Abstract
Diabetic Retinopathy (DR), a debilitating microvascular complication of diabetes, is one of the leading cause of blindness. However, the pathogenesis of this disease is not fully understood. Few Studies have reported the role of MicroRNA (miRNA), which is deregulated or altered in many diseases. Further, few pathways linked genes which have been suggested to be regulated by miRNAs, may play an important role in the regulation of glucose homeostasis and eventually may contribute to the establishment of DR. However, the roles of microRNAs (miRNAs) in DR are still not very clear. In current review, we explored various findings of scientific database demonstrating the role of miRNA in the progression and development of Diabetic Retinopathy.
Keywords: Mi RNA, Diabetic retinopathy, Glucose
Introduction
The occurrence of diabetes, a prolonged metabolic disorder, has been growing at worrying rates all over the world and is estimated to rise to 552 million adults by 2030. Upswing in cases and results in death, makes it one of the important health challenges in our society [1, 2]. In the absence of physical activity, people become overweight and obese and interactions with genetic predisposition, significantly modify the incidence of diabetes [1, 3]. Now Type 2 diabetes mellitus (T2DM), is affecting not only adults but also it is seen at an early age in children and adolescents [1, 2]. The rising incidence of obesity in children increase the T2DM burden in population. Hyperglycemia is the main characteristics in all diabetic patients. This is, due to lack of insulin, or loss of insulin sensitivity in the target tissue in the presence of normal insulin secretion, or a combination of both. While in type 1 diabetes, there is complete lack of insulin, due to beta cell malfunctioning. “Insulin resistance, which is the impaired ability of insulin to elicit its metabolic effects in the target tissues, particularly, in fat, liver and skeletal muscle, is one of the important causes of T2DM and cardiovascular disease”[4, 5]. Moreover, longstanding hyperglycemia can be root for life threatening macrovascular and microvascular problems. In addition, macrovascular problems like coronary heart disease and stroke are accelerated by atherosclerosis and hypertension and it is most common in diabetic patients [6, 7]. On the other hand, diabetic retinopathy is caused by microvascular episode [8, 9], where it is major cause of blindness, and diabetic nephropathy, affected tubules, glomeruli, interstitium and the vessels, causing malfunctioning in renal and finally leading to end-stage renal disease [10, 11]. In addition, diabetic neuropathy is also caused by microvascular disease [2, 12, 13], affecting specially the nervous system, eventually heading to the development of chronic diabetic foot ulcers, among other problems.
T2DM is characterised by the chronic inflammatory state, leading to chronic complications. These can be prevented by adequate nutrition and regular physical exercise, at an initial stage. However, our aim to get better and more effective treatment solutions for these long-term complications in patients remains of importance. Many clinical trials have addressed few of these questions [14–16]. The results of these studies express that even though glucose-lowering treatment decreases the risk of cardiovascular diseases, the risks of macrovascular and microvascular complications still persist, and the development of new therapeutic strategies is required. It has been advised that the main reason is that the interventions are implemented too late after the diagnosis of the disease. UKPDS and STENO-2 studies backed this work, wherein treatment of chronic hyperglycemia was undertaken at the initial stages of the disease [17, 18]. Lower glycemia and long-term reduction of the risk of macrovascular and microvascular complications is resulted by these interventions. However, most of the prevailing method used for therapies are not fully efficacious and therefore, there is a serious requirement for a better interpretation of the molecular mechanisms in T2DM development and its issues and eventually identify preferably therapeutic targets.
Small endogenous non-coding RNA molecules, 18–25 nucleotides in length are highly conserved, known as micro RNAs (miRNAs or miR). Their role in regulation of many key biological functions including the maintenance of cellular signalling in both physiological and pathophysiological states and the regulation of entire pathways have been reported. When miRNA regulation is altered, it may result into serious physiological complications, including chronic disorder, like diabetes and its associated complications. miRNA regulates post-transcriptional gene expression by binding to their target messenger RNAs (mRNAs), resulting in mRNA degradation or suppression of translation [19–21]. It has been proven that these molecules can be very encouraging remedial goal and have substantial promise for diagnostic biomarkers for the various diabetic problems. Thus, in this review we summarizes the most of the advanced reporting concerning the task of miRNA in the widespread pathophysiology of diabetic retinopathy.
