Summary
Recent work has found increasing evidence of mitigated, incompletely penetrant phenotypes in heterozygous carriers of recessive Mendelian disease variants. We leveraged whole-exome imputation within the full UK Biobank cohort (n ∼ 500K) to extend such analyses to 3,475 rare variants curated from ClinVar and OMIM. Testing these variants for association with 58 quantitative traits yielded 102 significant associations involving variants previously implicated in 34 different diseases. Notable examples included a POR missense variant implicated in Antley-Bixler syndrome that associated with a 1.76 (SE 0.27) cm increase in height and an ABCA3 missense variant implicated in interstitial lung disease that associated with reduced FEV1/FVC ratio. Association analyses with 1,134 disease traits yielded five additional variant-disease associations. We also observed contrasting levels of recessiveness between two more-common, classical Mendelian diseases. Carriers of cystic fibrosis variants exhibited increased risk of several mitigated disease phenotypes, whereas carriers of spinal muscular atrophy alleles showed no evidence of altered phenotypes. Incomplete penetrance of cystic fibrosis carrier phenotypes did not appear to be mediated by common allelic variation on the functional haplotype. Our results show that many disease-associated recessive variants can produce mitigated phenotypes in heterozygous carriers and motivate further work exploring penetrance mechanisms.
Keywords: Mendelian disease, recessive disease, penetrance, association study
Introduction
Since the advent of next-generation sequencing, the number of variants identified as contributing to Mendelian disease has grown rapidly.1 Roughly 20% of all protein-coding genes in humans have been associated with at least one Mendelian disease.2 Increasingly, studies of recessive disease variants have begun observing that these variants can sometimes cause mitigated phenotypes in heterozygous carriers, thereby contributing to population variation in complex traits and disease susceptibility.3, 4, 5, 6, 7, 8, 9, 10, 11 However, the rarity of most such variants together with their unavailability in most SNP-array-based genotyping studies has limited attempts to explore this phenomenon at scale. Early work focused on smaller cohorts recruited for specific diseases, such as a series of studies that demonstrated increased risk of male infertility,12,13 bronchiectasis,14, 15, 16 and asthma15,17 among other phenotypes in cystic fibrosis (CF [MIM: 219700]) carriers. More recently, larger data sets have enabled extending the breadth of such analyses to more phenotypes4,5 and to more recessive disease variants.3
With increasing exome sequencing of population biobank cohorts,18, 19, 20 a new opportunity to search for carrier phenotypes in a phenome-wide, exome-wide manner has emerged. Furthermore, biobank datasets present an opportunity to ameliorate ascertainment biases by assessing phenotypes in population cohorts, complementing analyses of affected individuals and their families. Family-based studies have been observed to be susceptible to ascertainment biases that inflate observed effects,21 while the opposite “healthy volunteer” phenomenon has been observed in biobank cohorts.22 Genome-wide genotyping and imputation in biobank datasets also provide opportunities to investigate potential genetic modifiers of incompletely penetrant carrier phenotypes.23
Here, we leveraged exome-wide imputation within the UK Biobank cohort24 to power a broad investigation of quantitative and disease phenotypes amongst carriers of recessive disease variants. Next, we performed a focused analysis of two relatively more common severe recessive Mendelian diseases, using the power afforded by high carrier frequencies to characterize carrier phenotypes or establish a truly recessive pattern of phenotypes. Finally, we considered the molecular mechanisms underlying incomplete penetrance observed amongst carriers, evaluating a previously proposed model of modified penetrance.23
Subjects and methods
Imputed carriers of recessive Mendelian disease variants in UK Biobank
We previously used the first tranche of whole-exome sequencing (WES) data released by the UK Biobank (n = 49,960)18 to impute coding variants into SNP-array data available for n = 487,409 participants in the full UK Biobank cohort,25 achieving accurate imputation of rare variant genotypes at minor allele frequencies (MAFs) down to ∼0.00005.24 Here, we analyzed a subset of imputed variants that were annotated in ClinVar26 as “pathogenic” or “likely pathogenic” for diseases annotated in OMIM2 as “autosomal recessive.” We further restricted to rare variants (MAF < 0.01) with a minimum MAF of 0.00001 and estimated imputation accuracy of R2 > 0.5, leaving 3,475 variants for analysis.
Association tests with quantitative traits
We tested imputed genotype dosages for association with 58 quantitative traits by using linear mixed models implemented in BOLT-LMM v2.3.4. These traits included the 54 quantitative traits we previously analyzed (which included blood count traits, serum biomarker traits, and other commonly studied traits with SNP heritability > 0.2 that were phenotyped in at least half of UK Biobank participants)24 and four additional traits (age of menarche, skin pigmentation, tanning ability, and hair color). We performed quantitative trait association analyses on n = 459,327 UK Biobank participants who reported European ancestry and had not withdrawn from the study at the time of analysis. We did not attempt to filter homozygotes or compound heterozygotes from these analyses, reasoning that such individuals would account for negligible numbers of carriers of the rare variants we analyzed (both based on allele frequencies and on the “healthy volunteer” ascertainment bias of UK Biobank).
Association tests with binary traits
We tested the same imputed variants for association with 1,134 binary disease phenotypes curated by UK Biobank. These consisted of the complete set of “first-occurrence” of disease traits in the UK Biobank converted to simple case and control status as well as the set of eight “algorithmically defined health outcomes” disease categories provided by the UK Biobank. We tested variants for association with binary traits by using the BinomiRare test27 to obtain p values robust to case-control imbalance while adjusting for age (stratified into 5-year tranches) and sex. For computational efficiency, we re-implemented the BinomiRare test and applied a binomial approximation when the number of observed cases among carriers exceeded 100. We estimated odds ratios (ORs) as xw/yz, where x, y, z, and w denote ratios of observed versus expected cases among carriers, cases among noncarriers, controls among carriers, and controls among noncarriers, respectively. We estimated 95% confidence intervals by using a normal approximation, i.e., converting p values to Z scores and then taking the 95% CI of the log odds ratio (OR) to be log(OR) ± 1.96 × log(OR)/z. We performed association analyses on an unrelated subset of n = 415,291 UK Biobank participants who reported European ancestry and had not withdrawn from the study.28
Analyses of cystic fibrosis carriers
We identified cystic fibrosis carriers in UK Biobank by using SNP-array genotypes for the p.Phe508del-encoding variant and (in auxiliary analyses) the missense SNP rs78655421, excluding participants with a cystic fibrosis report (according to the “first occurrences” data field). We verified that p.Phe508del-encoding genotypes called with the SNP-array were highly concordant (Pearson R = 0.996) with imputation of exome-sequencing-based genotypes. We applied the same analysis pipeline as above to test for associations with the 1,134 binary traits and applied a significance threshold of FDR < 5% (q value < 0.05).
