family members with FH to optimize the FST and cascade chatbot for use in a prospective, pragmatic trial to determine whether an optimized chatbot improved based on patient input can increase cascade genetic testing for FH.44 Enhancements have also been made to our follow-up chatbot with the goal of increasing engagement, including shortening the bot overall while adding new response language for common barriers that patients have indicated to us keep them from scheduling GC (i.e., plans to follow-up with doctor instead, no time, can’t afford screening and visits, anxious/scared, result isn’t important to me, other health issues to focus on, etc.). In response to these indicated barriers, Gia now provides a scripted response whose goal is to help the individual understand why scheduling a GC appointment could still be beneficial to them and their family.
Overall, initial data indicate chatbots may have utility across many aspects of genomics service delivery, including use cases to augment GC.43,45 Multiple additional potential use cases exist, including point-of-care triage based on testing and/or referral criteria for a wider array of hereditary conditions, pre-visit data collection and risk assessment, informed consent, results return, and the provision of longitudinal surveillance recommendations, among others. However, in the development and deployment of chatbots in the practice of GC, there are multiple issues to consider. Previous chatbot service research has identified potential concerns regarding accuracy, cyber-security, and the inability of AI-led services to empathize, with acceptability correlated negatively with poorer IT skills.46
In addition, what impact will chatbots have on practice in both positive and, potentially, negative ways? Do genetic counselors have angst about being replaced by bots? Is there wariness regarding certain use cases, such as return of genomic risk results? How might the use of chatbots impact the patient-provider relationship? Further, what languages are available? Will chatbots be accessible to underserved and underrepresented populations? Patient-centered user design, incorporation of health communication theories, ongoing iterations based on user feedback, and effectiveness and implementation research are necessary to ensure the development of chatbots accessible to broad, diverse populations and effective in facilitating genomics service delivery and GC, as described in the research agenda below.
Digital delivery of comprehensive genetic services
“Genome-first” testing, wherein a clinical genetic test may be deployed as a screening tool to identify otherwise healthy individuals who are at increased risk for specific hereditary conditions, has necessitated a different model for the generation and delivery of clinical genetic test results. Specifically, the process of returning genetic results by a genetic counselor, or trained care provider, needs to be robust and efficient in order to scale with the demand.
Previous analyses of time-based effort have determined that clinical genetic services are time consuming and labor intensive, with as little as 25%–41% of a genetic counselor’s time spent on direct patient care and up to 3.5–7 total hours spent per client in certain types of clinical encounters.47, 48, 49, 50, 51 Since 2015, Color Health has implemented a digitally enabled novel service-delivery model that utilizes a software and technology platform to deliver streamlined and efficient GC services to its clinical genetic-testing patients. The platform includes an online health history collection tool, an automated pedigree creation tool, an automated risk model calculation tool, online scheduling and rescheduling tools, and templatized summary notes. This service model and platform has been used to deliver results to >250,000 patients, has been utilized within health system populations52 and family member cascade testing programs,53 and will be leveraged to deliver the GC services for the All of Us Research Program (https://grantome.com/grant/NIH/OT2-OD028251-01).
Implementation assessment survey findings of 70 primary care providers and over 1,600 patients in one health system revealed that both providers and patients found value in utility in returning/receiving results via this type of testing model.54,55 Through this platform, genetic counselors spend much less time than industry averages conducting non-direct patient care activities—such as patient scheduling, clinical history intake, pedigree generation, and session note logging—thus increasing the proportion of time spent on direct care. The Color digitally enabled GC platform implements custom software tools to create time savings. As part of this platform, one major area of time savings is the replacement of the pre-test GC session with a series of web-based educational modules and a pre-test educational video that cover many of these concepts in a self-paced way. In addition, a novel protocol was implemented through the platform, in which telephone-based GC is offered as an optional service to individuals who receive negative clinical genetic results, but in which GC is required for individuals who receive a positive genetic result.