Diabetic Retinopathy
Diabetic retinopathy is one of the important complications of diabetic patients, eventually leading to visual impairment in aged 40–74 years. Main feature is spectrum of lesions within the retina [22, 23]. The reason behind DR are capillary degeneration, vascular permeability, capillary microaneurysms and excessive neovascularization. Death of some cells leads to neural retina dysfunctional, which changes retinal electrophysiology and results in an inability to discriminate between colons. Clinically, diabetic retinopathy has been divided into non-proliferative and proliferative disease stages. In the preliminary phase, hyperglycaemia may result into intramural pericyte death and thickening of the basement membrane, which made changes in the integrity of blood vessels within the retina, altering the blood-retinal barrier and vascular permeability [22]. In condition of non-proliferative diabetic retinopathy, most people do not observe any visual impairment.
Occlusions or degeneration of retinal capillaries are correlated with deteriorating prognosis24, which is most likely the result of ischemia followed by immediate secretion of angiogenic factors related to hypoxia. These conditions develop the disease into the proliferative phase where neovascularization and accumulation of fluid within the retina, termed macula edema, causes visual impairment. There is bleeding with linked distorting of the retinal architecture in severe cases, including development of a fibrovascular membrane which can leads to retinal detachment [22].
Diabetic retinopathy develops in many years, and about all patients with type 1 diabetes [23, 25], and most of the having type 2 diabetes [26], presents some retinal lesions after 15–20 years of disease. Moreover, the major vision threatening retinal disorder in type 1 diabetes appears to be proliferative retinopathy [27], while in type 2 diabetes there is a higher incidence of macula edema. Nevertheless, only a few of such cases will have progression leading into diminished vision.
Micro RNA and Dysregulation
In 1993, miRNAs were first identified in the nematode organism (Caenorhabditis elegans) [28–30]. After a long research lin-4 became first reported miRNA. It (lin-4) was successfully downregulated a nuclear protein called lin-14, thus initiating the second stage in larval development [30, 31]. After some time a second miRNA, let-7, had been identified in C. elegans that apparently to be highly conserved among species including humans [32, 33]. Now 35,828 mature miRNAs including 2000 in human being, have been reported across all species and registered in miRbase [35, 36].
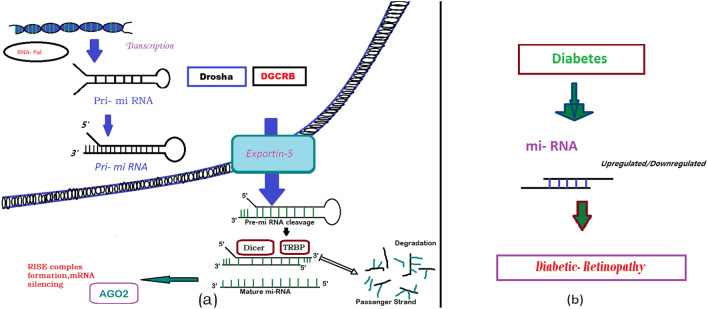
“Mature miRNAs are small noncoding ribonucleotides (~ 22 nt) capable of recognizing and binding to partially complementary sequences within the 3’ untranslated region (UTR) of specific mRNAs”[36]. Most miRNAs function by mediating the degradation or translational inhibition of mRNAs. [36, 37] (Fig. 1) Many authors have reported that “miRNAs also can affect translation and gene expression in a positive manner” [38–40], by pairing with the target mRNA through their seed region at the 5’ end. Now, miRNA synthesis and processing start as, miRNAs are transcribed as pri-miRNAs and processed to pre-miRNAs by the set of enzyme Drosha and DGCR8 inside the nucleus. [41] After transport into the cytoplasm pre-miRNAs are processed to mature miRNAs by the Dicer complex, and the obtained double-stranded RNA associates with the RNA-induced silencing complex that mediates the interaction of miRNAs with target mRNAs. In addition, most of the preliminary studies on miRNA function used deletions of Dicer [42], Drosha [43], DGCR8 [44], and Ago2 [45] genes. While the homozygous deletion of Dicer is embryonic lethal in mice and zebrafish [46] and tissue specific deletions of Dicer have been used to study the role of miRNAs in various cell types.