Analyses of spinal muscular atrophy carriers
We identified spinal muscular atrophy carriers (SMA1 [MIM: 253300], SMA2 [MIM: 253550], SMA3 [MIM: 253400], SMA4 [MIM: 271150]) in the UK Biobank n = 200K exome sequencing release as individuals with evidence of only one functional copy of SMN1 (MIM: 600354). We estimated the number of functional copies of each of SMN1 and SMN2 (MIM: 601627) on the basis of depth of coverage of exome-sequencing reads that mapped uniquely to the exon 7-intron 7 region of each gene (chr5: 70,951,800–70,952,600 for SMN1 and chr5: 70,076,400–70,077,100 for SMN2 in hg38 coordinates; these regions contain four paralogous sequence variants that distinguish the highly homologous genes and were captured by exome sequencing). This approach accounted for deletions of exons 7–8 that commonly inactivate copies of SMN2 and occasionally SMN1.29 We computed exome-sequencing read-depth by using mosdepth v0.3.130 and normalized each sample’s read-depth measurements against corresponding measurements from other samples with similar exome-wide sequencing depth profiles by using a pipeline we recently described.28
We validated the accuracy of this method for estimating SMN1 and SMN2 functional copy number by examining the dataset for individuals estimated to have 0 functional copies of SMN1. We found one individual with this genotype and confirmed that this individual had a diagnosis of spinal muscular atrophy (in additional to an extremely low serum creatinine measurement—11th-lowest in the entire UK Biobank cohort—suggesting very limited muscular function). This individual was also estimated to have an SMN2 copy number of 4, known to mitigate the SMA phenotype and to allow individuals to live into adulthood.
We analyzed SMA carriers for evidence of changes in three traits related to neuromuscular function: walking speed, hand grip strength (maximum of left- and right-hand measurements), and FEV1/FVC ratio (a measure of lung function). Using age, age squared, and sex as covariates, we performed linear regressions to test for an association between SMA carrier status and each trait.
Testing a model of modified penetrance in carriers of loss-of-function variants
To further investigate potential molecular mechanisms underlying why some recessive variant carriers display mild phenotypes but others do not, we considered a model of modified penetrance proposed by Castel et al. (2018).23 This model proposes that the penetrance of a deleterious variant can be affected by variants on the allele on the homologous chromosome, particularly in the case of common cis-eQTLs that modulate expression of the functional copy of the gene. To evaluate this model, we analyzed heterozygous carriers of relatively common disease variants in two genes, FLG (MIM: 135940) and CFTR (MIM: 602421). To perform association tests on variants carried on the haplotypes opposite the disease variants, we imputed variants from the Haplotype Reference Consortium panel (r1.1) by using Minimac3 v2.0.1 (run on genomic windows including 3 Mb flanks of each gene) and analyzed these variants together with the variants we previously imputed from WES.24 Next, we extracted the genotypes for these carriers at variants within 1 Mb up- and downstream of each gene. We then recoded the genotypes for each carrier to be hemizygous for the alleles sitting on the haplotype opposite from the deleterious variant. We performed association tests on these recoded hemizygous variants by using the Fisher’s exact test implemented in PLINK (v1.9)31 (--fisher-midp) (which could perform this analysis after we recoded the chromosome as “X” and coded all individuals as male). We assessed the power of these analyses to detect associations between common variants on the opposite haplotype by using the wp.logistic function in the WebPower R package.
Results
Quantitative phenotypes in carriers of recessive disease variants
Testing 3,475 rare recessive disease variants for association with 58 quantitative traits measured in the UK Biobank identified 102 significant (p < 2.5 × 10−7; Bonferroni-corrected) variant-trait associations (Figures 1 and S1 and Table S1). These associations involved variants reported to be pathogenic for 34 distinct recessive diseases. For many of these diseases (19/34 diseases), carriers exhibited significant deviations in multiple quantitative traits. Some of these multiple associations partly reflected correlated measurements of blood, lipid, or pigmentation traits, such as associations between a variant believed to cause Bernard-Soulier syndrome type C (BSS [MIM: 231200]) (a recessive bleeding disorder) and mean platelet volume (0.68 ± 0.03 SD), platelet distribution width (0.54 ± 0.03 SD), and platelet count (−0.65 ± 0.03 SD). However, others pointed to distinct manifestations of pleiotropy, such as associations of a variant for McArdle disease (GSD5 [MIM: 232600]) (a recessive glycogen storage disorder that interferes with muscle function) with both increased urate levels (0.15 ± 0.02 SD) and increased waist-hip ratio (0.10 ± 0.02 SD). The fraction of variants that associated with at least one quantitative trait varied by MAF, ranging from 22% (15 of 69 variants with MAF > 0.1%) to 0.5% (14 of 2,735 variants with MAF < 0.01%), probably reflecting reduced power to detect effects of very rare variants due to a combination of small sample size and reduced imputation accuracy (Table S2).