In order to quantify the time-savings of implementing this digital-tool-enhanced GC service, genetic counselors tracked time spent conducting direct and non-direct patient care activities over a period of 8 months. During this time, the team conducted over 1,800 post-test telephone GC sessions for patients who received a multi-gene panel for hereditary cancer risk. More than 50% of the genetic counselors’ time spent with a patient was used to provide direct care—the average total time spent per patient was 37 minutes. In order to quantify the impact of these sessions, post-counseling surveys were deployed to assess patient comprehension and satisfaction with their GC sessions. Overall, satisfaction with the GC, as delivered through the digitally enabled GC platform, was high for individuals who received both a positive (4.8 out of 5) and negative result (4.9 out of 5). Responses ranged from 4.9 to 5.0 for questions that addressed patient comprehension of their results. In addition, the use of a digital scheduling tool achieved a missed-appointment rate below the industry average. One study of failed-appointment rates, including no-shows and cancellations, at genetics clinics found the average failed-appointment rate to be 12%.56 The rate of missed appointments measured in this study was 3.1%. Allowing patients to schedule and reschedule appointments, as well as the flexibility inherent to telephone-based counseling, likely contributed to this low no-show rate. Broader implementation of similar software tools for all genetic counselors providing clinical care may improve efficiency and time available for both direct and non-direct patient care.
It should be noted that this study was conducted as a retrospective description of the processes and practices utilized by genetic counselors in a commercial setting. As such, no direct comparative analyses of time savings were possible. This digitally enabled GC platform has been used to deliver genetic testing results to patients, largely in patient cohorts that are composed mostly of healthy individuals. Further, it should be noted that a more in-depth series of follow-up GC encounters are necessary for patients who are found to carry a pathogenic or likely pathogenic variant, which is not delivered through this platform. These follow-up sessions often need to be traditional in-person appointments, since there are additional tests and screens that must be performed for such patients. As such, this digitally enabled GC platform is less well suited for the delivery of GC in high-risk clinical settings, where many patients are likely to be found to carry a pathogenic or likely pathogenic variant. Indeed, one of the biggest gaps that must be closed for digitally enabled GC delivery is how to ensure that patients who carry an actionable variant are actually seen in-person for their follow-up care and are not lost to follow-up. This is an area that is currently not addressed through the platform.
The digital GC platform has aided the return of GC results to many patients; however, additional studies are necessary to understand the long-term health outcomes and efficacy of GC delivery in this fashion. In addition, studies are needed to understand the cost-effectiveness of this type of delivery. Finally, to ensure that digital platform-enabled GC is able to serve a diverse demographic population equitably, translation of the digital experience and availability of real-time medical translation for the GC sessions should be implemented. Non-digital workflows should also be made available to ensure equitable delivery of services for individuals with lower digital literacy. As described in the research agenda below, user-centered design with a focus toward equitable access is an important factor towards ensuring adoption across a diverse population.
A research agenda for digital health-enabled genomics
The case studies above highlight key gaps in the current path for digital health technologies to enable genomic medicine. Here we highlight the imperative for a strategic approach to the development and delivery of these highly specialized digital health tools. To become mainstream in the application of genomics to medicine, a robust digital health research agenda is necessary across the translational continuum to demonstrate effectiveness and fill knowledge gaps. For optimal implementation and adoption of digital health tools in genomic medicine, these technologies will need to be supported by the payer community. In concert with technical, clinical, and data science considerations, a policy agenda that considers the challenges from participants to health systems will be a critical success factor (Box 1).
Box 1. A research agenda for digital health-enabled genomics.