Fig. 1.
a primary miRNA (pri-miRNA) transcript is encoded in the cell's DNA and transcribed in the nucleus, processed by an enzyme Dosha and exported into the cytoplasm where it is further processed by Dicer. After strand separation, the mature miRNA represses protein production either by blocking translation or causing transcript degradation [88]. b Upregulated or down regulated miRNA causes diabetic retinopathy
As of now 2500 mature miRNA sequences has been identified which signify greater than 5% of all genes. Although many miRNAs be present in miRNA families with similar seed sequences [41]. Now it has been considered that each miRNA family regulates more than 300 different target mRNAs and close to 50% of target mRNAs have binding sites for two or more miRNAs [41, 47, 48], regulate more than 75% of mRNAs in a cell [49]. So, entire protein networks and signalling pathways are regulated by miRNAs. They play important role in development, neuronal cell fate, apoptosis, and cell proliferation. Many altered miRNAs is responsible in disease progression including cancer, diabetes, neurological disorders, autoimmune and cardiovascular diseases. Even though miRNAs play important roles in diverse aspects of signalling and metabolic control while their exact function and targets of most of the identified miRNAs remain unknown.
Similarly other biomarkers, miRNAs are now increasingly accepted as biomarkers of disease progression. Keeping plasma glucose level normal, requires the production and secretion of insulin from pancreas, which then acts on insulin-sensitive tissues, including muscle, liver, and adipocytes. The first miRNA was identified in pancreatic islets as micro-RNA (miR)-375, control glucose homeostasis [50]. Afterward, many miRNAs have been recognized with key functions in pancreatic β-cells [51, 52]. Now, it is regarded as true that circulating miRNAs may represent a sensitive and more accurate assessment of diabetic retinopathy progression when compared to conventional clinical examination and/or analysis of retinal images.
McArthur et al. reported that “miR-200b, which targets VEGF-A, was found down regulated in the retinas of diabetic rats”. Moreover, “in vitro exposure of HUVECs and bovine retinal capillary endothelial cells (BRECs) to HG resulted in a downregulation of miR-200b and an upregulation in VEGF-A mRNA” [53]. As we know that NF-kB is a potent regulator of the immune response and play a very important role in the prior pathogenesis of DR by activating a pro-apoptotic instance in retinal pericytes [54]. miRNAs (miR-146, miR-155, and miR-21) which are regarded as transcriptionally regulated by NF-kB [55–57] were also revealed to be upregulated in the retinal ECs of diabetic rats [58].
Another group reported that “by using microarray screening assays, found that 304 miRNAs were differentially expressed in the transforming growth factor (TGF)-2-induced epithelial-mesenchymal transition of human retinal pigment epithelium cells”. It is an important finding because this incident is essential during the progress of proliferative vitreoretinopathy. Moreover, TGF-2 can induce cell migration as well as moderate fibrosis [59] and thus it will be important to pinpoint the targets for these miRNAs. So, endothelial cell damage is one of the important key mechanisms which causing retinal microvascular injury in diabetes.
Recent results have identified a novel mechanism by which “miR-195 regulates sirtuin (SIRT)-1 mediated tissue damage in diabetic retinopathy”. The expression of miR-195 was found to be upregulated in retinas of diabetic rats while intra-vitreal injection of antagomiR-195 ameliorated levels of SIRT-1. Hyperglycemia has been suggested as the cause for high miR-195 expression levels found (Table 1) [60]. In addition, the pathogenesis of diabetic retinopathy is affected by two very important factor, hypoxia-inducible factor-1α (HIF1) and VEGF [61]. Ling and colleagues recently found that “there is a cross-talk between HIF1 and VEGF through interaction with their common miRNAs, such as miR-106a, silencing either HIF1 or VEGF increased the availabilities of the shared miRNAs” [62]. Furthermore, “it has been shown that miR-126 is not only downregulated under hypoxic conditions in vivo and in vitro, but it can also modulate the expression of VEGF and MMP-9 protein levels in monkey chorioretinal vessel endothelial cells (RF/6A)” [63]. Moreover, it has been noticed that miR-200b, implicated in the controlling of oxidation resistance (Oxr)-1, a protein that regulate the sensitivity of neuronal cells to oxidative stress, in the retinas of Akita mice (a model of type 1 diabetes] wherein it appears to be upregulated [65), whereas in another study the “same miRNA has been found downregulated upon high glycemia in diabetic retinas and endothelial cells, having VEGF as a possible target”[53]. In an in-vitro study, a cell line of retinal pigment epithelial (RPE) cells and human retinal endothelial cells (HRECs) after exposure of higher glucose mimicked the DR progression and resulted in hyperglycemia-induced HREC/RPE apoptosis. They found miR-124/-125b, miR-135b/-199a levels decreased with DR progression in vitro. Moreover, miR-145/-146a expression decreased by degrees in high-glucose-treated HRECs, but increased in hyperglycemia-exposed RPE cells [65] Zhou et al. [66] reported that “protein levels of PPARa were altered by transfection of miR-21 mimic or miR-21 inhibitor in cultured cells and that the deletion of miR-21 binding sites abolished its regulatory effects on PPARγ, suggesting miR-21 regulates PPARγ by targeting its 3’UTRs in DR” [87]. These results could be very helpful from a therapeutic perspective, since miRNAs are responsible for neovascularization, matrix protein accumulation and vascular permeability, all are cause for loss of vision. More work needs to be done in order to recognise and validate the specific targets and cascades that can be modulated by some of these differentially expressed biomarkers.
