Abstract
Proteins are some of the most fascinating and challenging molecules in the universe, and they pose a big challenge for artificial intelligence. The implementation of machine learning/AI in protein science gives rise to a world of knowledge adventures in the workhorse of the cell and proteome homeostasis, which are essential for making life possible. This opens up epistemic horizons thanks to a coupling of human tacit–explicit knowledge with machine learning power, the benefits of which are already tangible, such as important advances in protein structure prediction. Moreover, the driving force behind the protein processes of self-organization, adjustment, and fitness requires a space corresponding to gigabytes of life data in its order of magnitude. There are many tasks such as novel protein design, protein folding pathways, and synthetic metabolic routes, as well as protein-aggregation mechanisms, pathogenesis of protein misfolding and disease, and proteostasis networks that are currently unexplored or unrevealed. In this systematic review and biochemical meta-analysis, we aim to contribute to bridging the gap between what we call binomial artificial intelligence (AI) and protein science (PS), a growing research enterprise with exciting and promising biotechnological and biomedical applications. We undertake our task by exploring “the state of the art” in AI and machine learning (ML) applications to protein science in the scientific literature to address some critical research questions in this domain, including What kind of tasks are already explored by ML approaches to protein sciences? What are the most common ML algorithms and databases used? What is the situational diagnostic of the AI–PS inter-field? What do ML processing steps have in common? We also formulate novel questions such as Is it possible to discover what the rules of protein evolution are with the binomial AI–PS? How do protein folding pathways evolve? What are the rules that dictate the folds? What are the minimal nuclear protein structures? How do protein aggregates form and why do they exhibit different toxicities? What are the structural properties of amyloid proteins? How can we design an effective proteostasis network to deal with misfolded proteins? We are a cross-functional group of scientists from several academic disciplines, and we have conducted the systematic review using a variant of the PICO and PRISMA approaches. The search was carried out in four databases (PubMed, Bireme, OVID, and EBSCO Web of Science), resulting in 144 research articles. After three rounds of quality screening, 93 articles were finally selected for further analysis. A summary of our findings is as follows: regarding AI applications, there are mainly four types: 1) genomics, 2) protein structure and function, 3) protein design and evolution, and 4) drug design. In terms of the ML algorithms and databases used, supervised learning was the most common approach (85%). As for the databases used for the ML models, PDB and UniprotKB/Swissprot were the most common ones (21 and 8%, respectively). Moreover, we identified that approximately 63% of the articles organized their results into three steps, which we labeled pre-process, process, and post-process. A few studies combined data from several databases or created their own databases after the pre-process. Our main finding is that, as of today, there are no research road maps serving as guides to address gaps in our knowledge of the AI–PS binomial. All research efforts to collect, integrate multidimensional data features, and then analyze and validate them are, so far, uncoordinated and scattered throughout the scientific literature without a clear epistemic goal or connection between the studies. Therefore, our main contribution to the scientific literature is to offer a road map to help solve problems in drug design, protein structures, design, and function prediction while also presenting the “state of the art” on research in the AI–PS binomial until February 2021. Thus, we pave the way toward future advances in the synthetic redesign of novel proteins and protein networks and artificial metabolic pathways, learning lessons from nature for the welfare of humankind. Many of the novel proteins and metabolic pathways are currently non-existent in nature, nor are they used in the chemical industry or biomedical field.
Keywords: artificial intelligence, proteins, protein design and engineering, machine learning, deep learning, protein prediction, protein classification, drug design
Introduction
Protein science witnesses the most exciting and demanding revolution of its own field; the magnitude of its genetic–epigenetic—molecular networks, inhibitors, activators, modulators, and metabolite information—is astronomical. It is organized in an open “protein self-organize, adjustment and fitness space”; for example, a protein of 100 amino acids would contain 20100 variants, and a process of searching–finding conformations in a protein of 100 amino acids can adopt ∼1046 conformation and a unique native state, the protein data exceeding many petabytes (1 petabyte is 1 million gigabytes) (Kauffman, 1992).
Therefore, the use of artificial intelligence in protein science is creating new avenues for understanding the ways of organizing and classifying life within its organisms to eventually design, control, and improve this organization. In this respect, protein synthesis is a case in point. Indeed, the discovery of the underlying mechanism of protein synthesis is an inter-field discovery, that is, “a significant achievement of 20th century biology that integrated results from two fields: molecular biology and biochemistry” (Baetu, 2015). More recently, the field of protein science is, in turn, another inter-field enterprise, this time between molecular biology and computer science, or better said, between a cross-functional team of researchers (biochemists, protein scientists, protein engineers, system biology scientists, bioinformatics, between others). Nowadays, it is possible to classify, share, and use a significant number of structural biology databases helping researchers throughout the world. Once the mechanism of DNA for protein synthesis is deduced, it will then be possible to replicate it via computational strategies through artificial intelligence (AI) and machine learning (ML) algorithms that can provide important information such as pattern recognition, nearest neighbors, vector profiles, back propagation, among others. AI has been used to exploit this novel knowledge to predict, design, classify, and evolve known proteins with improved and enhanced properties and applications in protein science (Paladino et al., 2017; Wardah et al., 2019;Cheng et al., 2008; Bernardes and Pedreira, 2013), which, in turn, makes its way to solve complex problems in the “fourth industrial revolution” and open new areas of protein research, growing at a very fast speed.
The techniques of machine learning are a subfield of AI, which has become popular due to the linear and non-linear processed data and the large amount of available combinatorial spaces. As a result, sophisticated algorithms have emerged, promoting the use of neural networks (Gainza et al., 2016) However, in spite of the large amount of research done in protein science, as far as we know, there are neither systematic reviews nor any biochemical meta-analysis in the scientific literature informing, illuminating, and guiding researchers on the best available ML techniques for particular tasks in protein science; albeit there have been recent reviews such as the work of AlQuraishi (2021), Dara et al. (2021), and Hie and Yang (2022), which prove that this inter-field is on evolution. By a biochemical meta-analysis, we mean an analysis resulting from two processes: identification and prediction. The former consists of identifying AI applications into the protein field where we classify and identify active and allosteric sites, molecular signatures, and molecular scaffolding not yet described in nature.
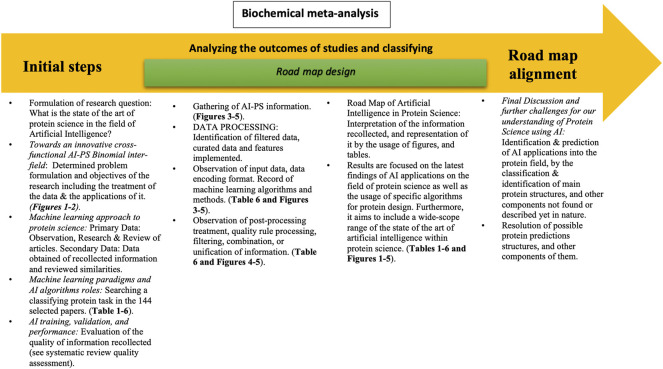
Each structural signature, pattern, or profile constitutes a singular part of the whole “lego-structure-kit” that is the protein space that includes the catalytic task space and shape space, which Kauffman (1992) defines as an abstract representation or mapping of all shapes and chemical reactions that can be catalyzed onto a space of task. The latter process is an analysis of the resulting predictions of structures, molecular signatures, regulatory sites, and ligand sites. Both processes are related to each other in the sense that the proteins in the identification process are searching targets of the 3D-structure for the prediction process that predicts the protein conformation multiple times from a template family or using model-free approach. The biochemical meta-analysis includes formulating the research question, searching and classifying protein tasks in the selected studies, gathering AI–PS information from the studies, evaluating the quality of the studies, analyzing and classifying the outcomes of studies, building up tables and figures for the interpretation of evidence, and presenting the results.
This study puts forward the use of ML classes and methods to address complex problems in protein science. Our point of departure is the state of the art of the AI–PS binomial; by binomial, we mean a biological name consisting of two terms that are partners in computational science as well as in biomedical or biotechnological science as a “two-feet principle” in order to understand, enhance, and control protein science development from an artificial intelligence perspective. Our cross-functional team aims at accelerating the steps of translating the basic scientific knowledge from protein science laboratories into AI applications. Here, we report a comprehensive, balanced systematic review for the literature in the inter-field and a biochemical meta-analysis, which includes a classification of screened articles: 1) by the ML techniques, they use and narrowing down the subareas, 2) by the classes, methods, algorithms, prediction type and programming language, 3) by some protein science queries, 4) by protein science applications, and 5) by protein science problems. Moreover, we present the main contributions of AI in several tasks, as well as a general outline of the processes that are carried out throughout the construction of the models and their applications. We outline a discussion on the best practices of validation, cross-validation, and individual control of testing ML models in order to assess the role that they play in the progress of ML techniques, integrating several data types and developing novel interpretations of computational methodology, thus enabling a wider range of protein’s-universe impacts. Finally, we provide future direction for machine learning approaches in the design of novel proteins, metabolic pathways, and synthetic redesign of protein networks.
Materials and Methods
A systematic review of the scientific literature found in the period (until February 2021) was carried out for this study (Figures 1–3) following the PIO (participants/intervention/outcome) approach and according to PRISMA declaration (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) Supplementary. No ethical approval or letter of individual consent was required for this research.
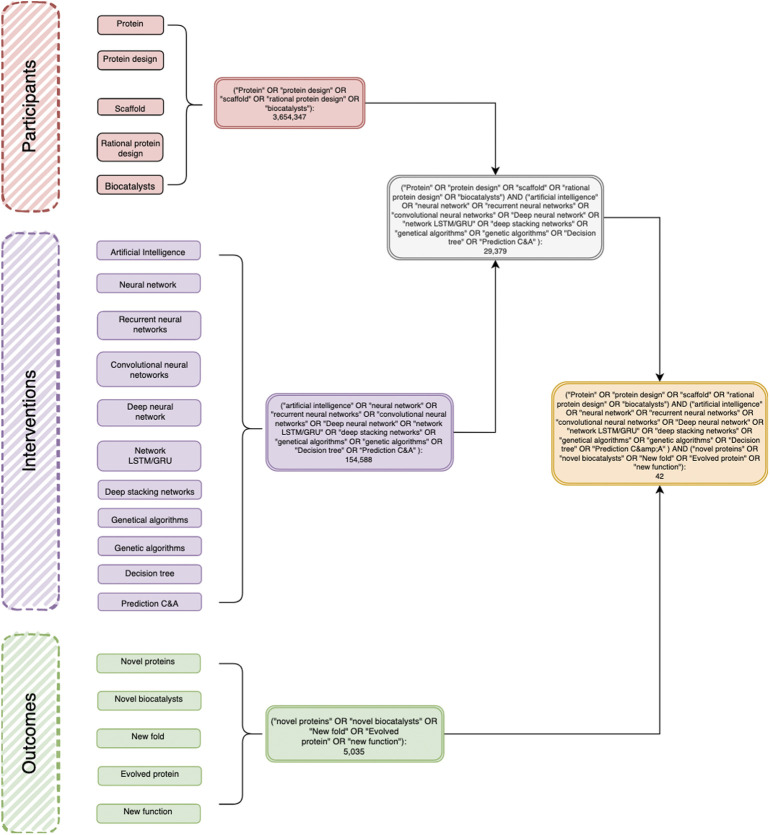
FIGURE 1.
A representative decision diagram showing the articles retrieved using the PIO strategy in the PubMed database. P (participants): Protein, Protein Design, Scaffold, Rational protein design, Biocatalysts. I (intervention): Networks: Neural networks, Recurrent neural networks, Networks LSTM/GRU, Convolutional neural network, Deep belief networks, Deep stacking networks C5.0; Genetic algorithms; Artificial intelligence; Decision trees; Classification; Prediction C&A; Software: Weka, RapidMiner, IBM Modeler; Programming Languages: Python, Java, OpenGL, C++ Shell; Development platform: Caffe Deep Learning, TensorFlow, IBM Distributed Deep Learning (DDL); Paradigm: Supervised Learning, Unsupervised learning, Reinforced learning, new function.
FIGURE 3.
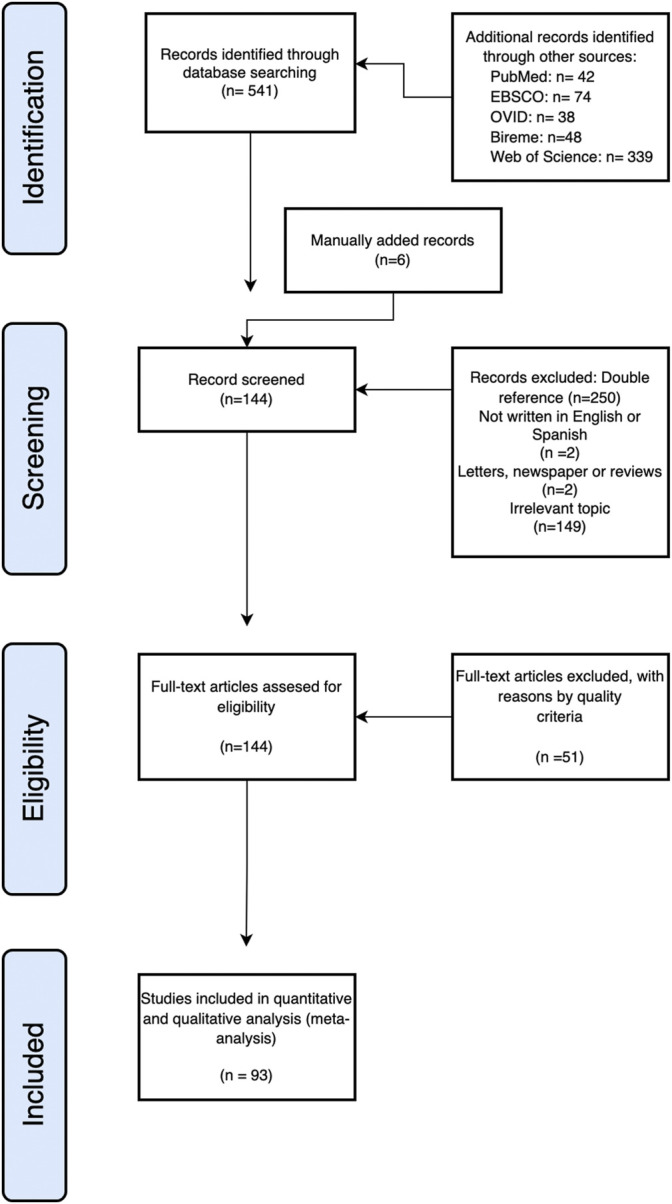
Flowchart of the review process. A PRISMA flowchart of the systematic review on AI for protein sciences.
PIO Strategy
One of the main objectives is to discuss new information in the latest findings about the functions of AI in protein design. Furthermore, this review and meta-analysis intend to include a wide scope of the status of artificial intelligence in protein science. The PIO (participants, intervention, and outcome) strategy was used to systematically search all databases and was the methodology to address the following research questions: What is the state of art in the use of artificial intelligence in the protein science field? What is the use of neural networks in the rational design of proteins? Which neural networks are used in the rational design of proteins? Protein design is currently considered a challenge. As artificial intelligence makes progress, this is presented as a solution to various issues toward addressing how this new branch can be used for the creation of high precision models in protein design. Following the PIO strategy, the next terms were used for the research.
Participants: articles about proteins and their MeSH terms in general were considered for inclusion; we gave special consideration to protein design and their related terms such as scaffold (as a main structure or template), rational design, and biocatalysts (as a main task target for protein evolution and design in the chemical–biotechnological industry and biomedical field):
• protein
• protein design
• scaffold
• rational protein design
• biocatalysts
Intervention: studies with any types of algorithms, software, programming language, platform, or paradigm using alone or in combination were selected.
Types of algorithms:
• neural networks
• recurrent neural networks
• network LSTM/GRU
• convolutional neural network
• deep belief networks
• deep stacking networks C5.0
• genetic algorithms
• artificial intelligence
• decision trees
• classification
• prediction C&A
Software:
• Weka
• RapidMiner
• IBM Modeler
Programming languages:
• Python
• Java
• OpenGL
• C++
• Shell
Development platform:
• Caffe
• DeepLearning4j
• TensorFlow
• IBM distributed deep learning (DDL)
Paradigm:
• supervised learning
• unsupervised learning
• reinforced learning
Outcomes:
• novel proteins
• protein structure prediction
• novel biocatalysts
• new fold
• evolved protein
• new function
Databases and Searches
The electronic databases used were PubMed, Bireme, EBSCO, and OVID. The concepts with similarity were searched with “OR,” and within the groups of each element of the PIO research, they were searched with the word “AND.” Next, a diagram was constructed in order to show the history of searches and concepts used (figure tree diagram). This figure describes in full detail the searching strategy in the PubMed database as well as all keywords used. Moreover, it includes the number of resulting articles. Subsequently, the results obtained from these searches were recorded. The references themselves were then downloaded into the Mendeley database. All references were taken, organized, and saved in Mendeley, eliminating duplicates for the final result.
Biochemical Meta-analysis
The biochemical meta-analysis included formulating the research question, searching and classifying protein tasks in the 144 selected studies, gathering AI–PS information from the 144 studies, evaluating the quality of the studies (as described in the systematic review, see flowchart of PRISMA), analyzing and classifying the intervention and outcome of studies (networks, software, programming languages, development platforms, paradigms, novel proteins, novel scaffold, new fold, etc.), and building up tables and figures for the interpretation of evidence and presenting the results.
By a biochemical meta-analysis, we mean an analysis resulting from two processes: identification and prediction. The former consists of identifying AI applications into the protein field: classify and identify active and allosteric sites, molecular signatures, and molecular scaffolding not yet described in nature, each of which constitute a single part of a grand-type Lego structure. The latter is an analysis of resulting predictions: structures, molecular signatures, regulatory and ligand sites, etc.
Biochemical Meta-Analysis and Designing the Road Map
PRELIMINARY: we determined the formulation of the problem and objectives of the research within the figure, which includes the treatment of the data and their applications. Note: the information was acquired from a list of various databases from which data were analyzed.
DATA COLLECTION: primary data: observation, research and review of articles. Secondary data: data of the reviewed articles and information shared among keywords.
DATA PRE-PROCESSING (ETL and training): identification of filtered data, curated data, and features implemented; machine learning input relationship with protein science servers.
DATA PROCESSING (training data and feature extraction): observation of input data and data encoding format. Record of machine learning algorithms and methods. Recognition of key information for processing data within databases.
DATA POST-PROCESSING: observation of post-processing treatment, rule quality processing, filtering, combination, or unification of information.
MEASURE: explanation of the process, the values of different metrics for the quantification of magnitudes, and the contribution for the completion within the process of information.
ANALYZE: identify the application of machine learning algorithm in which the input of the dataset to process data format, training set, and 3D structures.
IMPROVE: determine the set to whom these new forms will be applied in models of the researched data and contribute to future implementations in protein science.
Concerning the computational aspects as to how articles were classified, three initial divisions were made and are displayed in Table 1: Pre-process, process, and post-process, each of which contain, in turn, the following items:
TABLE 1.
An overview of the included articles on study and algorithm features based in their characteristics, strengths, limitations, and measure of precision.