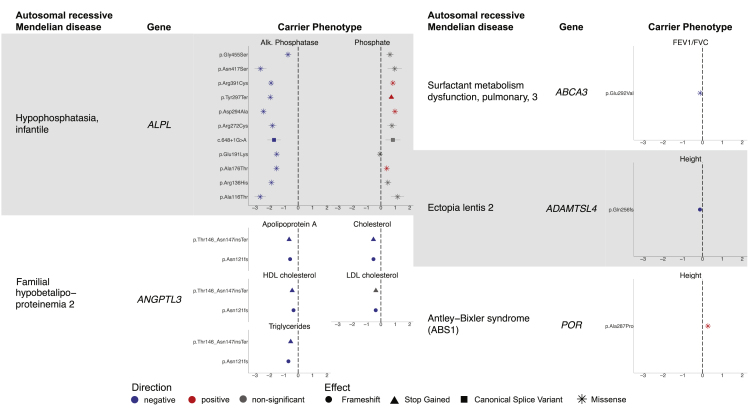
Figure 1.
Carriers of recessive Mendelian disease variants display quantitative phenotypes
Mendelian diseases and their associated genes are listed to the left in each column, and effect size is plotted on the right for each associated quantitative trait in units of standard deviation (error bars, 95% CIs). Positive-effect variants are shown in red, negative-effect variants in blue, and variants not Bonferroni significant for one of the displayed traits in gray. Marker shapes correspond to effects on the gene and gene product as reported in ClinVar.
For some of the diseases, carriers exhibited traits that might be expected based on the known biological mechanisms of the disease, supporting the validity of our analytical approach. For example, when considering Mendelian disorders where the production of a particular protein or compound is altered, one might expect a carrier to have reduced levels of that same molecule (Figure 1, left). We observed this phenomenon with infantile hypophosphatasia, which is defined by errors in alkaline phosphatase.32 In UK Biobank, carriers of several variants in ALPL (MIM: 171760) reported as pathogenic for recessive infantile hypophosphatasia (HPPI [MIM: 241500]) exhibited decreased alkaline phosphatase (ranging from −2.63 ± 0.20 SD to −0.71 ± 0.12 SD) and increased phosphate, as might be expected. Another example involved two variants in ANGPTL3 (MIM: 604774) that have been implicated in familial hypobetalipoproteinemia 2 (FHBL2 [MIM: 605019]), a recessive disorder in which individuals experience low levels of several lipid biomarkers.33 Carriers showed decreases in apolipoprotein A levels (−0.56 ± 0.04 SD; −0.63 ± 0.07 SD), cholesterol levels (−0.52 ± 0.04 SD; −0.52 ± 0.07 SD), and triglyceride levels (−0.67 ± 0.04 SD; −0.52 ± 0.07 SD).
Other diseases with more complex biological mechanisms yielded less straightforward carrier phenotypes. Here, we highlight three such examples (Figure 1, right). First, we highlight a missense variant in POR (MIM: 124015) implicated in Antley-Bixler syndrome (ABS1 [MIM: 201750]),34 a recessive skeletal disorder in which bones fuse prematurely, associated with a 0.27 ± 0.04 SD increase in height (i.e., 1.76 ± 0.27 cm). Second, we highlight a frameshift variant in ADAMTSL4 (MIM: 610113) implicated in ectopia lentis 2 (ECTOL2 [MIM: 225100]), a recessive disorder of the fibers in the eyes that can lead to vision problems, associated with a 0.13 ± 0.02 SD (0.84 ± 0.12 cm) decrease in height. Decreased height has been observed in individuals with ectopia lentis 2, but the mechanism by which ADAMTSL4 variants cause this change has not been extensively examined.35 Third, we highlight a missense variant in ABCA3 (MIM: 601615) implicated in pulmonary surfactant metabolism dysfunction 3 (SMDP3 [MIM: 610921]), a recessive interstitial lung disease caused by disruptions in the surface tension of lung surfactant,36 associated with a 0.12 ± 0.01 SD decrease in FEV1/FVC ratio, a measure of lung function. These examples add to the growing body of evidence that rare variants that cause severe disease in homozygotes or compound heterozygotes can often produce mild, subclinical phenotypes in heterozygous carriers.3,4
Disease phenotypes in heterozygous carriers of recessive variants
We next tested the same set of 3,475 rare recessive disease variants for association with 1,134 binary traits in UK Biobank, identifying five associations that reached significance (p < 1.27 × 10−8; Bonferroni corrected) (Table 1). As with the quantitative traits, some associations were expected from previous literature. Carriers of a frameshift variant in HBB (MIM: 141900) reported to be causal for beta-zero-thalassemia (MIM: 613985) exhibited increased risk of thalassemia,37 and carriers of a stop-gain variant in COL4A4 (MIM: 120131) implicated in Alport syndrome 2 (ATS2 [MIM: 203780]), a recessive disorder that involves kidney dysfunction, exhibited increased risk of hematuria (OR = 10.5; 95% CI, 5.2–21.2),38,39 as we and others have recently reported.24,40,41 Carriers of a missense variant in TYR (MIM: 606933) (tyrosinase) implicated in recessive oculocutaneous albinism type IA (OCA1A [MIM: 203100]) exhibited increased risk of disorders of aromatic amino acid metabolism (OR = 63.3; 95% CI, 16.3–245.1).42 A missense variant in TG (MIM: 188450) (thyroglobulin) implicated in recessive thyroid dyshormonogenesis 3 (TDH3 [MIM: 274700]) increased risk of hypothyroidism in carriers (OR = 2.20; 95% CI, 1.68–2.88).43,44
Table 1.
Carriers of Mendelian recessive disease variants exhibit increased risk of less-severe disease phenotypes
| Recessive disease association |
ClinVar reported variant |
Disease association in carriers | Trait category | OR (95% CI) | p value | |||
|---|---|---|---|---|---|---|---|---|
| Gene | Variant | Variant impact | MAF | |||||
| Alport syndrome 2 | COL4A4 | 2:227917083 G>C | p.Ser969Ter | 6.16E−4 | recurrent and persistent haematuria (N02) | genitourinary system disorders | 10.47 (5.17–21.2) | 6.75E−11 |
| Thyroid dyshormono-genesis 3 | TG | 8:133894854 C>T | p.Arg296Ter | 6.38E−4 | other hypothyroidism (E03) | endocrine, nutritional, and metabolic diseases | 2.20 (1.68–2.88) | 9.72E−9 |
| Beta-0 thalassaemia | HBB | 11:5248233 CAG>C | p.Pro6ArgfsTer17 | 2.15E−5 | thalassaemia (D56) | blood, blood-forming organs, and certain immune disorders | 3,183 (440–23,030) | 1.36E−15 |
| Albinism, oculo-cutaneous, type IA | TYR | 11:88961072 C>A | p.Thr373Lys | 1.10E−3 | disorders of aromatic amino acid metabolism (E70) | endocrine, nutritional, and metabolic diseases | 63.27 (16.33–245.10) | 1.94E−9 |
| Retinitis pigmentosa 80 and short rib thoracic dysplasia 9 | IFT140 | 16:1607935 C>A | c.2399+1G>T | 5.11E−4 | cystic kidney disease (Q61) | congenital disruptions and chromosomal abnormalities | 18.42 (8.47–40.05) | 1.98E−13 |
Odds ratios and p values are reported for the five associations that reached Bonferroni significance.
