Abstract
Ensifer adhaerens is a soil bacterium that attaches to other bacteria and may cause lysis of these other bacteria. Based on the sequence of its small-subunit rRNA gene, E. adhaerens is related to Sinorhizobium spp. E. adhaerens ATCC 33499 did not nodulate Phaseolus vulgaris (bean) or Leucaena leucocephala, but with symbiotic plasmids from Rhizobium tropici CFN299 it formed nitrogen-fixing nodules on both hosts. The nodule isolates were identified as E. adhaerens isolates by growth on selective media.
Rhizobia (Sinorhizobium, Allorhizobium, Mesorhizobium, Bradyrhizobium, Rhizobium, and Azorhizobium) form nitrogen-fixing nodules on the roots and stems of legumes. The genetic information for symbiosis is plasmid borne in Rhizobium and Sinorhizobium. Symbiotic plasmids may be eliminated, rendering the bacteria nonsymbiotic. Nonsymbiotic rhizobia exist naturally and can be more numerous than their symbiotic counterparts (16, 27).
Ensifer adhaerens strains are gram-negative soil bacteria that attach endwise to various living gram-positive and gram-negative bacteria and may cause lysis of these bacteria. E. adhaerens is a participant in a predatory chain involving other bacteria; however, it is an obligate predator only under nutrient limitation conditions. Its 16S rRNA gene sequence places E. adhaerens in the sinorhizobia (1).
Phaseolus vulgaris (bean), Vigna, and Macroptilium have been reported to be highly promiscuous hosts and are nodulated with a large range of rhizobia (18, 22, 25). We found that E. adhaerens ATCC 33499 did not form nodules on bean, Leucaena leucocephala, Vigna mungo, Macroptilium atropurpureum, or alfalfa when 10 plants of each species were grown in flasks with cotton, vermiculite, or agar as the support, as previously described (20). Thus, we wondered if E. adhaerens could become a bean and Leucaena nitrogen-fixing symbiont by acquiring symbiotic plasmids from a Rhizobium species. We chose Rhizobium tropici as the donor because R. tropici sym plasmids conferred on Agrobacterium tumefaciens the capacity to form nitrogen-fixing nodules on P. vulgaris and L. leucocephala (17).
R. tropici CFN299 and E. adhaerens ATCC 33499 are easily distinguishable by colony morphology on PY agar (5 g of peptone per liter, 3 g of yeast extract per liter, 0.7 g of calcium chloride per liter, 1.5% agar) plates; E. adhaerens produces larger amounts of slime and forms colonies faster than R. tropici. CFN299 does not grow in Luria broth (LB), while strain ATCC 33499 does. E. adhaerens ATCC 33499 is also resistant to 5 mg of gentamicin per liter, 100 mg of streptomycin per liter, 5 mg of chloramphenicol per liter, and 300 mg of erythromycin per liter, while R. tropici CFN299 is sensitive to all of these antibiotics.
No nifH genes were detected in Ensifer either by Southern blot hybridization or by PCR performed with nifH primers (6) (Table 1). Additionally, no nod gene products were obtained with E. adhaerens ATCC 33499 in a PCR with nodC primers 251F and 566R (28) or with nodBC primers (nodB 31 [TACCTGACSTTVGACGACGGTCC] and nodC RR [GAGACGGCGRCRRTGCTGGTTG]) that we have used to amplify nodBC or nodC gene sequences from Sinorhizobium meliloti, Sinorhizobium medicae, Sinorhizobium arboris, Sinorhizobium terangae, Sinorhizobium kostiense, Sinorhizobium saheli, Sinorhizobium fredii, R. tropici, and Rhizobium etli. The nucleotide sequences of the PCR products obtained with R. etli strains were determined and corresponded to the nodBC gene sequences (Claudia Silva, personal communication). No hybridization was obtained when the S. meliloti nodC PCR product was used as a probe in Southern blot hybridization with E. adhaerens ATCC 33499 total restricted DNA.