| Author/Year of Publication/Setting | Classes of machine learning | Methods | Algorithms | Protein Query | Characteristics | Strengths | Limitations | Validation and performance |
|---|---|---|---|---|---|---|---|---|
| Study characteristics and algorithm aspects | ||||||||
| Song J., 2021, China (Song et al., 2021) | Connectionist and Symbolist | An ensemble predictor with a deep convolutional neural network and LightGBM with ensemble learning algorithm | CNN, LightGBM | A sequence-based prediction method for protein–ATP-binding residues, including, PSSM, the predicted secondary structure, and one-hot encoding | The CNN frameworks are proposed as a multi-incepResNet-based predictor architecture and a multi-Xception-based predictor architecture. LightGBM, as a Gradient Boosting Decision Tree (GBDT) for classification and regression merged by an ensemble learning algorithm | The model enriches the protein–ATP-binding residue prediction ability using sequence information. Outstanding performance using ensemble learning algorithm in combination with a deep convolutional neural network and LightGBM as an ATP-binding tool | Distribution of the specific weights was calculated according to the ratio between the positive instances and the negative instances to solve the imbalance problem. The sensitivity prediction was only 0.189. This can be attributed by its very limited prediction coverage and the limited number of sequences in the training set | AUC (0.922 and 0.902), MCC (0.639 and 0.0642), and 5-fold cross-validation |
| Verma N., 2021, US (Verma et al., 2021) | Connectionist | A DNN framework (Ssnet), for the protein–ligand interaction prediction, which utilize the secondary structure of proteins extracted as a 1D representation based on the curvature and torsion of the protein backbone | DNN | Information about locations in a protein where a ligand can bind, including binding sites, allosteric sites, and cryptic sites, independently of the conformation | Curvature and torsion of protein backbone, feature vector for ligand. Multiple convolution networks with varying window sizes as branch convolution | The model does not show biases in the physicochemical properties and necessity of accurate 3D conformation while requiring significantly less computing time. Fast computation once the model is trained with weights bare fixed. No requirement of high-resolution structural data | Ssnet being blind to conformation limits its capability to account for mutations resulting from the same fold but significant difference in binding affinity. Ssnet should be treated as a tool to cull millions of drug-like molecules and not as an exact binding affinity prediction tool | AUC, ROC, and EF scores |
| Bond. S, 2020, US (Bond et al., 2020) | Connectionist | CCP4i2 Buccaneer automated model-building pipeline | PDB | Correctness of protein residues | Visual examination by the crystallographer. Coot provides validation tools to identify Ramachandran outliers, unusual rotamers, and other potential errors, as well as an interface to some tools from MolProbity | No cutoff has to be chosen | It may also have difficulties in that a residue built into the solvent 5 A° away from the structure is no different than one 10 A° away | COD for main chain 0.751; COD for side chain 0.613 |
| Kwon Y., 2020, Korea (Kwon et al., 2020) | Connectionist | A new neural network model for binding affinity prediction of a protein–ligand complex structure | 3D-CNN | Protein–protein complexes in a 3D structure | Ensemble of multiple independently trained networks that consist of multiple channels of 3D CNN layers. Protein–ligand complexes were represented as 3D grids voxelized binding pocket and ligand | Higher Pearson coefficient (0.827) than the state-of-the-art binding affinity prediction scoring functions. Accurate ranking of the relative binding affinities of possible multiple binders of a protein, comparable to the other scoring functions | For docking power, the Ak-score-single model is not as prominent as the other criteria models | Spearman and Pearson correlation coefficients |
| Li H., 2020, France, Hong Kong (Hongjian et al., 2021) | Connectionist, Symbolist and Analogist | Analyzed machine learning scoring functions for structure-based virtual screening | RF, BRT, kNN, NN, SVM, GBDT, multi-task DNN XGBoost | Comparison and review of machine learning scoring functions and classical scoring functions | Machine learning-based scoring function performs better than classical scoring functions, outperforming the average classical methods | Machine learning-based scoring function has introduced strong improvements over classical scoring functions, benchmarks for SBVS. | Current SBVS benchmarks do not actually mimic real test sets, and thus their ability to anticipate prospective performance is uncertain | N/A |
| Liang M., 2020, China (Liang and Nie, 2020) | Connectionist | Method that uses the relation between amino acids directly to predict enzyme function | RN, LSTM | State description matrix containing structural information by four parts, amino acid name (N), angles φ and ψ(A), relative distance (RD), and relative angle γ (RA) | A three-layer MLP; a four-layer MLP; a three-layer MLP, all with ReLU nonlinearities. The final layer was a linear layer that produced logits for optimization with a softmax loss function | Structural relationship information of amino acids and the relationship inference model can achieve good results in the protein functional classification | The model is currently only for single-label classification rather than multi-label classification and only predicts proteins approximately into six major classes. The training has a considerable time during the entire experiment; further optimization is necessary to improve performance | Accuracy, ROC curve, AUC, 3-fold cross-validation |
| Nie J., 2020, Singapore, Taiwan (Sua et al., 2020) | Probabilistic inference, symbolist, and analogist | Identification of lysine PTM site from a convolutional neural network and sequence graph transform techniques | RF, SVM, MNB, LR, Max Entropy, KNN, CNN, MLP | A computational technique to improve the identification of reaction sites for multiple lysine PTM sites in a protein sample | Improves the performance of identifying lysine PTM sites by using a novel combination with convolutional neural networks and sequence graph transform | As the current model that we are proposing is a multilabel model, it is very generalizable, especially when it comes to combinations of multilabel that the dataset does not have. In addition, such combinations of multilabel will increase the test sample size and provide a better idea of the accuracy of the model | Deep learning models are black-box models and may not be very useful for trying to understand the causes of PTMs and how to affect them. We gather that scientists would like to know the cause and effect in order to propose disease modification methods, rather than just pure identification of PTM’s | Cross-validation, precision accuracy, recall, Hamming-loss |
| Qin Z., 2020, US (Qin et al., 2020) | Connectionist | Learn method on amino acid sequence folds into a protein structure, along with the phi–psi angle information for high resolution of protein structure | MNNN | Prediction with only primary amino acid sequence without any template or co-evolutional information | Performs labeling of dihedral angles, combined with the sequence information, allowing the phi–psi angle prediction and building the atomic structure | Prediction consumes less than six orders of magnitude time. Prediction of the structure of an unknown protein is achieved, showing great advantage in the rational design of de novo proteins | Prediction accuracy can be further improved by incorporating new structure to refine the model | Prediction accuracy (85%) |
| Savojardo C., 2020, Italy (Savojardo et al., 2020a) | Connectionist | A method for protein subcellular localization prediction | DeepMITO, 1D-CNN | Performing proteome-wide prediction of sub-mitochondrial localization on representative proteomes | Its major characteristics is to combine proteome-wide experimental data with the predicted annotation of subcellular localization at submitochondrial level and complementary functional characterization in terms of biological processes and molecular functions. Evolutionary information, in the form of Position-Specific Scoring Matrices (PSSM) | The model allows users to search for proteins by organisms, mitochondrial compartment, biological process, or molecular function and to quickly retrieve and download results in different formats, including JSON and CSV | N/A | MCC coefficient |
| Wang M., 2020, US (Wang M. et al., 2020a) | Symbolist | A topology-based network tree, constructed by integrating the topological representation and NetTree for predicting protein–protein interaction (PPI) | TopNetTree, CNN, GBT | Protein structures, protein mutation, and mutation type | Convolutional Neural Networks, used in their Top Net Tree model, as a second module: consisting of the CNN-assisted GBT model | The proposed model achieved significantly better Rp than those of other existing methods, indicating that the topology-based machine learning methods have a better predictive power for PPI systems | Both GBTs and neural networks are quite sensitive to system errors of training of a model The ΔΔG of 27 non-binders (–8 kcal mol–1) did not follow the distribution of the whole dataset. | Person coefficient (Rp) = 0.65/0.68 and 10-fold cross-validation |
| Wardah W., 2020, Australia, Fiji, Japan, US (Wardah et al., 2020) | Pattern recognition | A convolutional neural network to identify the peptide-binding sites in proteins | CNN | Amino acid residues to create the image-like representations by feature vectors | Sets of convolution layers for image operations, followed by a pooling layer and a fully connected layer. The internal weights of the network were adjusted using the Adam optimizer. Bayesian optimization uses calculated values for configuring the model’s hyper-parameters based on prior observations | The model is able to predict a protein sequence with the highest sensitivity compared to any other tool | Improvement and especially in reducing the number of non-binding residues that get falsely classified as binding sites. Better feature engineering to produce better protein–peptide-binding site prediction results. More advanced computing environment | Sensitivity, specificity, AUC, ROC curve, and MCC coefficient |
| Yu C., 2020, Taiwan, US (Yu and Buehler, 2020) | Connectionist | A deep neural network model is based on translating protein sequences and structural information into a musical score, reflecting secondary structure information and information about the chain length and different protein molecules | RNN, LSTM | A vibrational spectrum of the amino acid, comprising amino acid sequence, fold geometry, or secondary structure | The RNN layers, Long Short-Term Memory Units are for time sequence features, alongside a dynamical conditioning. The attention dynamical conditioning model monitors the note velocity changes of the note sequences | The deep neural network is capable of training, classifying, and generating new protein sequences, reproducing existing sequences, and completely new sequences that do not exist yet. The model generates new proteins with an embedded secondary structure approach | The method could be extended to address folded structures of proteins by including more spatial information (relative distance of residuals, angles, or contact information). As well as the addition of combined optimization algorithms, like genetic algorithms | Molecular dynamics equilibration with normal mode analysis |
| Cui Y., 2019, China (Cui et al., 2019) | Pattern recognition | A deep learning model sequence-based for ab initio protein–ligand-binding residue prediction | DCNN | Protein sequences in order to construct several features for the input feature map | First representation, an amino acid sequence by m x d. First convolutional layer with k x d kernel size. Stage 1, with Plain(k x 1,2c) the same as for Block(k x 1,2c). Stage 2, with a Block(k x 1,2c) and Layer normalization-GLU-Conv block | The convolutional architecture provides the ability to process variable-length inputs. The hierarchical structure of the architecture enables us to capture long-distance dependencies between the residue and those that are precisely controlled. Augmentation of the training sets slightly improves the performance | The computational cost for training increases several times. Due to the considerable data skew, the training algorithm tends to fall into a local minimum where the network predicts all inputs as negative examples | Precision, Recall, MCC |
| Degiacomi M., 2019, UK (Degiacomi, 2019) | Deep machine learning | Conformational space generator | Molecular dynamics, random forests and autoencoder algorithms | Generative neural network trained on protein structures produced by molecular simulation can generate plausible conformations | Generative neural networks for the characterization of the conformational space of proteins featuring domain-level dynamics | The auto encoder does great at describing concerted motions (e.g., hinge motions) than at capturing subtle local fluctuations; it is most suitable to handle cases featuring domain-level rearrangements | This generative neural network model yet lies incapable of reproducing non-diversity-related cases, which is a subject of active research in the machine learning community | Performance assessed using different sizes of latent vector and optimizer |
| Fang C., 2019, China, Japan (Fang et al., 2019) | Connectionist | Protein sequence descriptor, position-specific scoring matrix, en DCNNMoRF | DCNN | Pinpoint molecular recognition features, which are key regions of intrinsically disordered proteins by machine learning methods | Ensemble deep convolutional neural network-based molecular recognition feature prediction. It does not incorporate any predicted features from other classifiers | The proposed method is highly performant for proteome-wide MoRF prediction without any protein category bias | It is yet difficult to predict if the new models will perform better only on the results, referring to the use of a new dataset. | Sensitivity, Specificity, Accuracy, AUC, ROC curve, MCC coefficient |
| Fang C., 2019, US (Fang et al., 2020) | Connectionist | Deep dense inception network for beta-turn prediction | DeepDIN | Protein sequence by creating four sets of features: physicochemical, HHBlits, predicted shape string and predicted eight-state secondary structure | Concatenate four convolved feature maps along the feature dimension. Feed the concatenated feature map into the stringed dense inception blocks. Dense layer, with Softmax function | Proposed process for beta-turn prediction outperforms the previous authors | Of the nine cases used, the amount of data belonging to each class may not produce a model with the ability to extract features or to be well generalized. Combined features improve prediction results than those features used alone | MCC and 5-fold cross-validation |
| Fu H., 2019, China (Fu et al., 2019) | Analogist | Classification Natural language prediction (NLP) task | CNN DL | Predict Lysine ubiquitination sites in large-scale | Input fragment. Multi-convolution-pooling layers. Fully connected layers | Extract features from the original protein fragments. First used in the prediction of ubiquitination | DeepUbi is not too deep. Only two convolution-pooling structures | 4-, 6-, 8-, and 10-fold cross-validation Sensitivity, Specificity, Acc, AUC, MCC, Acc >85% AUC = 0.9066/MCC= 0.78 |
| Guo Y., 2019, US (Guo et al., 2019) | Connectionist and Symbolist | Asymmetric Convolutional neural networks and bidirectional long short-term memory | ACNNs, BLSTM, DeepACLSTM | Sequence-based prediction for Protein Secondary Structure (P.S.S.) | The DeepACLSTM method is proposed to predict an 8-category PSS from protein sequence features and profile features | The method efficiently combines ACNN with BLSTM neural networks for the PPS prediction. Leveraging the feature vector dimension of the protein feature matrix | Expensive and time consuming | CB6133 0.742 CB513 0.705 |
| Haberal I., 2019, Norway, Turkey (Haberal and Ogul, 2019) | Connectionist | Three different deep learning architectures for prediction of metal-binding of Histidine (HIS) and Cysteine (CYS) amino acids | 2D CNN, LSTM, RNN | Three methods, PAM, ProCos, and BR to create the feature set from the frame vector; applying directly to raw protein sequences without any extensive feature engineering, while optimizing the model for predicting metal-binding site | The model is a 2D-CNN with four convolution layers, two pooling, two dropout, and two multi-layer perceptron layers. Each convolution layer has 3 × 3 pixel filters | The good performance of the model demonstrates the potential application for protein metal-binding site prediction. A competitive tool for future metal-binding studies, protein metal -interaction, protein secondary structure prediction, and protein function prediction. The CNN method provides better results for the prediction of protein metal binding using PAM attributes | The overall best results were obtained for a window of size 15. The lowest result was obtained in windows of size 101. The lowest result for the ProCos was obtained with the CNN model | Precision, Accuracy, Recall F-Measures K-fold (k = 3,5) cross-validation |
| Heinzinger M., 2019, Germany (Heinzinger et al., 2019) | Connectionist | Natural language processing with Deep learning | ELMo CharCNN LSTM | Protein function and structure prediction via analysis of unlabeled big data and deep learning processing | Novel representation of protein sequences as continuous vectors using language model ELMo, using NLP. | The approach improved over some popular methods using evolutionary information, and for some proteins even did beat the best. Thus, they prove to condense the underlying principles of protein sequences. Overall, the important novelty is speed | Although SeqVec embeddings generated the best predictions from single sequences, no solution improved over the best existing method using evolutionary information | Predictions of intrinsic disorder were evaluated through Matthew’s correlation coefficient and the False- Positive Rate. Also, the Gorodkin measure was used |
| Kaleel M., 2019, Ireland (Kaleel et al., 2019) | Connectionist and Symbolist | Deep neural network architecture composed of stacks of bidirectional recurrent neural networks and convolutional layers | RSA. | Three-dimensional structure of protein prediction | Predicting relative solvent accessibility (RSA) of amino acids within a protein is a significant step toward resolving the protein structure prediction challenge, especially in cases in which structural information about a protein is not available by homology transfer | High accuracy in four different classes (75% average). They performed all the training and testing in 5-fold cross-validation on a very large, state-of-the-art redundancy reduced set containing over 15,000 experimentally resolved proteins | The protein structure prediction challenge especially in cases in which structural information about a protein is not available by homology transfer | 2-class ACC 0.805 2-class F1 0.80 3-class ACC 0.664 3-class F1 0.66 4-class ACC 0.565 4-class F1 0.56 |
| Karimi M., 2019, US (Karimi et al., 2019) | Pattern recognition | Interpretable deep learning of compound–protein affinity | RNN–CNN models | Development of accurate deep learning models for predicting compound–protein affinity using only compound identities and protein sequences | Using only compound identities and protein sequences, and taking massive protein and compound data, RNN–CNN, and GCNN trained models outperform baseline models | Compared to conventional compound or protein representations using molecular descriptors or Pfam domains, the encoded representations learned from novel structurally annotated SPS sequences and SMILES strings improve both predictive power and training efficiency for various machine learning models | The resulting unified RNN/GCNN–CNN model did not improve against unified RNNCNN | Inferior relative error in IC50 within 5-fold for test cases and 20-fold for protein classes not included for training |
| Li C., 2019, China (Li and Liu, 2020) | Constrained optimization and Connectionist | Feature extractor techniques for protein-fold recognition | MotifCNN and MotifDCNN SVM CNN | Fold-specific features with biological attributes considering the evolutionary information from position-specific frequency matrices (PSFMs) considering the structure information from residue–residue | The predictor called MotifCNN-fold combines SVMs with the pairwise sequence similarity scores based on fold-specific features | The model incorporates the structural motifs into the CNNs, aiming to extract the more discriminative fold-specific features with biological attributes, considering the evolutionary information from PSFMs and the structure information from CCMs | Existing fold-specific features lack biological evidences and interpretability, the feature extraction method is still the bottleneck for the performance improvement of the machine learning-based methods | 2-fold cross-validation, Accuracy |
| Lin J., 2019, China (Lin et al., 2019) | Analogist and evolving structures | A drug target prediction method based on genetic algorithm and Bagging-SVM ensemble classifier | GA, SVM | Protein sequences by combining pseudo amino acid, dipeptide composition, and reduced sequence algorithms | GA is used to select the druggable protein dataset. The optimal feature vectors are for the SVM classifier. Bagging-SVM ensemble is for positive and negative sample sets | The method has a high reference value for the prediction of potential drug targets. An improvement over previous methods | N/A | Acc, MCC, Sn, Sp, AUC, PPV, NPV, F1-score,ROC curve and 5-fold cross-validation |
| Pagès G., 2019, France (Pagès et al., 2019) | Connectionist | Regression structure atomic depiction with a density function | 3D CNN | Protein model quality assessment | Three convolutional layers. Fully connected layers. Use of ELU as activation function | Competitivity with single-model protein model quality assessment. Trained to match CAD-score, on stage 2 of CASP 11 | Ornate does not reach the accuracy of the best meta-methods. Scoring time about 1 s for mid-size proteins | Network running using a GeForce GTX 680 GPU |
| Picart-Armada S., 2019, Belgium, UK, Spain (Picart-Armada et al., 2019) | Pattern recognition | Network propagation machine learning methods | PR, Random Randomraw EGAD, PPR, Raw, GM, MC, Z-scores, KNN, WSLD, COSNet, bagSVM, RF, SVM | Assess performance of several network propagation algorithms to find sensible gene targets for 22 common non-cancerous diseases | Two biological networks, six performance metrics, and compared two types of input gene-disease association scores. The impact of the design factors in performance was quantified through additive explanatory models | Network propagation seems effective for drug target discovery, reflecting the fact that drug targets tend to cluster within the network | Choice of the input network and the seed scores on the genes need careful consideration due to possibility of overestimation in performance indicators | There was a dramatic reduction in performance for most methods when using a complex-aware cross-validation strategy. Three cross-validation schemes were used |
| Savojardo C., 2019, Italy (Savojardo et al., 2020b) | Connectionist | A convolutional neural network architecture to extract relevant patterns from primary features | CNN | High prediction on discriminating four mitochondrial compartments (matrix, outer, inner, intermembrane) | Two pooling layers concatenated into a single vector with four independent output units with sigmoid activation function quantifying the membership of each considered mitochondrial compartment | Model has a robust approach with respect to class imbalance and accurate predictions for the four classification compartments | Adoption of more complex architecture, like recurrent layers can improve performance. However, the use of recurrent models leads to bad performance. Impossibility to predict multiple localization for a single protein sequence | 10-fold cross-validation, MCC from 0.45 to 0.65 |
| Schantz M., 2019, Argentina, Denmark, Malaysia (Klausen et al., 2019) | Connectionist | NetSurfP-2.0 | NetSurfP-2.0 | Predict local structural features of a protein from the primary sequence | A novel tool that can predict the most important local structural features with unprecedented accuracy and runtime. Is sequence-based and uses an architecture composed of convolutional and long short-term memory neural networks trained on solved protein structures. | Predicts solvent accessibility, secondary structure, structural disorder, and backbone dihedral angles for each residue of the input sequences | The models are presented with cases that are neither physically nor biologically meaningful | CASP12 0.726 TS115 0.778 CB513 0.794 |
| Taherzadeh G., 2019, Australia, US (Taherzadeh et al., 2019) | Constrained optimization and Connectionist | Predictor method of N- and mucin-type O-linked glycosylation sites in mammalian glycoproteins | DNN, SVM | An amino acid sequence binary vector, evolutionary information, physicochemical properties | DNN uses deep architectures of fully connected artificial neural networks. And SVM linear kernel for classification techniques to predict O-linked glycosylation sites | N-glycosylation model performs equally well for intra or cross-species datasets | Limitation to typical N-linked and mucin-type O-linked glycosylation sites due to lack of data for atypical N-linked and other types of O-linked glycosylation sites | AUC MCC, accuracy, sensitivity, specificity, ROC curve, 10-fold cross-validation |
| Torng W., 2019, US (Torng and Altman, 2019) | Analogist | Classification Softmax classifier for class probabilities | 3D CNN SVM | Protein functional site detection | Protein site representation as four atom channels and supervised labels | Achieved an average of 0.955 at a threshold of 0.99 on PROSITE families. Good performance where sequence motifs are absent, but a function is known | Loss of specific orientation data. NOS structures 1TLL and 1F20 and catalytic sites in TRYPSIN-like enzymes not detected | 5-fold cross-validation Precision, Recall Precision = 0.99 Recall = 0.955 |
| Wan C., 2019, UK (Wan et al., 2019) | Connectionist | A novel method (STRING2GO), with a deep maxout neural networks for protein functional predictive information | DMNN, SVM | Protein functional biological network node neighborhoods and co-occurrence function information | The network architecture consists of three fully connected hidden layers, followed by an output layer with as many neurons as the numbers of terms selected for the biological process functional domain. A sigmoid function is used as activation function and the AdaGrad optimizer is implemented | Successful learning of the functional representation classifiers for making predictions | Potential improvement of predictive accuracy by integrating representations from other data sources with the current PPI network embedding representations | AUC, ROC, MCC |
| Wang D., 2019, China (Wang D. et al., 2020) | Evolutionary | An Artificial Intelligence-based protein structure Refinement method | Multi-objective PSO | Query sequence structures as the initial particle selection for conformation representation | Use of multiple energy functions as multi-objectives. Initialization, energy map of the initial particles. Iteration, energy landscape of the 4th iteration. Selection of non-dominated solutions and added to the Pareto set. And selection of the global best position and the best position every swarm has had by the use of the dominance relationship of swarms, moving to the optimal direction | Success of AIR can be attributed to three main aspects: the first is the anisotropy of multiple templates. The complementarity of multi-objective energy functions and the swarm intelligence of the PSO algorithm, for effective search of good solutions. The larger number of iterations allows the algorithm to perform a more detailed search on the search space, which can improve the quality of the output models | Restriction of the velocity of the dihedral angles in each iteration to a reasonable range for balancing the accuracy and the searching conformation. There are still some unreasonable solutions in the Pareto set. The final step, which ranks the structures in Pareto set, needs more studies | RMSD value |
| Yu C., 2019, US (Yu et al., 2019) | Connectionist | Regression musical patterns by the extension of protein designed | RNN LSTM | Generation of audible sound from amino acid sequence for application on designer materials | An RNN utilized for melody generation. (LSTM) for time sequence featuring | Mechanism to explain the importance of protein sequences. 4.- It can be applied to express the structure of other nanostructures | N/A | N/A |
| Zhang D., 2019, US (Zhang and Kabuka, 2019) | Connectionist | Protein sequence pre-processing, unsupervised learning, supervised, and deep feature extraction | Multimodal DNN | Identify protein–protein interactions and classify families via deep learning models | Multi-modal deep representation learning structure by incorporating the protein physicochemical features with the graph topological features from the PPI networks | The model outperforms most of the baseline machine learning models analyzed by the authors, using the same reference datasets | If there is a certain type of PPI that previous models cannot handle, the article will not say if the new model can | PPI prediction accuracy for eight species ranged from 96.76 to 99.77%, which implies the multi-modal deep representation-learning framework achieves superior performance compared to other computational methods |
| Zhang Y., 2019, China (Zhang et al., 2019) | Connectionist | A new prediction approach appropriate for imbalanced DNA–protein-binding sites data | ADASYN | Employment of PSSM feature and sequence feature for predicting DNA-binding sites in proteins | Introduction of new feature representation approach by combining position-specific scoring matrix, one-hot encoding and predicted solvent accessibility features. Apply adaptive synthetic sampling to oversample the minority class and Bootstrap strategy for a majority class to deal with the imbalance problem | Demonstration that the method achieves a high prediction performance and outperforms the state-of-the-art sequence-based DNA–protein-binding site predictors | Consideration of some other physicochemical features to construct the model and try to explain the biological meaning of CNN filters | Sensitivity, Specificity, Accuracy, Precision, and MCC coefficient |
| Zheng W., 2019, US (Zheng et al., 2019) | Probabilistic inference, Symbolist | Two fully deep learning automated structure prediction pipelines for guided protein structure prediction | Zhang-Server and QUARK | Starting from a full-length query sequence structure | Three core modules: multiple sequence alignment (MSA) generation protocol to construct deep sequence-profiles for contact prediction; an improved meta- method, NeBcon, which combines multiple contact predictors, including ResPRE that predicts contact-maps by coupling precision-matrices with deep residual convolutional neural networks; an optimized contact potential to guide structure assembly simulations | Improvement of the accuracy of protein structure prediction for both FM and TBM targets. Accurate evolutionary coupling information for contact prediction, thus improving the performance of structure prediction. And properly balancing the components of the energy function was vital for accurate structure prediction | Incorrect prediction of contacts between the N- and C- terminal protein regions. Low accuracy of contact prediction in the Terminal regions due to MSAs with many gaps in these regions, as the accuracy of contact-map prediction and FM target modeling is highly influenced by the number of effective sequences in the MSA. | TM-score and p-values |
| Cuperus J., 2018, US (Cuperus et al., 2017) | Connectionist | Regression dropout probability distribution | DNN, CNN, LSTM | Predict protein expression | Hierarchical representation of image features from data | Prediction and visualization of transcription factor binding, Dnase I hypersensitivity sites, enhancers, and DNA methylation sites | Measurement of protein expression with yeast possessing only 5000 genes | k-mer feature, Cross-validation, Held-out R2 = 0.61 |