Table 1.
Summary of miRNAs that are involved in "diabetic retinopathy"
| S. No | miRNA | Model | Technique | miRNA function |
|---|---|---|---|---|
| 1 | miR-132 [58] | Animal (Sprague–Dawley rats) | miRNA RT-PCR arrays (Taqman; Applied Biosystem) | Upregulated with increased NF-kB, ICAM-1 and MCP-1, in diabetic retinal endothelial cells and retina |
| miR-146 [58] | Animal (Sprague–Dawley rats) | miRNA RT-PCR arrays (Taqman; Applied Biosystem) | Upregulated and ativated by NF-κB in the Retinal endothelial cell of diabetic rats | |
| miR-155 and miR-21 [58] | Animal (Sprague–Dawley rats) | miRNA RT-PCR arrays (Taqman; Applied Biosystem) | Upregulated and activated by both VEGF and NF-κB in the Retinal endothelial cell of diabetic rats | |
| 2 | miR-34 family [58] | Animal (Sprague–Dawley rats) | miRNA RT-PCR arrays (Taqman; Applied Biosystem) | Upregulated in diabetic rats upon VEGF and p53 responses in retinas |
| 3 | miR-34a [67] | Cell Line | MicroRNA Assay (TaqMan; Applied Biosystems) | Downregulated and can inhibit proloiferation of retinal pigment epithelial cells |
| 4 | miR-29b [68] | Animal (Wistar rats) | MicroRNA Assay (TaqMan; Applied Biosystems) | Upregulated, potential target is cellular activator of x cellular activator of PKR, RAX (PKR activator X], in retinal ganglion cells |
| 5 | miR-195 [69] | Cell Line (Human retinal pigment and dermal microvascular epithelium) | One Step RT-PCR | Upregulated in retinas of diabetic rats. Regulates sirtuin 1 mediated tissue damage, in human retinal and dermal microvascular endothelial cells |
| 6 | miR-200b [53, 64] | Animal (Akita mice) | Microarray and quantitative RT-PCR | Downregulated upon high glycemia with VEGF as a direct target, in diabetic retinas and endothelial cells. Upregulated in Akita mouse retinas. Regulates the expression of oxidation resistance-1 |
| 7 | miR-126 [63] | Animal (Monkey chorioretinal vessel endothelial cells and Wistar rats) | Real time PCR (SYBR Green) | Downregulated by hypoxia and reduced in the retinal tissue of streptozotocin-induced diabetic rats. Inhibition of VEGF and MMP-9 |
| 8 | miR-27b and miR-320a [70] | Human | TaqMan miRNA assays | Downregulated and upregulated, involve in progression of retinopathy in patients with T1D |
| 9 | miR-21, miR-181c, miR-1179 [71] | Human | TaqMan Low Density Array and real-time PCR | Upregulated and act as biomarker for proliferative daibetic retinopathy |
| 10 | miR-21 [72] | Human | Quantitative real-time PCR | Upregulated and involved in beta cell dysfunction |
| 11 | miR-93 [73] | Human | qRT-PCR | Upregulated and promote the tumor progression |
Future Perspective
In diabetes mellitus specially in type 2, the research on miRNA are still debatable. Substantially, research on miRNA biomarkers in DR has been hypothesis driven and non-hypothesis-driven research is still in its early phase in the area of diabetes. Though, both upgraded accessibility of biobanks that store samples properly from clinical cohorts, and increasing experience with the use of both cognitive genomics and biostatistics is likely to change this scenario in the next future. An open omic approach to get “Molecular signatures,” are likely to better research based on individual biomarkers as single biomarkers can hardly reflect the biological complexity of the primary microvascular injury.