-
•
Design: Use biodesign principles in developing research programs—understand the problem we are trying to solve
-
•
Clinical: Utilize implementation science principles to guide digital health tool integration and evaluation in healthcare systems
-
•
Data science: Apply FAIR principles to the data; ensure that technology is validated against an acceptable clinical standard
-
•
Policy: Conduct outcomes research to establish utility and economic justification for adoption; understand what safeguards on privacy and cybersecurity should be deployed to assure trust
-
•
Equity: To close the digital divide, address access, literacy, awareness, ethnocultural, and knowledge gaps in the application of digital health technologies
Design
Digital technology developers often ideate in the absence of the end user. We recommend that biodesign principles be rigorously applied as product developers consider the technology by asking up front: "what is the problem we are trying to solve?" Needs-driven approaches are vital, and early engagement of the consumer, participant, patients, and provider communities—who are all end users—will ensure that features that are most desired by diverse users are considered for the product up front.57
Key technical and research questions for genomic digital health tool and platform development are: (1) Does the technology reproduce across a range of diverse populations (ancestry, ethnicity, age, sex, gender, socioeconomic status, language)? (2) Do the measures correlate with “ground truth” (clinically accepted benchmarks or standards; for example, correlating a smart watch that measures heart rate over time with a Holter monitor)? That is, can we be certain about the phenomena or phenotypes that the device is reporting? (3) How can these data be interoperable with other data streams? (4) What human-factors research and implementation research is needed to sustain user engagement with apps and devices?
Clinical
For digital health technologies to be fully integrated into healthcare systems using genomics, an understanding of the workflows and data flows of systems is necessary. An implementation science strategy will enable understanding of (1) the environments in which the technology is being considered for use, (2) how the end user wishes to interact with the technology, (3) how best to deliver the information from the technology, and (4) how to iteratively integrate the technology into workflows to ensure its sustainable use.
To be acceptable to the clinical community, these technologies will require a clear agenda to validate the technology’s accuracy (analytic validity), its ability to reproducibly measure the clinical features of phenotype it purports to measure (clinical validity), and that it provides value by delivering information that changes behaviors or outcomes (clinical utility).
Data science
Digital health data are computationally intense and constitute “big data.” As such, increasing machine actionability by human users will be required. In 2016, FAIR principles (findable, accessible, interoperable, and reusable) were published and are now being widely adopted to promote optimal quality and reuse of data. Reuse and reanalysis should be the norm given the inherently noisy nature of digital health data, particularly from wearable devices; therefore, multiple and diverse data science strategies should be applied to optimize abilities to detect signals from noise.
Policy
Paramount to the utility of digital health is the evidence that it favorably changes health outcomes and the economics of healthcare. Research on the cost benefit and effectiveness of these technologies in concert with the payer community will enable adoption.
A significant challenge to digital health is privacy and cybersecurity. End users will have varying degrees of tolerance for data sharing and fears of data breaches. These issues will need to be better understood such that the infrastructures and protections at various levels, depending on tolerance, can be established for regular and sustainable use.
The regulatory pathways for digital devices and algorithms will need clear definition, particularly as developers and investors will require an understanding of what it will take for regulatory approval. Risk categories for digital devices and the care decisions they impact will likely determine aspects of the regulatory approval pathway.
Equity
Digital tools are not equitably accessible and affordable. Research and policy agendas will need to address digital and data literacy, the languages the devices use, awareness, and knowledge as well as how to create low-cost options and provide for universal access. Learning how best to achieve the trust, engagement, and value that digital health technologies bring to underserved and underrepresented communities should be of paramount importance for the ongoing research agenda for digital health and genomics.
While many of these areas of research apply more broadly to digital health and healthcare delivery, it is imperative that the genomic medicine community develop this agenda in the context of optimizing the use, integration, and interpretation of genomic data to optimize the health of individuals and their treatment.
Conclusions
Digital health applications have shown great promise in terms of improving outcomes of genetic healthcare, as well as improving access, quality, and efficiency of genetic services. Yet digital tools can also exacerbate disparities if digital models do not represent broad populations or fail to be used by diverse groups. The call to action for the genetics community includes not only developing scalable and efficient tools, but also designing equitable digital genomic solutions accessible for all potential users, conducting comparative effectiveness research, and addressing the policy agenda that will enable the evidence-based use of digital genomics in medical practice.