| Fang C.,US, 2018 (Fang et al., 2018) | Pattern recognition | A deep learning network architecture for both local and global interactions between amino acids for secondary structure prediction | Deep3I | A protein secondary structure prediction model | A designed feature matrix corresponding to the primary amino acid sequence of a protein, which consists of a rich set of information derived from individual amino acid, as well as the context of the protein sequence | This model uses a more sophisticated, yet efficient, deep learning architecture. The model utilizes hierarchical deep inception blocks to effectively process local and nonlocal interactions of residues | Further application of the model to predict other protein structure-related properties, such as backbone torsion angles, solvent accessibility, contact number, and protein order/disorder region, will be done in the future | Accuracy, p-value |
| Feinberg E., 2018, China, US (Feinberg et al., 2018) | Connectionist | A PotentialNet family of graph convolutions | GCNN | A generalized graph convolution to include intramolecular interactions and noncovalent interactions between different molecules | First: graph convolutions over only bonds, which derives new node feature maps. Second: entails both bond-based and spatial distance-based propagations of information. Third: a graph gather operation is conducted over the ligand atoms, whose feature maps are derived from bonded ligand information and spatial proximity to protein atoms | Statistically significant performance increases were observed for all three prediction tasks, electronic property (multitask), solubility (single task), and toxicity prediction (multitask). Spatial graph convolutions can learn an accurate mapping of protein−ligand structures to binding free energies using the same relatively low amount of data | Drawback to train−test split is possible overfitting to the test set through hyperparameter searching. Another limitation is that train and test sets will contain similar examples | Regression enrichment factor (EF), Pearson, and Spearman coefficient, R-squared, MUE (mean-unsigned error) |
| Frasca M., 2018, Italia (Frasca et al., 2018) | Analogist | Clustering Hopfield model | COSNet ParCOSNet HNN | AFP (Automated Protein Function Prediction) | Network parameters are learned to cope with the label imbalance | Advantage of the sparsity of input graphs and the scarcity of positive proteins in characterizing data in the AFP. | Time execution increased less than the density, and more than the number of nodes | 5-fold cross-validation Implementation and execution in a Nvidia GeForce GTX980 GPU target Precision, Recall, F-score, AUPRC |
| Hanson J., Australia, China, 2018 (Hanson et al., 2019) | Pattern recognition | A sequence-based prediction of one- dimensional structural properties of proteins | CNN, LSTM-BRNN | Improving the prediction of protein secondary structure, backbone angles, solvent accessibility | The model leverages an ensemble of LSTM-BRNN and ResNet models, together with predicted residue–residue contact maps, to continue the push toward the attainable limit of prediction for 3- and 8-state secondary structures, backbone angles (h, s, and w), half-sphere exposure, contact numbers and solvent accessible surface area (ASA) | The large improvement of fragment structural accuracy. A new method for predicting one-dimensional structural properties of proteins based on an ensemble of different types of neural networks (LSTM-BRNN, ResNet, and FC-NN) with predicted contact map input from SPOT-contact. The employment of an ensemble of different types of neural networks contributes another 0.5% improvement | Long proteins are also shown to take extensive time, especially for 2D analysis tools. The use of CPU and GPU is shown to not make a major difference in the time taken, as the speed increase introduced by GPU acceleration mainly comes during training | 10-fold cross-validation, Accuracy |
| Hanson J., Australia, China, 2018 (Hanson et al., 2018) | Connectionist | Method by stacking residual 2D-CNN with residual bidirectional recurrent LSTM networks, with 2D evolutionary coupling-based information | CNN, 2D-BRLSTM | Protein contact map prediction | Transformation of sequence-based 1D features into a 2D representation (outer concatenation function). ResNet, 2D-BRLSTM and FullyConnected (FC) | Method achieves a robust performance. The model is more accurate in contact prediction across different sequence separations, proteins with a different number of homologous sequences and residues with a different number of contacts | Coding limitation environment imposed by the 2D-BRLSTM model; training and testing input is limited to proteins of length 300 and 700 residues | AUC >0.95, ROC curve, precision |
| Huang L., 2018, US (Huang et al., 2008) | Connectionist | A novel PPI prediction method based on deep learning neural network and regularized Laplacian kernel | ENN-RL | Protein–protein interaction network | Contains five layers including the input layer, three hidden layers, and the output layer. Sigmoid is adopted as the activation function for each neuron, and layers are connected with dropouts. Regularized Laplacian kernel applied to the transition matrix built upon that evolved the PPI network | The transition matrix learned from our evolution neural network can also help build optimized kernel fusion, which effectively overcome the limitation of the traditional WOLP method that needs a relatively large and connected training network to obtain the optimal weights | The results show that our method can further improve the prediction performance by up to 2%, which is very close to an upper bound that is obtained by an approximate Bayesian computation-based sampling method | Cross-validation, AUC, sensitivity |
| Khurana S., 2018,Qatar, USA (Khurana et al., 2018) | Analogist | Clustering Natural language processing task | CNN FFNN | Solubility prediction | Use additional biological features from external feature extraction tool kits from the protein sequences | DeepSol is at least 3.5% more accurate than PaRSnIP and 15% than PROSO II. DeepSol is superior to all the current sequence-based protein solubility predictors | DeepSol S2 model was surpassed by PaRSnIP on sensitivity for soluble proteins | 10-fold cross-validation Acc, MCC 15% MCC = 0.55 3.5% DeepSol S1- 69 DeepSol S2- 69% |
| Le N., 2018, Taiwan (Le et al., 2018) | Analogist | Regression Softmax layer for classification | CNN | Classify Rab protein molecules | 2D-CNN and position-specific scoring matrices. PSSM profiles of 20 × 20 matrices | Construct a robust deep neural network for classifying each of four specific molecular functions. Powerful model for discovering new proteins that belong to Rab molecular functions | Consideration of the potential effects of more rigorous classification tests | 5-fold cross-validation Sensitivity, Specificity, Acc, AUC, F-score, MCC Acc = 99, 99.5, 96.3, 97.6% |
| Li H., 2018, China (Huang et al., 2018) | Constrained optimization | Regression Adam optimizer | DNN CNN LSTM | Prediction of protein interactions | Machine learning approach for computational methods for the prediction of PPIs | Insight into the identification of protein–protein interactions (PPIs) into protein functions | Manual input of features into the networks | Hold-out testing set model validation Acc, recall, precision, F-score, MCC Acc = 0.9878 Recall= 0.9891 Precision = 0.9861 F-score= 0.9876 MCC= 0.9757 |
| Long H., 2018, China, US (Long et al., 2018) | Connectionist | Classification sigmoid function | HDL CNN LSTM RNN | Predicting hydroxylation sites | CNN deep learning model. Convolution layer consists of a set of filters through dimensions of input data | p-values between AUCs of other methods are less than 0.000001 | Comparative results for CNN and iHyd-PseCp networks | 5-fold cross-validation Sn, Sp, Acc, MCC, TPR, FPR, Precision, recall |
| Makrodimitris S., 2018, Netherlands (Makrodimitris et al., 2019) | Analogist | Clustering constrained optimization | KNN LSDR | Protein function prediction | Transformation of the GO terms into a lower-dimensional space | GO-aware LSDR has slightly better performance on SDp. LSDR reduces the number of dimensions in the label-space. Improve power of the term-specific predictors | LSDR generates inconsistent parent–child pairs. GO-aware terms have a higher inconsistencies | 3-fold cross-validation Fp, AUPRCp, SDp, Ft, AUCRPCt |
| Popova M., 2018, Russia, US (Popova et al., 2018) | Constrained optimization | Regression Stack-RNN as a generative model | Stack-RNN LSTM. | De novo drug design | Deep neural network generative novel molecules (G) and predictive novel compounds (P) | The ReLeaSe method does not rely on predefined chemical descriptors No manual feature engineering for input representation | Extension of the system to afford multi-objective optimization of several target properties | 5-fold cross-validation (5CV) model trained using a GPU Acc R2, RMSE Acc R2 = 0.91 RMSE = 0.53 |
| Sunseri J., 2018, US (Sunseri et al., 2019) | Connectionist | Regression distributed atom densities | CNN | Cathepsin S model ligand protein | CNN based on scoring functions | CNN scoring function outperforms Vina on most tasks without manual intervention | Difficulties with Cathepsin S, for de novo docking | AUC, ROC, MCC |
| Zhang B., 2018, China (Zhang B. et al., 2018) | Connectionist | A novel deep learning architecture to improve synergy protein secondary structure prediction | CNN, RNN, BRNN | Four input features; position-specific scoring matrix, protein coding features, physical properties, characterization of protein sequence | A local block comprising two 1D convolutional networks with 100 kernels, and the concatenation of their outputs. BGRU block, the concatenation of input from the previous layer and before the previous layer is fed to the 1D convolutional filter. After reducing the dimensionality, the 500-dimensional data are transferred to the next BGRU layer | The CNN was successful at feature extraction, and the RNN was successful at sequence processing. The residual network connected the interval BGRU network to improve modeling long-range dependencies. When the staked layers were increased to two layers, the performance increased to 70.5%, and three-layer networks increased further to 71.4% accuracy | When the recurrent neural network was constructed by unidirectional GRU, the performance dropped to 67.2%. The unidirectional GRU network was ineffective at capturing contextual dependencies | Precision, Recall, F1-score, macro-F1, Accuracy |
| Zhang L., 2018, China (Zhang L. et al., 2018) | Connectionist | Two novel approaches that separately generate reliable noninteracting pairs, based on sequence similarity and on random walk in the PPI network | DNN, Adam algorithm | Use of auto-covariance descriptor to extract the features from amino acid sequences and deep neural networks to predict PPIs | The feature vectors of two individual proteins extracted by AC are employed as the inputs for these two DNNs, respectively. Adam algorithm is applied to speed up training. The dropout technique is employed to avoid overfitting. The ReLU activation function and cross-entropy loss are employed, since they can both accelerate the model training and obtain better prediction results | To reduce the bias and enhance the generalization ability of the generated negative dataset, these two strategies separately adjust the degree of the non-interacting proteins and approximate the degree to that of the positive dataset. | NIP-SS is competent on all datasets and hold a good performance, whereas NIP-RW can only obtain a good performance on small dataset (positive samples ≤6000) because of the restriction of random walk and the results of extensive experiments | Precision, Accuracy, Recall, Specificity, MCC coefficient, F1-score, AUC, Sensitivity |
| Zhao X., 2018, China (Zhao et al., 2018) | Connectionist | Bi-modal deep architecture with sub-nets handling two parts (raw protein sequence and physicochemical properties) | CNN and DNN | Raw sequence and physicochemical properties of protein for characterization of the acetylated fragments | Multi-layer 1D CNN for feature extractor and DNN with attention layer with a softmax layer | Capability of transfer learning for species-specific model, combining raw protein sequence and physicochemical information | Interpretation of biological aspect, overfitting problems on small-scale data | 10-fold cross-validation; ACC = 0.708, sensitivity (SEN) = 0.723, specificity (SPE) = 0.707, AUC = 0.783, MCC = 0.251 |
| Armenteros J., 2017, Denmark (Almagro Armenteros et al., 2017) | Analogist | Classification optimization | CNN RNN BLSTM FFNN Attention models | Predict protein subcellular localization | CNN extracts motif information using different motif sizes. Recurrent neural network scans the sequence in both directions | A-BLSTM and the CONV A-BLSTM models achieved the highest performance | Training time for the full ensemble was 80 h, approximately 5 h per model | Nested cross-validation and held-out set for testing models Gorodkin, Acc, MCC 72.90% 72.89% |
| Jimenez J., 2017, Spain (Jiménez et al., 2017) | Bayesian | Regression sigmoid activation function, depicting the probability | 3D CNN | Predict protein–ligand-binding sites Drug design | Fully connected networks. Hierarchical organized layers | Four convolutional layers with max pooling and dropout after every two convolutional layers, followed by one regular fully connected layer | Demand of significant computational resources than other methods for ligand-binding prediction | 10-fold cross-validation Using Nvidia GeForce GTX 1080 GPU for accelerated computing DCC, DVO AUC, ROC, Sn, SP, Precision, F1-score, MCC, Cohen’s Kappa coefficient |
| Müller A., 2017, Switzerland (Müller et al., 2018) | Analogist | Regression SoftMax function for temperature-controlled probability | RNN LSTM | Design of new peptide combinatorial de novo peptide design | The computed output y is compared to the actual amino acid to calculate the categorical cross-entropy loss | The network models were shown to generate peptide libraries of a desired size within the applicability domain of the model | Increasing the network size to more than two layers with 256 neurons led to rapid over-fitting of the training data distribution | 5-fold cross-validation Network training and generated sequences on a Nvidia GeForce GTX 1080 Ti GPU |
| Ragoza M., 2017, US (Ragoza et al., 2017) | Connectionist | Classification distributed atom densities | CNN SGD | Protein-ligand score for drug discovery | CNN architecture: construction using simple parameterization and serve as a starting point for optimization | On a per-target basis, CNN scoring outperforms Vina scoring for 90% of the DUD-E targets | CNN performance is worse at intra-target pose ranking, which is more relevant to molecular docking | 3-fold cross-validation ROC, AUC, FPR, TPR, RF-score, NNScore. CNN-0.815 Vina-0.645 |
| Szalkai B., 2017, Hungary (Szalkai and Grolmusz, 2018a) | Pattern recognition | A classification by amino acid sequence multi-label classification ability | ANN | Protein classification by amino acid sequence | The convolutional architecture with 1D spatial pyramid pooling and fully connected layers. The network has six one-dimensional convolution layers with kernel sizes [6,6,5,5,5,5] and depths (filter counts) [128,128,256,256, 512,512], with parametric rectified linear unit activation. Each max pooling layer was followed by a batch normalization layer | The model outperformed the existing solutions and have attained a near 100% of accuracy in multi-label, multi-family classification | Network variants without batch normalization and five (instead of six) layers showed a performance drop of several percentage points. With more GPU RAM available, one can further improve upon the performance of our neural network by simply increasing the number of convolutional or fully connected layers | Precision, Recall, F1-value, AUC, ROC curve |
| Szalkai B., 2017, Hungary (Szalkai and Grolmusz, 2018b) | Logical Inference | Classification Hierarchical classification tree | ANN | Hierarchical biological sequence classification | SECLAF implements a multi-label binary cross-entropy classification loss on the output neurons | SECLAF produces the most accurate artificial neural network for residue sequence classification to date | Preparation of the input data must be done by the user | AUC |
| Vang Y., 2017, US (Vang and Xie, 2017) | Analogist | Regression Distributed representation with NLP | CNN | HLA class I-peptide-binding prediction | The CNN architecture: convolutional and fully connected dense layers | Effective for validation, distribution, and representation for automatic encoding with no handcrafted encode construction | Provided sufficient data, the method is able to make prediction for any length peptides or allele subtype | 70% training set and 30% validation set (Hold-out) and 10-fold cross-validation GPU for faster computation of model SRCC, AUC SRCC = 0.521, 0.521, 0.513 AUC= 0.836, 0.819, 0.818 66.7% |
| Wang S., 2017, US (Wang et al., 2017) | Analogist | Classification Regression Regularization and optimization | UDNN RNN 2 | Prediction of Protein Contact Map | Consists of two major modules, each being a residual neural network | 3D models built from contact prediction have Tm score >0.5 for 208 of the 398 membrane proteins | No recognition of predict contact maps from PDB. | Algorithm runs on GPU card. Acc L/k (k= 10, 5, 2, 1) Long-range 47% CCMpred- 21% CASP11–30% |
| Yeh C., 2017, UK, US (Yeh et al., 2018) | Evolving structures | Optimization GA | GA multithreaded processing | Designed helical repeat proteins (DHRs) | Iterates through mutation, scoring, ranking, and selection | Aims to control the overall shape and size of a protein using existing blocks | First workload imbalance, less efficient work sharing and overheads in scheduling | RMSD value |
| Simha R., 2015, Canada, Germany, US (Simha et al., 2015) | Bayesian | Classification Probabilistic generative model Bayesian networks | MDLoc BN | Protein multi-location prediction | Each iteration of the learning process obtains a Bayesian network structure of locations using the software package BANJO. | Improvement of MDLoc over preliminary methods with Bayesian network classifiers | MDLoc’s precision values are lower than those of BNCs, MDLoc’s | 5-fold cross-validation Presi, Recsi, Acc, F1-scoresi |
| Yang J., 2015 China, US (Yang et al., 2015) | Analogist | Regression hierarchical order reduction | SVR | Structure prediction of cysteine-rich proteins | Position-specific scoring matrix (PSSM): each oxidized cysteine residue is represented as a vector of 20 elements | Cyscon improved the average accuracy of connectivity pattern prediction | Contact information must be predicted from sequence either by feature-based training or by correlated mutations | 10-fold cross-validation and 20-fold cross-validation QC, QP 21.9% |
| Folkman L., 2014, Australia (Folkman et al., 2014) | Bayesian Constrained optimization | Classification predicted probability of the mutation | SFFS SVM EASE-MM | Model designed for a specific type of mutation | Feature-based multiple models with each model designed for a specific type of mutations | EASE-MM archived balanced results for different types of mutations based on the accessible surface area, secondary structure, or magnitude of stability changes | Using an independent test set of 238 mutations, results were compared in with related work | 10-fold cross-validation ROC, AUC, MCC, Q2, Sn, Sp, PVV, NPV AUC = 0.82 MCC = 0.44 Q2 = 74.71 Sn = 73.14 Sp = 75.28 PVV = 52.30 NPV = 88.33 |
| Li Z., 2014, US (Li et al., 2014) | Bayesian | Classification Probability output prediction | SPIN NN | Sequence profile prediction | Sequence Profiles by Integrated Neural network based on fragment-derived Sequence profiles and structure-derived energy profiles | SPIN improves over the fragment-derived profile by 6.7% (from 23.6 to 30.3%) in sequence identity between predicted and raw sequences | Minor improvement in the core of proteins, which have 10% less hydrophilic residues in predicted sequences than raw sequences | 10-fold cross-validation MSE, Precision, Recovery rate |
| Eisenbeis S., 2012, Germany (Eisenbeis et al., 2012) | N/A | N/A | N/A | Enzyme design | No network | No network | No network | — |
| Qi Y., 2012, US (Qi et al., 2012) | Connectionist | Classification Back propagation in deep layers | DNN | Prediction of local properties in proteins | An amino acid feature extraction layer. A sequential feature extraction layer. A series of classical neural network layers | For the prediction of coiled coil regions, our performance of 97.4% beats the best result (94%) on the same dataset from using the same evaluation setup | The largest improvement is observed for relative solvent accessibility prediction, from 79.2 to 81.0% in the multitask setting | 3- and 10-fold cross-validation Acc, precision, recall, F1 80.3% |
| Ebina T., 2011, Japan (Ebina et al., 2011) | Analogist | Classification Domain linker prediction SVM | DROP SVM RF | Domain predictor | Vector encoding. Random Forest feature selection. SVM parameter optimization. Prediction assessment | Advantage for testing several averaging windows, 600 properties encoded, averaged with five different windows into a 3000-dimensional vector | Computational time required for performing an exhaustive search | 5-fold cross-validation AUC, Sn, Precision, NDO, AOS |
| Yang Y., 2011, US (Yang et al., 2011) | Probability Inference | Regression probabilistic-based matching | SPARKS-X Algorithm | Single-method fold recognition | The model is built by modeller9v7 using the alignment generated by SPARKS-X | SPAKRS-X performs significantly better in recognizing structurally similar proteins (3%) and in building better models (3%) | HHPRED improve 3% over SPARKS-X due to significantly more sophisticated model building techniques | ROC, TPR, FPR |
| Briesemeister S., 2010, Germany (Briesemeister et al., 2010) | Bayesian | Classification probabilistic approach | NB | Predict protein subcellular localization | Yloc, based on the simple naive Bayes classifier | Small number of features and the simple architecture guarantee interpretable predictions | Returns in confidence estimates that rate predictions are reliable or not | 5-fold cross-validation Acc, F1-score, precision, recall |
| Lin G., 2010, US (Lin et al., 2010) | Analogist | Classification Optimization | SVM SVR | Protein folding kinetic rate and real-value folding rate | SVM classifier to classify folding types based on binary kinetic mechanism (two-state or multi-state), instead of using structural classes of all-α-class, all-β-class and α/β-class | The accuracy of fold rate prediction is improved over previous sequence-based prediction methods | Performance can be further enhanced with additional information | Leave-one-out cross-validation (LOOCV) Classification accuracy surface, Predicted precision |
| Tian J., 2010, China (Tian et al., 2010) | Analogist | Classification Optimization | RFR SVM RF | Effect on single or multi-site mutation on protein thermostability | Random forest includes bootstrap re-sampling, random feature selection, in-depth decision, tree construction, and out-of-bag error estimates | Overall accuracy of classification and the Pearson correlation coefficient of regression were 79.2% and 0.72 | Direct comparison of Prethermut with the other published predictor was not performed as a result of data limitation and differences | 10-fold cross-validation Overall accuracy (Q2), MCC, Sn, Sp, Pearson correlation coefficient ® Acc = 79.2% r = 0.72 |
| Zhao F., 2010, US (Zhao et al., 2010) | Bayesian | Classification probabilistic graphical model | CNF SVM | Protein folding | Conformations of a residue in the protein backbone is described as a probabilistic distribution of (θ, τ) | The method generates conformations by restricting the local conformations of a protein | CNF can generate decoys with lower energy but not improve decoy quality | 5-, 7-, and 10-fold cross-validation Accuracy (Q3) Q3 = 80.1% |
| Hong E., 2009, US (Hong et al., 2009) | Symbolist | Classification Branch and bound tree Logical inference | BroMap | Tenth human fibronectin, D44.1 and DI.3 antibodies, Human erythropoietin | BroMAP attempts the reduction of the problem size within each node through DEE and elimination | Lower bounds are exploited in branching and subproblem selection for fast discovery of strong upper bounds | BroMAP is particularly applicable to large protein design problems where DEE/A∗ struggles and can also substitute for DEE/A∗ in general GMEC search | N/A |
| Özen A., 2009, Turkey (Özen et al., 2009) | Analogists | Classification Regression Constrained optimization | SVM KNN DT SVR | Single-site amino acid substitution | Early Integration. Intermediate Integration. Late Integration | Possible combination including new feature set, new kernel, or a learning method to improve accuracy. | Training any classifier with an unbalanced dataset in favor of negative instances makes it difficult to learn the positive instances | 20-fold cross-validation Acc, Error rate, Precision, Recall, FP rate Acc= 0.842, 0.835 |
| Ebrahimpour A., 2008, Malaysia [(Ebrahimpour et al., 2008) | Connectionist | Classification Back and batch back propagation | ANN FFNN IBP BBP QP GA LM | Lipase production Syncephalastrum racemosum, Pseudomonas sp. strain S5 and Pseudomonas aeruginosa | ANN architecture: input layer with six neurons, an output layer with one neuron, and a hidden layer. Transfer functions of hidden and output layers are iteratively determined | Maximum predicted values by ANN (0.47 Uml -1) and RSM (0.476 U–l - 1), whereas R2 and AAD were determined as 0.989 and 0.059% for ANN and 0.95 and 0.078% for RSM, respectively | ANN has the disadvantage of requiring large amounts of training data | RMSE, R2, AAD RMSE<0.0001 R2 = 0.9998 |
| Huang W., 2008, Taiwan (Huang et al., 2008) | Analogist | Clustering Combinatorial optimization | GA SVM KNN | Prediction method for predicting subcellular localization of novel proteins | Preparation of SVM, binary classifiers of LIBSVM. Sequence representation. Inclusion of essential GO terms | Bias-free estimation of the accuracy reduces computational cost | Computational demand is impractical for large datasets | 10-fold cross-validation and leave-one-out cross-validation (LOOCV) Accuracy, MCC Acc= 90.6–85.7% |
| Katzman S., 2008, US (Katzman et al., 2008) | Bayesian | Classification Probabilistic | MUSTER SVM | Local structure prediction | Calculation of output of each unit in each layer. Soft max function to all outputs of a given layer represents valid probability distribution | Accurate predictions of novel alphabets for extending the performance | Smaller windows and number of units, the network has fewer total degrees of freedom | 3-fold cross-validation, Qn |
| Liao J., 2007, US (Liao et al., 2007) | Supervised Learning | Classification Regression | RR Lasso PLSR SVMR LPSVMR LPBoosR MR ORMR | Proteinase K variants | Design of protein variants. Expression of the protein variants. Analysis of protein variant sequences and activities to assess the contribution of each amino acid substitution | Machine learning algorithms make it possible to use more complex and expensive tests to only protein properties | Computational resources are cheap; we instead used the 1000 subsamples of the training sets | Cross-validation |
| Raveh B., 2007, Israel (Raveh et al., 2007) | Connectionist | Clustering Pattern recognition | K-means Clustering | Existence of α-helices, parallel β-sheets, anti-parallel sheets and loops. Non-conventional hybrid structures | Network motif vector (k means of motif vector). Enriched Interaction graphs | Rediscovery existence of conventional a-helices, parallel b-sheets, anti-parallel sheets and loops, and non-conventional hybrid structures | Limitation to backbone interactions, the degree of each node in the network was bounded from above by two covalent and two possible hydrogen bonds | 10-fold cross-validation |
| Shamim M., 2007, India (Shamim et al., 2007) | Analogist | Classification Regression | SVM | Protein-fold prediction | LIBSVM provides a choice of in-built kernels, such as Linear, Polynomial, Radial basis function (RBF), and Gaussian, we use RBF kernel | Overall accuracy of 65.2% for fold discrimination and individual propensities, which is better than those from the literature | Incrementation of backbone conformation results in the reduction on accuracy prediction | 2-fold cross-validation 5-fold cross-validation Accuracy (Q), Sn, Sp Q= 65.2% >70% |
| Hung C., 2006, Taiwan (Hung et al., 2006) | Symbolist | Regression Genetic algorithm casual tree | DFS HMM GA AGCT | Predict protein functions | AGCT study applies a hybrid methodology based on genetic programming with a causal tree model to predicting protein function | The model is developed to exploit global search capabilities in genetic programming for predicting protein functions of a distantly related protein family that has difficulties in the conserved domain identification | Ratios of comparison between the heuristic signal match and exhaustive sequence alignment are low | Cross-validation |
| Sidhu A., 2006, UK (Sidhu and Zheng, 2006) | Symbolist | Classification Logical Inference | BBFNN NN DT | Predict signal peptide | BBFNN Characteristics: Mutation matrix for protein sequence encoding. BBFNN is a linear combination of K bio-bases with the bio-basis function | The BBFNN has improved the accuracy by a further 5%. Most cost-effective and efficient way of predicting signal peptides | Size of the positive examples in the dataset reduces prediction accuracy | 5-fold cross-validation Accuracy Acc >90%, 97.16% for BBFNN 97.63% for C4.5 |
| Zimmermann O., 2006, Germany, US (Zimmermann and Hansmann, 2006) | Analogist | Classification | SVM C-SVM algorithm implementation | Prediction of dihedral regions | Implementation of the sequence window of length seven and three separate predictions: helix, extended beta, and outliers | Profile-only SVM classifiers show a prediction performance of 80% | The approach is based on sequence profiles only. Models show a tendency to over-predict extended residues and under-predict residues in the helical state | Acc, MCC, Sn, Sp Acc = 93.3%, 93.4% MCC = 0.645, 0.671 |
| Capriotti E., 2005, Italy (Capriotti et al., 2005) | Analogist | Classification | SVM | Protein stability prediction | Prediction of the direction of the protein stability changes upon single-point mutation from the protein tertiary structure | Large extent protein stability can be evaluated with specific interactions in the sequence neighbors captured | Correlation of predicted with expected/experimental values is 0.71 with a standard error of 1.30 kcal/mol and 0.62 with an SE of 1.45 kcal/mol | Cross-validation Accuracy, MCC, Q2 = 0.80, 0.77 MCC = 0.51, 0.42 |
| Rossi A., 2001, Italy (Rossi et al., 2001) | Connectionist | Regression Perceptron algorithm | NN | Barnase and chymotrypsin inhibitor | Two- and three-body energy functions. Partitioning the 20 amino acids into classes (Hydrophobic, Neutral, Charged) | The method is able to identify crucial sites for folding process: for 2ci2 and barnase and shows a very good agreement with experimental results | No improvement on success rate by introducing more sophisticated energy functions. Important features of real proteins are neglected by short-range Hamiltonians | N/A |
pre-process
database, pretreatment, and input
process
machine learning paradigm and input, algorithm and development software, three aspects of the neural network used (characteristics, strengths, and limitations) and output.
post-process
input and web server when applied.
Most of the research reported in these articles performs a pretreatment over the protein database used, that is, processes of randomization and training, in order to leave the data prepared for the computational process itself, for when the algorithm is to be executed on a software platform and within a particular machine learning paradigm (mostly supervised, unsupervised, and deep learning, as shown in Figure 4). We also reported special characteristics as well as strengths and limitations of the neural networks used. Finally, part of the post-process, when applied, concerns the web server where research results are stored. Moreover, some of these aspects are also registered in Tables 2–6 as well as some others (programming language and software license type).
FIGURE 4.
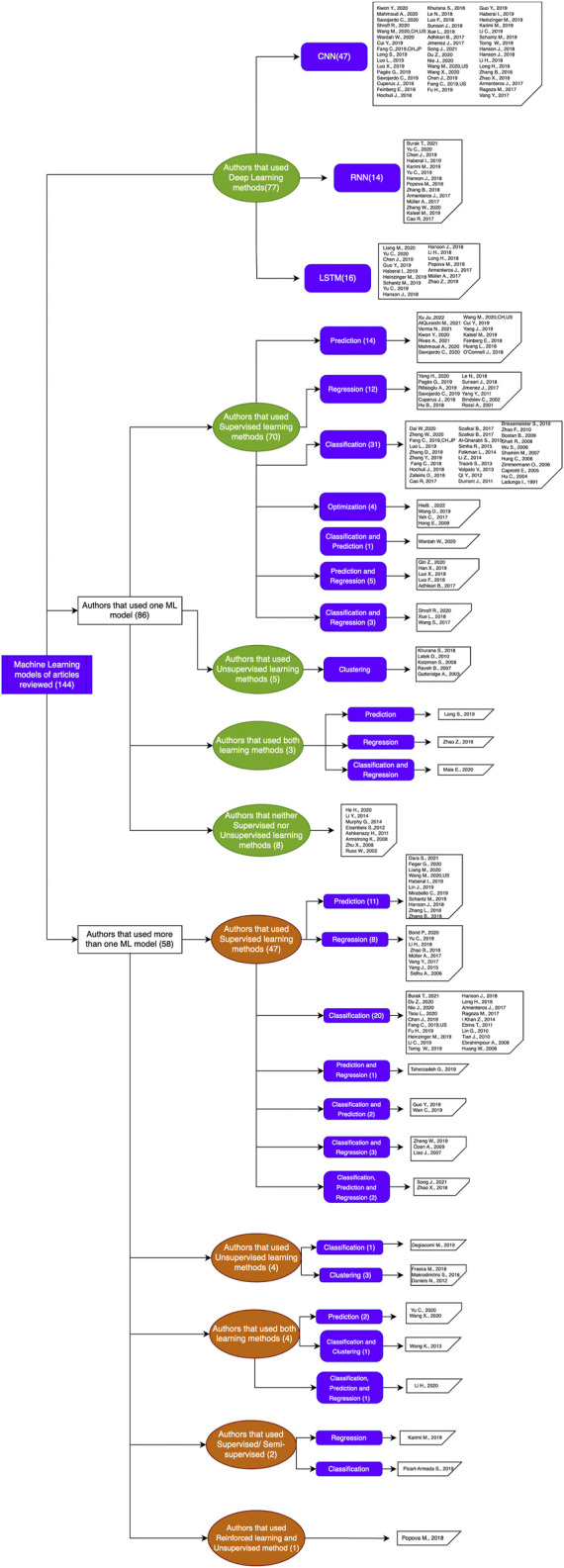
Machine Learning paradigms: superviser learning, unsupervised learning, reinforcement learning.
TABLE 2.