As miRNAs can regulate gene expression using different strategy, together with mRNA cleavage, translational repression and de-adenylation. As we know, miRNAs have swiftly come up as promising targets for the development of novel therapeutics. Most prevailing technique to target specific miRNA are Double-stranded (ds) miRNA mimics and anti-mRNA antisense oligo deoxyribonucleotide. Though miRNAs have a specific and defined target in the pathogenic mechanism of the disease but miRNA-based therapy comes with the trump card that they point many genes engaged in the similar pathway process [74]. Now, one of the big alarms with the miRNA-based therapy is their proper and safe delivery, as these modulators must leave the circulatory system to get into the target tissue and should be able to cross blood-retina barrier. In addition, the second relevant issue is their half-life in circulation. In this modern age with advanced technology in drug delivery system, could open up the use of miRNAs for diabetic retinopathy.
Silencing DR-Inducing miRNAs
Now four ways open the door to silence the miRNA, (1) anti-miRNA oligonucleotides (AMOs), (2) miRNA-inhibiting natural agents, (3) miRNA sponges, (4) gene knockout [75]. All four methods are in a nutshell introduced below.
Anti-miRNA Oligonucleotides (AMOs)
AMOs are designed in a such manner to complement miRNAs that are stopped from binding to their target and specific sequences [75]. Nevertheless, There is significant hurdle in delivery of AMOs in vivo to their successful implementation in therapeutics. Authors reported that “Chemical modification of AMOs can be beneficial by improving hybridization affinity for the target mRNA, resistance to nuclease degradation, or activation of RNaseH or other proteins involved in the terminating mechanism”[76]. 2’-O-Me modification as well as the 2’- O-methoxyethyl (2’-MOE) and 2’-fluoro (2’-F) chemistries is modified at the 2’ position of the sugar moiety, while LNA comprises a group of bicyclic RNA analogues in which the furanose ring in the sugar-phosphate backbone is chemically locked in an RNA mimicking N-type (C3’-endo) conformation by the introduction of a 2’-O,4’-C methylene bridge [77–81]. LNA has shown the highest affinity towards complementary in amid these chemical modifying methods [82, 83].
miRNA-Inhibiting Natural Agents
Some of the very important natural agents (product) derived from food material are illustrated to have miRNA inhibiting effect. Curcumin and its analogue CDF were found to downregulate miR-21, which is a key miRNA in tumour aggressiveness. Curcumin (diferuloylmethane), a natural compound, is a bright yellow chemical produced by plants of the Curcuma longa species, commonly used as a kitchen spice, is exhibited to modulate a number of histone modifying enzymes and miRNAs” [84], and author’s earlier work has shown that “curcumin ameliorates retinal metabolic abnormalities postulated to be important in the development of diabetic retinopathy” [85]. Thus, natural compounds (product) could be very useful in curbing the progression of retinopathy in diabetic subjects via regulating both metabolic abnormalities and epigenetic modifications.
miRNA Sponges
The microRNA (miRNA) “sponge” method was presented three years ago as a means to form continuous miRNA loss of function in cell lines and transgenic organisms. As we know, miRNA sponges contain complementary binding sites to the seed region of the miRNA of interest, which permits them to obstruct an entire group of associated miRNAs. Thus, subcloning the miRNA binding site region into a vector containing a U6 small nuclear RNA promoter with 50 and 30 stem-loop elements are used to transfer sponges in to cell [86].
Conclusion
In conclusion, our understanding of the importance of miRNAs in the pathogenesis of DR has grown substantially, and intervention studies in experimental animals indicate that treatments targeting miRNAs can be beneficial. Furthermore, the progress of a highly sensitive, reliable and less invasive test with high predictive power for clinical stratification of DR will provide a substitute marker for clinical trials, give better prognosis information and steer development of future therapies focused towards preventing the vascular complications of diabetes.
Acknowledgements
Funding was provided by Science and Engineering Research Board (SERB) Start Up Research Grant (Young Scientist) (Sanction order No. YSS/2015/000054).
Author Contribution
Study concept and design: Sadashiv; drafting of the manuscript: Praveen Sharma; Checked and corrected: Sunita Tiwari, Shailendra Dwivedi, Amit Pal & Pankaj Kumar Singh.
Declarations
Conflict of interest
There is no conflict of interest.