TABLE 1.
Strains and characteristics
| Straina | Small-subunit rRNA patternsb | R. tropici plasmidsc | nifH gened | Acetylene reduction activity with P. vulgaris nodules (nmol of ethylene/h/plant)e
|
|
|---|---|---|---|---|---|
| 14 days postinoculation | 18 days postinoculation | ||||
| E. adhaerens transconjugants | |||||
| CFNEA40 | ACEGIK | a, cf | + | 102.7 | NDg |
| CFNEA41 | ACEGIK | a, b, c | + | 104.8 | ND |
| CFNEA50 | ACEGIK | a, c | + | 108.0 | 84.9 |
| CFNEA51 | ACEGIK | a, c | + | 106.9 | 85.9 |
| CFNEA52 | ACEGIK | a, b, cf | + | 103.8 | 84.9 |
| CFNEA53 | ACEGIK | a, b, c | + | 115.3 | 96.4 |
| CFNEA54 | ACEGIK | b, c | + | 113.2 | 94.3 |
| CFNEA55 | ACEGIK | b, c | + | 126.9 | 104.8 |
| CFNEA56 | ACEGIK | b, c | + | 114.3 | 94.3 |
| E. adhaerens ATCC 33499 | ACEGIK | ||||
| R. tropici strains | |||||
| CFN299 | BDFHJL | + | |||
| CFN299 Tn5-mob-6 | 166.8 | ||||
| CFN299 Tn5-mob-7 | 182.4 | ||||
Only transconjugant CFNEA41 was derived from CFN299 Tn5-mob-7; all other transconjugants were derived from CFN299 Tn5-mob-6. R. etli CFN42 was also used in this study.
The different letters represent distinct patterns obtained with restriction enzymes HinfI, MspI, RsaI, HhaI, Sau3A1, and DdeI; each letter position refers to a different restriction enzyme used for PCR-synthesized 16S rRNA genes (15) obtained with primers rD1 and fD1 (29).
Eckhardt gel analysis was performed as modified (23) with liquid early-exponential-phase cultures in horizontal gels with sodium dodecyl sulfate in the agarose. The plasmid sizes were as follows: plasmid a, 185 kb; plasmid b, 220 kb; and plasmid c, 500 kb (nod and nif plasmid).
The presence of nifH was determined by Southern blot hybridization or by PCR synthesis with nif primers as described previously (6).
Acetylene reduction activity was determined 14 and 18 days postinoculation. The data are averages for five plants from one experiment. The plant-to-plant variation was not more than 10% for each transconjugant. The levels of nitrogen fixation with the E. adhaerens transconjugants were around 40% of the levels with the parental CFN299 strains in the four other experiments. Plants were maintained in a growth chamber at 28°C.
Plasmid cointegration or rearrangement occurred, which led to an approximately 700-kb plasmid or to an additional 300-kb plasmid.
ND, not determined.
R. tropici CFN299 Tn5-mob-6 and CFN299 Tn5-mob-7 were obtained by mating CFN299 and S17-1(pSUP5011) and were selected on the basis of their ability to transfer to Agrobacterium tumefaciens GMI9023 the capacity to form nodules on bean as previously described (17). R. tropici CFN299 Tn5-mob-6 and CFN299 Tn5-mob-7 were able to form nitrogen-fixing nodules when they were tested individually with bean plants (Table 1). R. tropici CFN299 Tn5-mob-6 and CFN299 Tn5-mob-7 were shown to have Tn5-mob in the nod-nif plasmid by hybridization of Eckhardt gels with Tn5 (data not shown).
E. adhaerens transconjugants obtained from matings on PY agar plates with R. tropici CFN299 Tn5-mob-6 and CFN299 Tn5-mob-7 were selected on LB containing 200 mg of neomycin per liter because E. adhaerens grows on LB containing 100 mg of neomycin per liter. Transconjugants grew in the presence of up to 800 mg of neomycin per liter, while the recipient E. adhaerens ATCC 33499 strain was sensitive to neomycin at concentrations greater than 100 mg per liter. Transconjugants CFNEA40 and CFNEA50 (from R. tropici CFN299 Tn5-mob-6) and CFNEA41 (from R. tropici CFN299 Tn5-mob-7) were selected.