An overview of the protein and drug design articles with the quality assessment.
| First Author/Year of Publication/Country | Database | Initial scaffold (ID) | Designed Protein | ML model | Software/Sever | Programming language/Platform | License | Quality (%) | Machine learning | Protein application |
|---|---|---|---|---|---|---|---|---|---|---|
| Protein and drug design | ||||||||||
| Hie B., 2022, USA (Hie and Yang, 2022) | N/A | Sequence-to-function machine learning surrogate model t | Protein engineering design | Machine learning optimization | N/A | N/A | N/A | 50% | Supervised learning: optimization | Protein design |
| Dara S., 2021, India (Dara et al., 2021) | ZINC, BindingDB, PUBCHEM, Drugbank, REAL, Genomic Database, Adaptable Clinical Trail Database, DataFoundry, SWISS-PROT, SCoP, dbEST. Genome Information Management System, BIOMOLQUEST, PDB, SWISS-PORT, ENZIME | Target identification, hit discovery, hit to lead, lead optimization | PPI prediction, protein folding, drug repurposing, virtual screening, activity scoring, QSAR, drug design, evaluation of ADME/T properties | AutoEncoder, ANN,CNN, DL, MLP,NB, RF, RNN, CNN, SVM, LR | N/A | N/A | N/A | 50% | Supervised learning: prediction | Drug discovery |
| Feger G., 2020, Czech Republic, France (Feger et al., 2020) | PDB | Peptide amphiphile scaffolds | Amphiphilic peptide scaffold design | SVM, RF | SasFit | C | Open source | 60 | Supervised Learning: Prediction | Protein design |
| He H., 2020, China (He et al., 2020) | Multiple databases | Multiple organisms | Review of novel drug discovery techniques | Multiple methods for structure prediction, ligand-binding site, undruggable to drug rabble targets, hidden allosteric site | N/A | N/A | N/A | 50 | N/A | Drug discovery |
| Maia E., 2020, Brazil (Maia et al., 2020) | Multiple databases | structure-based virtual screening (SBVS) | Drug development | VSA | N/A | Multiple languages | N/A | 60 | Supervised Learning: Unsupervised Learning | Drug development design |
| Qin Z., 2020, US (Qin et al., 2020) | PDB | Phi–psi angle and sequence of natural protein, only of standard amino acids | Protein design of fold alpha-helical structure | MNNN | Tensorflow https://github.com/IBM/mnnn | Python | Open Source | 95 | Supervised Learning: Prediction Regression | Protein design |
| Tsou L., 2020, Taiwan (Tsou et al., 2020) | ChEMBL | In-house database of 165,000 compounds | TNBC inhibitors and GPCR classification prediction | DNN, RF | N/A | N/A | N/A | 60 | Supervised Learning: Classification | Drug design |
| Wang X., 2020, China (Wang X. et al., 2020) | KIBA, Davis dataset | Kinase protein family | Predict drug-target-binding affinity | CNN, GCN | N/A | N/A | N/A | 60 | Supervised Learning, Semi-Supervised Learning: Prediction | Drug-target binding-affinity |
| Yu C., 2020, Taiwan, US (Yu and Buehler, 2020) | PDB | α-helix-rich proteins | De novo protein design | RNN, LSTM | TensorFlow, https://github.com/tensorflow/magenta/issues/1438 | Python | Open Source | 90 | Supervised Learning: Unsupervised Learning: Prediction | Protein design |
| Fang C., 2019, US (Fang et al., 2020) | UniProt | Proteins from datasets BT426 and BT6376 containing at least one beta-turn | Beta-turn prediction | HMM, CNN, DeepDIN | Tensorflow, Keras http://dslsrv8.cs.missouri.edu/∼cf797/MUFoldBetaTurn/download.html | Python | Open Source | 90 | Supervised Learning: Classification | Protein design |
| Karimi M., 2019, US (Karimi et al., 2019) | BindingDB, STITCH, Uniref | Various protein classes | Compound–protein affinity prediction | RNN, CNN | https://github.com/ShenLab/DeepAffinity | N/A | N/A | 75 | Semi-supervised, Unsupervised Learning: Regression | Drug design |
| Lin J., 2019, China (Lin et al., 2019) | DrugBank | Druggable proteins and non-druggable proteins | Drug target prediction | SVM, GA | https://github.com/QUST-AIBBDRC/GA-Bagging-SVM | Matlab | MathWorks | 90 | Supervised Learning: Prediction | Drug design |
| Hu B., 2018, China (Hu et al., 2018) | DDI, SIDER, TWOSIDES, HPRD, Drug Bank, Offsides PubChem | Semantic meta-paths ADR | meta-path-based proximities ADR | SDHINE, Network embedding | TensorFlow, N/A | C, C++, Python | Apache 2.0 | 65 | Supervised Learning: Regression | Drug design |
| Popova M., 2018, Russia, US (Popova et al., 2018) | PHYSPROP, ChEMBL, KKB | SMILE string | Drug design (de novo design) | Stack-RNN, LSTM, ReLeaSE | PyTorch, TensorFlow ReLeaSE https://github.com/isayev/ReLeaSE | Python, CUDA | Open Source | 75 | Reinforced Learning, Unsupervised Learning: Regression | Drug design |
| Zafeiris D., 2018, UK (Zafeiris et al., 2018) | GEO, Array Expression | Amyloid beta-precursor protein, microtubule-associated protein tau, apolipoprotein E | Biomarker discovery for Alzheimer’s disease | ANN | N/A | N/A | N/A | 50 | Supervised Learning: Classification | Enzyme design |
| Jimenez J., 2017, Spain (Jiménez et al., 2017) | scPDB | PDB ID File or PDB file | Predict protein–ligand-binding sites Drug design | 3D-DCNN | Keras, Theano www.playmolecule.org | Python | Open Source | 90 | Supervised Learning: Regression | Drug design |
| Müller A., 2017, Switzerland (Müller et al., 2018) | ADAM, APD DADP | Antimicrobial peptide Amino acid sequences | Design of new peptide combinatorial de novo peptide design | RNN, LSTM | modlAMP Python package https://github.com/alexarnimueller/LSTM_peptides | Python | Open Source | 100 | Supervised Learning: Regression | Drug design |
| Ragoza M., 2017, US (Ragoza et al., 2017) | PDB ChEMBL | Spatial and chemical features of protein–ligand complex | Protein–ligand score for drug discovery | CNN, SGD | Gnina Caffe https://github.com/gnina | C++ | Open Source | 85 | Supervised Learning: Classification | Drug design |
| Yeh C., 2017, UK, US (Yeh et al., 2018) | JSON database: centers of mass and geometric relationship data | Helical repeat proteins, Center of mass (CoM) using C-α protein sequence | Designed helical repeat proteins (DHRs) | GA multithreaded processing | ELFIN https://github.com/joy13975/elfin | Python, C++, MATLAB | Apache 2.0 open source 3-Clause BSD | 90 | Supervised Learning: Optimization | Drug design |
| Folkman L., 2014, Australia (Folkman et al., 2014) | ProTherm | Protein sequence and amino acid substitution | Model designed for a specific type of mutation | EASE-MM, SVM | EASE-MM LISVM http://www.ict.griffigr.edu.au/bioinf/ease | Python, Linux | Open Source | 75 | Supervised Learning: Classification | Model design |
| Khan Z., 2014, Pakistan (Khan et al., 2015) | BRENDA | Amino Acid sequence and alkaline enzyme | Enzyme catalysis | DT, KNN, MLP, PNN, SVM | MATLAB Bioweka Weka | Java | Open Source MathWorks | 50 | Supervised Learning: Classification | Drug design |
| Li Y., 2014, US (Li and Cirino, 2014) | PDB | E. coli | Designs of improved enzymes and enzymes with new functions and activities | Computational design and scaffolding and compartmentalization | N/A | N/A | N/A | 50 | N/A | Drug design |
| Murphy G., 2014, US (Murphy et al., 2015) | DND_4HB protein | DND_4HB protein | Design an up-down four-helix bundle | Computational folding | N/A | N/A | N/A | 50 | N/A | Drug design |
| Traoré S., 2013, France (Traoré et al., 2013) | PDB | 3D protein structure | Structure-based computational protein design framework | CFN | CPD http://genoweb.toulouse.inra.fr/tschiex/CPD | Perl | Open source | 65 | Supervised Learning: Classification | Protein design |
| Volpato V., 2013, Ireland (Volpato et al., 2013) | ENZYME UniProt | Oxidoreductase, transferase, hydrolase, lyase, isomerase, and ligase | Acid-residue frequency derived from multiple sequence alignments extracted from uniref90 | N-to-1 Neural Network | N/A | N/A | N/A | 65 | Supervised Learning: Classification | Drug design |
| Daniels N., 2012, US (Daniels et al., 2012) | SCOP | Protein sequence, 207 beta structural SCOP super families | Detection for beta-structural proteins into the twilight zone, make over a 100-new-fold prediction genome of T. maritima | HMM, MRF | SMURFLite http://smurf.cs.tufts.edu/smurflite/ | N/A | Open Source | 65 | Unsupervised Learning: Clustering | Drug design |
| Eisenbeis S., 2012, Germany (Eisenbeis et al., 2012) | PDB | (βα)8-barrel and the flavodoxin-like fold, CheY, HisF | Enzyme design | Rational recombination | http://pubs.acs.org Modeller, Rosetta | Python | IBM, Academic nonprofit freeware | 75 | N/A | Drug design |
| Ebina T., 2011, Japan (Ebina et al., 2011) | DS-All dataset | Protein sequence | Domain predictor | DROP, SVM, RF | DROP http://web.tuat.ac.jp/∼domserv/DROP.html | Bash script | Open source | 75 | Supervised Learning: Classification | Drug design |
| Bostan B., 2009, US (Bostan et al., 2009) | KEGG | Given a species proteome | Predict homologous signaling pathway | PSP | N/A | N/A | N/A | 50 | Supervised Learning: Classification | Model design |
| Hong E., 2009, US (Hong et al., 2009) | Standard rotamer library Expanded rotamer library | Fn3: Derived from protein Fn3, 10th human fibronectin-type III domain | Tenth human fibronectin, D44.1 and DI.3 antibodies, Human erythropoietin | BroMAP | BroMAP | C++, Linux | Open Source | 100 | Supervised Learning: Optimization | Drug design |
| Özen A., 2009, Turkey (Özen et al., 2009) | ProTherm | Structure-based features: amino acid substitution likelihood equilibrium fluctuations α, Cβ, packing density | Single-site amino acid substitution | SVM, KNN, DT, SVR | MOSEK http://www.prc.boun.edu.tr/appserv/prc/mlsta | N/A | Open Source | 85 | Supervised Learning: Classification Regression | Model design |
| Ebrahimpour A., 2008, Malaysia (Ebrahimpour et al., 2008) | GenBank | Geobacillus sp. Strain | Lipase production Syncephalastrum racemosum, Pseudomonas sp. Strain S5 and Pseudomonas aeruginosa | ANN, FFNN, IBP, BBP, QP, GA, LM | CPC-X Software N/A | Java | Neural Power version 2.5 | 75 | Supervised Learning: Classification | Protein design |
| Zhu X., 2008, China (Zhu and Lai, 2009) | PDB | 223 scaffold proteins | Pocket residues of ribose-binding protein (2dri), tyrosyl-t/RNA synthetase (4ts1), and tryptophan synthase (1a50). No metal ion-binding sites | Vector matching | N/A | N/A | N/A | 65 | N/A | Drug design |
| Liao J., 2007, US (Liao et al., 2007) | GenBank | Proteinase K-catalyzed hydrolysis of the tetrapeptide N-Succinyl-Ala-Ala-Pro-Leu p-nitroanilide | Proteinase K variants | RR, Lasso, PLSR, SVMR, LPSVMR, LPBoosR, MR, ORMR | N/A | Matlab | MathWorks | 75 | Supervised Learning: Classification Regression | Protein design |
| Raveh B., 2007, Israel (Raveh et al., 2007) | PDB | TIM-barrel fold 1YPI. Whole β-sheet global structures | Existence of α-helices, parallel β-sheets, anti-parallel sheets and loops. Non-conventional hybrid structures | K-means clustering | Matlab | Matlab | MathWorks | 75 | Unsupervised Learning: Clustering | Protein design |
| Zimmermann O., 2006, Germany (Zimmermann and Hansmann, 2006) | PDB | Protein sequence | Prediction of dihedral regions | C-SVM | LIBSVM-library DHPRED http://www.fz- juelich.de/nic/cbb | C, Python, Linux, Windows | Open source | 80 | Supervised Learning: Classification | Protein design |
| Russ W., 2002, US (Russ and Ranganathan, 2002) | N/A | SH3 domain GroEL minichaperone WW domain prototype | Thermostable consensus phytase, 84.5 kDa protein | Knowledge-base potential functions | N/A | N/A | N/A | 65 | N/A | Protein design |
| Rossi A., 2001, Italy (Rossi et al., 2001) | PDB, HSSP | 2ci2 Barnase | Barnase and chymotrypsin inhibitor | Perceptron | N/A | N/A | N/A | 90 | Supervised Learning: Regression | Drug design |
3D-CNN, Three-dimensional convolutional neural network; ANN, Artificial neural network; BBP, Back Back propagation; BroMap, Branch and bound map estimation; CFN, Cost function network; CNN, Convolutional neural network; DeepDIN, Deep dense inception network; DT, Decision tree; DROP, Domain linker prediction using optimal feature; EASE-MM, Evolutionary Amino acid, and Structural Encodings with Multiple Models; FFNN, Feed forward neural network; GA, Genetic algorithms; GCN, Graph convolutional network; HMM, Hidden Markov model; IBP, Incremental back propagation; KNN, k-nearest neighbor; Lasso, Least absolute shrinkage and selection operator; LM, Levenberg–Marquardt; LPBoostR, Linear programming boosting regression; LPSVMR, Linear programming support vector machine regression; LSTM, Long short-term memory; MLP, Multilayer perceptron; MR, Matching loss regression; MRF, Markov random forest; MNNN, Multi-scale neighborhood-based neural network; ORMR, One-norm regularization matching-loss regression; PLSR, Partial least-squares regression; PNN, Probabilistic neural network; PSP, Predict Signal Pathway; QP, quick prob; ReLeaSE, Reinforcement Learning for Structural Evolution; RF, Random forest; RNN, Recurrent neural network; RR, Ridge regression; SDHINE, Meta path-based heterogeneous information embedding approach; SFFS, Sequential forward floating selection; SGD, Stochastic gradient descent; SVM, Support vector machine; SVMR, Support vector machine regression; SVR, Support vector regression; VSA, Virtual screening algorithms.
TABLE 6.
An overview of the protein contact map prediction, protein-binding prediction, protein site prediction, and genomics articles with the quality assessment.
| First Author/Year of Publication/Country | Database | Initial scaffold (ID) | Designed Protein | ML model | Software/Sever | Programming language/Platform | License | Quality (%) | Machine learning | Protein application |
|---|---|---|---|---|---|---|---|---|---|---|
| Protein Contact Map Prediction | ||||||||||
| Yang H., 2020, China (Yang H. et al., 2020) | SCOPe 2.07 | N/A | Contact map protein prediction | GAN | Keras, Tensorflow https://github.com/melissaya/GANcon | Python | Open Source | 70 | Supervised Learning: Regression | Contact map prediction |
| Hanson J., 2018, Australia, China (Hanson et al., 2018) | PDB UniProt | Primary amino acid sequence, proteins from CASP12 | Protein contact map prediction | CNN, 2D-BRLSTM | http://sparks-lab.org/jack/server/SPOTContact/ | N/A | N/A | 95 | Supervised Learning: Prediction | Protein contact map prediction |
| Ashkenazy H., 2011, Israel (Ashkenazy et al., 2011) | PDB | 3D protein structure | Contact map prediction | WMC | http://tau.ac.il/∼haimash/WMC | Perl | Open Source | 45 | N/A | Protein map prediction |
| Durrant J., 2011, US (Durrant and McCammon, 2011) | PDB MOAD | Crystal structure data | Identification of small-molecule ligands | ANN scoring function map | NNScore 2.0 http://www.nbcr.net/software/nnscore/ | Python | Open Source | 50 | Supervised Learning: Classification | Protein map prediction |
| Lin G., 2010, US (Lin et al., 2010) | PDB | Protein Folding Rates. Predicting protein folding rates from geometric contact and amino acid sequence | Protein folding kinetic rate and real-value folding rate | SVM, SVR | SeqRate http://casp.rnet.missouri.edu/fold_rate/index.html | Java | Open Source | 75 | Supervised Learning: Classification | Protein map prediction |
| Protein-Binding Prediction | ||||||||||
| Song J., 2021, China (Song et al., 2021) | PDB Swiss-Prot | ATP-binding proteins | Protein–ATP-Binding Residues | DCNN, LightGBM | TensorFlow, Keras https://github.com/tlsjz/ATPensemble | Python | Open Source | 80 | Supervised Learning: Regression Prediction Classification | Prediction of Protein–ATP Binding Residues |
| Kwon Y., 2020, Korea (Kwon et al., 2020) | PDBind-2016 | VEGFR2 kinase domain and adenosine deaminase | Prediction of affinity-binding of a protein–ligand complex | 3D-CNN | Keras, Tensorflow | Python | Open Source | 85 | Supervised Learning: Prediction | Protein affinity-binding prediction |
| Mahmoud A., 2020, Switzerland, US (Mahmoud et al., 2020) | PDB | HIV-1 protease, dihydrofolate reductase | Hydration site occupancy and thermodynamics predictions | CNN | https://hub.docker.com/r/lilllab/watsite3 | N/A | Open Source | 65 | Supervised Learning: Regression Classification | Protein–ligand-binding prediction |
| Wang M., 2020, US (Wang M. et al., 2020a) | SKEMPI 1.0, 2.0 dataset AB-Bind S645 dataset | Protein–protein complexes | Protein–ligand-binding affinity predictions | Site-specific persistent homology, CNN, GBT | TopNetTree, Keras https://doi.org/10.24433/CO.0537487.v1 | Matlab, java, python | Open Source | 90 | Supervised Learning: Prediction | Protein–protein-binding affinity |
| Luo X., 2019, China (Luo et al., 2020) | Transfac | DNA sequences | predicting DNA–protein binding | CNN | Keras, Tensorflow https://github.com/gao-lab/ePooling | Python | Open Source | 70 | Supervised Learning: Regression Prediction | Protein-binding prediction |
| Protein Site Prediction | ||||||||||
| Zheng W., 2020, China, US (Zheng et al., 2020) | SCOPe2.07 | N/A | Protein domain boundaries | DRNN | https://zhanglab.ccmb.med.umich.edu/FUpred/ | N/A | Open Source | 60 | Supervised Learning: Classification | Protein domain identification |
| Cui Y., 2019, China (Cui et al., 2019) | BioLip | Fourteen binding residues | Protein–ligand-binding residue prediction | DCNN | TensorFlow, https://github.com/yfCuiFaith/DeepCSeqSite | Python | Open Source | 100 | Supervised Learning: Prediction | Protein site prediction |
| Fu H., 2019, China (Fu et al., 2019) | PLMD | Sequences and physicochemical properties of protein | Predict Lysine ubiquitination sites in large scale | CNN, DL DeepUbi | TensorFlow, DeepUbi: https://github.com/Sunmile/DeepUbi | Python, MATLAB, Linux | Open Source | 100 | Supervised Learning: Classification | Protein site prediction |
| Haberal I., 2019,, Norway, Turkey (Haberal and Ogul, 2019) | PDB | Metal binding of histidine and Cysteine amino acids | Prediction of metal binding in proteins | 2D-CNN, LSTM, RNN | Keras, TensorFlow | Python | Open Source | 100 | Supervised Learning: Prediction | Protein site prediction |
| Savojardo C., 2019, Italy (Savojardo et al., 2020b) | UniprotKB/Swiss-Prot | Mitochondrial proteins | Sub-mitochondrial cellular localization | CNN | http://busca.biocomp.unibo.it/deepmito | Python | Open Source | 75 | Supervised Learning: Regression | Protein sub-mitochondrial site prediction |
| Simha R., 2015, Canada, Germany, US (Simha et al., 2015) | DBMLoc dataset | N/A | Protein multi-location prediction | MDLoc, BN | MDLoc http://www.eecis.udel.edu/compbio/mdloc | Python | Open Source | 75 | Supervised Learning: Classification | Protein site prediction |
| Briesemeister S., 2010, Germany (Briesemeister et al., 2010) | UniProt | Protein sequence | Predict protein subcellular localization | NB | Yloc Weka www.multiloc.org/YLoc | Python, Java, Linux | Open source | 85 | Supervised Learning: Classification | Protein site prediction |
| Huang W., 2008, Taiwan (Huang et al., 2008) | UniProt GO | SCL12, SCL16 Sequence-based, GO terms, protein sequence | Prediction method for predicting subcellular localization of novel proteins | GA, SVM | LIBSVM ProlocGO http://iclab.life.nctu.edu.tw/prolocgo | N/A | N/A | 75 | Supervised Learning: Classification | Protein site prediction |
| Ladunga I., 1991, Hungary (Ladunga et al., 1991) | UniProt | Signal peptide | Novel predicted signal peptides | NN (Tiling algorithm) | N/A | C | N/A | 50 | Supervised Learning: Classification | Protein site prediction |
| Genomics | ||||||||||
| Dai W., China, 2020 (Dai et al., 2020) | Reactome DB and InBio Map DB | Human essential gene | Predict human essential genes | Network embedding, SVM | N/A | N/A | N/A | 50 | Supervised Learning: Classification | Human gene prediction |
| Picart-Armada S., 2019, Belguim, UK, Spain (Picart-Armada et al., 2019) | STRING | Gene-disease data from 22 common non-cancerous diseases | Target disease gene identification | PR, Random Randomraw EGAD, PPR, Raw, GM, MC, Z-scores, KNN, WSLD, COSNet, bagSVM, RF, SVM | https://github.com/b2slab/genedise | R | Open Source | 80 | Semi-supervised, Supervised Learning: Classification | Target gene identification, target drug discovery |
2D-BRLSTM, two-dimensional bidirectional Res-long short-term memory; 2D-CNN, Two-dimensional convolutional neural ubcell; 3D-CNN, Three-dimensional convolutional neural ubcell; ANN, Artificial neural network; BN, Bayesian Network; CNN, Convolutional neural network; DCNN, Deep Convolutional neural network; DL, Deep learning; GAs, Genetic algorithms; GBT, Gradient boost tree; KNN, k-nearest neighbor; LightGBM, Light Gradient Boosting Machine; LSTM, Long short-term memory; NB, Naïve Bayes; NN, Neural network; RNN, Recurrent neural network; SVM, Support vector machine; SVR, Support vector regression; WMC, Weighted multiple conformation.
Results
Article Scaffolding
This article is arranged as follows (Figure 2): first, we provide a representation of the process in designing, preparing, and describing of the guideline throughout the article. Secondly, we review the presented formulation of the research question toward the determined problem formulation and objectives of the research, including the treatment of the data and the applications of it. Thirdly, the article processes the observation, research, and review of a series of articles to further study the data obtained and review similarities. Furthermore, the gathering of AI–PS information, within this processing of the identification of filtered data, curated data and features implemented, the observation of input data, data encoding format, recording of machine learning algorithms and methods, as so the post-processing treatment, quality rule processing, filtering, combination, or unification of information, which passes into the interpretation of the information recollected, and representation of it by the usage of figures and tables, portrays the results, which are focused on the latest findings of AI applications in the field of protein science as well as the usage of specific algorithms for protein design. Therefore, this aims to include a wide-scope range of the state of the art of artificial intelligence within protein science; this leads us to a latter analysis and discussion regarding the identification and prediction of AI applications into the protein field, by classification and identification of main protein structures, and other components not found or described yet in nature, and the resolution of possible protein prediction structures and other components of them are plausible outcomes of future research.
FIGURE 2.
Flowchart of article scaffold. Representation of the process throughout the entire article. The biochemical meta-analysis consists of three main steps: the systematic review, the road map design, and the road map alignment. In the systematic review, the research question is formulated in order to set the basis and objectives of the project. It also includes the observation and synthesis of information obtained from a variety of articles and the correlation made between them. The latter followed by the quality evaluation of the collected information. The road map design consists of analyzing the outcome of the studies and classifying them, thus being able to interpret the information recollected and represent it through the usage of figures and tables. This aims to include a wide range of the state of the art or artificial intelligence. Finally, the road map alignment includes the final discussion and further changes for our understanding of protein science using AI and the resolution of possible protein science application targets.
Toward an Innovative Cross-Functional AI–PS Binomial Inter-field
This systematic review and meta-analysis are focused on the latest findings of AI applications to the field of protein science as well as specific algorithms used for protein design. Furthermore, it aims to include a wide scope of the state of the art of artificial intelligence in protein science. PIO is the methodology used to address the following research question: What is the state of the art in the use of artificial intelligence in the protein science field? Figure 1 shows the total number of articles retrieved using the PIO strategy in the PubMed database.
The systematic review process began with 541 references obtained from five electronic databases: 42 were from PubMed, 74 were from Ebsco, 48 were from Bireme, 38 were from OVID, and 339 were from Web of Science. In the first screening, 403 articles were removed: 250 articles with a double reference; 2 not written in Spanish or English; 149 whose topic was irrelevant to the review; and two newspapers, letters, or reviews. This election process left 138 references, and manually we added 6, thus getting a total of 144 articles for the review (Figure 3).
A second screening (eligibility) was performed using the following set of quality criteria:
1. Clear research questions and objectives.
2. Definition of the measured concepts.
3. Reliability and feasibility of the instruments to be measured.
4. Detailed description of the method.
5. Scaffolding and enhanced protein information.
6. Characteristics of scaffolding and its realization.
7. Appropriate system and learning approach.
8. Journal impact.
A total of 93 articles were included for further analysis, and 51 studies were removed based on quality criteria.
Machine Learning Approach to Protein Science
Proteins are influenced by epigenetic phenomena (cellular stress, aging, etc.) because of their multiple structure-folding-function within protein science (PS), phenomena that can be challenged through the use of artificial intelligence (AI).There are several questions within this interdisciplinary approach such as How do proteins evolve? How do proteins fold and get their tridimensional structure? What are their networks within proteins? Given the astronomical numbers of possibilities for protein structures, configurations, and functions that require the use of AI as a tool to fully understand protein behavior.
A total of 144 articles were assessed for quality (Tables 2–6) resulting in 93 articles (Table 1), those articles that were greater or equal to 75 in the quality percentage qualifications were kept for the final biochemical meta-analysis. For this review and meta-analysis, we identified five main applications of AI into PS (Tables 2–6 and Figures 4–6)
-
I. Protein design and drug design (Table 2)
a) De novo protein design.
b) Novel biocatalyst design.
c) Novel function and ligand interaction.
d) Evolution of non-existent proteins in nature.
e) Chemical structure and properties.
f) Drug–drug interaction.
g) Drug–receptor interaction.
h) Drug effects.
-
II. Protein function, function prediction, and novel function (Table 3)
a) Protein–ligand interactions.
b) Hydroxylation site prediction.
c) Prediction of the local properties in proteins.
d) Enzymatic function prediction.
e) Predicting protein–protein interactions.
f) Function prediction.
g) Molecular property prediction.