Ethical Approval
This article doesn’t require ethical clearance certificate as it is a review article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus-present and future perspectives. Nat Rev Endocrinol. 2012;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- 2.Moura LI, Dias AM, Carvalho E, de Sousa HC. Recent advances on the development of wound dressings for diabetic foot ulcer treatment: a review. Acta Biomater. 2013;9:7093–7114. doi: 10.1016/j.actbio.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 3.Whiting DR, Guariguata L, Weil C, Shaw J. Idf diabetes atlas: Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94:311–321. doi: 10.1016/j.diabres.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 4.Reaven GM. Pathophysiology of insulin resistance in human disease. Physiol Rev. 1995;75:473–486. doi: 10.1152/physrev.1995.75.3.473. [DOI] [PubMed] [Google Scholar]
- 5.DeFronzo RA, Bonadonna RC, Ferrannini E. Pathogenesis of niddm: a balanced overview. Diabetes Care. 1992;15:318–368. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- 6.Fontbonne A, Eschwege E, Cambien F, Richard JL, et al. Hypertriglyceridaemia as a risk factor of coronary heart disease mortality in subjects with impaired glucose tolerance or diabetes. Results from the 11-year follow-up of the paris prospective study. Diabetologia. 1989;32:300–304. doi: 10.1007/BF00265546. [DOI] [PubMed] [Google Scholar]
- 7.Srikanth S, Deedwania P. Primary and secondary prevention strategy for cardiovascular disease in diabetes mellitus. Cardiol Clin. 2011;29:47–70. doi: 10.1016/j.ccl.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Yun JS, Ko SH, Kim JH, Moon KW, Park YM, Yoo KD, et al. Diabetic retinopathy and endothelial dysfunction in patients with type 2 diabetes mellitus. Diabetes Metab J. 2013;37:262–269. doi: 10.4093/dmj.2013.37.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein R, Moss SE, Klein BE, Davis MD, DeMets DL. Wisconsin epidemiologic study of diabetic retinopathy. XII. Relationship of c-peptide and diabetic retinopathy. Diabetes. 1990;39:1445–1450. doi: 10.2337/diab.39.11.1445. [DOI] [PubMed] [Google Scholar]
- 10.Andersen AR, Christiansen JS, Andersen JK, Kreiner S, Deckert T. Diabetic nephropathy in type 1 (insulin-dependent) diabetes: an epidemiological study. Diabetologia. 1983;25:496–501. doi: 10.1007/BF00284458. [DOI] [PubMed] [Google Scholar]
- 11.Higgins GC, Coughlan MT. Mitochondrial dysfunction and mitophagy: The beginning and end to diabetic nephropathy? Br J Pharmacol. 2014;171:1917–1942. doi: 10.1111/bph.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SK, Lee KJ, Hahm JR, Lee SM, Jung TS, et al. Clinical significance of the presence of autonomic and vestibular dysfunction in diabetic patients with peripheral neuropathy. Diabetes Metab J. 2012;36:64–69. doi: 10.4093/dmj.2012.36.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da Silva L, Carvalho E, Cruz MT. Role of neuropeptides in skin inflammation and its involvement in diabetic wound healing. Exp Opin Biol Ther. 2010;10:1427–1439. doi: 10.1517/14712598.2010.515207. [DOI] [PubMed] [Google Scholar]
- 14.Hata J, Arima H, Zoungas S, Fulcher G, Pollock C, et al. Effects of the endpoint adjudication process on the results of a randomised controlled trial: the advance trial. PLoS ONE. 2013;8:e55807. doi: 10.1371/journal.pone.0055807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor PJ, Ismail-Beigi F. Near-normalization of glucose and microvascular diabetes complications: data from accord and advance. Ther Adv Endocrinol Metab. 2011;2:17–26. doi: 10.1177/2042018810390545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchi C, Del Prato S. Metabolic memory and individual treatment aims in type 2 diabetes-outcome-lessons learned from large clinical trials. Rev Diabet Stud. 2011;8:432–440. doi: 10.1900/RDS.2011.8.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008;359:1577–1589. doi: 10.1056/NEJMoa0806470. [DOI] [PubMed] [Google Scholar]
- 18.Gaede P, Lund-Andersen H, Parving HH, Pedersen O. Effect of a multifactorial intervention on mortality in type 2 diabetes. N Engl J Med. 2008;358:580–591. doi: 10.1056/NEJMoa0706245. [DOI] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maqbool R, Hussain MU. MicroRNAs and human diseases: diagnostic and therapeutic potential. Cell Tissue Res. 2014;013:1787–1793. doi: 10.1007/s00441-013-1787-3. [DOI] [PubMed] [Google Scholar]
- 22.Frank RN. Diabetic retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 23.Hirai FE, Tielsch JM, Klein BE, Klein R. Ten-year change in vision-related quality of life in type 1 diabetes: Wisconsin epidemiologic study of diabetic retinopathy. Ophthalmology. 2011;118:353–358. doi: 10.1016/j.ophtha.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresnick GH, Engerman R, Davis MD, de Venecia G, Myers FL. Patterns of ischemia in diabetic retinopathy. Trans Sect Ophthalmol Am Acad Ophthalmol Otolaryngol. 1976;81:694–709. [PubMed] [Google Scholar]
- 25.Roy MS, Klein R, O’Colmain BJ, Klein BE, Moss SE, et al. The prevalence of diabetic retinopathy among adult type 1 diabetic persons in the United States. Arch Ophthalmol. 2004;122:546–551. doi: 10.1001/archopht.122.4.546. [DOI] [PubMed] [Google Scholar]
- 26.Kempen JH, O’Colmain BJ, Leske MC, Haffner SM, Klein R, et al. The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol. 2004;122:552–563. doi: 10.1001/archopht.122.4.552. [DOI] [PubMed] [Google Scholar]
- 27.Klein R, Knudtson MD, Lee KE, Gangnon R, Klein BE. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: XXII. the twenty-five-year progression of retinopathy in persons with type 1 diabetes. Ophthalmology 2008;115: 1859–1868. [DOI] [PMC free article] [PubMed]
- 28.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 1989;57:49–57. [DOI] [PubMed]
- 29.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 1981; 24:59–69. [DOI] [PubMed]
- 30.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843–54. [DOI] [PubMed]
- 31.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 32.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 33.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 34.XXX. http://mirbase.org, Release 21, accessed June 2014.
- 35.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 37.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 38.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 41.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205(9):2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21(16):1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35(3):217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 47.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 48.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 49.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 50.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 51.Dumortier O, Van Obberghen E. MicroRNAs in pancreas development. Diabetes Obes Metab. 2012;14(suppl 3):22–28. doi: 10.1111/j.1463-1326.2012.01656.x. [DOI] [PubMed] [Google Scholar]
- 52.Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, et al. Emerging roles of non-coding RNAs in pancreatic β-cell function and dysfunction. Diabetes Obes Metab. 2012;14(suppl 3):12–21. doi: 10.1111/j.1463-1326.2012.01654.x. [DOI] [PubMed] [Google Scholar]
- 53.McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011;60:1314–1323. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kowluru RA, Koppolu P, Chakrabarti S, Chen S. Diabetes-induced activation of nuclear transcriptional factor in the retina, and its inhibition by antioxidants. Free Radic Res. 2003;37:1169–1180. doi: 10.1080/10715760310001604189. [DOI] [PubMed] [Google Scholar]
- 55.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, et al. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 58.Kovacs B, Lumayag S, Cowan C, Xu S. microRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Ye S, Xiao W, Luo L, Liu Y. Differentially expressed microRNAs in tgfbeta2-induced epithelial-mesenchymal transition in retinal pigment epithelium cells. Int J Mol Med. 2014;33:1195–1200. doi: 10.3892/ijmm.2014.1688. [DOI] [PubMed] [Google Scholar]
- 60.Mortuza R, Feng B, Chakrabarti S. Mir-195 regulates sirt1-mediated changes in diabetic retinopathy. Diabetologia. 2014;57:1037–1046. doi: 10.1007/s00125-014-3197-9. [DOI] [PubMed] [Google Scholar]
- 61.Bento CF, Fernandes R, Matafome P, Sena C, Seica R, et al. Methylglyoxal-induced imbalance in the ratio of vascular endothelial growth factor to angiopoietin 2 secreted by retinal pigment epithelial cells leads to endothelial dysfunction. Exp Physiol. 2010;95:955–970. doi: 10.1113/expphysiol.2010.053561. [DOI] [PubMed] [Google Scholar]
- 62.Ling S, Birnbaum Y, Nanhwan MK, Thomas B, Bajaj M, et al. MicroRNA-dependent cross-talk between vegf and hif1alpha in the diabetic retina. Cell Signal. 2013;25:2840–2847. doi: 10.1016/j.cellsig.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 63.Ye P, Liu J, He F, Xu W, Yao K. Hypoxia-induced deregulation of mir-126 and its regulative effect on vegf and mmp-9 expression. Int J Med Sci. 2014;11:17–23. doi: 10.7150/ijms.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murray AR, Chen Q, Takahashi Y, Zhou KK, Park K, et al. MicroRNA-200b downregulates oxidation resistance 1 (oxr1) expression in the retina of type 1 diabetes model. Invest Ophthalmol Vis Sci. 2013;54:1689–1697. doi: 10.1167/iovs.12-10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiaoyun G, Jianan X, Yang L, Ying L, Guanfang S. Differentially expressed MicroRNAs in the development of early diabetic retinopathy. J Diabetes Res. 2017;2017:4727–4942. doi: 10.1155/2017/4727942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JYJ, et al. MicroRNA-21 targets peroxisome proliferatorsactivated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci USA. 2011;108:10355–10360. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou Q, Tang J, Wang Z, Wang C, Chen X, et al. Inhibitory effect of microRNA-34a on retinal pigment epithelial cell proliferation and migration. Invest Ophthalmol Vis Sci. 2013;54:6481–6488. doi: 10.1167/iovs.13-11873. [DOI] [PubMed] [Google Scholar]
- 68.Silva VA, Polesskaya A, Sousa TA, Correa VM, Andre ND, et al. Expression and cellular localization of microRNA-29b and rax, an activator of the RNA-dependent protein kinase (pkr), in the retina of streptozotocin-induced diabetic rats. Mol Vis. 2011;17:2228–2240. [PMC free article] [PubMed] [Google Scholar]
- 69.Mortuza R, Feng B, Chakrabarti S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia. 2014;57(5):1037–1046. doi: 10.1007/s00125-014-3197-9. [DOI] [PubMed] [Google Scholar]
- 70.Zampetaki A, Willeit P, Burr S, Yin X, Langley SR, et al. Angiogenic micro- RNAs linked to incidence and progression of diabetic retinopathy in type 1 diabetes. Diabetes. 2016;65(1):216–227. doi: 10.2337/db15-0389. [DOI] [PubMed] [Google Scholar]
- 71.Qing S, Yuan S, Yun C, Hui H, Mao P, et al. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem. 2014;34:1733–1740. doi: 10.1159/000366374. [DOI] [PubMed] [Google Scholar]
- 72.Jiang Q, Lyu XM, Yuan Y, Wang L. Plasma miR-21 expression: an indicator for the severity of type 2 diabetes with diabetic retinopathy. Biosci Rep. 2017;37:2. doi: 10.1042/BSR20160589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou HL, Wang Y, Gang Q, Zhang Y, Sun Y. Plasma level of miR-93 is associated with higher risk to develop type 2 diabetic retinopathy. Graefe's Arch Clin Exp Ophthalmol. 2017;255(6):1159–1166. doi: 10.1007/s00417-017-3638-5. [DOI] [PubMed] [Google Scholar]
- 74.Caroli A, Cardillo MT, Galea R, Biasucci LM. Potential therapeutic role of microRNAs in ischemic heart disease. J Cardiol. 2013;61(5):315–320. doi: 10.1016/j.jjcc.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 75.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16(11):2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3(1):1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44(1):55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, et al. Potent inhibition of microRNA in vivo without degradation. Nucl Acids Res. 2009;37(1):70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esau CC, Monia BP. Therapeutic potential for microRNAs. Adv Drug Deliv Rev. 2007;59:101–114. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 80.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21(2):74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 81.Stenvang J, Kauppinen S. MicroRNAs as targets for antisense-based therapeutics. Exp Opin Biol Therapy. 2008;8(1):59–68. doi: 10.1517/14712598.8.1.59. [DOI] [PubMed] [Google Scholar]
- 82.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8(1):1–7. doi: 10.1016/S1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 83.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucl Acids Res. 2006;34(8):2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Howell JC, Chun E, Farrell AN, Hur EY, Caroti CM, et al. Global microRNA expression profiling: curcumin (diferuloylmethane) alters oxidative stress-responsive microRNAs in human ARPE-19 cells. Mol Vis. 2013;19:544–560. [PMC free article] [PubMed] [Google Scholar]
- 85.Kowluru RA, Kanwar M. Effects of curcumin on retinal oxidative stress and inflammation in diabetes. Nutri Metab. 2007;4:8. doi: 10.1186/1743-7075-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in ammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J, Wang KC, Wu W, et al. MicroRNA-21 targets peroxisome proliferators activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Nat Acad Sci USA. 2011;108:10355–10360. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.George SM. MicroRNA gets down to business. Nat Biotechnol. 2007;25(6):631–638. doi: 10.1038/nbt0607-631. [DOI] [PubMed] [Google Scholar]