Additionally, a mixture of E. adhaerens transconjugants derived from CFN299 Tn5-mob-6 and CFN299 Tn5-mob-7 was inoculated onto plants, and transconjugants CFNEA51 to CFNEA56 were selected from well-developed red nodules as follows. All bacteria isolated from bean nodules were recovered on PY medium, and 10 individual colonies per nodule were then tested for growth in LB containing 200 mg of neomycin per liter. All isolates on PY agar had the colony morphology of E. adhaerens ATCC 33499, not the colony morphology of R. tropici CFN299. All of the isolated colonies tested grew in LB containing 200 mg of neomycin per liter. One isolated colony from a nodule from each of six different plants was purified further by five serial steps involving colony isolation on LB containing 200 mg of neomycin per liter. The parental strain of transconjugants CFNEA51 to CFNEA56 was recognized on the basis of the size of the band hybridizing to a Tn5 probe in each Ensifer transconjugant. The transconjugants were all derived from CFN299 Tn5-mob-6, perhaps indicating that the Tn5-mob-7 insertion had affected some loci involved in competition for nodule formation. The gene interrupted by Tn5-mob-7 will be described elsewhere.
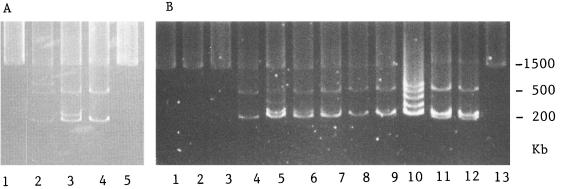
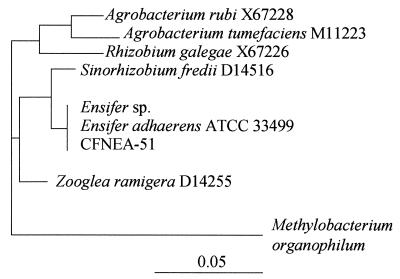
E. adhaerens ATCC 33499 harbors two megaplasmids, as revealed by the Eckhardt procedure. Ensifer transconjugants acquired two or three plasmids from the R. tropici donor strain (Table 1; Fig. 1). nif genes were detected in the Ensifer transconjugants by using nifH primers in a PCR (Table 1), and the total DNA restriction fingerprints of all transconjugants were identical to the E. adhaerens ATCC 33499 fingerprint (data not shown). Ribosomal fingerprints (15) were determined by 16S rRNA gene restriction enzyme digestion with HinfI, MspI, RsaI, HhaI, Sau3A1, and DdeI of PCR products generated with primers fD1 and rD1 (29) from all E. adhaerens transconjugants (Table 1), and the 16S rRNA gene sequence of E. adhaerens transconjugant CFNEA51 was determined (2). Almost the entire 16S rRNA gene was sequenced with an automated sequencer. The resulting sequence was hand aligned with selected comparison sequences, taking into consideration the secondary structure of the 16S rRNA molecule. The aligned sequences were then analyzed by the distance matrix method by using the FITCH option of the PHYLIP program (7). Distances were corrected by the method of Jukes and Cantor (14). Phylogenetic analysis of E. adhaerens transconjugant CFNEA51 (Fig. 2) indicated that the 16S rRNA gene sequence was identical to the original E. adhaerens ATCC 33499 sequence and the sequence of another strain of E. adhaerens included for comparison. As reported previously (1), the Ensifer strains were most closely related to Sinorhizobium spp.
FIG. 1.