-
III. Fold ID, physicochemical properties, and protein classification (Table 4)
a) Fold Id.
b) Glycation site predictor.
c) Phosphorylation site predictor.
d) Protein–protein interaction.
e) Intrinsically disordered protein prediction.
-
IV. Protein structure prediction (Table 5)
a) Protein structure prediction: primary, secondary, and 3D-structures; domains, active sites, allosteric sites, and structural feature prediction.
b) Protein structure classification: folds, structural families, intrinsically disorder proteins, etc.
c) Protein–protein interactions and protein networks.
d) Protein–ligand interactions: substrates, inhibitors, activators, ions, etc.
V. Protein contact map prediction, protein-binding prediction, protein site prediction, and genomics (Table 6)
1) Contact map prediction.
2) Protein sub-mitochondrial site prediction.
3) Genomics.
FIGURE 6.
Representation of the specific case for protein structure prediction in the supervised learning framework. Revealing the most common flow followed by the studies analyzed. From extraction, training data, feature extraction procedures and data continuity. Including the PDB database, the most common supervised algorithms, SVM, SVR, 3DCNN.
TABLE 3.
An overview of the protein function prediction, function prediction, and novel function articles with the quality assessment.
| First Author/Year of Publication/Country | Database | Initial scaffold (ID) | Designed Protein | ML model | Software/Sever | Programming language/Platform | License | Quality (%) | Machine learning | Protein application | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein function prediction | ||||||||||||
| Verma N., 2021, US (Verma et al., 2021) | DrugBank matador PDB | Human, C. Elegans | Protein–ligand interactions | DNN | GitHub (https://github.com/ekraka/SSnet) | Python | Open source | 75 | Supervised learning: Prediction | Protein–ligand interaction prediction | ||
| Du Z., 2020, China, Russia, US (Du et al., 2020) | CAFA3, SwissProt | Human, C. Elegans | Automated function prediction | NLP, CNN | Keras, TensorFlow | Python | Open Source | 70 | Supervised Learning: Classification | Protein function prediction | ||
| Liang M., 2020, China (Liang and Nie, 2020) | PDB | Relative angle of (C – Cα – C) principal plane | Enzymatic function prediction | RN, LSTM | TensorFlow | Python | Open Source | 90 | Supervised Learning: Prediction | Protein function prediction, Function ID | ||
| Rifaioglu A., 2019, Turkey, UK (Rifaioglu et al., 2019) | UniProtKB/Swiss-Prot | N/A | GO term prediction | DNN | Tensorflow, https://github.com/cansyl/DEEPred | Python | Open Source | 70 | Supervised Learning: Regression | Protein function prediction | ||
| Torng W., 2019, US (Torng and Altman, 2019) | PROSITE NOS dataset | Protein structure as 3D images | Protein functional site detection | DL, 3D-CNN, SVM | N/A https://simtk.org/projects/fscnn | Python | N/A | 75 | Supervised Learning: Classification | Protein function prediction | ||
| Wan C., 2019, UK (Wan et al., 2019) | UniProtKB/Swiss-Prot | Human proteins | Function prediction | DMNN, SVM | Keras, https://github.com/psipred/STRING2GO | Python | Open Source | 80 | Supervised Learning: Prediction Classification | Protein function prediction | ||
| Feinberg E., 2018, China, US (Feinberg et al., 2018) | PDB Bind 2007 | Scaffold split for grouping ligands in common frameworks | Molecular Property Prediction | GCNN | PyTorch, NumPy and SciPy | Python | Open Source | 100 | Supervised Learning: Prediction | Protein function prediction | ||
| Frasca M., 2018, Italy (Frasca et al., 2018) | STRING GO | Organisms: Homo sapiens (human) S. cerevisiae (yeast) Mus musculus (mouse) | AFP (Automated Protein Function Prediction) | COSNet, ParCOSNet, HNN | COSNet, ParCOSNet | C, C++, R, CUDA | Open Source | 75 | Unsupervised Learning: Clustering | Protein function prediction | ||
| Khurana S., 2018, Qatar, US (Khurana et al., 2018) | pepcDB database | k-mer structure and additional sequence and structural features extracted from the protein sequence | Solubility prediction | CNN, DL, FFNN | PROSO II https://zenodo.org/record/1162886#.XSP26ffPzOQ DeepSol: https://github.com/sameerkhurana10/DSOL_rv0.2 | Python, Linux | Open source | 95 | Unsupervised Learning: Clustering | Protein function prediction | ||
| Li H., 2018, China (Li et al., 2018) | HPRD DIP HIPPIE | Primary sequence Escherichia coli, Drosophila, Caenorhabditis elegans, Pan’s PPI datasets | Prediction of protein interactions | DNN, CNN, LSTM | Keras, Theano, TensorFlow, N/A | Python | Open Source | 85 | Supervised Learning: Regression | Protein function prediction | ||
| Long H., 2018, China, US (Long et al., 2018) | UniProt | PseAAC Hydroxyproline and hydroxylysine | Predicting hydroxylation sites | CNN, LSTM | MXNet, N/A | R | Apache 2.0 | 85 | Supervised Learning: Classification | Protein function prediction | ||
| Makrodimitris S., 2018, Netherlands (Makrodimitris et al., 2019) | Arabidopsis thaliana proteins | Arabidopsis thaliana protein | Protein function prediction | KNN, LSDR | SciPy https://github. Com/stamakro/SSP-LSDR. | Python, MATLAB Bioinformatics toolbox | Open source, Mathworks | 80 | Unsupervised Learning: Clustering | Protein function prediction | ||
| Zhang L., 2018, China (Zhang L. et al., 2018) | UniProt, DIP | S. cerevisiae, H. sapiens, and M. musculus | Predicting Protein–Protein interactions | DNN, Adam Algorithm | TensorFlow | Python | Open Source | 100 | Supervised Learning: Prediction | Protein function prediction | ||
| Adhikari B., 2017, US (Adhikari et al., 2018) | DNCON Dataset | N/A | Contact map protein prediction | CNN | TensorFlow, Keras http://sysbio.rnet.missouri.edu/dncon2/ | Python | Open Source | 65 | Supervised Learning: Regression Predection | Protein residue–residue contacts | ||
| Cao R, 2017, US (Cao et al., 2017) | UniProt | Protein sequence | Protein function prediction | RNN | ProLanGO Model N/A | N/A | N/A | 50 | Supervised Learning: Classification | Protein function prediction | ||
| Al-Gharabli S., 2015, Jordan (Al-Gharabli et al., 2015) | PDB | Amino acid sequence hydrophobicity | Prediction of dihedral angles physiochemical properties, enzyme loops | ANN | N/A | N/A | N/A | 50 | Supervised Learning: Classification | Protein function prediction | ||
| Qi Y., 2012, US (Qi et al., 2012) | Standard benchmark, CB513 DSSP | PSI-BLAST amino acid embedding | Prediction of the local properties in proteins | DNN | Torch5 | C | Open Source | 100 | Supervised Learning: Classification | Protein function prediction | ||
| Yang Y., 2011, US (Yang et al., 2011) | SPINE | Protein sequence | Single-method fold recognition | SPARKS-X Algorithm | SPARKS-X https://sparks-lab.org/server/sparks-x/ | Shell script | Open Source | 75 | Supervised Learning: Regression | Protein function prediction | ||
| Latek D., 2010, Poland (Latek and Kolinski, 2011) | 10 globular proteins, 216 residues, and S100A1 protein | 10 globular proteins and S100A1 protein | Predicted Nuclear Overhauser Effect signals on the basis of low-energy structures from CABS-NMR | CABS, MC | CABS- NMR toolkit http://biocomp.chem.uw.edu.pl/services.php | N/A | N/A | 70 | Unsupervised Learning: Clustering | Protein function prediction | ||
| Tian J., 2010, China, US (Tian et al., 2010) | ProTherm PDB | 3D structures | Effect on single- or multi-site mutation on protein thermostability | RFR, RF, SVM | Prethermut http://www.mobioinfor.cn/prethermut/ | R, Perl, Linux | Open Source | 75 | Supervised Learning: Classification | Protein function prediction | ||
| Wu S., 2008, US (Wu and Zhang, 2008) | PDB | PDB protein sequence | Protein contact predictor | MUSTER | MUSTER http://zhang.bioinformatics.ku.edu/MUSTER | N/A | N/A | 50 | Supervised Learning: Classification | Protein function prediction | ||
| Hung C., 2006, Taiwan (Hung et al., 2006) | NCBI | Nucleocapsid (nsp1) of a coronavirus family | Predict protein functions | AGCT | N/A | N/A | N/A | 75 | Supervised Learning: Classification | Protein function prediction | ||
| Sidhu A., 2006, UK (Sidhu and Zheng, 2006) | Swiss-Prot | Signal peptides and non-secretory proteins from Human, E. coli, prokaryotic | Predict signal peptide | BBFNN, DT | N/A | N/A | N/A | 75 | Supervised Learning: Regression | Protein function prediction | ||
| Capriotti E., 2005, Italy (Capriotti et al., 2005) | ProTherm | Protein tertiary structure | Protein stability prediction | SVM | I-Mutant2.0 http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant2.0/I-Mutant2.0.cgi | Python | Open Source | 75 | Supervised Learning: Classification | Protein function prediction | ||
| Hu C., 2004, US (Hu et al., 2004) | WhatIF database UniProt | 3D coarse-grained structure from protein sequences | Optimal non-linear scoring | SVM non-linear Gaussian kernel functions | N/A | N/A | N/A | 65 | Supervised Learning: Classification | Protein function prediction | ||
| Gutteridge A., 2003, UK (Gutteridge et al., 2003) | PDB | Amino acid sequence of quinolate phosphoribosyl transferase | Predict active site | FFNN | N/A | N/A | N/A | 50 | Unsupervised Learning: Clustering | Protein function prediction | ||
| Function Prediction and Novel Function | ||||||||||||
| Nie J., Singapore 2020 (Sua et al., 2020) | UniProt | acetyl-lysine (S1), “crotonyl-lysine” (S2), “methyl-lysine” (S3), or “succinyl-lysine” (S4) | Identification of Lysine PTM sites | RF, SVM, MNB, LR, ME, KNN, CNN, MLP | Tensorflow, https://github.com/khanhlee/lysineSGT | Python | N/A | 100 | Supervised Learning: Classification | Function ID | ||
| Savojardo C.., 2020, Italy (Savojardo et al., 2020a) | UniProtKB GOA, DeepMitoDB | Human, mouse, fly, yeast, and Arabidopsis thaliana | protein sub-mitochondrial localization | DeepMito, 1D-CNN | N/A | N/A | N/A | 75 | Supervised learning: Prediction | Function ID | ||
| Fang C., 2019, China, Japan (Fang et al., 2019) | PDB | MoRF-containing membrane protein chains | Molecular recognition features MoRFs prediction | DCNN | N/A | N/A | N/A | 75 | Supervised Learning: Classification | Function ID and Fold ID | ||
| Zhang Y., 2019, China (Zhang et al., 2019) | PDB | PDNA-543, PDNA-224 and PDNA-316 | Identification of DNA–protein-binding site | ADASYN | Theano | Python | Open Source | 85 | Supervised Learning: Classification | Function ID and Fold ID | ||
| Hanson J., 2018, Australia, China (Hanson et al., 2019) | PISCES CASP12 PDB | 5N5EA 6FI2A 6FQ3A | Sequence-based prediction of one-dimensional structural properties of proteins | CNN, 2D-BRLSTM | N/A | N/A | N/A | 80 | Supervised Learning: Classification | Function ID | ||
| Shah R., 2008, US (Shah et al., 2008) | D Dataset | Protein sequence | Homology detection | SVM | SVM-HUSTLE http://www.sysbio.org/sysbio/networkbio/svm_hustl | N/A | N/A | 70 | Supervised Learning: Classification | Function ID and Fold ID | ||
1D-CNN, one-dimensional convolutional neural network; 2D-BRLSTM, two-dimensional bidirectional recurrent long short-term memory; 3D-CNN, three-dimensional convolutional neural network; ADASYN, Adaptive Synthetic Sampling; ANN, Artificial neural network; AGCT, Alignment genetic causal tree; BBFNN, Biobasis function neural network; CABS, C-alpha-beta side; CNN, Convolutional neural network; COSNet, Cost-sensitive neural network; DCNN, Deep Convolutional neural network; DMNN, Deep mahout neural network; DFS, Depth first search; DL, Deep learning; DNN, Deep neural network; DTNN, Deep tensor neural network; FFNN, Feed forward neural network; GA, Genetic algorithms; HDL, Hybrid Deep learning; HMM, Hidden Markov model; HNN, Hopfield neural network; KNN, k-nearest neighbor; LR, Logistic regression; LSDR, Label-Space dimensionality reduction; LSTM, Long short-term memory; MC, Monte Carlo; ME, Max Entropy; MLP, Multilayer; MNB, Multinomial Naïve Bayes; MNPP, Message passing neural network; NLP, Natural language processing; NN, Neural network; ParCOSNet, Parallel COSNet; RF, Random forest; RN, Relational network; RNN, Recurrent neural network; SPARK-X, Probabilistic-based matching; SVM, Support vector machine.
TABLE 4.
An overview of the fold id, physicochemical properties, and protein classification articles with the quality assessment.
| First Author/Year of Publication/Country | Database | Initial scaffold (ID) | Designed Protein | ML model | Software/Sever | Programming language/Platform | License | Quality (%) | Machine learning | Protein application |
|---|---|---|---|---|---|---|---|---|---|---|
| Fold ID and physicochemical properties | ||||||||||
| Rives A., 2020, UK, USA (Rives et al., 2021) | SCOPe | Protein data in the form of unlabeled amino acid sequences. Small vocabulary of 20 canonical elements | Predicted model contains information about biological properties in its representations | Deep contextual language model | https://github.com/facebookresearch/esm | Python | Open source | 70 | Supervised learning; prediction | Physicochemical and biological properties |
| Li H., 2020, France, Hong Kong (Hongjian et al., 2021) | PDB, PubChem, ZINC, ChEMB,L BindingDB, HTS | Chemical Estrogen receptor α (Erα) Anaplastic lymphoma kinase Neuraminidase (NA) Reducing the level of Dmiro protein in flies Acetylcholinesterase (AchE) | Protein–ligand complex | RF, BRT, kNN, NN, SVM, GBDT, multi-task DNN XGBoost | Descriptor data bank ODDT BINANA RF-Score-v1 RF-Score-v3 MIEC-SVM | Python | Open Source | 100 | Supervised Learning; Unsupervised Learning; Prediction Classification Regression | Physicochemical properties |
| Shroff R., 2020, US (Shroff et al., 2020) | PDB | N/A | amino acid association guide mutation | 3D CNN | Theano www. Mutcompute.com | Python | Open Source | 70 | Supervised Learning: Class Prediction | Microenvironment mutation identification |
| Wang M., 2020, China, US (Wang M. et al., 2020b) | UniProt | E. coli, M. musculus, H. sapiens | Protein malonylation site prediction | DL-CNN | Keras, https://github.com/QUST-AIBBDRC/DeepMal/ | Python, Matlab | Open Source | 80 | Supervised Learning: Classification | Malonylation site prediction |
| Chen J., 2019, China (Chen et al., 2019) | Datasets A(CPLM),B,C | Proteins and reducing sugars | Glycation product prediction | RNN, CNN | N/A | N/A | N/A | 60 | Supervised Learning: Classification | Glycation site predictor |
| Han X., 2019, Singapore, US (Han et al., 2019) | eSol | Cell-free protein expression from E. coli | Protein solubility | GAN | N/A | N/A | N/A | 60 | Supervised Learning: Regression Prediction | Protein solubility prediction |
| Heinzinger M., 2019, Germany (Heinzinger et al., 2019) | UniProt, PDB | TS115 CB513 CASP12 | Protein sequence representation | NLP, ELMo | Pytorch, https://embed.protein.properties/ | Python | Open Source | 80 | Supervised Learning: Classification | Fold ID |
| Kaleel M., 2019, Ireland (Kaleel et al., 2019) | PDB | Amino acids are subcellular into four classes involving RSA | Prediction of relative solvent accessibility | BRNN | http://distilldeep.ucd.ie/paleale/ | Python | Open Source | 90 | Supervised Learning: Prediction | Protein relative solvent accessibility prediction |
| Li C., 2019, China (Li and Liu, 2020) | LE dataset from SCOP | Multiple superfamilies | Detect the structural motifs related with the protein folds | MotifCNN and MotifDCNN SVM CNN | TensorFlow | Python | Open source | 100 | Supervised Learning: Classification | Fold ID |
| Luo L., 2019, China (Luo L. et al., 2019) | BioCreative II, BioCreative III, BioCreative II.5 | PPI protein articles | Protein–protein interaction | KeSACNN | Keras | Python | Open Source | 50 | Supervised Learning: Classification | Physicochemical properties |
| Taherzadeh T., 2019, Australia, US (Taherzadeh et al., 2019) | Uniprot, dbPTM, Uniprep, UnicarKB, GlycoProtDB | Glycoprotein | N- and O-linked glycosylation | DNN SVM | TensorFlow, https://sparks-lab.org/server/sprint-gly/ | Python | Open Source | 80 | Supervised Learning: Regression Prediction | Glycosylation site identification |
| Zhang D., 2019, US (Zhang and Kabuka, 2019) | DIP, HPRD, UniProt | D. melanogaster, S. cerevisiae, E. coli, C. elegans, H. sapiens, H. pylori, M. musculus, R. norvegicus | Protein–protein interactions and protein family prediction | Multimodal DNN | N/A | N/A | N/A | 75 | Supervised Learning: Classification | Physicochemical properties |
| Cuperus J., 2018, US (Cuperus et al., 2017) | 5′ UTR library of 50-nt-long random sequences | Yeast Saccharomyces cerevisiae | Predict protein expression | CNN | Keras, Theano, https://github.com/Seeliglab/2017---Deep-learning-yeast-UTRs | Python | Open Source | 85 | Supervised Learning: Regression | Fold ID |
| Hochuli J., 2018, US (Hochuli et al., 2018) | PDB | Ligands SMILE Protein FASTA | Identify protein–ligand scoring | CNN | Gnina, Caffe Github.com/gnina | C++, Python | Open source | 50 | Supervised Learning: Classification | Protein Scoring |
| Luo F., 2018, China (Luo F. et al., 2019) | Phospho.ELM, PhosphositePlus, HPRD, dbPTM, SysPTM | Kinase protein family | Protein phosphorylation | CNN | https://github.com/USTCHIlab/DeepPhos | N/A | N/A | 60 | Supervised Learning: Regression Prediction | Phosphorylation site predictor |
| Zhao X., 2018, China (Zhao et al., 2018) | PLMD | Lysine | Lysine acetylation sites | CNN DNN | Keras, Theano, https://github.com/jiagenlee/DeepAce | Python | Open Source | 80 | Supervised Learning: Regression Classification Prediction | Acetylation site prediction |
| Zhao F., 2010, US (Zhao et al., 2010) | CASP | (PSSM) Position-specific scoring matrix generated by PSI-BLAST | Protein folding | CNF | CNF | N/A | N/A | 80 | Supervised Learning: Classification | Fold ID |
| Armstrong K., 2008, US (Armstrong and Tidor, 2008) | PDB | Protein sequence | Protein engineering space of foldable sequences | Computational mapping | N/A | C++ | Open source | 50 | N/A | Fold ID |
| Shamim M., 2007, India (Shamim et al., 2007) | D-B dataset Ext. D-B dataset | Structural information of amino acid residue and amino acid residue pairs | Protein fold prediction | SVM | LIBSVM-library | C++, Java, Python Windows, Linux | Open source | 80 | Supervised Learning: Classification | Fold ID |
| Protein Classification | ||||||||||
| Burak T., 2021, Turkey (Alakuş and Türkoğlu, 2021) | UniProt | Protein sequence from 60 different families | Protein family classification/identification | FIBHASH | N/A | N/A | N/A | 70 | Supervised Learning: Classification | Protein classification |
| Zhao Z., 2019, China (Zhao and Gong, 2019) | Monomers and dimers from the author | Monomers and dimers from the author | Protein–protein interaction | LSTM | N/A | N/A | N/A | 60 | Supervised Learning: Unsupervised Learning: Regression | Interface residue pair prediction |
| Huang L., 2018, US (Huang et al., 2018) | DIP, HPRD | PPI network graph | Protein–protein interaction | ENN-RL | TensorFlow, https://www.eecis.udel.edu/∼lliao/enn/ | Python | Open Source | 75 | Supervised Learning: Prediction | Protein–protein interaction |
| Le N., 2018, Taiwan (Le et al., 2018) | UniProt GO | Rab GGT activity Rab GDI activity Rab GTPase binding Rab GEF activity | Classify Rab protein molecules | 2D-CNN | Keras, Theano DeepRab; http://bio216.bioinfo.yzu.edu.tw/deeprab/ | Python | Open Source | 90 | Supervised Learning: Regression | Protein Classification |
| Xue L., 2018, China, US (Xue et al., 2019) | Swiss-Prot, TrEMBL | Secretory protein | Protein sequence into T3Ses or non-T3Ses | DCNN | Keras, https://github.com/lje00006/DeepT3 | Python | Open Source | 60 | Supervised Learning: Regression Classification | Protein classification |
| Zhao B., 2018, US (Zhao and Xue, 2018) | DisProt PDB | Intrinsically disordered proteins (IDPs), intrinsically disordered regions (IDRs), and intrinsically disordered amino acids (IDAAs) | N/A | ANN, DT | DisEMBL, IUPred, VSL2, Dbann, and Espritz | N/A | N/A | 50 | Supervised Learning: Regression | Intrinsically disordered protein prediction |
| Szalkai B., 2017, Hungary(Szalkai and Grolmusz, 2018a) | Swiss-Prot, UniProt, GO | Thyroid hormone, phenol-containing compound, cellular modified amino acid, protein kinase superfamily | protein classification by amino acid sequence | ANN | TensorFlow | Python | Open Source | 90 | Supervised Learning: Classification | Protein Classification |
| Szalkai B., 2017, Hungary (Szalkai and Grolmusz, 2018b) | UniProt GO | Classes.tre | Hierarchical Biological Sequence Classification | DNN | SECLAF, TensorFlow https://pitgroup.org/seclaf/ | Python | Open Source | 85 | Supervised Learning: Classification | Protein Classification |
3D-CNN, three-dimensional convolutional neural network; ANN, Artificial neural network; BLSTM, Bidirectional long short-term memory; BRNN, Bidirectional recurrent neural network; BRT, Booster regression tree; CNF, Conditional neural filed; DNN, Deep neural network; DT, Decision Tree; ELMO, Embeddings from language models; ENN-RL, Evolution neural network-based Regularized Laplacian kernel; FIBHASH, Fibonacci numbers and hashing table; GAN, Generative adversarial network; GBDT, Gradient boosted decision tree; GR, Genetic recombination; KNN, k-nearest neighbor; KeSCANN, Knowledge-enriched Self-Attention convolutional neural network; LSTM, Long short-term memory; MotifCNN, Motif convolutional neural network; Motif DNN, Motif deep neural network; Multimodal DNN, Multimodal deep neural network; NLP, Natural language processing; NN, Neural network; RF, Random forest; RNN, Recurrent neural network; SPARK-X, Probabilistic-based matching; SVM, Support vector machine.
TABLE 5.