Acknowledgments
A.C.S. receives NIH funding for chatbot-related research via NHLBI 1R01HL148246. Y.B. receives funding from the Canadian Institutes of Health Research (CIHR) for "Genomics ADvISER" and "Genetics Adviser" via CIHR grant numbers 143310 and 165963.
Declaration of interests
G.S.G. is an employee of the National Institutes of Health, the Department of Health and Human Services, and the United States government. The opinions expressed in this article are the author’s own and do not reflect the view of these organizations. This article was prepared while G.S.G. was employed at Duke University. At that time, G.S.G. was a consultant for KonicaMinolta and Fabric Genomics. G.S.G. was an owner of Peer Medical, Origin Commercial Advisors, Predigen, MeTree&You, and Coprata. G.S.G. received royalties from Elsevier. A.Y.Z. is a full-time employee and shareholder of Color Health, Inc. A.C.S. is an employee of 23andMe. The article was prepared while A.C.S. was employed at Geisinger. At that time, A.C.S. was a consultant for Invitae and 23andMe.
Web resources
Genetics Adviser, https://www.geneticsadviser.com
Genomics ADvISER, https://www.genomicsadviser.com
References
- 1.Bombard Y., Hayeems R.Z. How digital tools can advance quality and equity in genomic medicine. Nat. Rev. Genet. 2020;21:505–506. doi: 10.1038/s41576-020-0260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birch P.H. Interactive e-counselling for genetics pre-test decisions: where are we now? Clin. Genet. 2015;87:209–217. doi: 10.1111/cge.12430. [DOI] [PubMed] [Google Scholar]
- 3.Griffith J.M., Sorenson J.R., Bowling J.M., Jennings-Grant T. Assessment of an interactive computer-based patient prenatal genetic screening and testing education tool. Health Educ. Behav. 2005;32:613–626. doi: 10.1177/1090198105278747. [DOI] [PubMed] [Google Scholar]
- 4.Adam S., Birch P.H., Coe R.R., Bansback N., Jones A.L., Connolly M.B., Demos M.K., Toyota E.B., Farrer M.J., Friedman J.M. Assessing an interactive online tool to support parents’ genomic testing decisions. J. Genet. Couns. 2018;28:10–17. doi: 10.1007/s10897-018-0281-1. [DOI] [PubMed] [Google Scholar]
- 5.Rupert D.J., Squiers L.B., Renaud J.M., Whitehead N.S., Osborn R.J., Furberg R.D., Squire C.M., Tzeng J.P. Communicating risk of hereditary breast and ovarian cancer with an interactive decision support tool. Patient Educ. Couns. 2013;92:188–196. doi: 10.1016/j.pec.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Biesecker B.B., Lewis K.L., Biesecker L.G. Web-based platform vs genetic counselors in educating patients about carrier results from exome sequencing-reply. JAMA Intern. Med. 2018;178:999. doi: 10.1001/jamainternmed.2018.2236. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz M.D., Valdimarsdottir H.B., DeMarco T.A., Peshkin B.N., Lawrence W., Rispoli J., Brown K., Isaacs C., O’Neill S., Shelby R., et al. Randomized trial of a decision aid for BRCA1/BRCA2 mutation carriers: impact on measures of decision making and satisfaction. Health Psychol. 2009;28:11–19. doi: 10.1037/a0013147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooker G.W., Leventhal K.-G., DeMarco T., Peshkin B.N., Finch C., Wahl E., Joines J.R., Brown K., Valdimarsdottir H., Schwartz M.D. Longitudinal changes in patient distress following interactive decision aid use among BRCA1/2 carriers: a randomized trial. Med. Decis. Making. 2011;31:412–421. doi: 10.1177/0272989x10381283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bombard Y., Clausen M., Shickh S., Mighton C., Casalino S., Kim T.H.M., Muir S.M., Carlsson L., Baxter N., Scheer A., et al. Effectiveness of the Genomics ADvISER decision aid for the selection of secondary findings from genomic sequencing: a randomized clinical trial. Genet. Med. 2020;22:727–735. doi: 10.1038/s41436-019-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C., Gonzalez R., Milliron K.J., Strecher V.J., Merajver S.D. Genetic counseling for BRCA1/2: a randomized controlled trial of two strategies to facilitate the education and counseling process. Am. J. Med. Genet. A. 2005;134A:66–73. doi: 10.1002/ajmg.a.30577. [DOI] [PubMed] [Google Scholar]
- 11.Yee L.M., Wolf M., Mullen R., Bergeron A.R., Cooper Bailey S., Levine R., Grobman W.A. A randomized trial of a prenatal genetic testing interactive computerized information aid. Prenat. Diagn. 2014;34:552–557. doi: 10.1002/pd.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellani C., Perobelli S., Bianchi V., Seia M., Melotti P., Zanolla L., Assael B.M., Lalatta F. An interactive computer program can effectively educate potential users of cystic fibrosis carrier tests. Am. J. Med. Genet. A. 2011;66:406–407. doi: 10.1097/ogx.0b013e31823385c2. [DOI] [PubMed] [Google Scholar]
- 13.Gason A.A., Aitken M., Delatycki M.B., Sheffield E., Metcalfe S.A. Multimedia messages in genetics: design, development, and evaluation of a computer-based instructional resource for secondary school students in a Tay Sachs disease carrier screening program. Genet. Med. 2004;6:226–231. doi: 10.1097/01.gim.0000132681.36771.63. [DOI] [PubMed] [Google Scholar]
- 14.Green M.J., Peterson S.K., Baker M.W., Harper G.R., Friedman L.C., Rubinstein W.S., Mauger D.T. Effect of a computer-based decision aid on knowledge, perceptions, and intentions about genetic testing for breast cancer susceptibility: a randomized controlled trial. JAMA. 2004;292:442–452. doi: 10.1001/jama.292.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green M.J., Peterson S.K., Baker M.W., Friedman L.C., Harper G.R., Rubinstein W.S., Peters J.A., Mauger D.T. Use of an educational computer program before genetic counseling for breast cancer susceptibility: effects on duration and content of counseling sessions. Genet. Med. 2005;7:221–229. doi: 10.1097/01.gim.0000159905.13125.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albada A., Ausems M.G.E.M., Otten R., Bensing J.M., van Dulmen S. Use and evaluation of an individually tailored website for counselees prior to breast cancer genetic counseling. J. Cancer Educ. 2011;26:670–681. doi: 10.1007/s13187-011-0227-x. [DOI] [PubMed] [Google Scholar]
- 17.Stacey D., Légaré F., Lewis K., Col N.F., Barry M.J., Bennett C.L., Eden K.B., Holmes-Rovner M., Llewellyn-Thomas H., Lyddiatt A., et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst. Rev. 2017;4:CD001431. doi: 10.1002/14651858.CD001431.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy A.D.M., Sculpher M.J., Coulter A., Dwyer N., Rees M., Abrams K.R., Horsley S., Cowley D., Kidson C., Kirwin C., et al. Effects of decision aids for menorrhagia on treatment choices, health outcomes, and costs: a randomized controlled trial. JAMA. 2002;288:2701–2708. doi: 10.1001/jama.288.21.2701. [DOI] [PubMed] [Google Scholar]
- 19.Vuorma S., Rissanen P., Aalto A.M., Kujansuu E., Hurskainen R., Teperi J. Factors predicting choice of treatment for menorrhagia in gynaecology outpatient clinics. Soc. Sci. Med. 2003;56:1653–1660. doi: 10.1016/s0277-9536(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 20.Shickh S., Rafferty S.A., Clausen M., Kodida R., Mighton C., Panchal S., Lorentz J., Ward T., Watkins N., Elser C., et al. Incidental Genomics Study Team The role of digital tools in the delivery of genomic medicine: enhancing patient-centered care. Genet. Med. 2021;23:1086–1094. doi: 10.1038/s41436-021-01112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bombard Y., Clausen M., Mighton C., Carlsson L., Casalino S., Glogowski E., Schrader K., Evans M., Scheer A., Baxter N., et al. The Genomics ADvISER: development and usability testing of a decision aid for the selection of incidental sequencing results. Eur. J. Hum. Genet. 2018;26:984–995. doi: 10.1038/s41431-018-0144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Légaré F., O’Connor A.C., Graham I., Saucier D., Côté L., Cauchon M., Paré L. Supporting patients facing difficult health care decisions: use of the Ottawa Decision Support Framework. Can. Fam. Physician. 2006;52:476–477. [PMC free article] [PubMed] [Google Scholar]
- 23.Shickh S., Clausen M., Mighton C., Casalino S., Joshi E., Glogowski E., Schrader K.A., Scheer A., Elser C., Panchal S., et al. Evaluation of a decision aid for incidental genomic results, the Genomics ADvISER: protocol for a mixed methods randomised controlled trial. BMJ Open. 2018;8:e021876. doi: 10.1136/bmjopen-2018-021876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reble E., Gutierrez Salazar M., Zakoor K.-R., Khalouei S., Clausen M., Kodida R., Shickh S., Mighton C., Cohn I., Schrader K.A., et al. Beyond medically actionable results: an analytical pipeline for decreasing the burden of returning all clinically significant secondary findings. Hum. Genet. 2021;140:493–504. doi: 10.1007/s00439-020-02220-9. [DOI] [PubMed] [Google Scholar]
- 25.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R., et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 26.Jarvik G.P., Amendola L.M., Berg J.S., Brothers K., Clayton E.W., Chung W., Evans B.J., Evans J.P., Fullerton S.M., Gallego C.J., et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am. J. Hum. Genet. 2014;94:818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg J.S., Adams M., Nassar N., Bizon C., Lee K., Schmitt C.P., Wilhelmsen K.C., Evans J.P. An informatics approach to analyzing the incidentalome. Genet. Med. 2013;15:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg J.S., Khoury M.J., Evans J.P. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet. Med. 2011;13:499–504. doi: 10.1097/gim.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 29.Mighton C., Carlsson L., Clausen M., Casalino S., Shickh S., McCuaig L., Joshi E., Panchal S., Graham T., Aronson M., et al. Development of patient “profiles” to tailor counseling for incidental genomic sequencing results. Eur. J. Hum. Genet. 2019;27:1008–1017. doi: 10.1038/s41431-019-0352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snir M., Nazareth S., Simmons E., Hayward L., Ashcraft K., Bristow S.L., Esplin E.D., Aradhya S. Democratizing genomics: leveraging software to make genetics an integral part of routine care. Am. J. Med. Genet. C Semin. Med. Genet. 2021;187:14–27. doi: 10.1002/ajmg.c.31866. [DOI] [PubMed] [Google Scholar]
- 31.Zarouali B., Van den Broeck E., Walrave M., Poels K. Predicting consumer responses to a chatbot on facebook. Cyberpsychol. Behav. Soc. Netw. 2018;21:491–497. doi: 10.1089/cyber.2017.0518. [DOI] [PubMed] [Google Scholar]
- 32.Maeda E., Miyata A., Boivin J., Nomura K., Kumazawa Y., Shirasawa H., Saito H., Terada Y. Promoting fertility awareness and preconception health using a chatbot: a randomized controlled trial. Reprod. Biomed. Online. 2020;41:1133–1143. doi: 10.1016/j.rbmo.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S., Bhatia S., Bhatia A. Quro: facilitating user symptom check using a personalised chatbot-oriented dialogue system. Stud. Health Technol. Inform. 2018;252:51–56. [PubMed] [Google Scholar]
- 34.Fitzpatrick K.K., Darcy A., Vierhile M. Delivering cognitive behavior therapy to young adults with symptoms of depression and anxiety using a fully automated conversational agent (woebot): a randomized controlled trial. JMIR Ment. Health. 2017;4:e19. doi: 10.2196/mental.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein N., Brooks K. A fully automated conversational artificial intelligence for weight loss: longitudinal observational study among overweight and obese adults. JMIR Diabetes. 2017;2:e28. doi: 10.2196/diabetes.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crutzen R., Peters G.-J.Y., Portugal S.D., Fisser E.M., Grolleman J.J. An artificially intelligent chat agent that answers adolescents’ questions related to sex, drugs, and alcohol: an exploratory study. J. Adolesc. Health. 2011;48:514–519. doi: 10.1016/j.jadohealth.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Ponathil A., Ozkan F., Welch B., Bertrand J., Chalil Madathil K. Family health history collected by virtual conversational agents: an empirical study to investigate the efficacy of this approach. J. Genet. Couns. 2020;29:1081–1092. doi: 10.1002/jgc4.1239. [DOI] [PubMed] [Google Scholar]
- 38.Heald B., Keel E., Marquard J., Burke C.A., Kalady M.F., Church J.M., Liska D., Mankaney G., Hurley K., Eng C. Using chatbots to screen for heritable cancer syndromes in patients undergoing routine colonoscopy. J. Med. Genet. 2021;58:807–814. doi: 10.1136/jmedgenet-2020-107294. [DOI] [PubMed] [Google Scholar]
- 39.Nazareth S., Hayward L., Simmons E., Snir M., Hatchell K.E., Rojahn S., Slotnick R.N., Nussbaum R.L. Hereditary cancer risk using a genetic chatbot before routine care visits. Obstet. Gynecol. 2021;138:860–870. doi: 10.1097/aog.0000000000004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siglen E., Vetti H.H., Lunde A.B.F., Hatlebrekke T.A., Strømsvik N., Hamang A., Hovland S.T., Rettberg J.W., Steen V.M., Bjorvatn C. Ask Rosa - the making of a digital genetic conversation tool, a chatbot, about hereditary breast and ovarian cancer. Patient Educ. Couns. 2021 doi: 10.1016/j.pec.2021.09.027. [DOI] [PubMed] [Google Scholar]
- 41.Ireland D., Bradford D., Szepe E., Lynch E., Martyn M., Hansen D., Gaff C. Introducing Edna: a trainee chatbot designed to support communication about additional (secondary) genomic findings. Patient Educ. Couns. 2021;104:739–749. doi: 10.1016/j.pec.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz M.L.B., McCormick C.Z., Lazzeri A.L., Lindbuchler D.M., Hallquist M.L.G., Manickam K., Buchanan A.H., Rahm A.K., Giovanni M.A., Frisbie L., et al. A model for genome-first care: returning secondary genomic findings to participants and their healthcare providers in a large research cohort. Am. J. Hum. Genet. 2018;103:328–337. doi: 10.1016/j.ajhg.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidlen T., Schwartz M., DiLoreto K., Kirchner H.L., Sturm A.C. Patient assessment of chatbots for the scalable delivery of genetic counseling. J. Genet. Couns. 2019;28:1166–1177. doi: 10.1002/jgc4.1169. [DOI] [PubMed] [Google Scholar]
- 44.Campbell-Salome G., Jones L.K., Masnick M.F., Walton N.A., Ahmed C.D., Buchanan A.H., Brangan A., Esplin E.D., Kann D.G., Ladd I.G., et al. Developing and optimizing innovative tools to address familial hypercholesterolemia underdiagnosis: identification methods, patient Activation, and cascade testing for familial hypercholesterolemia. Circ. Genom Precis Med. 2021;14:e003120. doi: 10.1161/circgen.120.003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nazareth S., Nussbaum R.L., Siglen E., Wicklund C.A. Chatbots & artificial intelligence to scale genetic information delivery. J. Genet. Couns. 2021;30:7–10. doi: 10.1002/jgc4.1359. [DOI] [PubMed] [Google Scholar]
- 46.Nadarzynski T., Miles O., Cowie A., Ridge D. Acceptability of artificial intelligence (AI)-led chatbot services in healthcare: a mixed-methods study. Digit Health. 2019;5 doi: 10.1177/2055207619871808. 205520761987180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernhardt B.A., Weiner J., Foster E.C., Tumpson J.E., Pyeritz R.E. The economics of clinical genetics services. II. A time analysis of a medical genetics clinic. Am. J. Hum. Genet. 1987;41:559–565. [PMC free article] [PubMed] [Google Scholar]
- 48.Cooksey J.A., Forte G., Flanagan P.A., Benkendorf J., Blitzer M.G. The medical genetics workforce: an analysis of clinical geneticist subgroups. Genet. Med. 2006;8:603–614. doi: 10.1097/01.gim.0000242307.83900.77. [DOI] [PubMed] [Google Scholar]
- 49.Mahon S.M. Allocation of work activities in a comprehensive cancer genetics program. Clin. J. Oncol. Nurs. 2013;17:397–404. doi: 10.1188/13.cjon.397-404. [DOI] [PubMed] [Google Scholar]
- 50.Heald B., Gustafson S., Mester J., Arscott P., Lynch K., Moline J., Eng C. A time study of cancer genetic counselors using a genetic counselor-only patient care model versus a traditional combined genetic counselor plus medical geneticist care model. J. Natl. Compr. Canc. Netw. 2013;11:1076–1081. doi: 10.6004/jnccn.2013.0129. [DOI] [PubMed] [Google Scholar]
- 51.McPherson E., Zaleski C., Benishek K., McCarty C.A., Giampietro P.F., Reynolds K., Rasmussen K. Clinical genetics provider real-time workflow study. Genet. Med. 2008;10:699–706. doi: 10.1097/gim.0b013e31818fd617. [DOI] [PubMed] [Google Scholar]
- 52.David S.P., Dunnenberger H.M., Ali R., Matsil A., Lemke A.A., Singh L., Zimmer A., Hulick P.J. Implementing primary care mediated population genetic screening within an integrated health system. J. Am. Board Fam. Med. 2021;34:861–865. doi: 10.3122/jabfm.2021.04.200381. [DOI] [PubMed] [Google Scholar]
- 53.Caswell-Jin J.L., Zimmer A.D., Stedden W., Kingham K.E., Zhou A.Y., Kurian A.W. Cascade genetic testing of relatives for hereditary cancer risk: results of an online initiative. J. Natl. Cancer Inst. 2019;111:95–98. doi: 10.1093/jnci/djy147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemke A.A., Amendola L.M., Kuchta K., Dunnenberger H.M., Thompson J., Johnson C., Ilbawi N., Oshman L., Hulick P.J. Primary care physician experiences with integrated population-scale genetic testing: a mixed-methods assessment. J. Pers Med. 2020;10:165. doi: 10.3390/jpm10040165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemke A.A., Amendola L.M., Thompson J., Dunnenberger H.M., Kuchta K., Wang C., Dilzell-Yu K., Hulick P.J. Patient-reported outcomes and experiences with population genetic testing offered through a primary care network. Genet. Test. Mol. Biomarkers. 2021;25:152–160. doi: 10.1089/gtmb.2020.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphreys L., Hunter A.G., Zimak A., O’Brien A., Korneluk Y., Cappelli M. Why patients do not attend for their appointments at a genetics clinic. J. Med. Genet. 2000;37:810–815. doi: 10.1136/jmg.37.10.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yock P.G., Zenios S., Makower J., Brinton T.J., Kumar U.N., Jay Watkins F.T., Denend L., Krummel T.M., Kurihara C.Q. Cambridge University Press; 2015. Biodesign: The Process of Innovating Medical Technologies. [Google Scholar]