Plasmids visualized by a modified Eckhardt procedure. (A) Lanes 1 and 5, E. adhaerens ATCC 33499; lane 2, CFNEA40; lane 3, CFNEA41; lane 4, CFNEA50. (B) Lanes 1 to 3 and 13, E. adhaerens ATCC 33499; lane 4, CFNEA51; lane 5, CFNEA52; lane 6, CFNEA53; lane 7, CFNEA54; lane 8, CFNEA55; lane 9, CFNEA56; lane 10, CFN42; lane 11, CFN299 Tn5-mob-6; lane 12, CFN299 Tn5-mob-7.
FIG. 2.
Phylogenetic tree based on the 16S rRNA gene sequence.
Since it has been found that E. adhaerens sticks very tightly to other bacteria and that separation from these bacteria is difficult (1, 4), great effort was expended to purify E. adhaerens transconjugants prior to inoculation of plants. The procedure used to purify all E. adhaerens transconjugants included several steps consisting of dilution with Tween 40 and plating on LB containing 200 mg of neomycin per liter for single-colony isolation before the transconjugants were tested in plant nodulation assays to determine levels of nitrogen fixation.
E. adhaerens transconjugants (Table 1) were found to form nitrogen-fixing nodules in five independent experiments with bean (three to five plants per strain were tested in each experiment). CFNEA50 to CFNEA56 also formed nitrogen-fixing nodules on L. leucocephala plants, which were green (Fig. 3), while all uninoculated control plants lacked nodules and were yellow. Leucaena plant development indicated that nitrogen was transferred to the plants. The identities of the strains in all bean and L. leucocephala nodules were verified by growing colonies isolated from nodules on LB containing 200 mg of neomycin per liter, and isolates from the more than 800 nodules tested corresponded to the Ensifer transconjugants. Tests for nodule surface sterility were performed for all nodules as described previously (17).
FIG. 3.
Leucaena plants (45 days old) inoculated with R. tropici CFN299 (plant 1), E. adhaerens transconjugant CFNEA51 (plant 2), and E. adhaerens ATCC 33499 (plant 3). Plant 4 was an uninoculated control plant.
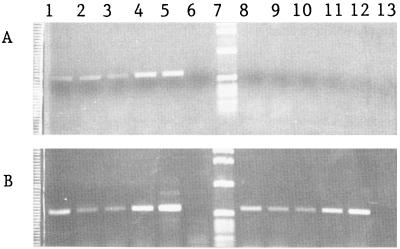
Twenty nodules recovered from different nitrogen-fixing plants inoculated with CFNEA51 and CFNEA53 were surface sterilized and individually macerated, and 1 drop of each preparation was placed on PY agar and analyzed for resistance to antibiotics as described above. The remainder of the nodule extract was used for PCR with nifH primers (6) or with R. tropici citrate synthase gene (11) primers P231p (AAGAAGCCCATTTGCTTCC) and P2318 (TTAACCCTTTGGCGCTTTTT), which yielded a 624-bp product. While PCR products were obtained with nifH primers from nodules formed by either R. tropici or E. adhaerens transconjugants, PCR products were obtained with citrate synthase gene primers from R. tropici nodules but not from E. adhaerens CFNEA51 or CFNEA53 nodule extracts or E. adhaerens ATCC 33499 purified DNA (Fig. 4). Citrate synthase gene PCR products were digested with MspI which yielded the expected digestion fragments. These results support the result that R. tropici CFN299 parental strains were not present as contaminants in E. adhaerens transconjugant nodules.
FIG. 4.
PCR products obtained directly from nodule extracts. Each surface-sterilized nodule was squeezed in 0.1 ml of sterilized water and extracted twice with phenol-chloroform-isoamyl alcohol and once with chloroform; the resulting preparation was precipitated with ethanol and resuspended in 15 μl of H2O, and 5-μl aliquots were used either with R. tropici citrate synthase gene primers (see text) (A) or with nifH gene primers (6) (B). Lanes 1 to 4, nodules formed by R. tropici strain CFN299; lane 5, R. tropici CFN299 DNA; lane 6, no-DNA control; lane 7, 1-kb DNA ladder marker; lanes 8 to 12, nodules from E. adhaerens transconjugants (lanes 8 and 9, CFNEA51; lane 10, CFNEA53; lanes 11 and 12, CFNEA51), lane 13, E. adhaerens ATCC 33499 DNA.