An overview of the protein structure prediction articles with the quality assessment.
| First Author/Year of Publication/Country | Database | Initial scaffold (ID) | Designed Protein | ML model | Software/Sever | Programming language/Platform | License | Quality (%) | Machine learning | Protein application |
|---|---|---|---|---|---|---|---|---|---|---|
| Protein Structure Prediction | ||||||||||
| Xu J., 2022, USA (Xu et al., 2021) | CASP13, PDB, PISCES, CATH | Discrete probability over distance for three backbone atom pair and inter-residue orientation | Structure prediction | Convolutional residual neural network | https://github.com/j3xugit/RaptorX-3DModeling/ | python | Open source | 70 | Supervised Learning; Prediction | Protein structure prediction |
| ALQuraishi M., 2021, USA (AlQuraishi, 2021) | PDB, CASP14 | Primary protein sequence | Structure prediction | Markov random field, Attention networks | N/A | N/A | N/A | 50% | Supervised Learning: Prediction | Protein structure prediction |
| Bond P., 2020, UK (Bond et al., 2020) | PDB | Only residues with side chains longer than beta-carbon | Predicting the correctness of protein residues | NN, MLP | CCP4 | C++, Python | Open Source | 60 | Supervised Learning: Regression | Protein structure prediction |
| Wardah W., 2020, Australia, Fiji, Japan, US (Wardah et al., 2020) | BioLiP | Positive (binding) or negative (non-binding), protein sequence classification | Predicting Protein-peptide-binding sites | CNN | PyTorch, https://github.com/WafaaWardah/Visual | Python | Open Source | 100 | Supervised Learning: Prediction Classification | Protein structure prediction |
| Yang J., 2019, China, USA (Yang J. et al., 2020) | CASP13, Uniclust30 | Representation of the rigid-body transform from one residue to another; angles and distances | Predicted inter-residue orientations | Deep residual convolutional neural network | https://yanglab.nankai.edu.cn/trRosetta/ | Python | Open source | 70 | Supervised Learning; Prediction | Protein structure prediction |
| Degiacomi M., 2019, UK (Degiacomi, 2019) | PDB | Malate dehydrogenase (1MLD), αB crystallin (2WJ7) Phospholipase A2 (1POA), Envelope glycoprotein (1SVB), MurD, closed (3UAG), MurD, open (1E0D), MurD, closed + open (3UAG,1E0D), HIV-1 (1E6J) | Enhancement of molecular conformational space generator | Molecular dynamics, RF, auto encoder | Keras, Tensorflow | Python | Open Source | 80 | Unsupervised Learning: Classification | Protein conformational space |
| Guo Y., 2019, US (Guo et al., 2019) | CB513, CASP10, CASP11 | Protein sequences | Protein secondary structure | ACNN, BLSTM | Keras, Tensorflow, https://github.com/GYBTA/DALSTM/ | Python | Open Source | 80 | Supervised Learning: Prediction Classification | Protein secondary structure prediction |
| Long S., 2019, China (Long and Tian, 2019) | Jpred dataset cullpdb dataset UniRef90 UniProt | Multiple superfamilies | Protein secondary structure prediction | CNN | TensorFlow N/A | Python | Open Source | 60 | Supervised Learning; Unsupervised Learning; Prediction | Protein structure prediction |
| Mirabello C., 2019, Sweden (Mirabello and Wallner, 2019) | PDB | N/A | Method prediction | NLP, DNN | Keras, TensorFlow https://bitbucket.org/clami66/rawmsa | Python | Open Source | 70 | Supervised Learning: Prediction | Protein structure prediction |
| Pagès G., 2019, France (Pagès et al., 2019) | CASP | Model QA | Protein model quality assessment | 3D CNN | TensorFlow, Ornate https://team.inria.fr/nanod/software/Ornate/ | C++, Python | Open Source | 85 | Supervised Learning: Regression | Model protein prediction |
| Schantz M., 2019, Argentina, Denmark, Malaysia (Klausen et al., 2019) | PDB, PISCES | Crystal structures | Prediction of protein structural features | CNN, LSTM | Keras | Python | Open source | 100 | Supervised Learning: Prediction | Protein structure prediction |
| Wang D., 2019, China (Wang D. et al., 2020) | CASP11, 12 | Caspase 14 | Protein structure refinement | Multi-objective PSO | AIR 2.0 www.csbio.sjtu.edu.cn/bioinf/AIR/ | Python | Open Source | 95 | Supervised Learning: Optimization | Protein structure prediction |
| Yu C., 2019, US (Yu et al., 2019) | PDB | 194l (lysozyme), 107m (myoglobin), 6cgz (β-barrel), a silk protein, amyloid protein, and others | Generation of audible sound from amino acid sequence for application on designer materials | RNN, LSTM | Magenta TensorFlow, Melody RNN | Java, Python | Open Source | 100 | Supervised Learning: Regression | Protein sequence prediction |
| Zheng W., 2019, US (Zheng et al., 2019) | CASP13 | Query sequence profiles | Automated structure prediction pipeline | ZhangServer and QUARK pipelines | Zhang and Quark server | N/A | Open Source | 85 | Supervised Learning: Classification Regression | Protein structure prediction |
| Fang C., 2018, US (Fang et al., 2018) | PDB JPRED CASP CB513 | Different super-families, CASP10, 11, 12 | Protein secondary structure prediction | Deep3I network | MUFOLD-SS TensorFlow and Keras | Python | Open Source | 80 | Supervised Learning: Classification | Protein structure prediction |
| O’Connell J., 2018, Australia, China, US (O’Connell et al., 2018) | SPIN dataset | N/A | Sequence profile compatible | DNN | http://sparks-lab.org. SPIN | N/A | Open Source | 65 | Supervised Learning: Prediction | Protein sequence prediction |
| Sunseri J., 2018, US (Sunseri et al., 2019) | D3R Grand challenge 3 Grand challenge 3 | Input ligand SMILES protein FASTA CSAR | Cathepsin S model ligand protein | CNN | Gnina, Caffe, https://github.com/gnina | C++, Python | Open Source | 100 | Supervised Learning: Regression | Protein model prediction |
| Zhang B., 2018, China (Zhang B. et al., 2018) | PDB, PISCES, TR5534 Dataset | CASP10, 11, 12 and 13 | Prediction of performance of protein | CNN, RNN, BRNN | Keras | Python | Open Source | 100 | Supervised Learning, Prediction | Protein structure prediction |
| Armenteros J., 2017, Denmark (Almagro Armenteros et al., 2017) | UniProt | Protein sequence, Sequence information | Predict protein subcellular localization | CNN, RNN BLSTM, FFNN, Attention models | Lasagne, Theano, Deep Loc: http://www.cbs.dtu.dk/services/DeepLoc | Python | License MIT | 90 | Supervised Learning: Classification | Protein structure prediction |
| Vang Y., 2017, US (Vang and Xie, 2017) | IEDB MHCBN SYFPEITHI | Human leukocyte antigen (HLA) complex | HLA class I-peptide-binding prediction | NLP, CNN | Keras, Theano, https://github.com/uci-cbcl/HLA-bind | Python | Open Source | 100 | Supervised Learning: Regression | Protein structure prediction |
| Wang S., 2017, US (Wang et al., 2017) | Pfam CASP CAMEO | 150 Pfam families 105 CASP11 test proteins 76 hard CAMEO | 5f5pH | DRNN | TensorFlow, Theano http://raptorx.uchicago.edu/ContactMap/ | Python | Apache 2.0 | 75 | Supervised Learning: Classification Regression | Protein structure prediction |
| Yang J., 2015, China, US (Yang et al., 2015) | PDB SPx dataset PDBCYS dataset | Amino acid sequence | Structure prediction of cysteine-rich proteins | HMM, SVR | CYSCON http://www.csbio.sjtu.edu.cn/bioinf/Cyscon/ | N/A | N/A | 75 | Supervised Learning: Regression | Protein structure prediction |
| Li Z., 2014, US (Li et al., 2014) | PISCES | TL2282 dataset TS500 dataset TR1532 dataset | Sequence profile prediction | SPIN, NN | SPIN http://sparks-lab.org | Python, Linux | Open Source | 85 | Supervised Learning: Classification | Protein structure prediction |
| Wong K., 2013, Canada, US, Saudi Arabia (Wong et al., 2013) | Protein-Binding Microarray dataset | DNA sequence | DNA-motif discovery | Kmer-HMM | kmerHMM http://www.cs.toronto.edu/wkc/kmerHMM | N/A | N/A | 50 | Supervised Learning: Classification. Unsupervised Learning: Clustering | Model Discovery |
| Katzman S., 2008, US (Katzman et al., 2008) | PDB PISCES | Amino acid sequence of a protein of unknown structure | Local structure prediction | Multi-layer NN | PREDICT-2ND http://www.soe.ucsc.edu/∼karplus/predict-2nd/ | C++ | Open source | 80 | Unsupervised Learning: Clustering | Protein structure prediction |
| Bindslev C., 2002, Denmark (Bindslev-Jensen et al., 2003) | 20 Patients with allergy to Macrozoarces americanus | Macrozoarces americanus | Investigate potential allergenicity of Ice Structuring Protein (ISP) | DT | N/A | N/A | N/A | 45 | Supervised Learning: Regression | Protein structure prediction |
3D-CNN, three-dimensional convolutional neural network; ACNN, Asymmetric convolutional neural network; BLSTM, Bidirectional long short-term memory; BRNN, Bidirectional recurrent neural network; CNN, Convolutional neural network; Deep3I, Deep inception-inside-inception network; DNN, Deep neural network; DRNN, Deep residual neural network; DT, Decision Tree; FFNN, Feed forward neural network; HMM, Hidden Markov model; K-merHMM, K.mer Hidden Markov model; LSTM, Long short-term memory; MC, Monte Carlo; ML, Model; MLP, Multilayer perceptron; NN, Neural network; PSO, Particle swarm optimization; RNN, Recurrent neural network; RNN 2, Residual neural network; SPIN, Sequence Profiles by Integrated Neural network; SVR, Support vector regression; UDNN, Ultradeep neural network.
The 40% (57/144) of the protein studies by AI applications were the following ones: myoglobin, silk protein, amyloid proteins, Rab family, cathepsin S family, kinases family, K proteinase, barnase, apolipoprotein family, protein DND_4HB, and antimicrobial peptides. Studies in enzymes should be pointed out, oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases, NOS (nitric oxide synthase), lysozyme, which are included in the columns of the initial scaffold (Tables 2–6). These proteins are very useful in the industry as well as in the biomedical fields. With respect to the type of organisms, the more explored are the following ones: E. coli, Drosophila, Caenorhabditis elegans, Homo sapiens, S. cerevisiae yeast, Mus musculus (mouse), Geobacillus, and Coronavirus.
Tables 2–6 present the lists of the most commonly used databases in AI applications on PS. Of all the studies reviewed, the single use of main databases and datasets used is as follows:
1) PDB (30/144) 21%.
2) Author’s dataset construction (21/144)15%.
3) UniProt either UniProtKB or UniProtKB/SwissProt (12/144)8%.
4) CASP (critical assessment of protein structure prediction) database (5/144)3%.
5) SCOP (structural classification of proteins) (4/144)3%.
6) N/A, GenBank (4/144) 3%.
7) Protherm (3/144) 2%.
8) BioLip (biologically relevant ligand–protein) (2/144) 1%.
9) PLMD (protein lysine modifications database) (2/144) 1%.
10) And each of the next databases ChEMBL, eSol, GEO, DSSP, Drugbank, BioCreative, Transfac, STRING, BRENDA, SPINE, PISCES, NCBI, D3R Grand challenge 3, and KEGG with a (1/144)1%.
From the studies reviewed, (23/144), 16% use two databases. Of these, the latter (11/23) 48% uses a combination of the PDB and HSPP, PISCES, ProTherm, MOAD, SPx dataset, ChEMBL, DisProt, and UniProt/SwissProt; (4/23)17% use a combination of the GO database with UniProt or STRING; (4/23)17% uses a combination of the UniProt/SwissProt database with ENZYME, DIP, TrEMBL, and CAFA database; and a (2/23)9% combination among DIP, HPRD, SKEMPI database, and SPx dataset. The rest (24/144)17% belongs to a combination of three or more databases with PDB, UniProt, among others.
Moreover, several authors (Shamim et al., 2007; Simha et al., 2015; Yang et al., 2015; Li et al., 2018; Torng and Altman, 2019) focused on using previously constructed datasets, while others chose the creation of their own, based on their own design and outcome, for example, NOS, PPI’s, SPX, DBMLoc, D-B, and Extended D-B (Tables 2–6 and Figure 5).
FIGURE 5.
Machine learning and artificial intelligence applications to protein sciences. Information includes the number of studies, applications, databases, methods, and validation used.
The following tables show the principal protein categories that were found in this study. Table 2 shows the result of each of the 38 articles that were considered in the protein and drug design category.
Table 3 shows 26 studies that are related to protein function prediction and 6 studies related to function prediction and novel function.
Table 4 shows 19 studies that are related to fold ID and physicochemical properties and 8 studies related to protein classification.
Table 5 shows 26 studies that are related to protein structure prediction.
Table 6 shows five studies for protein contact map prediction, five studies for protein-binding prediction, nine studies for protein site prediction, and two studies for genomics.
Table 1 shows the overview of the extracted information of the selected studies based on the quality criteria.
Machine Learning Paradigms and AI Algorithm Roles
The most applied approach we found as a result of our review and meta-analysis corresponds to supervised learning (123/144)85%, which focuses on classification algorithms (CNN, NB, KNN, RF, SVM, etc.) and regression algorithms (SVR, RFR, DT, ANN, DNN, etc.) that are used for a variety of tasks: detection of functional sites, hydroxylation sites, amino acid composition, DNA expression sequences, protein interaction, biomarker finding, protein design, drug design, 3D structure prediction, and protein folding (Tables 2–6 and Figures 4, 5). Within supervised machine learning (123), we found that classification techniques overrule, by far, regression ones (31/123) (for reference, see Tables 2–6). On a closer look, we see that these methods are generally very good at prediction tasks, although complexity may be significantly increased by the execution time required, something that is often reported as a drawback of this method (AlQuraishi, 2021).
In contrast to supervised learning, it is only (17/144)12% focusing on unsupervised learning, using clustering algorithms (CNN, FFNN, LSDR, DL, HMM, MRF, NN, etc.) for various purposes, such as protein solubility prediction, protein prediction of new functions, discovery of DNA motifs, detection of protein structures, and prediction of the nuclear Overhauser effect at low energies. Of the eight articles using this approach, two of them report an improvement in performance as an advantage, one of them in time reduction (Frasca et al., 2018) and the other one in the acceleration of automated protein function prediction methods in general (Makrodimitris et al., 2019). At the same time, however, a disadvantage reported is that time execution may be increased, a fact that should not surprise us, for it is well known that unsupervised learning algorithms are characterized by being computationally very complex methods (Table 1 and Figures 4–7).
FIGURE 7.
Representation of the whole AI process based on the selected protein application. The process amalgams several steps: protein application (protein design, protein classification, protein prediction, etc.), extraction (selection of database), transform (code development and filtering), and load (input of the training data) (ETL) for the training data and the feature extraction procedure is the building of the machine learning network. Outcome step and a proposal server application.
On the other hand, supervised machine learning is used just a little more than deep learning techniques. Moreover, it is interesting to note that roughly (77/144)53% of the deep learning articles combine two clustering algorithms: CNN (47/77)61% and LSTM (16/77)21%. Of course, some articles put forward optimization procedures in an algorithmic genetic fashion (Figures 4–7).
Regarding hybrid algorithms using neural networks, we found that all 11 articles explicitly stating their use of hybrid algorithms belong to the deep learning paradigm, combining CNN and LSTM or RNN and CNN. One of them (Almagro Armenteros et al., 2017) goes even further; in that, it uses a combination of these two neural networks to predict protein subcellular localization and then an attention mechanism to identify protein regions important for subcellular localization (Table 1 and Figures 4–6).
It is interesting to note as well that nine articles are used for prediction (glycation product prediction (Chen et al., 2019), protein secondary structure (Guo et al., 2019), prediction of metal binding in proteins (Haberal and Ogul, 2019), compound–protein affinity prediction (Karimi et al., 2019), prediction of protein structural features (Klausen et al., 2019), protein contact map prediction (Hanson et al., 2018), prediction of protein interactions (Huang et al., 2018), predicting hydroxylation sites (Long et al., 2018), and predicting protein subcellular localization (Almagro Armenteros et al., 2017)), of which two perform prediction from original sequences (Almagro Armenteros et al., 2017;Li et al., 2018).
Moreover, one of them highlights that one of its applications is for the design of new drugs and one of them performs this task (Karimi et al., 2019).
It is tempting to put forward the claim that hybrid algorithms in deep learning are very good for prediction tasks as well as for applications in the new drug design. It is noteworthy to mention that these articles belong to the last 3 years of our revision, something that suggests that there is a tendency for the use of hybrid methods in the near future (Table 1).
AI Training, Validation, and Performance
Validation process allows obtaining a quantitative measure of the models’ efficiency. In this systematic review, several methodologies were used to train and validate in the machine and deep learning proposed by means of hold-out and k-fold cross-validation; The most utilized was the k-fold cross-validation, each one with a different folding proposal, e.g., 2-, 3-, 5-, and 10-fold (Szalkai and Grolmusz, 2018a), trained and validated its algorithm utilizing two validations: 3- and 5-fold cross-validations. Several articles used a graphics processing unit (GPU) that was employed to accelerate the deep learning training and validation process. The most utilized AI algorithm in these articles was CNN, with a 33% occurrence, followed by DNN with 9%, both programmed with Python. The performance of the AI algorithms for protein design was evaluated using parameters such as sensitivity, specificity, true-positive rate, false-positive rate, accuracy, recall, precision, F1-score, area under the curve (AUC), receiver operating characteristic (ROC) curve, and Matthew’s correlation coefficient (MCC). For the case of the hold-out validation, a percentage of the data that is taken and that percentage is randomly removed from the dataset is selected. This methodology, in particular, is computationally very simple; however, it suffers from a high variance because it is not known that data will end up in the test set or in the training one and of the importance that these data might have. In hold-out validation, datasets, which for this review are the databases of proteins, genes, peptides, etc. (see Tables 2–6 and Figures 4–6), are randomly divided into two partitions with different proportions (50, 70, or 75% training—50, 30, or 25% validation), which are mutually exclusive. The first part of the database is used to feed the input vectors of the methods and train the machine or deep learning algorithms, while the rest is used to evaluate and validate the results obtained with their proposed algorithms. In contrast, with this type of validation technique, hold-out takes a long time for computational processing, especially for large datasets, in particular case, the large protein databases. As a result of our meta-analysis, we found the use of the hold-out methodology to train and validate their AI proposals, as CNN, RNN, LSTM, and FFNN (Tables 1–6 and Figures 4–6) in the prediction of expressions, interactions, and subcellular localization of proteins and also in the prediction of the peptide binding.
Another technique for evaluating the performance of AI methods, particularly for large databases such as protein design, is cross-validation. Cross-validation is a technique used to (generally) obtain the ability of a model to fit an unknown dataset given a collected dataset. In this context, the k-fold cross-validation is an iterative process that consists of dividing the dataset randomly into k groups of approximately the same size. In this sense, although not all possible combinations of sets are examined, an estimate of the average accuracy more than acceptable can be obtained by training the model only k-fold. The first set is used to train the AI models and the other is used to test and validate them, doing this process k times using a different group for validation in the iteration. Although cross-validation is computationally an intensive method of training and validation, its advantages are the reduction of computational time because the process is repeated k times, where all the data are tested once and used for training, maintaining a reduced variance and bias. Of the total 93 articles in this review, 41 of them (47%) used the following cross-validation schemes: leave-one-out, 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 10-fold, and 20-fold cross-validations. For most of them, the use of 5-fold and 10-fold cross-validations to analyze the performance of their AI proposals predominated, with 16 and 17 articles, respectively. This method was preferred for the evaluation to the performance of CNN and SVM algorithms, with databases such as PBD, ProTherm, UniProt, GO, and ChEMBL. Additionally, in seven articles (17%), they carried out various types of cross-validations to obtain more information on the performance of their proposals. Another variant to evaluate performance was observed in three articles (7%), which combined the use of both hold-out and cross-validation methodologies in their proposals, which provide them more effective comparison of results in terms of validation schemes.
In contrast, in 22 articles of this review, 25% did not mention neither their training methods nor the validation performed to evaluate the performance of their algorithms used. Likewise, 7% of the articles evaluated their methods using various types of cross-validations at the same time to obtain more information on the performance of their proposals, e.g., 4-fold, 6-fold, 8-fold, and 1-fold, or 3-fold, 5-fold, 7-fold, and 1-fold, or 10- and 20-fold, for databases of PDB, UniProt, GO, ChEMBL, ProTherm, PISCES, GenBank, STRING, and new databases as NOS, SPx, D-B, and Ext D-B.
In general, the performance of all proposed AI algorithms was evaluated using several parameters such as sensitivity, specificity, true-positive rate, false-positive rate, accuracy, recall, precision, root-mean-square error (RMSE), R 2, F1-score, area under the curve (AUC), receiver operating characteristic (ROC) curve, and Matthew’s correlation coefficient (MCC) (Table 1).
Of the 87 articles selected as finalists, we have the following: 32 use one single algorithm and 55 use a combination of two or three algorithms sequentially. In machine learning, we found 30; in deep learning, we found 20 applying machine learning (SVM); 11 deep learning (RNN); and 6 using optimization through genetic algorithms.
Regarding the programming language in which each study was developed, we found 47 articles do not specify what language they are based on, 75 articles are based on the Python language, of which 57 are based entirely on Python and 18 are in combination with other software; see Tables 2–6.
Twelve articles are based on the C++ language of which only three are based exclusively on that language and nine in combination with Python, with C, R, and CUDA and C++ language in the Linux environment.
Other nine articles are based on MATLAB of which only four are based exclusively on that language and five in combination in conjunction with Python and Bioinformatics and with Python and C++.
Six articles are based on the C language of which three are based exclusively on that language and three in combination in conjunction with C++, R, and CUDA, with Java and Python and one with Linux and Windows environment.
Finally, seven articles are based on the Java language of which two are written exclusively in this language and five in combination with TensorFlow and with C and Python.
Regarding software licenses, 90 articles were found to be Open Source. An article is licensed by Neural Power version 2.5. One article specifies an open license type belonging to IBM and GNU, respectively. Unfortunately, 45 items did not specify the type of license they own.
Road Map of Artificial Intelligence in Protein Science
The goal of this analysis is to provide a road map to apply machine learning and AI techniques in protein science. One of the results of our meta-analysis, for example, in protein structure prediction, is shown in Figure 6 in which we can observe the two main strategies for protein structure prediction. In Figure 2, we show the scaffold-template-based modeling that is the most commonly used for the scientist in this field with very good results. However, recently Senior and collaborators using a free modeling approach successfully developed an AlphaFold algorithm using a deep neural network. They generated an outstanding accuracy of the 3D structure of a protein with an unknown fold in CASP14 (Senior et al., 2020). This led to an unsolved big question about the importance of the starting point in protein structure prediction, in particular, and in protein science, in general.
The road map of this research is an evolving and a dynamic process (Figure 7). It begins by obtaining information from a list of several databases, followed by a pre-treatment step over the extracted data, including those steps for eliminating redundancies within sequences, structure threshold based on RMSD values, and the like. Further steps contribute to the required pre-processing to complete the reporting process, and then proceed to the data process of the information itself, which includes the input data and the application of the machine learning algorithm, in which the input data are set to be processed into FASTA sequences, training sets, or 3D structures, depending on the function of algorithm in turn. The algorithms used fall into four categories: supervised learning, unsupervised learning, deep learning, and optimization, where each of these categories include a set of their own subparts, which are then combined and configured to predict new ways to model previous data and contribute to future implementations in protein science. The post-processing of data and the support of the new data acquired are made up of models and sequences that were loaded on the platforms to servers such as “DeepUbi, DeepSol, COSNet, Gnina, among others”, in which these servers are used for the storage or implementation of their respective methods. Figure 7 shows that more than half of the reported research completed the three pre-process, process and post-process steps we set forward, so this sequence may be applied to protein science including protein design, classification, physicochemical properties, functionalities, folding properties, and new functions such as homology prediction, domain prediction, subcellular localization, drug design, sensitivity, and other enhancers that can provide new catalysts and new functions, all of which provide any future development for biomolecular enhancement within protein science through machine learning. Model development is intrinsically related to the protein application to be developed. Data extraction varies depending on the architecture of the model to be developed since the data become more complex as the transformation, training, and feature extraction process unfold. The extraction ranges from obtaining the amino acid sequence, secondary structure to the 3D atomic model, using the atomic coordinates. Transforming data emphasizes on performing an adequate filtering for the use of the information for the training of the model, which leads to the feature extraction for the use of machine learning model and finally generating a final output. The process road map includes the fusion of these different applied AI learnings, models, and classifications into a connected deep learning layer that will be included in future research and test datasets to cover the terms of AI science, proteins, and their applications.
Final Discussion and Further Challenges for Our Understanding of Protein Science Using AI
Novelties and Future Direction in the Binomial PS-IA Research
The protein science field has great expectations on ML methods as indispensable tools for the biomedical sciences as well as for the chemical and biotechnology industry, for applied research is moving toward synthetic organisms with artificial metabolic networks, regulators, and so on, creating synthetic molecular factories. The binomial PS-IA research is evolving and strengthening, as shown in the Results section (Tables 1–6 and Figures 4–7). Our research reveals that road maps are most needed to solve complex problems in PS, guiding the exploration into the protein universe. As depicted in Table 1, ML techniques, which are used nowadays, are tailored to the expected results; Tables 1–6 display an array of networks of several solving problem methods, hence showing that guidance is needed in the form of road maps.
It is important to emphasize that in order to design a model algorithm bank functioning as a kit-tool, it is essential to understand the source from which the data are obtained and then used to train each model. The studies analyzed solve classification, regression, and optimization problems. As depicted in Table 1, models providing a solution make use of probabilistic inference, functions, activation functions, reduction of the hierarchical order, and logical inference. These results support the fact that machine learning models are heterogeneous, time demanding to design, and correctly evaluate complex models—since the result may not always be as expected or the method may not be carried out successfully. As illustrated in Table 3, there are some physical limitations blocking the full execution of the various models or algorithms, for example, when there is no appropriate computational equipment. Not surprisingly, several authors report that executing a model requires a high demand on execution time, computational power, extensive time to correctly evaluate the model, large memory consumption, and optimization toward GPUs (Frasca et al., 2018; Almagro Armenteros et al., 2017; Yeh et al., 2018; Jiménez et al., 2017; Lin et al., 2010). Another crucial aspect mentioned in Table 1 is the lack of input data to train the model, something that influences the model’s precision and accuracy (Pagès et al., 2019; Cuperus et al., 2017; Folkman et al., 2014; Qi et al., 2012). Moreover, there are also limitations in model construction, such as errors in the training process, manual intervention of data, overadjustment of the model, and an inadequate algorithm construction. In the studies analyzed, there are cases in which there is no description regarding the performance of the comprehensive models, generating gaps in the understanding of the behavior of the algorithms or models, like whether they are deterministic (Long et al., 2018; Ragoza et al., 2017; Makrodimitris et al., 2019). As stated in the ML and AI Algorithm section, supervised learning is the most used method, something that highlights the use of classification algorithms. Moreover, there seems to be a current trend to solve problems in protein science using techniques that require a cross-functional group of scientists, something that, in turn, highlights the fact that there is plenty of unexplored terrain in the use of unsupervised machine learning.
An interesting finding is the implementation of free code and software, as shown in the AI Training, Validation, and Performance section. Our results exhibit a tendency to create models with transparency, which means that every study implemented in a public server has access to all new models created. Another crucial result is the one depicted in the Road Map of Artificial Intelligence in Protein Science section, which is an abstraction that reduces the design of an artificial intelligence model to be used in the resolution of a specific problem in protein science. The whole process follows three steps directed to build a competent model; these steps are 1) the procedure to obtain raw data and which type of processing should be followed for the model to be adequate, 2) the type of algorithm that may be used depending on the complexity of the problem, and finally, 3) the interpretation of results.
Overall, AI displays a window of opportunities to solve complex problems in PS because of its potential in finding patterns and correlating information that requires the integration of protein data exceeding many petabytes. However, we are still far away from solving all the protein tasks computationally. As a result of our biochemical meta-analysis, we showed that AI applications are strongly directed to function identification and protein classification (Tables 1–6), for machine learning models and methods are heterogeneous and do not always draw a clear line as to whether a process should go in a certain sequence (Table 1 and Figures 4–7). It should also be noted that there is no optimal method, which is why applications have different purposes and conditions, suggesting that algorithms must be customized based on the expected outcome or query (Table 1).
The evaluation accuracy horizon is an open epistemic horizon, as shown in Table 1: the metrics for ML methods used in several applications are limited; there are no reported research articles using random forest, in which the cross-validation is unnecessary. In summary, none of the studies reported explicitly use robustly validated methods.