A more intriguing association involved a splice donor variant in IFT140 (MIM: 614620) previously implicated in recessive short-rib thoracic dysplasia 9 (SRTD9 [MIM: 266920]) and retinitis pigmentosa 80 (RP80 [MIM: 617781]), often with accompanying renal disease.45,46 Carriers of this variant exhibited increased risk of cystic kidney disease (OR = 18.4; 95% CI, 8.5–40.1), corroborating recent findings from analyses of directly sequenced individuals and imputation with the TOPMed reference panel.47,48 Loss of function of both copies of IFT140 appears to be inviable based on murine studies,49 such that this canonical splice variant has been observed in cases of recessive disease only in compound heterozygotes with partial function of the other copy of the gene. While retinitis pigmentosa 80 primarily manifests in visual symptoms and recessive short-rib thoracic dysplasia 9 in skeletal symptoms, IFT140 encodes a protein related to cilia function that also is expressed in the kidney, and renal symptoms have been noted in both diseases. The observed association between carriers of the splice variant and cystic kidney disease suggests partial haploinsufficiency of IFT140 in its role in the kidney.
Contrasting recessiveness of cystic fibrosis and spinal muscular atrophy
In light of the diversity of autosomal recessive Mendelian diseases for which we observed mitigated phenotypes in carriers, we decided to more closely investigate two relatively common recessive Mendelian diseases to ask whether mitigated phenotypes were a ubiquitous feature of recessive disease carriers. To do so, we identified diseases with sufficiently high carrier frequencies in UK Biobank that we would be well-powered to identify mitigated carrier phenotypes or lack thereof. The two diseases we identified on the basis of these criteria were cystic fibrosis (CF) and spinal muscular atrophy (SMA).
Previous studies have identified mitigated phenotypes in carriers of cystic fibrosis variants related to phenotypic manifestations of the disease.4,5 To further explore the extent of this phenomenon utilizing the deep phenotyping of UK Biobank, we tested our full set of quantitative and binary traits for associations with carriers of the most common CF pathogenic variant, CFTR p.Phe508del (MAF = 1.6%), which was directly genotyped by UK Biobank SNP arrays. Carriers of this variant showed significant associations (q value < 0.05) with asthma (OR = 1.12; 95% CI, 1.06–1.17), aspergillosis (OR = 2.60; 95% CI, 1.63–4.13), bronchiectasis (OR = 1.40; 95% CI, 1.20–1.61), and duodenal ulcer (OR = 1.30; 95% CI, 1.15–1.45) (Figure 2A and Table S3). Four additional associations reached significance at a relaxed FDR threshold of 10%: COPD (OR = 1.17; 95% CI, 1.07–1.27), cholelithiasis (OR = 1.13; 95% CI, 1.06–1.22), male infertility (OR = 2.10; 95% CI, 1.40–3.15), and other prostate disorders (OR = 1.39; 95% CI, 1.15–1.67) (Table S3). We also tested carriers of the next most common cystic fibrosis pathogenic variant, CFTR p.Arg117His (MAF = 0.2% in UK Biobank) but concluded that power was insufficient (Table S3).
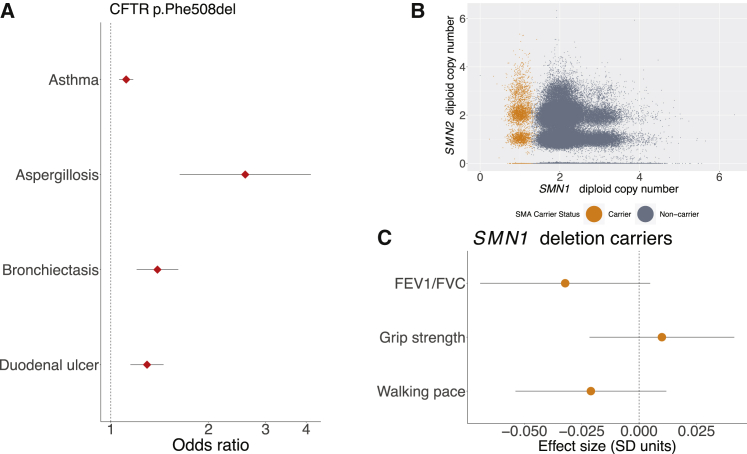
Figure 2.
Cystic fibrosis carriers show mitigated phenotypes, but spinal muscular atrophy carriers do not
(A) Carriers with the CFTR p.Phe508del inframe deletion exhibit increased risk of several mitigated disease phenotypes (data points, odds ratios; error bars, 95% CIs).
(B) Genotyping for SMA carrier status using exome sequencing data from the UK Biobank. SMN1 and SMN2 copy numbers were estimated on the basis sequencing read depth, and SMA carriers (orange) were identified as those individuals estimated to have one functional copy of SMN1 (with a deletion allele on the homologous chromosome).
(C) SMN1 deletion carriers did not display evidence of changes in any of three traits related to neuromuscular function (data points, mean values in units of standard deviations; error bars, 95% CIs).