E. adhaerens ATCC 33499 was a suitable recipient for rhizobial symbiotic plasmids. Sym plasmid stability was assessed by using isolated colonies of CFNEA50 and CFNEA55 after growth for 100 generations in PY liquid medium without antibiotics. All 600 colonies tested (300 colonies of each transconjugant) were resistant to neomycin (200 mg/liter), suggesting that they harbored the R. tropici CFN299 symbiotic plasmid, but two of six E. adhaerens transconjugants lost the symbiotic plasmid after 2 months of storage at 4°C, indicating that there was some instability.
There was a short delay (1 or 2 days) in the onset of bean nodulation with Ensifer transconjugants CFNEA52 to CFNEA56 compared to the R. tropici mutant CFN299 Tn5-mob-6. At 16 days postinoculation, the average number of nodules elicited by transconjugants CFNEA52 to CFNEA56 was 80% of the number of nodules obtained with donor strain CFN299 Tn5-mob-6. The nodules formed by the transconjugants were red and large. There were no nodules on any of the control (uninoculated) bean plants.
In competition experiments, when a 1:1 or 1:1,000 ratio of R. tropici CFN299 to E. adhaerens transconjugant CFNEA53 was used to inoculate bean plants (a total of 106 bacteria were inoculated per plant), no nodules were formed by the E. adhaerens transconjugants since only R. tropici CFN299 was recovered from inside nodules. The nodule isolates did not form colonies on LB containing 200 mg of neomycin per liter. PCR analysis of some of the nodules revealed only CFN299 with the R. tropici ccsA gene primers. In the experiments in which we deliberatedly mixed R. tropici CFN299 with E. adhaerens transconjugants, we were able to easily distinguish nodules formed by R. tropici CFN299. This finding supports the notion that in the experiments described above the E. adhaerens transconjugants, and not a contaminating parental strain, really formed the nodules.
To test if E. adhaerens ATCC 33499 could adhere to and be introduced into nodules together with R. tropici CFN299, mixtures of the two bacteria in various proportions were inoculated onto roots of bean seedlings. R. tropici was found to be the sole occupant of the bean nodules. It has been reported that E. adhaerens does not attack A. tumefaciens, Rhizobium leguminosarum, or S. meliloti (9, 31). When mixtures of E. adhaerens and R. tropici were coinoculated onto bean plants, the numbers of nodules obtained were similar to the numbers of nodules obtained with R. tropici CFN299 alone, and all of the nodules were formed by CFN299. A similar result was obtained with an inoculum prepared from a coculture of R. tropici CFN299 and E. adhaerens grown in PY medium for 24 h. The ability of E. adhaerens to attack other bacteria has been reported to be dependent on the growth conditions (3).
Genes involved in uptake of bean root exudates have been located on plasmid c (which carries the nod-nif genes) and on plasmid a (180 kb) of R. tropici CFN299, and these uptake genes have a role in symbiosis (26). Plasmid b was found to contain symbiotic determinants that conferred a symbiotic advantage to A. tumefaciens harboring only plasmid c (17). We found that plasmids a and b were cotransferred from R. tropici CFN299, along with the nod-nif plasmid, into A. tumefaciens (17) or into Ensifer (this study). The A. tumefaciens transconjugant containing all three plasmids nodulated better and fixed more nitrogen than transconjugants containing only plasmid c (17). Nevertheless, in competition experiments with R. tropici CFN299, A. tumefaciens harboring R. tropici plasmids a, b, and c was not recovered from the nodules, indicating that this transconjugant was not as competitive for nodule formation as the wild-type R. tropici strain (20). Similarly, in this study we found that Ensifer transconjugant CFN299 containing R. tropici plasmids b and c was not as competitive as R. tropici wild-type strain CFN299.
Several reports have addressed the role of plasmids in rhizobia with regard to symbiosis and metabolism (24; reviewed in references 8 and 21). Catabolic genes (23) and genes for lipopolysaccharide (5) or exopolysaccharide (10, 13) biosynthesis are plasmid borne. Rhizobial plasmids have been transferred between different strains and species in the laboratory (reviewed in reference 19). S. meliloti transconjugants that acquired the R. leguminosarum nod-nif plasmids either formed ineffective nodules (non-nitrogen-fixing nodules) or no nodules on pea or vetch (12, 30). In these examples, S. meliloti contained its normal complement of symbiotic megaplasmids, and functional incompatibility of plasmids may have occurred.
Levels of DNA-DNA homology greater than 30% between E. adhaerens ATCC 33499 and all Sinorhizobium species (unpublished data) also support the hypothesis that these bacteria are closely related. Taken together, our data suggest that E. adhaerens might be a misclassified bacterium, seemingly a nonsymbiotic bacterium, but comprehensive polyphasic taxonomic characterization of E. adhaerens is required to clarify the taxonomic position of this organism. Additionally, it would be interesting to search for predatory activities in Sinorhizobium species.
Acknowledgments
We thank Julio Martínez for technical support, Claudia Silva and Valeria Souza for providing nodBC primers, and Michael Dunn for reading the manuscript.
This study was supported by PAPIIT DGAPA-UNAM grant IN201600.
REFERENCES
- 1.Balkwill, D. L. Ensifer. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. The proteobacteria, in press. Springer-Verlag, New York, N.Y.
- 2.Balkwill D L, Drake G R, Reeves R H, Fredrickson J K, White D C, Ringelberg D B, Chandler D P, Romine M F, Kennedy D W, Spadoni C M. Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int J Syst Bacteriol. 1997;47:191–201. doi: 10.1099/00207713-47-1-191. [DOI] [PubMed] [Google Scholar]
- 3.Casida L E., Jr Bacterial predators of Micrococcus luteus in soil. Appl Environ Microbiol. 1980;39:1035–1041. doi: 10.1128/aem.39.5.1035-1041.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casida L E., Jr Ensifer adhaerens gen. nov., sp. nov.: a bacterial predator of bacteria in soil. Int J Syst Bacteriol. 1982;32:339–345. [Google Scholar]
- 5.Cava J R, Elias P M, Turowski D A, Noel K D. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J Bacteriol. 1989;171:8–15. doi: 10.1128/jb.171.1.8-15.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eardly B D, Young J P W, Selander R K. Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl Environ Microbiol. 1992;58:1809–1815. doi: 10.1128/aem.58.6.1809-1815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1993 PHYLIP (phylogeny inference package), version 3.5c University of Washington, Seattle.
- 8.García-de los Santos A, Brom S, Romero D. Rhizobium plasmids in bacteria-legume interactions. World J Microbiol Biotechnol. 1996;12:119–125. doi: 10.1007/BF00364676. [DOI] [PubMed] [Google Scholar]
- 9.Germida J J, Casida L E., Jr Ensifer adhaerens predatory activity against other soil bacteria in soil, as monitored by indirect phage analysis. Appl Environ Microbiol. 1983;45:1380–1388. doi: 10.1128/aem.45.4.1380-1388.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 11.Hernández-Lucas I, Pardo M A, Segovia L, Miranda J, Martínez-Romero E. Rhizobium tropici chromosomal citrate synthase gene. Appl Environ Microbiol. 1995;61:3992–3997. doi: 10.1128/aem.61.11.3992-3997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooykaas P J J, Snijdewint F G M, Schilperoort R A. Identification of the sym plasmid of Rhizobium leguminosarum strain 1001 and its transfer to and expression in other rhizobia and Agrobacterium tumefaciens. Plasmid. 1982;8:73–82. doi: 10.1016/0147-619x(82)90042-7. [DOI] [PubMed] [Google Scholar]
- 13.Hynes M F, Simon R, Müller P, Niehaus K, Labes M, Pühler A. The two megaplasmids of Rhizobium meliloti are involved in the effective nodulation of alfalfa. Mol Gen Genet. 1986;202:356–362. [Google Scholar]
- 14.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 15.Laguerre G, Allard M-R, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laguerre G, Bardin M, Amarger N. Isolation from soil of symbiotic and nonsymbiotic Rhizobium leguminosarum by DNA hybridization. Can J Microbiol. 1993;39:1142–1149. [Google Scholar]
- 17.Martínez E, Palacios R, Sánchez F. Nitrogen-fixing nodules induced by Agrobacterium tumefaciens harboring Rhizobium phaseoli plasmids. J Bacteriol. 1987;169:2828–2834. doi: 10.1128/jb.169.6.2828-2834.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez E, Pardo M A, Palacios R, Cevallos M A. Reiteration of nitrogen fixation gene sequences and specificity of Rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris. J Gen Microbiol. 1985;131:1779–1786. [Google Scholar]
- 19.Martínez E, Romero D, Palacios R. The Rhizobium genome. Crit Rev Plant Sci. 1990;9:59–93. [Google Scholar]
- 20.Martínez-Romero E, Rosenblueth M. Increased bean (Phaseolus vulgaris L.) nodulation competitiveness of genetically modified Rhizobium strains. Appl Environ Microbiol. 1990;56:2384–2388. doi: 10.1128/aem.56.8.2384-2388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercado-Blanco J, Toro N. Plasmids in rhizobia: the role of nonsymbiotic plasmids. Mol Plant-Microbe Interact. 1996;9:535–545. [Google Scholar]
- 22.Michiels J, Dombrecht B, Vermeiren N, Xi C, Luyten E, Vanderleyden J. Phaseolus vulgaris is a non-selective host for nodulation. FEMS Microbiol Ecol. 1998;26:193–205. [Google Scholar]
- 23.Oresnik I J, Pacarynuk L A, O'Brien S A P, Yost C K, Hynes M F. Plasmid-encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol Plant-Microbe Interact. 1998;11:1175–1185. [Google Scholar]
- 24.Perret X, Freiberg C, Rosenthal A, Broughton W J, Fellay R. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol Microbiol. 1999;32:415–425. doi: 10.1046/j.1365-2958.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- 25.Perret X, Staehelin C, Broughton W J. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenblueth M, Hynes M F, Martínez-Romero E. Rhizobium tropici teu genes involved in specific uptake of Phaseolus vulgaris is bean-exudate compounds. Mol Gen Genet. 1998;258:587–598. doi: 10.1007/s004380050772. [DOI] [PubMed] [Google Scholar]
- 27.Segovia L, Piñero D, Palacios R, Martínez-Romero E. Genetic structure of a soil population of nonsymbiotic Rhizobium leguminosarum. Appl Environ Microbiol. 1991;57:426–433. doi: 10.1128/aem.57.2.426-433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Phylogeny of Sym plasmids of rhizobia by PCR-based sequencing of a nodC segment. J Bacteriol. 1995;177:468–472. doi: 10.1128/jb.177.2.468-472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijffelman C A, Pees E, Van Brussel A N N, Hooykaas P J J. Repression of small bacteriocin excretion in Rhizobium leguminosarum and Rhizobium trifolii by transmissible plasmids. Mol Gen Genet. 1983;192:171–176. [Google Scholar]
- 31.Zeph L R, Casida L E., Jr Gram-negative versus gram-positive (actinomycete) nonobligate bacterial predators of bacteria in soil. Appl Environ Microbiol. 1986;52:819–823. doi: 10.1128/aem.52.4.819-823.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]