We end by commenting on a key problem in the binomial AI–PS. As well known, it is not possible to work directly with the protein sequences. To tackle this challenge, several studies address this limitation by representing the sequence of a protein as an input to the deep learning model (Almagro Armenteros et al., 2017; Long et al., 2018; Fu et al., 2019). Moreover, given some featured procedures comprising what may be called the coding architecture, which is based on creating a specific-weight matrix or a bit vector that represents the sample. This practice was observed in some articles (Cuperus et al., 2017; Jiménez et al., 2017; Khurana et al., 2018; Le et al., 2018) that work with 2D convolutional neural networks in which the authors reported an increase in sensitivity and precision when using indexed datasets. A similar abstraction was observed in 3D convolutional neural networks since the structural representation of a protein is not a rotational invariant; several authors (Jiménez et al., 2017; Ragoza et al., 2017; Hochuli et al., 2018; Pagès et al., 2019; Sunseri et al., 2019; Torng and Altman, 2019) propose using a volumetric map divided into voxels centered on the backbone atoms, representing the physicochemical properties of proteins.
Regarding other review articles along the lines we have followed, the closest we found is the one by Dara et al. (2021). This review article is restricted to drug discovery, one of the five applications we analyzed (genomics, protein structure and function, protein design and evolution, and drug design).
Of a total of 38 articles we presented in Table 2 concerning protein and drug design, only 11 of them were about protein design, so the comparison is not at all fair between these two articles, as far as the analysis of the bibliography analyzed is concerned. However, we share with these authors part of the challenges for researchers in this area: data quality as well as the heterogeneity of databases to be searched for.
Optimization and the characteristics of a prediction must be carried out with a few design considerations, including how to represent the protein data and what type of learning algorithm to use. These form the establishment of a priority acquisition, standard acquisition, etc., and the generation of a protein based on a base model, with the aim that one day it would be possible to have controllable predictive models that can read and generate outputs in a consensual terminology, as revised in Hie and Yang (2022). Clearly showing a replacement of conventional methods to the use of machine learning algorithms (neural networks), attributed to improvements in design, computational power, etc., the result of a machine learning algorithm is not deterministic, but rather, it is intended to perform transformation functions in relation to the complexity of the data, as depicted in AlQuraishi (2021). There are volumes and volumes of empirical protein data. It is extremely difficult to synthesize such data for correct use in existing algorithms; however, machine learning has helped to compile a large number of methodologies, considering specific assumptions. Nevertheless, most of the empirical methodologies to demonstrate that drugs are safe and effectively continue to be used since there is a gap in the understanding of how the learning transmission of the data to the model is carried out (Dara et al., 2021).
In order to close our reflection as a research team, we believe that a landmark for the epistemic horizon in research is the reassurance that cross-functional groups of scientists from several academic disciplines, in this case including the participation of experts from the natural sciences (organic chemistry, physics and chemistry of proteins, molecular and structural biology, protein engineering, systems biology, microfluid chip engineering, and nanobiotechnology), together with those in computer science (artificial intelligence, knowledge engineering) promote the innovation process in tecno-sciences by combining tacit and explicit knowledge, sharing skills, methodologies, tools, ideas, concepts, experiences, and challenges to fully explore the binomial AI–PS promising area of research (Hey et al., 2019; Mataeimoghadam et al., 2020; Senior et al., 2020; Tsuchiya and Tomii, 2020). A very recent successful case study that highlights this approach is the team of creators of system Alphafold (Senior et al., 2020; AlQuraishi, 2021), one which in the CASP (Critical Assessment of Protein Structure Prediction) competition of three-dimensional protein structure modeling were able to determine the 3D structure of a protein from its amino acid sequence. By doing so, this group of researchers solved one of natural science’s open (until now) and most challenging problems using a deep learning approach combining template-based modeling (TBM) and free modeling (FM). The key point is that the neural network prediction encompasses backbone torsion angles and pairwise distances between residues (Senior et al., 2020). At the dawn of the year 2021, this peak of the iceberg brings fresh air and a great power to the protein science field, in particular, and to the life-sciences more broadly, encouraging the new generation of scientists to work as cross-functional teams in order to tackle novel tasks toward the understanding of nature.
One challenge for the binomial AI–PS research area is to tackle the representation of tacit knowledge and include it in the ML algorithms. The relevance of tacit knowledge in the building up of protein science knowledge has come a long way since Polanyi first noted it, extending to different fields in the search for an improvement of their practical skills. In AI, the predominant way of knowledge acquisition and performance is a formal one in which the machine learns and expresses explicitly through guidelines and that works in a focalized mean; the new task alludes to a tacit dimension (Polanyi, 1962), which remains in the edge of attention and incorporates aspects that are taught and learned mostly through practice and in a comprehensive manner (it is context-specific, spreads in the laboratory environment, and comes into play in decision-making.
Some Conclusions
To sum up, the systematic review and the biochemical meta-analysis offered in this article focused on the enormous innovation that has been made in the binomial AI–PS research, both in its applications and its road maps to solve protein structures and function prediction, protein and drug design, among other tasks. The contribution of this study is 3-fold: firstly, the setup of a cross-functional group in which computer scientists, professionals in biomedicine, and a philosopher constructed a common language and together identified relevant literature in the inter-field of AI–PS and constructed a bridge between the two fields, which can serve as a framework for further research in either area.
Secondly, we stressed the importance of a finer-grain understanding of training and validation methods of ML models and their outcomes, combining databases from several areas of knowledge (life-science experiments, in silico simulations, ML, direct evolution approach, etc.) that allowed us to classify, stratify, and contribute to the evolving protein science field. Thirdly, we showed that the binomial AI–PS, a progressive research program, as Lakatos would say and has still several challenges to tackle, such as the development of a comprehensive machine learning benchmarking enterprise, the experimental confirmation of the structure of the 3D modeling in laboratories, the classification, etc., controls the vulnerability of the neural networks, the development of a tool-kit to design novel biocatalysts not found in nature using reverse engineering, human-made metabolic routes, the design of new antibody molecular factory, novel proteostasis systems, the understanding of protein folding and protein-aggregation mechanisms, etc. Finally, we suggested that there may be a paradigm shift in the AI–PS research as a result of the recent great outcome of Alphafold, encouraging its use to the new generation of scientists.
In any case, what is clear is that a cross-functional group of scientists from several knowledge domains is required to work in coordination for sharing ideas, methodologies, and challenges toward the development of road maps and computational tools, paradigms, tacit, and explicit knowledge to fully explore and close the gap of the binomial AI–PS, a promising research area.
Acknowledgments
The authors would like to acknowledge the experimental support and fruitful discussions provided by Dr. Elsa de la Chesnaye. We also wish to thank Dr. Laura Bonifaz for her support. The contributions made by the assigned pre-graduate research fellows at the Universidad lberoamericana and UNAM are greatly appreciated. We are also thankful for the contributions of Perla Sueiras, Daniela Monroy, Maria Fernanda Frlas, Pablo Cardenas and Mattea Cussel for translation and proofread the manuscript, and Rogelio Ezequiel and Alonso Loyo for the artwork.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation.
Author Contributions
Conceived and designed the experiments: MA-B and NA-B. Performed the systematic review: JV-A, MVA, FO-F, RZ-S, NA-B, and MA-B. Analyzed the data: JV-A, LO-T, MVA, FP-E, AA, FO-F, RZ-S, NA-B, NK-V, SVA, and MA-B. Contributed to reagents/materials/analysis tools: NK-V, NA-B, CR-M, and MA-B. Wrote the article: JV-A, LO-T, MVA, AA, FP-E, NA-B, and MA-B. Contributed to helpful discussions: JV-A, LO-T, MVA, FP-E, AA, FO-F, RZ-S, NA-B, NK-V, CR-M, SVA, and MA-B.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- 1D-CNN
one-dimensional convolutional neural network
- 2D-BRLSTM
two-dimensional bidirectional recurrent long short-term memory
- 2D-CNN
Two-dimensional convolutional neural network
- 3D-CNN
Three-dimensional convolutional neural network
- ACNN
Asymmetric convolutional neural network
- ADASYN
Adaptive synthetic sampling
- AGCT
Alignment genetic causal tree
- ANN
Artificial neural network
- BBFNN
Biobasis function neural network
- BBP
Back back propagation
- BLSTM
Bidirectional long short-term memory
- BN
Bayesian network
- BRNN
Bidirectional recurrent neural network
- BroMap
Branch and bound map estimation
- BRT
Booster regression tree
- CABS
C-alpha–beta side
- CFN
Cost function network
- CNF
Conditional neural field
- CNN
Convolutional neural network
- COSNet
Cost-sensitive neural network
- DCNN
Deep convolutional neural network
- DeepDIN
Deep dense inception network
- Deep3I
Deep inception-inside-inception network
- DFS
Depth first search
- DL
Deep learning
- DMNN
Deep mahout neural network
- DNN
Deep neural network
- DRNN
Deep residual neural network
- DROP
Domain linker prediction using optimal feature
- DT
Decision tree
- DTNN
Deep tensor neural network
- EASE-MM
Evolutionary amino acid and structural encodings with multiple models
- ELMO
Embeddings from language models
- ENN-RL
Evolution neural network-based regularized Laplacian kernel
- FFNN
Feed forward neural network
- FIBHASH
Fibonacci numbers and hashing table
- GA
Genetic algorithms
- GAN
Generative adversarial network
- GBT
Gradient boost tree
- GBDT
Gradient boosted decision tree
- GCN
Graph convolutional network
- GR
Genetic recombination
- HDL
Hybrid deep learning
- HMM
Hidden Markov model
- HNN
Hopfield neural network
- IBP
Incremental back propagation
- KeSCANN
Knowledge-enriched self-attention convolutional neural network
- K-merHMM
K.mer Hidden Markov model
- KNN
k-nearest neighbor
- Lasso
Least absolute shrinkage and selection operator
- LightGBM
Light gradient boosting machine
- LM
Levenberg–Marquardt
- LPBoostR
Linear programming boosting regression
- LPSVMR
Linear programming support vector machine regression
- LR
Logistic regression
- LSDR
Label-space dimensionality reduction
- LSTM
Long short-term memory
- MC
Monte Carlo
- ME
Max entropy
- ML
Model
- MLP
Multilayer perceptron
- MNB
Multinomial naïve bayes
- MNNN
Multi-scale neighborhood-based neural network
- MNPP
Message passing neural network
- MotifCNN
Motif convolutional neural network
- Motif DNN
Motif deep neural network
- MR
Matching loss regression
- MRF
Markov random forest
- Multimodal DNN
Multimodal deep neural network
- NB
Naïve Bayes
- NLP
Natural language processing
- ORMR
One-norm regularization matching-loss regression
- ParCOSNet
Parallel COSNet
- PLSR
Partial least-squares regression
- PNN
Probabilistic neural network
- PS
Protein science
- PSO
Particle swarm optimization
- PSP
Predict signal pathway
- QP
quick prob
- ReLeaSE
Reinforcement learning for structural evolution
- RF
Random forest
- RN
Relational network
- RNN
Recurrent neural network
- RNN 2
Residual neural network
- RR
Ridge regression
- SDHINE
Meta path-based heterogeneous information embedding approach
- SFFS
Sequential forward floating selection
- SGD
Stochastic gradient descent
- SPARK-X
Probabilistic-based matching
- SPIN
Sequence profiles by integrated neural network
- SVM
Support vector machine
- SVMR
Support vector machine regression
- SVR
Support vector regression
- UDNN
Ultradeep neural network
- VSA
Virtual screening algorithms
- WMC
Weighted multiple conformations
References
- Adhikari B., Hou J., Cheng J. (2018). DNCON2: Improved Protein Contact Prediction Using Two-Level Deep Convolutional Neural Networks. BioInformatics 34, 1466–1472. 10.1093/bioinformatics/btx781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Gharabli S. I., Agtash S. A., Rawashdeh N. A., Barqawi K. R. (2015). Artificial Neural Networks for Dihedral Angles Prediction in Enzyme Loops: A Novel Approach. Ijbra 11, 153–161. 10.1504/IJBRA.2015.068090 [DOI] [PubMed] [Google Scholar]
- Alakuş T. B., Türkoğlu İ. (2021). A Novel Fibonacci Hash Method for Protein Family Identification by Using Recurrent Neural Networks. Turk. J. Electr. Eng. Comput. Sci. 29, 370–386. Available at: http://10.0.15.66/elk-2003-116 . 10.0.15.66/elk-2003-116 [DOI] [Google Scholar]
- Almagro Armenteros J. J., Sønderby C. K., Sønderby S. K., Nielsen H., Winther O. (2017). DeepLoc: Prediction of Protein Subcellular Localization Using Deep Learning. Bioinformatics 33, 3387–3395. 10.1093/bioinformatics/btx431 [DOI] [PubMed] [Google Scholar]
- AlQuraishi M. (2021). Machine Learning in Protein Structure Prediction. Curr. Opin. Chem. Biol. 65, 1–8. 10.1016/j.cbpa.2021.04.005 [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Tidor B. (2008). Computationally Mapping Sequence Space to Understand Evolutionary Protein Engineering. Biotechnol. Prog. 24, 62–73. 10.1021/bp070134h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Unger R., Kliger Y. (2011). Hidden Conformations in Protein Structures. Bioinformatics 27, 1941–1947. 10.1093/bioinformatics/btr292 [DOI] [PubMed] [Google Scholar]
- Baetu T. (2015). Carl F, Craver and Lindley Darden: In Search of Mechanisms: Discoveries across the Life Sciences. Hpls 36, 459–461. 10.1007/s40656-014-0038-6 [DOI] [Google Scholar]
- Bernardes J., Pedreira C. (2013). A Review of Protein Function Prediction under Machine Learning Perspective. Biot 7, 122–141. 10.2174/18722083113079990006 [DOI] [PubMed] [Google Scholar]
- Bindslev-Jensen C., Sten E., Earl L. K., Crevel R. W. R., Bindslev-Jensen U., Hansen T. K., et al. (2003). Assessment of the Potential Allergenicity of Ice Structuring Protein Type III HPLC 12 Using the FAO/WHO 2001 Decision Tree for Novel Foods. Food Chem. Toxicol. 41, 81–87. 10.1016/S0278-6915(02)00212-0 [DOI] [PubMed] [Google Scholar]
- Bond P. S., Wilson K. S., Cowtan K. D. (2020). Predicting Protein Model Correctness in Coot Using Machine Learning. Acta Cryst. Sect. D. Struct. Biol. 76, 713–723. 10.1107/S2059798320009080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostan B., Greiner R., Szafron D., Lu P. (2009). Predicting Homologous Signaling Pathways Using Machine Learning. Bioinformatics 25, 2913–2920. 10.1093/bioinformatics/btp532 [DOI] [PubMed] [Google Scholar]
- Briesemeister S., Rahnenführer J., Kohlbacher O. (2010). Going from where to Why-Interpretable Prediction of Protein Subcellular Localization. Bioinformatics 26, 1232–1238. 10.1093/bioinformatics/btq115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Freitas C., Chan L., Sun M., Jiang H., Chen Z. (2017). ProLanGO: Protein Function Prediction Using Neural Machine Translation Based on a Recurrent Neural Network. Molecules 22, 1732. 10.3390/molecules22101732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. (2005). I-Mutant2.0: Predicting Stability Changes upon Mutation from the Protein Sequence or Structure. Nucleic Acids Res. 33, W306–W310. 10.1093/nar/gki375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yang R., Zhang C., Zhang L., Zhang Q. (2019). DeepGly: A Deep Learning Framework with Recurrent and Convolutional Neural Networks to Identify Protein Glycation Sites from Imbalanced Data. IEEE ACCESS 7, 142368–142378. 10.1109/ACCESS.2019.2944411 [DOI] [Google Scholar]
- Cheng J., Tegge A. N., Baldi P. (2008). Machine Learning Methods for Protein Structure Prediction. IEEE Rev. Biomed. Eng. 1, 41–49. 10.1109/RBME.2008.2008239 [DOI] [PubMed] [Google Scholar]
- Cui Y., Dong Q., Hong D., Wang X. (2019). Predicting Protein-Ligand Binding Residues with Deep Convolutional Neural Networks. BMC Bioinforma. 20, 93. 10.1186/s12859-019-2672-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J. T., Groves B., Kuchina A., Rosenberg A. B., Jojic N., Fields S., et al. (2017). Deep Learning of the Regulatory Grammar of Yeast 5′ Untranslated Regions from 500,000 Random Sequences. Genome Res. 27, 2015–2024. 10.1101/gr.224964.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Chang Q., Peng W., Zhong J., Li Y. (2020). Network Embedding the Protein-Protein Interaction Network for Human Essential Genes Identification. Genes. 11, 153. 10.3390/genes11020153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels N. M., Hosur R., Berger B., Cowen L. J. (2012). SMURFLite: Combining Simplified Markov Random Fields with Simulated Evolution Improves Remote Homology Detection for Beta-Structural Proteins into the Twilight Zone. Bioinformatics 28, 1216–1222. 10.1093/bioinformatics/bts110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dara S., Dhamercherla S., Jadav S. S., Babu C. H., Ahsan M. J. (2021). Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 55 (3), 1947–1999. 10.1007/s10462-021-10058-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degiacomi M. T. (2019). Coupling Molecular Dynamics and Deep Learning to Mine Protein Conformational Space. Structure 27, 1034–1040. 10.1016/j.str.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Du Z., He Y., Li J., Uversky V. N. (2020). DeepAdd: Protein Function Prediction from K-Mer Embedding and Additional Features. Comput. Biol. Chem. 89, 107379. N.PAG--N.PAG. Available at: http://10.0.3.248/j.compbiolchem.2020.107379 . 10.1016/j.compbiolchem.2020.107379 [DOI] [PubMed] [Google Scholar]
- Durrant J. D., McCammon J. A. (2011). NNScore 2.0: A Neural-Network Receptor-Ligand Scoring Function. J. Chem. Inf. Model.. 51, 2897–2903. 10.1021/ci2003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina T., Toh H., Kuroda Y. (2011). DROP: An SVM Domain Linker Predictor Trained with Optimal Features Selected by Random Forest. Bioinformatics 27, 487–494. 10.1093/bioinformatics/btq700 [DOI] [PubMed] [Google Scholar]
- Ebrahimpour A., Rahman R. N. Z. R. A., Ean Ch'ng D. H., Basri M., Salleh A. B. (2008). A Modeling Study by Response Surface Methodology and Artificial Neural Network on Culture Parameters Optimization for Thermostable Lipase Production from a Newly Isolated Thermophilic Geobacillus Sp. Strain ARM. BMC Biotechnol. 8, 96. 10.1186/1472-6750-8-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis S., Proffitt W., Coles M., Truffault V., Shanmugaratnam S., Meiler J., et al. (2012). Potential of Fragment Recombination for Rational Design of Proteins. J. Am. Chem. Soc. 134, 4019–4022. 10.1021/ja211657k [DOI] [PubMed] [Google Scholar]
- Fang C., Moriwaki Y., Tian A., Li C., Shimizu K. (2019). Identifying Short Disorder-To-Order Binding Regions in Disordered Proteins with a Deep Convolutional Neural Network Method. J. Bioinform. Comput. Biol. 17, 1950004. 10.1142/S0219720019500045 [DOI] [PubMed] [Google Scholar]
- Fang C., Shang Y., Xu D. (2020). A Deep Dense Inception Network for Protein Beta‐turn Prediction. Proteins 88, 143–151. 10.1002/prot.25780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C., Shang Y., Xu D. (2018). MUFOLD-SS: New Deep Inception-Inside-Inception Networks for Protein Secondary Structure Prediction. Proteins 86, 592–598. 10.1002/prot.25487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feger G., Angelov B., Angelova A. (2020). Prediction of Amphiphilic Cell-Penetrating Peptide Building Blocks from Protein-Derived Amino Acid Sequences for Engineering of Drug Delivery Nanoassemblies. J. Phys. Chem. B 124, 4069–4078. 10.1021/acs.jpcb.0c01618 [DOI] [PubMed] [Google Scholar]
- Feinberg E. N., Sur D., Wu Z., Husic B. E., Mai H., Li Y., et al. (2018). PotentialNet for Molecular Property Prediction. ACS Cent. Sci. 4, 1520–1530. 10.1021/acscentsci.8b00507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman L., Stantic B., Sattar A. (2014). Feature-based Multiple Models Improve Classification of Mutation-Induced Stability Changes. BMC Genomics 15, 96. 10.1186/1471-2164-15-S4-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca M., Grossi G., Gliozzo J., Mesiti M., Notaro M., Perlasca P., et al. (2018). A GPU-Based Algorithm for Fast Node Label Learning in Large and Unbalanced Biomolecular Networks. BMC Bioinforma. 19, 353. 10.1186/s12859-018-2301-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Yang Y., Wang X., Wang H., Xu Y. (2019). DeepUbi: A Deep Learning Framework for Prediction of Ubiquitination Sites in Proteins. BMC Bioinforma. 20, 86. 10.1186/s12859-019-2677-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainza P., Nisonoff H. M., Donald B. R. (2016). Algorithms for Protein Design. Curr. Opin. Struct. Biol. 39, 16–26. 10.1016/j.sbi.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Li W., Wang B., Liu H., Zhou D. (2019). DeepACLSTM: Deep Asymmetric Convolutional Long Short-Term Memory Neural Models for Protein Secondary Structure Prediction. BMC Bioinforma. 20, 341. 10.1186/s12859-019-2940-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge A., Bartlett G. J., Thornton J. M. (2003). Using a Neural Network and Spatial Clustering to Predict the Location of Active Sites in Enzymes. J. Mol. Biol. 330, 719–734. 10.1016/S0022-2836(03)00515-1 [DOI] [PubMed] [Google Scholar]
- Haberal İ., Oğul H. (2019). Prediction of Protein Metal Binding Sites Using Deep Neural Networks. Mol. Inf. 38, 1800169. 10.1002/minf.201800169 [DOI] [PubMed] [Google Scholar]
- Han X., Zhang L., Zhou K., Wang X. (2019). ProGAN: Protein Solubility Generative Adversarial Nets for Data Augmentation in DNN Framework. Comput. Chem. Eng. 131, 106533. N.PAG--N.PAG. Available at: http://10.0.3.248/j.compchemeng.2019.106533 . 10.1016/j.compchemeng.2019.106533 [DOI] [Google Scholar]
- Hanson J., Paliwal K., Litfin T., Yang Y., Zhou Y. (2018). Accurate Prediction of Protein Contact Maps by Coupling Residual Two-Dimensional Bidirectional Long Short-Term Memory with Convolutional Neural Networks. Bioinformatics 34, 4039–4045. Available at: http://10.0.4.69/bioinformatics/bty481 . 10.1093/bioinformatics/bty481 [DOI] [PubMed] [Google Scholar]
- Hanson J., Paliwal K., Litfin T., Yang Y., Zhou Y. (2019). Improving Prediction of Protein Secondary Structure, Backbone Angles, Solvent Accessibility and Contact Numbers by Using Predicted Contact Maps and an Ensemble of Recurrent and Residual Convolutional Neural Networks. Bioinformatics 35, 2403–2410. 10.1093/bioinformatics/bty1006 [DOI] [PubMed] [Google Scholar]
- He H., Liu B., Luo H., Zhang T., Jiang J. (2020). Big Data and Artificial Intelligence Discover Novel Drugs Targeting Proteins without 3D Structure and Overcome the Undruggable Targets. STROKE Vasc. Neurol. 5, 381–387. 10.1136/svn-2019-000323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzinger M., Elnaggar A., Wang Y., Dallago C., Nechaev D., Matthes F., et al. (2019). Modeling Aspects of the Language of Life through Transfer-Learning Protein Sequences. BMC Bioinforma. 20, 723. 10.1186/s12859-019-3220-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey T., Butler K., Jackson S., Thiyagalingam J. (2019). Machine Learning and Big Scientific Data. Philos. Trans. A Math. Phys. Eng. Sci. 378 (2166), 20190054. arXiv. Available at: file:///Users/Myriam/Documents/2020/manuscritos . 10.1098/rsta.2019.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hie B. L., Yang K. K. (2022). Adaptive Machine Learning for Protein Engineering. Curr. Opin. Struct. Biol. 72, 145–152. 10.1016/j.sbi.2021.11.002 [DOI] [PubMed] [Google Scholar]
- Hochuli J., Helbling A., Skaist T., Ragoza M., Koes D. R. (2018). Visualizing Convolutional Neural Network Protein-Ligand Scoring. J. Mol. Graph. Model. 84, 96–108. 10.1016/j.jmgm.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E.-J., Lippow S. M., Tidor B., Lozano-Pérez T. (2009). Rotamer Optimization for Protein Design through MAP Estimation and Problem-Size Reduction. J. Comput. Chem. 30, 1923–1945. 10.1002/jcc.21188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Wang H., Wang L., Yuan W. (2018). Adverse Drug Reaction Predictions Using Stacking Deep Heterogeneous Information Network Embedding Approach. Molecules 23, 3193. 10.3390/molecules23123193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Li X., Liang J. (2004). Developing Optimal Non-linear Scoring Function for Protein Design. Bioinformatics 20, 3080–3098. 10.1093/bioinformatics/bth369 [DOI] [PubMed] [Google Scholar]