The ORs we calculated for carriers of p.Phe508del, while significant, were much smaller than those recently reported in an analysis of CF carriers ascertained from a database of insurance claims from individuals who had been tested for carrier status4 (Figure S2). Furthermore, several reported associations did not replicate in our analysis of UK Biobank. For example, whereas the claims analysis showed a strong association between carrier status and short stature,4 we did not observe an association between p.Phe508del carrier status and height in UK Biobank despite ample power (effect size = −0.000 ± 0.006 SD). We verified that genotyping error in UK Biobank was minimal and unlikely to contribute to these differences (see subjects and methods). The ORs we computed were more consistent with those reported by Çolak et al. (2020) via p.Phe508del genotyping in the Copenhagen General Population Study.5 These results underscore the importance of understanding issues of ascertainment bias when studying penetrance in population studies.50, 51, 52
In contrast to CF, potential phenotypes of SMA carriers have not (to our knowledge) previously been explored, in part because of the difficulty of genotyping SMA carrier mutations, most of which arise from structural variation at the SMN1–SMN2 locus.29,53 SMA is usually caused by loss-of-function mutations in both copies of SMN1, and disease severity is then determined by the number of functional copies of the paralogous SMN2. The availability of WES data for n ∼ 200K UK Biobank participants19 enabled us to estimate the number of functional copies of SMN1 and SMN2 in each sequenced sample from WES depth-of-coverage (Figure 2B). We ascertained 3,462 SMA carriers (i.e., individuals likely to carry only one functional copy of SMN1) in this way from the set of whole-exome sequenced individuals of European ancestry (n = 187,720), consistent with previously reported SMN1 deletion frequency.29 Interestingly, we found no significant associations between SMA carrier status and potential manifestations of muscle weakness—walking speed, grip strength, and FEV1/FVC ratio (Figure 2C)—even when stratifying for SMN2 copy number (Table S4). These results suggest that SMA is a truly recessive disease in which muscle weakness phenotypes only manifest in individuals who carry two SMN1 alleles inactivated by loss-of-function variants.
Testing a model of modified penetrance
In all the instances in which we observed mitigated phenotypes in carriers of recessive disease variants, the associated phenotypes exhibited incomplete penetrance in heterozygotes. We therefore sought to explore the possible molecular mechanisms underlying this incomplete penetrance.
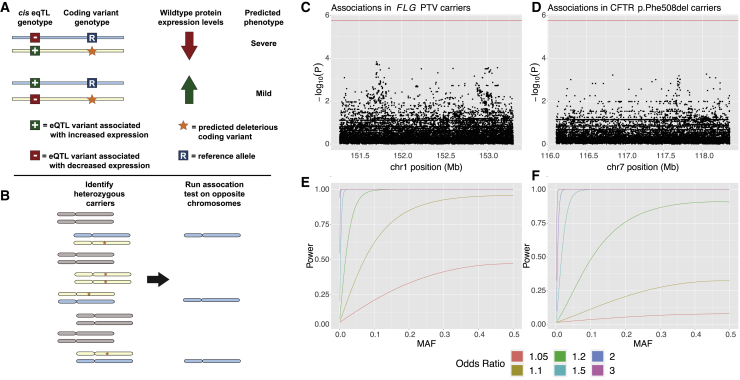
Castel et al. (2018) previously proposed a model of modified penetrance in which the haplotype arrangement of loss-of-function and expression-modifying variants in an individual might affect overall phenotype (Figure 3A).23 In this model, the phenotypic impact of a deleterious variant inactivating one copy of a gene is mediated by the amount of expression of the functional copy (on the opposite haplotype), such that a common cis-eQTL influencing expression of the functional allele can influence the severity of the phenotype. Explicitly, if the cis-eQTL increases expression of the functional, wild-type protein, this could partially ameliorate the loss of the other copy; in contrast, if the cis-eQTL decreases expression of the functional copy, one might expect the carrier to have a more severe phenotype.
Figure 3.
Common variants on haplotypes opposite recessive alleles in carriers of two Mendelian diseases do not appear to modify penetrance of carrier phenotypes
(A) The “modified penetrance” model of Castel et al. (2018)23 posits that cis-eQTLs can increase or decrease severity of a deleterious variant by modulating the quantity of functional protein produced by the opposite haplotype.
(B) To test this hypothesis, we examined opposite haplotypes (blue chromosomes) in heterozygous carriers of recessive disease variants (orange stars on yellow chromosomes). We tested variants carried on these opposite haplotypes for association with mitigated phenotypes observed in carriers.
(C and D) Manhattan plots showing association test results for variants on the opposite haplotype of deleterious variants in FLG and CFTR with asthma (the phenotype most strongly associated with carrier status in each case). No association reached Bonferroni significance (red line).
(E and F) Power analyses for the tests conducted in (C) and (D) indicate that these tests were well-powered to detect common variant effects with odds ratios > 1.2.
To explore this hypothesis, we considered two genes, FLG and CFTR, in which variants known to both cause recessive disease and produce mitigated phenotypes in carriers are sufficiently common to power analysis. Loss-of-function variants in FLG are known to cause ichthyosis vulgaris (MIM: 146700) in homozygotes or compound heterozygotes, and carrier status has been associated with asthma and atopic dermatitis.54,55 In UK Biobank, 10.3% of participants carried a loss-of-function variant in FLG that was associated with asthma or atopic dermatitis in heterozygotes. As discussed in the previous section, mutations in CFTR are responsible for CF, as well as several mitigated phenotypes in carriers. Approximately 3.1% of individuals in the UK Biobank are carriers for the p.Phe508del variant in CFTR that we considered for this analysis.
To determine whether variants on the opposite (putatively functional) haplotype in carriers might affect their susceptibility to mitigated phenotypes, we restricted our analysis just to carriers of these deleterious variants (Figure 3B). For each nearby variant at each locus, we then ran an association test between opposite-haplotype genotypes and mitigated phenotypes. No tested variant at either locus significantly associated with phenotype (Figure 3C). Given that we were well-powered to detect common variant associations with an OR > 1.2 in both scenarios (Figure 3D), these results suggest that the modified penetrance model is unlikely to underlie incomplete penetrance of these carrier phenotypes.
Discussion
Our results demonstrate that for a wide range of Mendelian diseases, variants traditionally considered to be recessive can cause milder phenotypes in heterozygous carriers. We also observed that entirely recessive effects do exist: heterozygous carriers for spinal muscular atrophy exhibited no evidence of even a subtle effect on phenotypes related to muscle strength. These observations suggest a spectrum of recessiveness that is now becoming visible in very large population cohorts.
Our study did have several limitations. First, even with the large sample size provided by exome sequencing in UK Biobank, we still lacked power to assess potential effects of many very rare variants that are known to cause Mendelian recessive diseases. Second, our examination of potential interactions between variants on opposite haplotypes was even more power constrained, such that we could only assess this model for two diseases involving common variants. Third, the effects we estimated are likely to be influenced by the “healthy volunteer” ascertainment bias observed in analyses of population biobank cohorts.22
As even larger, well-phenotyped cohorts with WES or whole-genome sequencing become available, our ability to determine the extent of mild carrier phenotypes will increase. More comprehensive phenome-wide and genome-wide studies will allow for an assessment of how common the phenomenon of incomplete recessivity is among severe Mendelian diseases and the spectrum of phenotypes that can manifest. Moreover, the higher power afforded by extremely large studies will also enable more extensive exploration of potential interactions between variants that could help to explain incomplete penetrance and shed light on the molecular mechanisms that underlie mitigated phenotypes.
Acknowledgments
We thank A. Gusev, A. Price, and S. Sunyaev for helpful discussions. This research was conducted with the UK Biobank Resource under application No. 10438. A.R.B. was supported by US NIH grant T32 HG229516 and fellowship F31 HL154537. M.L.A.H. was supported by US NIH fellowship F32 HL160061. M.A.S. was supported by the MIT John W. Jarve (1978) Seed Fund for Science Innovation and US NIH fellowship F31 MH124393. R.E.M. was supported by US NIH grant K25 HL150334 and NSF grant DMS-1939015. P.-R.L. was supported by US NIH grant DP2 ES030554, a Burroughs Wellcome Fund Career Award at the Scientific Interfaces, the Next Generation Fund at the Broad Institute of MIT and Harvard, and a Sloan Research Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Computational analyses were performed on the O2 High Performance Compute Cluster, supported by the Research Computing Group, at Harvard Medical School (http://rc.hms.harvard.edu).
Declaration of interests
The authors declare no competing interests.
Published: May 31, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.05.008.
Contributor Information
Alison R. Barton, Email: alisonbarton@g.harvard.edu.
Po-Ru Loh, Email: poruloh@broadinstitute.org.
Web resources
OMIM, https://omim.org/
Supplemental information
Data and code availability
Access to the UK Biobank Resource is available by application (http://www.ukbiobank.ac.uk/). BOLT-LMM (v2.3.4) is available at https://data.broadinstitute.org/alkesgroup/BOLT-LMM/. mosdepth (v0.3.1) is available at https://github.com/brentp/mosdepth. Minimac4 (v.1.0.1) is available at https://genome.sph.umich.edu/wiki/Minimac4. PLINK (v1.9) is available from https://www.cog-genomics.org/plink/1.9/.
References
- 1.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 2.Hamosh A., Amberger J.S., Bocchini C., Scott A.F., Rasmussen S.A. Online mendelian inheritance in man (OMIM®): victor McKusick’s magnum opus. Am. J. Med. Genet. 2021;185:3259–3265. doi: 10.1002/ajmg.a.62407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright C.F., West B., Tuke M., Jones S.E., Patel K., Laver T.W., Beaumont R.N., Tyrrell J., Wood A.R., Frayling T.M., et al. Assessing the pathogenicity, penetrance, and expressivity of putative disease-causing variants in a population setting. Am. J. Hum. Genet. 2019;104:275–286. doi: 10.1016/j.ajhg.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller A.C., Comellas A.P., Hornick D.B., Stoltz D.A., Cavanaugh J.E., Gerke A.K., Welsh M.J., Zabner J., Polgreen P.M. Cystic fibrosis carriers are at increased risk for a wide range of cystic fibrosis-related conditions. Proc. Natl. Acad. Sci. U S A. 2020;117:1621–1627. doi: 10.1073/pnas.1914912117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Çolak Y., Nordestgaard B.G., Afzal S. Morbidity and mortality in carriers of the cystic fibrosis mutation CFTR Phe508del in the general population. Eur. Respir. J. 2020;56:2000558. doi: 10.1183/13993003.00558-2020. [DOI] [PubMed] [Google Scholar]
- 6.Mäkitie O., Pereira R.C., Kaitila I., Turan S., Bastepe M., Laine T., Kröger H., Cole W.G., Jüppner H. Long-term clinical outcome and carrier phenotype in autosomal recessive hypophosphatemia caused by a novel DMP1 mutation. J. Bone Miner. Res. 2010;25:2165–2174. doi: 10.1002/jbmr.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagher H., Buzza M., Colville D., Jones C., Powell H., Fassett R., Wilson D., Agar J., Savige J. A comparison of the clinical, histopathologic, and ultrastructural phenotypes in carriers of X-linked and autosomal recessive Alport’s syndrome. Am. J. Kidney Dis. 2001;38:1217–1228. doi: 10.1053/ajkd.2001.29217. [DOI] [PubMed] [Google Scholar]
- 8.Watts J.A., Morley M., Burdick J.T., Fiori J.L., Ewens W.J., Spielman R.S., Cheung V.G. Gene expression phenotype in heterozygous carriers of ataxia telangiectasia. Am. J. Hum. Genet. 2002;71:791–800. doi: 10.1086/342974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidransky E., Nalls M.A., Aasly J.O., Aharon-Peretz J., Annesi G., Barbosa E.R., Bar-Shira A., Berg D., Bras J., Brice A., et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vieira S.R.L., Morris H.R. Neurodegenerative disease risk in carriers of autosomal recessive disease. Front. Neurol. 2021;12:679927. doi: 10.3389/fneur.2021.679927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez Y., Wormser O., Sadaka Y., Birk R., Narkis G., Birk O.S. A rare variant in PGAP2 causes autosomal recessive hyperphosphatasia with mental retardation syndrome, with a mild phenotype in heterozygous carriers. BioMed Res. Int. 2017;2017:3470234. doi: 10.1155/2017/3470234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chillón M., Casals T., Mercier B., Bassas L., Lissens W., Silber S., Romey M.-C., Ruiz-Romero J., Verlingue C., Claustres M., et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N. Engl. J. Med. 1995;332:1475–1480. doi: 10.1056/nejm199506013322204. [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Chen Z., Ni Y., Li Z. CFTR mutations in men with congenital bilateral absence of the vas deferens (CBAVD): a systemic review and meta-analysis. Hum. Reprod. 2012;27:25–35. doi: 10.1093/humrep/der377. [DOI] [PubMed] [Google Scholar]
- 14.Girodon E., Cazeneuve C., Lebargy F., Chinet T., Costes B., Ghanem N., Martin J., Lemay S., Scheid P., Housset B., et al. CFTR gene mutations in adults with disseminated bronchiectasis. Eur. J. Hum. Genet. 1997;5:149–155. doi: 10.1159/000484750. [DOI] [PubMed] [Google Scholar]
- 15.Tzetis M., Efthymiadou A., Strofalis S., Psychou P., Dimakou A., Pouliou E., Doudounakis S., Kanavakis E. CFTR gene mutations – including three novel nucleotide substitutions – and haplotype background in patients with asthma, disseminated bronchiectasis and chronic obstructive pulmonary disease. Hum. Genet. 2001;108:216–221. doi: 10.1007/s004390100467. [DOI] [PubMed] [Google Scholar]
- 16.Casals T., De-Gracia J., Gallego M., Dorca J., Rodríguez-Sanchón B., Ramos M., Giménez J., Cisteró-Bahima A., Olveira C., Estivill X. Bronchiectasis in adult patients: an expression of heterozygosity for CFTR gene mutations? Clin. Genet. 2004;65:490–495. doi: 10.1111/j.0009-9163.2004.00265.x. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen A.O., Qayum S., Bouchelouche P.N., Laursen L.C., Dahl R., Dahl M. Risk of asthma in heterozygous carriers for cystic fibrosis: a meta-analysis. J. Cyst. Fibros. 2016;15:563–567. doi: 10.1183/13993003.congress-2016.pa1258. [DOI] [PubMed] [Google Scholar]
- 18.Van Hout C.V., Tachmazidou I., Backman J.D., Hoffman J.D., Liu D., Pandey A.K., Gonzaga-Jauregui C., Khalid S., Ye B., Banerjee N., et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586:749–756. doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szustakowski J.D., Balasubramanian S., Kvikstad E., Khalid S., Bronson P.G., Sasson A., Wong E., Liu D., Wade Davis J., Haefliger C., et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 2021;53:942–948. doi: 10.1038/s41588-021-00885-0. [DOI] [PubMed] [Google Scholar]
- 20.Backman J.D., Li A.H., Marcketta A., Sun D., Mbatchou J., Kessler M.D., Benner C., Liu D., Locke A.E., Balasubramanian S., et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;599:628–634. doi: 10.1038/s41586-021-04103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper D.N., Krawczak M., Polychronakos C., Tyler-Smith C., Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry A., Littlejohns T.J., Sudlow C., Doherty N., Adamska L., Sprosen T., Collins R., Allen N.E. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am. J. Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castel S.E., Cervera A., Mohammadi P., Aguet F., Reverter F., Wolman A., Guigo R., Iossifov I., Vasileva A., Lappalainen T. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat. Genet. 2018;50:1327–1334. doi: 10.1038/s41588-018-0192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton A.R., Sherman M.A., Mukamel R.E., Loh P.-R. Whole-exome imputation within UK Biobank powers rare coding variant association and fine-mapping analyses. Nat. Genet. 2021;53:1260–1269. doi: 10.1038/s41588-021-00892-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sofer T. BinomiRare: a robust test of the association of a rare variant with a disease for pooled analysis and meta-analysis, with application to the HCHS/SOL. SOL. Genet. Epidemiol. 2017;41:388–395. doi: 10.1002/gepi.22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukamel R.E., Handsaker R.E., Sherman M.A., Barton A.R., Zheng Y., McCarroll S.A., Loh P.-R. Protein-coding repeat polymorphisms strongly shape diverse human phenotypes. Science. 2021;373:1499–1505. doi: 10.1126/science.abg8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Sanchis-Juan A., French C.E., Connell A.J., Delon I., Kingsbury Z., Chawla A., Halpern A.L., Taft R.J., Bentley D.R., et al. Spinal muscular atrophy diagnosis and carrier screening from genome sequencing data. Genet. Med. 2020;22:945–953. doi: 10.1038/s41436-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen B.S., Quinlan A.R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7–015. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss M.J., Cole D.E., Ray K., Whyte M.P., Lafferty M.A., Mulivor R.A., Harris H. A missense mutation in the human liver/bone/kidney alkaline phosphatase gene causing a lethal form of hypophosphatasia. Proc. Natl. Acad. Sci. U S A. 1988;85:7666–7669. doi: 10.1073/pnas.85.20.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musunuru K., Pirruccello J.P., Do R., Peloso G.M., Guiducci C., Sougnez C., Garimella K.V., Fisher S., Abreu J., Barry A.J., et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/nejmoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flück C.E., Tajima T., Pandey A.V., Arlt W., Okuhara K., Verge C.F., Jabs E.W., Mendonça B.B., Fujieda K., Miller W.L. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat. Genet. 2004;36:228–230. doi: 10.1038/ng1300. [DOI] [PubMed] [Google Scholar]
- 35.Neuhann T.M., Stegerer A., Riess A., Blair E., Martin T., Wieser S., Kläs R., Bouman A., Kuechler A., Rittinger O. ADAMTSL4-associated isolated ectopia lentis: further patients, novel mutations and a detailed phenotype description. Am. J. Med. Genet. 2015;167:2376–2381. doi: 10.1002/ajmg.a.37157. [DOI] [PubMed] [Google Scholar]
- 36.Shulenin S., Nogee L.M., Annilo T., Wert S.E., Whitsett J.A., Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N. Engl. J. Med. 2004;350:1296–1303. doi: 10.1056/nejmoa032178. [DOI] [PubMed] [Google Scholar]
- 37.Taher A.T., Musallam K.M., Cappellini M.D. β-Thalassemias. N. Engl. J. Med. 2021;384:727–743. doi: 10.1056/nejmra2021838. [DOI] [PubMed] [Google Scholar]
- 38.Longo I., Porcedda P., Mari F., Giachino D., Meloni I., Deplano C., Brusco A., Bosio M., Massella L., Lavoratti G., et al. COL4A3/COL4A4 mutations: from familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int. 2002;61:1947–1956. doi: 10.1046/j.1523-1755.2002.00379.x. [DOI] [PubMed] [Google Scholar]
- 39.Buzza M., Wang Y.Y., Dagher H., Babon J.J., Cotton R.G., Powell H., Dowling J., Savige J. COL4A4 mutation in thin basement membrane disease previously described in Alport syndrome1. Kidney Int. 2001;60:480–483. doi: 10.1046/j.1523-1755.2001.060002480.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang C., Song Y., Chen Z., Yuan X., Chen X., Ding G., Guan Y., McGrath M., Song C., Tong Y., Wang H. A nonsense mutation in COL4A4 gene causing isolated hematuria in either heterozygous or homozygous state. Front. Genet. 2019;10:628. doi: 10.3389/fgene.2019.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., Venkataraman G.R., Wainberg M., Ollila H.M., Kiiskinen T., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grønskov K., Ek J., Brondum-Nielsen K. Oculocutaneous albinism. Orphanet J. Rare Dis. 2007;2:43. doi: 10.1186/1750-1172-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ieiri T., Cochaux P., Targovnik H.M., Suzuki M., Shimoda S., Perret J., Vassart G. A 3’ splice site mutation in the thyroglobulin gene responsible for congenital goiter with hypothyroidism. J. Clin. Invest. 1991;88:1901–1905. doi: 10.1172/jci115513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grasberger H., Refetoff S. Genetic causes of congenital hypothyroidism due to dyshormonogenesis. Curr. Opin. Pediatr. 2011;23:421–428. doi: 10.1097/mop.0b013e32834726a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrault I., Saunier S., Hanein S., Filhol E., Bizet A.A., Collins F., Salih M.A.M., Gerber S., Delphin N., Bigot K., et al. Mainzer-Saldino syndrome is a ciliopathy caused by IFT140 mutations. Am. J. Hum. Genet. 2012;90:864–870. doi: 10.1016/j.ajhg.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidts M., Frank V., Eisenberger T., al Turki S., Bizet A.A., Antony D., Rix S., Decker C., Bachmann N., Bald M., et al. Combined NGS approaches identify mutations in the intraflagellar transport gene IFT140 in skeletal ciliopathies with early progressive kidney disease. Hum. Mutat. 2013;34:714–724. doi: 10.1002/humu.22294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. NHLBI Trans-Omics for Precision Medicine TOPMed Consortium Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senum S.R., Li Y., Sabrina) M., Benson K.A., Joli G., Olinger E., Lavu S., Madsen C.D., Gregory A.V., Neatu R., et al. Monoallelic IFT140 pathogenic variants are an important cause of the autosomal dominant polycystic kidney-spectrum phenotype. Am. J. Hum. Genet. 2021;109:136–156. doi: 10.1016/j.ajhg.2021.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jonassen J.A., SanAgustin J., Baker S.P., Pazour G.J. Disruption of IFT complex A causes cystic kidneys without mitotic spindle misorientation. J. Am. Soc. Nephrol. 2012;23:641–651. doi: 10.1681/asn.2011080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganguli M., Lytle M.E., Reynolds M.D., Dodge H.H. Random versus volunteer selection for a community-based study. J. Gerontol. A Biol. Sci. Med. Sci. 1998;53A:M39–M46. doi: 10.1093/gerona/53a.1.m39. [DOI] [PubMed] [Google Scholar]
- 51.Mirshahi U.L., Colclough K., Wright C.F., Wood A.R., Beaumont R.N., Tyrrell J., Laver T.W., Stahl R., Golden A., Goehringer J.M., et al. The penetrance of age-related monogenic disease depends on ascertainment context. Preprint at medRxiv. 2021 doi: 10.1101/2021.06.28.21259641. [DOI] [Google Scholar]
- 52.Forrest I.S., Chaudhary K., Vy H.M.T., Bafna S., Jordan D.M., Rocheleau G., Loos R.J.F., Cho J.H., Do R. Ancestrally and temporally diverse analysis of penetrance of clinical variants in 72,434 individuals. Preprint at medRxiv. 2021 doi: 10.1101/2021.03.11.21253430. [DOI] [Google Scholar]
- 53.Alías L., Bernal S., Fuentes-Prior P., Barceló M.J., Also E., Martínez-Hernández R., Rodríguez-Alvarez F.J., Martín Y., Aller E., Grau E., et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum. Genet. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 54.Weidinger S., O’Sullivan M., Illig T., Baurecht H., Depner M., Rodriguez E., Ruether A., Klopp N., Vogelberg C., Weiland S.K., et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J. Allergy Clin. Immunol. 2008;121:1203–1209.e1. doi: 10.1016/j.jaci.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 55.Wang Q., Dhindsa R.S., Carss K., Harper A.R., Nag A., Tachmazidou I., Vitsios D., Deevi S.V.V., Mackay A., Muthas D., et al. AstraZeneca Genomics Initiative Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature. 2021;597:527–532. doi: 10.1038/s41586-021-03855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to the UK Biobank Resource is available by application (http://www.ukbiobank.ac.uk/). BOLT-LMM (v2.3.4) is available at https://data.broadinstitute.org/alkesgroup/BOLT-LMM/. mosdepth (v0.3.1) is available at https://github.com/brentp/mosdepth. Minimac4 (v.1.0.1) is available at https://genome.sph.umich.edu/wiki/Minimac4. PLINK (v1.9) is available from https://www.cog-genomics.org/plink/1.9/.