- Huang L., Liao L., Wu C. H. (2018). Completing Sparse and Disconnected Protein-Protein Network by Deep Learning. BMC Bioinforma. 19, 103. 10.1186/s12859-018-2112-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.-L., Tung C.-W., Ho S.-W., Hwang S.-F., Ho S.-Y. (2008). ProLoc-GO: Utilizing Informative Gene Ontology Terms for Sequence-Based Prediction of Protein Subcellular Localization. BMC Bioinforma. 9, 80. 10.1186/1471-2105-9-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C.-M., Huang Y.-M., Chang M.-S. (2006). Alignment Using Genetic Programming with Causal Trees for Identification of Protein Functions. Nonlinear Analysis Theory, Methods & Appl. 65, 1070–1093. 10.1016/j.na.2005.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J., Doerr S., Martínez-Rosell G., Rose A. S., De Fabritiis G. (2017). DeepSite: Protein-Binding Site Predictor Using 3D-Convolutional Neural Networks. Bioinformatics 33, 3036–3042. 10.1093/bioinformatics/btx350 [DOI] [PubMed] [Google Scholar]
- Kaleel M., Torrisi M., Mooney C., Pollastri G. (2019). PaleAle 5.0: Prediction of Protein Relative Solvent Accessibility by Deep Learning. Amino Acids 51, 1289–1296. Available at: http://10.0.3.239/s00726-019-02767-6 . 10.1007/s00726-019-02767-6 [DOI] [PubMed] [Google Scholar]
- Karimi M., Wu D., Wang Z., Shen Y. (2019). DeepAffinity: Interpretable Deep Learning of Compound-Protein Affinity through Unified Recurrent and Convolutional Neural Networks. Bioinformatics 35, 3329–3338. Available at: http://10.0.4.69/bioinformatics/btz111 . 10.1093/bioinformatics/btz111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S., Barrett C., Thiltgen G., Karchin R., Karplus K. (2008). Predict-2nd: A Tool for Generalized Protein Local Structure Prediction. Bioinformatics 24, 2453–2459. 10.1093/bioinformatics/btn438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S. A. (1992). “Origins of Order in Evolution: Self-Organization and Selection,” in Understanding Origins (Netherlands: Springer; ), 153–181. 10.1007/978-94-015-8054-0_8 [DOI] [Google Scholar]
- Khan Z. U., Hayat M., Khan M. A. (2015). Discrimination of Acidic and Alkaline Enzyme Using Chou's Pseudo Amino Acid Composition in Conjunction with Probabilistic Neural Network Model. J. Theor. Biol. 365, 197–203. 10.1016/j.jtbi.2014.10.014 [DOI] [PubMed] [Google Scholar]
- Khurana S., Rawi R., Kunji K., Chuang G.-Y., Bensmail H., Mall R. (2018). DeepSol: A Deep Learning Framework for Sequence-Based Protein Solubility Prediction. Bioinformatics 34, 2605–2613. 10.1093/bioinformatics/bty166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen M. S., Jespersen M. C., Nielsen H., Jensen K. K., Jurtz V. I., Sønderby C. K., et al. (2019). NetSurfP‐2.0: Improved Prediction of Protein Structural Features by Integrated Deep Learning. Proteins 87, 520–527. 10.1002/prot.25674 [DOI] [PubMed] [Google Scholar]
- Kwon Y., Shin W.-H., Ko J., Lee J. (2020). AK-score: Accurate Protein-Ligand Binding Affinity Prediction Using an Ensemble of 3D-Convolutional Neural Networks. Ijms 21, 8424. 10.3390/ijms21228424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladunga I., Czakó F., Csabai I., Geszti T. (1991). Improving Signal Peptide Prediction Accuracy by Simulated Neural Network. Bioinformatics 7, 485–487. 10.1093/bioinformatics/7.4.485 [DOI] [PubMed] [Google Scholar]
- Latek D., Kolinski A. (2011). CABS-NMR-De Novo Tool for Rapid Global Fold Determination from Chemical Shifts, Residual Dipolar Couplings and Sparse Methyl-Methyl Noes. J. Comput. Chem. 32, 536–544. 10.1002/jcc.21640 [DOI] [PubMed] [Google Scholar]
- Le N.-Q. -K., Ho Q.-T., Ou Y.-Y. (2018). Classifying the Molecular Functions of Rab GTPases in Membrane Trafficking Using Deep Convolutional Neural Networks. Anal. Biochem. 555, 33–41. 10.1016/j.ab.2018.06.011 [DOI] [PubMed] [Google Scholar]
- Li C.-C., Liu B. (2020). MotifCNN-fold: Protein Fold Recognition Based on Fold-specific Features Extracted by Motif-Based Convolutional Neural Networks. Brief. Bioinform. 21, 2133–2141. 10.1093/bib/bbz133 [DOI] [PubMed] [Google Scholar]
- Li H., Gong X.-J., Yu H., Zhou C. (2018). Deep Neural Network Based Predictions of Protein Interactions Using Primary Sequences. Molecules 23, 1923. 10.3390/molecules23081923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Sze K. H., Lu G., Ballester P. J. (2021). Machine‐learning Scoring Functions for Structure‐based Virtual Screening. WIREs Comput. Mol. Sci. 11. 10.1002/wcms.1478 [DOI] [Google Scholar]
- Li Y., Cirino P. C. (2014). Recent Advances in Engineering Proteins for Biocatalysis. Biotechnol. Bioeng. 111, 1273–1287. 10.1002/bit.25240 [DOI] [PubMed] [Google Scholar]
- Li Z., Yang Y., Faraggi E., Zhan J., Zhou Y. (2014). Direct Prediction of Profiles of Sequences Compatible with a Protein Structure by Neural Networks with Fragment-Based Local and Energy-Based Nonlocal Profiles. Proteins 82, 2565–2573. 10.1002/prot.24620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Nie J. (2020). Prediction of Enzyme Function Based on a Structure Relation Network. IEEE ACCESS 8, 132360–132366. 10.1109/ACCESS.2020.3010028 [DOI] [Google Scholar]
- Liao J., Warmuth M. K., Govindarajan S., Ness J. E., Wang R. P., Gustafsson C., et al. (2007). Engineering Proteinase K Using Machine Learning and Synthetic Genes. BMC Biotechnol. 7, 16. 10.1186/1472-6750-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G. N., Wang Z., Xu D., Cheng J. (2010). SeqRate: Sequence-Based Protein Folding Type Classification and Rates Prediction. BMC Bioinforma. 11, S1. 10.1186/1471-2105-11-S3-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Chen H., Li S., Liu Y., Li X., Yu B. (2019). Accurate Prediction of Potential Druggable Proteins Based on Genetic Algorithm and Bagging-SVM Ensemble Classifier. Artif. Intell. Med. 98, 35–47. Available at: http://10.0.3.248/j.artmed.2019.07.005 . 10.1016/j.artmed.2019.07.005 [DOI] [PubMed] [Google Scholar]
- Long H., Liao B., Xu X., Yang J. (2018). A Hybrid Deep Learning Model for Predicting Protein Hydroxylation Sites. Ijms 19, 2817. 10.3390/ijms19092817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S., Tian P. (2019). Protein Secondary Structure Prediction with Context Convolutional Neural Network. RSC Adv. 9, 38391–38396. 10.1039/c9ra05218f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Wang M., Liu Y., Zhao X.-M., Li A. (2019). DeepPhos: Prediction of Protein Phosphorylation Sites with Deep Learning. Bioinformatics 35, 2766–2773. 10.1093/bioinformatics/bty1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Yang Z., Wang L., Zhang Y., Lin H., Wang J. (2019). KeSACNN: a Protein-Protein Interaction Article Classification Approach Based on Deep Neural Network. Ijdmb 22, 131–148. 10.1504/ijdmb.2019.099724 [DOI] [Google Scholar]
- Luo X., Tu X., Ding Y., Gao G., Deng M. (2020). Expectation Pooling: an Effective and Interpretable Pooling Method for Predicting DNA-Protein Binding. Bioinformatics 36, 1405–1412. 10.1093/bioinformatics/btz768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud A. H., Masters M. R., Yang Y., Lill M. A. (2020). Elucidating the Multiple Roles of Hydration for Accurate Protein-Ligand Binding Prediction via Deep Learning. Commun. Chem. 3, 19. 10.1038/s42004-020-0261-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia E. H. B., Assis L. C., de Oliveira T. A., da Silva A. M., Taranto A. G. (2020). Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 8. 10.3389/fchem.2020.00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrodimitris S., Van Ham R. C. H. J., Reinders M. J. T. (2019). Improving Protein Function Prediction Using Protein Sequence and GO-Term Similarities. Bioinformatics 35, 1116–1124. 10.1093/bioinformatics/bty751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataeimoghadam F., Newton M. A. H., Dehzangi A., Karim A., Jayaram B., Ranganathan S., et al. (2020). Enhancing Protein Backbone Angle Prediction by Using Simpler Models of Deep Neural Networks. Sci. Rep. 10, 1–12. 10.1038/s41598-020-76317-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello C., Wallner B. (2019). rawMSA: End-To-End Deep Learning Using Raw Multiple Sequence Alignments. PLoS One 14, e0220182. 10.1371/journal.pone.0220182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A. T., Hiss J. A., Schneider G. (2018). Recurrent Neural Network Model for Constructive Peptide Design. J. Chem. Inf. Model.. 58, 472–479. 10.1021/acs.jcim.7b00414 [DOI] [PubMed] [Google Scholar]
- Murphy G. S., Sathyamoorthy B., Der B. S., Machius M. C., Pulavarti S. V., Szyperski T., et al. (2015). Computational De Novo Design of a Four-Helix Bundle Protein-Dnd_4hb. Protein Sci. 24, 434–445. 10.1002/pro.2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell J., Li Z., Hanson J., Heffernan R., Lyons J., Paliwal K., et al. (2018). SPIN2: Predicting Sequence Profiles from Protein Structures Using Deep Neural Networks. Proteins 86, 629–633. 10.1002/prot.25489 [DOI] [PubMed] [Google Scholar]
- Özen A., Gönen M., Alpaydın E., Haliloğlu T. (2009). Machine Learning Integration for Predicting the Effect of Single Amino Acid Substitutions on Protein Stability. BMC Struct. Biol. 9. 10.1186/1472-6807-9-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès G., Charmettant B., Grudinin S., Valencia A. (2019). Protein Model Quality Assessment Using 3D Oriented Convolutional Neural Networks. Bioinformatics 35, 3313–3319. 10.1093/bioinformatics/btz122 [DOI] [PubMed] [Google Scholar]
- Paladino A., Marchetti F., Rinaldi S., Colombo G. (2017). Protein Design: from Computer Models to Artificial Intelligence. WIREs Comput. Mol. Sci. 7, e1318. 10.1002/wcms.1318 [DOI] [Google Scholar]
- Picart-Armada S., Barrett S. J., Willé D. R., Perera-Lluna A., Gutteridge A., Dessailly B. H. (2019). Benchmarking Network Propagation Methods for Disease Gene Identification. PLoS Comput. Biol. 15, e1007276–24. Available at: http://10.0.5.91/journal.pcbi.1007276 . 10.1371/journal.pcbi.1007276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanyi M. (1962). Personal Knowledge. Towards a Post-Critical Philosophy. 2nd ed.. London: Routledge & Kegan Paul. [Google Scholar]
- Popova M., Isayev O., Tropsha A. (2018). Deep Reinforcement Learning for De Novo Drug Design. Sci. Adv. 4, eaap7885. 10.1126/sciadv.aap7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Oja M., Weston J., Noble W. S. (2012). A Unified Multitask Architecture for Predicting Local Protein Properties. PLoS One 7, e32235. 10.1371/journal.pone.0032235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z., Wu L., Sun H., Huo S., Ma T., Lim E., et al. (2020). Artificial Intelligence Method to Design and Fold Alpha-Helical Structural Proteins from the Primary Amino Acid Sequence. Extreme Mech. Lett. 36, 100652. 10.1016/j.eml.2020.100652 [DOI] [Google Scholar]
- Ragoza M., Hochuli J., Idrobo E., Sunseri J., Koes D. R. (2017). Protein-Ligand Scoring with Convolutional Neural Networks. J. Chem. Inf. Model.. 57, 942–957. 10.1021/acs.jcim.6b00740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh B., Rahat O., Basri R., Schreiber G. (2007). Rediscovering Secondary Structures as Network Motifs-Aan Unsupervised Learning Approach. Bioinformatics 23, e163–e169. 10.1093/bioinformatics/btl290 [DOI] [PubMed] [Google Scholar]
- Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., et al. (2021). Biological Structure and Function Emerge from Scaling Unsupervised Learning to 250 Million Protein Sequences. Proc. Natl. Acad. Sci. U. S. A. 118, e2016239118. 10.1073/pnas.2016239118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Micheletti C., Seno F., Maritan A. (2001). A Self-Consistent Knowledge-Based Approach to Protein Design. Biophysical J. 80, 480–490. 10.1016/S0006-3495(01)76030-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ W. P., Ranganathan R. (2002). Knowledge-based Potential Functions in Protein Design. Curr. Opin. Struct. Biol. 12, 447–452. 10.1016/S0959-440X(02)00346-9 [DOI] [PubMed] [Google Scholar]
- Savojardo C., Martelli P. L., Tartari G., Casadio R. (2020b). Large-scale Prediction and Analysis of Protein Sub-mitochondrial Localization with DeepMito. BMC Bioinforma. 21, 266. N.PAG--N.PAG. Available at: http://10.0.4.162/s12859-020-03617-z . 10.1186/s12859-020-03617-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savojardo C., Bruciaferri N., Tartari G., Martelli P. L., Casadio R. (2020a). DeepMito: Accurate Prediction of Protein Sub-mitochondrial Localization Using Convolutional Neural Networks. Bioinformatics 36, 56–64. Available at: http://10.0.4.69/bioinformatics/btz512 . 10.1093/bioinformatics/btz512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., et al. (2020). Improved Protein Structure Prediction Using Potentials from Deep Learning. Nature 577, 706–710. 10.1038/s41586-019-1923-7 [DOI] [PubMed] [Google Scholar]
- Shah A. R., Oehmen C. S., Webb-Robertson B.-J. (2008). SVM-HUSTLE--an Iterative Semi-supervised Machine Learning Approach for Pairwise Protein Remote Homology Detection. Bioinformatics 24, 783–790. 10.1093/bioinformatics/btn028 [DOI] [PubMed] [Google Scholar]
- Shamim M. T. A., Anwaruddin M., Nagarajaram H. A. (2007). Support Vector Machine-Based Classification of Protein Folds Using the Structural Properties of Amino Acid Residues and Amino Acid Residue Pairs. Bioinformatics 23, 3320–3327. 10.1093/bioinformatics/btm527 [DOI] [PubMed] [Google Scholar]
- Shroff R., Cole A. W., Diaz D. J., Morrow B. R., Donnell I., Annapareddy A., et al. (2020). Discovery of Novel Gain-Of-Function Mutations Guided by Structure-Based Deep Learning. ACS Synth. Biol. 9, 2927–2935. 10.1021/acssynbio.0c00345 [DOI] [PubMed] [Google Scholar]
- Sidhu A., Yang Z. R. (2006). Prediction of Signal Peptides Using Bio-Basis Function Neural Networks and Decision Trees. Appl. Bioinforma. 5, 13–19. 10.2165/00822942-200605010-00002 [DOI] [PubMed] [Google Scholar]
- Simha R., Briesemeister S., Kohlbacher O., Shatkay H. (2015). Protein (Multi-)location Prediction: Utilizing Interdependencies via a Generative Model. Bioinformatics 31, i365–i374. 10.1093/bioinformatics/btv264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Liu G., Jiang J., Zhang P., Liang Y. (2021). Prediction of Protein-ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm. Ijms 22, 939. 10.3390/ijms22020939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sua J. N., Lim S. Y., Yulius M. H., Su X., Yapp E. K. Y., Le N. Q. K., et al. (2020). Incorporating Convolutional Neural Networks and Sequence Graph Transform for Identifying Multilabel Protein Lysine PTM Sites. Chemom. Intelligent Laboratory Syst. 206, 104171. 10.1016/j.chemolab.2020.104171 [DOI] [Google Scholar]
- Sunseri J., King J. E., Francoeur P. G., Koes D. R. (2019). Convolutional Neural Network Scoring and Minimization in the D3R 2017 Community Challenge. J. Comput. Aided. Mol. Des. 33, 19–34. 10.1007/s10822-018-0133-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureyya Rifaioglu A., Doğan T., Jesus Martin M., Cetin-Atalay R., Atalay V. (2019). DEEPred: Automated Protein Function Prediction with Multi-Task Feed-Forward Deep Neural Networks. Sci. Rep. 9, 7344. 10.1038/s41598-019-43708-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalkai B., Grolmusz V. (2018a). Near Perfect Protein Multi-Label Classification with Deep Neural Networks. METHODS 132, 50–56. 10.1016/j.ymeth.2017.06.034 [DOI] [PubMed] [Google Scholar]
- Szalkai B., Grolmusz V. (2018b). SECLAF: A Webserver and Deep Neural Network Design Tool for Hierarchical Biological Sequence Classification. Bioinformatics 34, 2487–2489. 10.1093/bioinformatics/bty116 [DOI] [PubMed] [Google Scholar]
- Taherzadeh G., Dehzangi A., Golchin M., Zhou Y., Campbell M. P. (2019). SPRINT-gly: Predicting N- and O-Linked Glycosylation Sites of Human and Mouse Proteins by Using Sequence and Predicted Structural Properties. Bioinformatics 35, 4140–4146. 10.1093/bioinformatics/btz215 [DOI] [PubMed] [Google Scholar]
- Tian J., Wu N., Chu X., Fan Y. (2010). Predicting Changes in Protein Thermostability Brought about by Single- or Multi-Site Mutations. BMC Bioinforma. 11, 370. 10.1186/1471-2105-11-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torng W., Altman R. B. (2019). High Precision Protein Functional Site Detection Using 3D Convolutional Neural Networks. Bioinformatics 35, 1503–1512. 10.1093/bioinformatics/bty813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traoré S., Allouche D., André I., De Givry S., Katsirelos G., Schiex T., et al. (2013). A New Framework for Computational Protein Design through Cost Function Network Optimization. Bioinformatics 29, 2129–2136. 10.1093/bioinformatics/btt374 [DOI] [PubMed] [Google Scholar]
- Tsou L. K., Yeh S.-H., Ueng S.-H., Chang C.-P., Song J.-S., Wu M.-H., et al. (2020). Comparative Study between Deep Learning and QSAR Classifications for TNBC Inhibitors and Novel GPCR Agonist Discovery. Sci. Rep. 10, 16771. 10.1038/s41598-020-73681-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y., Tomii K. (2020). Neural Networks for Protein Structure and Function Prediction and Dynamic Analysis. Biophys. Rev. 12, 569–573. 10.1007/s12551-020-00685-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang Y. S., Xie X. (2017). HLA Class I Binding Prediction via Convolutional Neural Networks. Bioinformatics 33, 2658–2665. 10.1093/bioinformatics/btx264 [DOI] [PubMed] [Google Scholar]
- Verma N., Qu X., Trozzi F., Elsaied M., Karki N., Tao Y., et al. (2021). SSnet: A Deep Learning Approach for Protein-Ligand Interaction Prediction. Ijms 22, 1392. 10.3390/ijms22031392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpato V., Adelfio A., Pollastri G. (2013). Accurate Prediction of Protein Enzymatic Class by N-To-1 Neural Networks. BMC Bioinforma. 14, S11. 10.1186/1471-2105-14-S1-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C., Cozzetto D., Fa R., Jones D. T. (2019). Using Deep Maxout Neural Networks to Improve the Accuracy of Function Prediction from Protein Interaction Networks. PLoS One 14, e0209958–21. Available at: http://10.0.5.91/journal.pone.0209958 . 10.1371/journal.pone.0209958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Geng L., Zhao Y.-J., Yang Y., Huang Y., Zhang Y., et al. (2020). Artificial Intelligence-Based Multi-Objective Optimization Protocol for Protein Structure Refinement. Bioinformatics 36, 437–448. 10.1093/bioinformatics/btz544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cang Z., Wei G.-W. (2020a). A Topology-Based Network Tree for the Prediction of Protein-Protein Binding Affinity Changes Following Mutation. Nat. Mach. Intell. 2, 116–123. 10.1038/s42256-020-0149-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cui X., Li S., Yang X., Ma A., Zhang Y., et al. (2020b). DeepMal: Accurate Prediction of Protein Malonylation Sites by Deep Neural Networks. Chemom. Intelligent Laboratory Syst. 207, 104175. 10.1016/j.chemolab.2020.104175 [DOI] [Google Scholar]
- Wang X., Liu Y., Lu F., Li H., Gao P., Wei D. (2020). Dipeptide Frequency of Word Frequency and Graph Convolutional Networks for DTA Prediction. Front. Bioeng. Biotechnol. 8. 10.3389/fbioe.2020.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sun S., Li Z., Zhang R., Xu J. (2017). Accurate De Novo Prediction of Protein Contact Map by Ultra-deep Learning Model. PLoS Comput. Biol. 13, e1005324. 10.1371/journal.pcbi.1005324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardah W., Dehzangi A., Taherzadeh G., Rashid M. A., Khan M. G. M., Tsunoda T., et al. (2020). Predicting Protein-Peptide Binding Sites with a Deep Convolutional Neural Network. J. Theor. Biol. 496, 110278. 10.1016/j.jtbi.2020.110278 [DOI] [PubMed] [Google Scholar]
- Wardah W., Khan M. G. M., Sharma A., Rashid M. A. (2019). Protein Secondary Structure Prediction Using Neural Networks and Deep Learning: A Review. Comput. Biol. Chem. 81, 1–8. 10.1016/j.compbiolchem.2019.107093 [DOI] [PubMed] [Google Scholar]
- Wong K.-C., Chan T.-M., Peng C., Li Y., Zhang Z. (2013). DNA Motif Elucidation Using Belief Propagation. Nucleic Acids Res. 41, e153. 10.1093/nar/gkt574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhang Y. (2008). A Comprehensive Assessment of Sequence-Based and Template-Based Methods for Protein Contact Prediction. Bioinformatics 24, 924–931. 10.1093/bioinformatics/btn069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Mcpartlon M., Li J. (2021). Improved Protein Structure Prediction by Deep Learning Irrespective of Co-evolution Information. Nat. Mach. Intell. 3, 601–609. 10.1038/s42256-021-00348-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Tang B., Chen W., Luo J. (2019). DeepT3: Deep Convolutional Neural Networks Accurately Identify Gram-Negative Bacterial Type III Secreted Effectors Using the N-Terminal Sequence. Bioinformatics 35, 2051–2057. 10.1093/bioinformatics/bty931 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang M., Yu Z., Zhao X.-M., Li A. (2020). GANcon: Protein Contact Map Prediction with Deep Generative Adversarial Network. IEEE ACCESS 8, 80899–80907. 10.1109/ACCESS.2020.2991605 [DOI] [Google Scholar]
- Yang J., Anishchenko I., Park H., Peng Z., Ovchinnikov S., Baker D. (2020). Improved Protein Structure Prediction Using Predicted Interresidue Orientations. Proc. Natl. Acad. Sci. U.S.A. 117, 1496–1503. 10.1073/pnas.1914677117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., He B.-J., Jang R., Zhang Y., Shen H.-B. (2015). Accurate Disulfide-Bonding Network Predictions Improveab Initiostructure Prediction of Cysteine-Rich Proteins. Bioinformatics 31, btv459–3781. 10.1093/bioinformatics/btv459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Faraggi E., Zhao H., Zhou Y. (2011). Improving Protein Fold Recognition and Template-Based Modeling by Employing Probabilistic-Based Matching between Predicted One-Dimensional Structural Properties of Query and Corresponding Native Properties of Templates. Bioinformatics 27, 2076–2082. 10.1093/bioinformatics/btr350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C.-T., Brunette T., Baker D., McIntosh-Smith S., Parmeggiani F. (2018). Elfin: An Algorithm for the Computational Design of Custom Three-Dimensional Structures from Modular Repeat Protein Building Blocks. J. Struct. Biol. 201, 100–107. 10.1016/j.jsb.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Yu C.-H., Buehler M. J. (2020). Sonification Based De Novo Protein Design Using Artificial Intelligence, Structure Prediction, and Analysis Using Molecular Modeling. Apl. Bioeng. 4, 016108. 10.1063/1.5133026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-H., Qin Z., Martin-Martinez F. J., Buehler M. J. (2019). A Self-Consistent Sonification Method to Translate Amino Acid Sequences into Musical Compositions and Application in Protein Design Using Artificial Intelligence. ACS Nano 13, 7471–7482. 10.1021/acsnano.9b02180 [DOI] [PubMed] [Google Scholar]
- Zafeiris D., Rutella S., Ball G. R. (2018). An Artificial Neural Network Integrated Pipeline for Biomarker Discovery Using Alzheimer's Disease as a Case Study. Comput. Struct. Biotechnol. J. 16, 77–87. 10.1016/j.csbj.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Li J., Lü Q. (2018). Prediction of 8-state Protein Secondary Structures by a Novel Deep Learning Architecture. BMC Bioinforma. 19, 293. Available at: http://10.0.4.162/s12859-018-2280-5 . 10.1186/s12859-018-2280-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Kabuka M. (2019). Multimodal Deep Representation Learning for Protein Interaction Identification and Protein Family Classification. BMC Bioinforma. 20, 531. 10.1186/s12859-019-3084-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu G., Guo M., Wang J. (2018). Predicting Protein-Protein Interactions Using High-Quality Non-interacting Pairs. BMC Bioinforma. 19, 525. 10.1186/s12859-018-2525-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qiao S., Ji S., Han N., Liu D., Zhou J. (2019). Identification of DNA-Protein Binding Sites by Bootstrap Multiple Convolutional Neural Networks on Sequence Information. Eng. Appl. Artif. Intell. 79, 58–66. 10.1016/j.engappai.2019.01.003 [DOI] [Google Scholar]
- Zhao B., Xue B. (2018). Decision-tree Based Meta-Strategy Improved Accuracy of Disorder Prediction and Identified Novel Disordered Residues inside Binding Motifs. Ijms 19, 3052. 10.3390/ijms19103052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Peng J., Xu J. (2010). Fragment-free Approach to Protein Folding Using Conditional Neural Fields. Bioinformatics 26, i310–i317. 10.1093/bioinformatics/btq193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Li J., Wang R., He F., Yue L., Yin M. (2018). General and Species-specific Lysine Acetylation Site Prediction Using a Bi-modal Deep Architecture. IEEE ACCESS 6, 63560–63569. 10.1109/ACCESS.2018.2874882 [DOI] [Google Scholar]
- Zhao Z., Gong X. (2019). Protein-Protein Interaction Interface Residue Pair Prediction Based on Deep Learning Architecture. IEEE/ACM Trans. Comput. Biol. Bioinf. 16, 1753–1759. 10.1109/TCBB.2017.2706682 [DOI] [PubMed] [Google Scholar]
- Zheng W., Li Y., Zhang C., Pearce R., Mortuza S. M., Zhang Y. (2019). Deep‐learning Contact‐map Guided Protein Structure Prediction in CASP13. Proteins 87, 1149–1164. 10.1002/prot.25792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zhou X., Wuyun Q., Pearce R., Li Y., Zhang Y. (2020). FUpred: Detecting Protein Domains through Deep-Learning-Based Contact Map Prediction. Bioinformatics 36, 3749–3757. 10.1093/bioinformatics/btaa217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Lai L. (2009). A Novel Method for Enzyme Design. J. Comput. Chem. 30, 256–267. 10.1002/jcc.21050 [DOI] [PubMed] [Google Scholar]
- Zimmermann O., Hansmann U. H. E. (2006). Support Vector Machines for Prediction of Dihedral Angle Regions. Bioinformatics 22, 3009–3015. 10.1093/bioinformatics/btl489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation.