Abstract
Despite the introduction of vaccines and drugs for SARS-CoV-2, the COVID-19 pandemic continues to spread throughout the world. In severe COVID-19 patients, elevated levels of proinflammatory cytokines have been detected in the blood, lung cells, and bronchoalveolar lavage, which is referred to as a cytokine storm, a consequence of overactivation of the NLR family pyrin domain-containing protein 3 (NLRP3) inflammasome and resultant excessive cytokine production. The hyperinflammatory response and cytokine storm cause multiorgan impairment including the central nervous system, in addition to a detriment to the respiratory system. Hyperactive NLRP3 inflammasome, due to dysregulated immune response, is the primary cause of COVID-19 severity. The severity could be enhanced due to viral evolution leading to the emergence of mutated variants of concern, such as delta and omicron. In this review, we elaborate on the inflammatory responses associated with the NLRP3 inflammasome activation in COVID-19 pathogenesis, the mechanisms for the NLRP3 inflammasome activation and pathway involved, cytokine storm, and neurological complications as long-term consequences of SARS-CoV-2 infection. Also discussed is the therapeutic potential of NLRP3 inflammasome inhibitors for the treatment of COVID-19.
Keywords: SARS-CoV-2, COVID-19, NLRP3 inflammasome, proinflammatory cytokines, cytokine storm, neuroinflammation
Introduction
Emerging and re-emerging pathogens are a massive threat to the world population and the primary global concern of the current public health [1,2]. The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported in Wuhan, China [3]. SARS-CoV-2 is the etiologic agent of coronavirus disease 2019 (COVID-19), the culprit behind the current pandemic [4,5]. This viral pathogen causes flu-like syndrome with mild symptoms in majority of infected individuals [6-8]. However, a subset of infected patients (~15% in the early phase of the COVID-19 pandemic) suffer from severe illnesses that require hospitalization and ventilation support [9,10]. SARS-CoV-2 has astounding infectivity due to global air connectivity, culminating in the worst pandemic ever [11-14]. According to the world health organization (WHO), as of February 23, 2022, there are more than 426 million confirmed cases and over 5 million deaths (https://covid19.who.int/). To treat COVID-19, drug repurposing for expeditious drug development against SARS-CoV-2 is unprecedented [15,16]. Meanwhile, work-speed vaccine pipelines and the application of modern biotechnology have yielded ever quicker vaccines [17,18]. Therefore, vaccination represents one of the most promising counter-pandemic measures to the COVID-19 [19]. However, COVID-19 remains massive and the need for effective therapies urgent, in part due to the emergence of mutated variants of concern (VOCs) [20-22].
Phylogenetically, SARS-CoV-2 is a member of the genus β-coronavirus, which includes 2003 SARS-CoV and 2012 the Middle East Respiratory Syndrome coronavirus (MERS-CoV) [23-25]. SARS-CoV-2 genome sequence is 80% similar to SARS-CoV [5,26,27]. Structurally, SARS-CoV-2 resembles other coronaviruses (CoVs), is spherical with ~100 nm in diameter, and has a single-stranded positive-sense RNA (ssRNA) [28,29]. It encodes 4-structural proteins: membrane glycoprotein (M), spike glycoprotein (S), envelope glycoprotein (E), and nucleocapsid (N) [30]. The N-protein conjugating with genomic RNA forms nucleocapsid, and to enclose nucleocapsid viral envelope assembled by three protein components S, M, and E [29]. The SARS-CoV-2 S-protein binds to the receptors: angiotensin-converting enzyme 2 (ACE2), and the viral entry is facilitated by transmembrane protein serine protease 2 (TMPRSS2) [31-35]. Identification of polybasic cleavage site in the S-protein and its enhanced affinity for ACE2 [26,36,37] may impart in greater transmissibility and increased virulence of SARS-CoV-2 [38,39]. The virus infects cells expressing ACE2, including monocytes, macrophages, alveolar cells, intestinal epithelial cells, endothelial cells, kidney cells, neurons, neuroepithelial cells and glial cells [36,40-42].
In the lungs of SARS-CoV-2 infected patients, an elevated level of cytokine release has been reported. The elevated levels of cytokines trigger an aberrant uncontrolled response known as cytokine storm, also referred to as cytokine release syndrome (CRS) [43-45]. The accumulating cytokines empower SARS-CoV-2 invasion by attracting immune cells, engaging aggressive inflammatory responses, and severe respiratory complications like acute respiratory distress syndrome (ARDS) [46-49]. The ARDS and related acute lung injury (ALI) occur due to the storm of inflammatory cytokines, notably interleukin-1β (IL-1β), IL-6, and tumor necrosis factor (TNF)-α [50]. Although the mechanisms underlying cytokine storm are multifaceted, accumulating evidence indicates that host cells necessitate inflammasome activation to produce inflammatory cytokines leading to the storm [45,51]. Inflammasomes are multiprotein cytosolic platforms with a tendency to aggregate in response to pathogen-induced or host-mediated assaults [52]. The NLR family pyrin domain-containing protein 3 (NLRP3) is a well-characterized inflammasome and initiates an inflammatory response by inducing excessive cytokine production against viral infection [53]. An involvement of NLRP3 inflammasome in SARS-CoV-2-associated intracellular signaling is shown in Figure 1.
Figure 1.
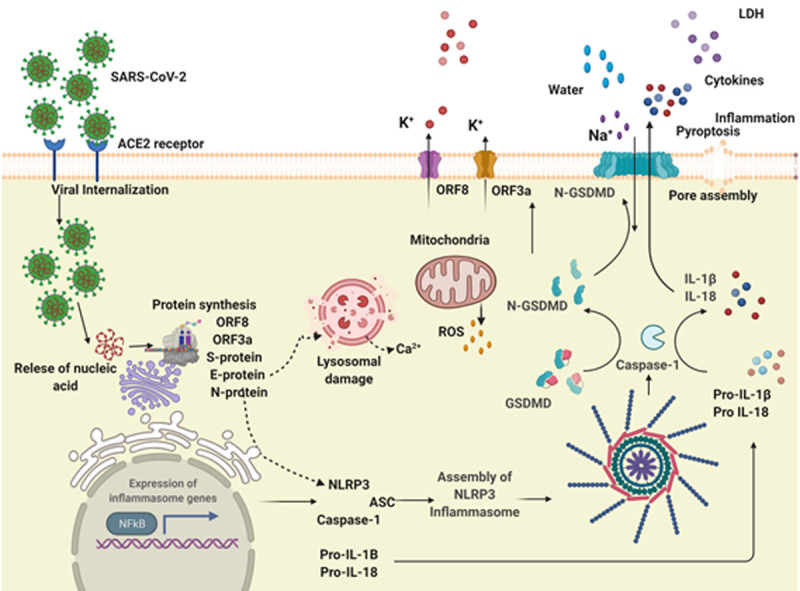
Activation of NLRP3 inflammasome by SARS-CoV-2. SARS-CoV-2 infection occurs by binding of Spike glycoprotein with cell surface receptor ACE2 leading to viral internalization, followed by the release of nucleic acid, viral replication and synthesis of viral proteins. Infection of SARS-CoV-2 upregulates the NFkB pathway leading to increased expression and synthesis of NLRP3 and IL-1β. Viral proteins including S, N, E and viroporins interact with NLRP3 and facilitate inflammasome assembly via oligomerization, the interaction of NLRP3 with ASC and cleavage of caspas1 leading to maturation and release of IL-18 and IL-1β. Ion channels and ion flux are also involved in NLRP3 inflammasome activation. Mitochondrial ROS and lysosomal degradation further impart NLRP3 inflammasome activation.
In addition to its attack on the respiratory system, SARS-CoV-2 was found to induce neurological syndromes including dizziness, hypersomnia, hypogeusia, headaches, myalgia, ataxia, seizures, and impaired consciousness in a high proportion of infected patients [54-56]. Indeed, COVID-19 patients exhibit diverse neurological symptoms that are similar to other respiratory viral infections [57-60]. These neurological symptoms in SARS-CoV-2-infected patients are disabling and quite frequent in their occurrence [61,62]. The neurodegenerative changes, brain edema, and even encephalitis were observed in the severe COVID-19 patients [33,63-65] and some of them were found positive for SARS-CoV-2 in the cerebrospinal fluid (CSF) and brain tissues [66-68]. The presence of SARS-CoV-2 in the CSF and brain tissues in postmortem cases demonstrated that this virus is not only restricted to the respiratory system but can enter the central nervous systems (CNS) inducing neurological manifestations [69]. The consequences of SARS-CoV-2 CNS infection are discussed in section titled NLRP3 inflammasome activation and COVID19-assoociated neurological symptoms and illustrated in Figure 2.
Figure 2.
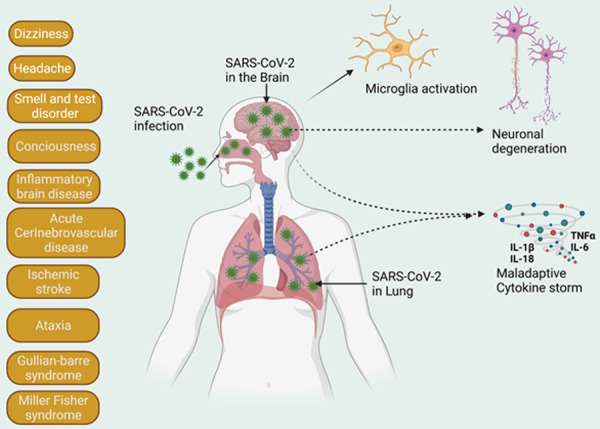
COVID-19-associated neurological consequences. The graphical representation of impacts of neurological outcomes in COVID-19 patients. These neurological syndromes are diverse in nature including dizziness, headache, loss of sense of taste and smell, brain fog and various inflammatory brain diseases.
Evidently, hyperinflammation and cytokine storm is the key pathophysiological processes leading to COVID-19 severity. This review tentatively covers the current progress on SARS-CoV-2-induced NLRP3 inflammasome activation, hyperinflammation, cytokine storm, and neurological consequences. We scrutinize the COVID-19-mediated neurological symptoms associated with NLRP3 inflammasome activation in microglia, astrocytes, and other CNS cells. Also discussed are the mechanistic pathways, checkpoints of inflammasome activation, cytokine storm, and pyroptosis, which could be the potential targets for intervening/ameliorating the severity of the COVID-19 pandemic.
Inflammasome complexes
The induction of the inflammatory process in cells is often mediated by inflammasomes which are multiprotein cytosolic platforms of the innate immune defense system [52]. Inflammasomes tend to aggregate in response to various microbe-associated and host-generated assaults and orchestrate the development of local and systemic inflammation [52,70]. These harmful threats are detected by components of the host innate immune system, the pattern-recognition receptors (PRRs). To trigger inflammatory pathways for the removal of microbial infection and repairment of tissue damage PRRs recognize pathogen-associated molecular patterns (PAMPs) or endogenous stress generated damage-associated molecular patterns (DAMPs). Inflammasomes are defined by their sensor proteins (PRRs) and get oligomerized in response to PAMPs and DAMPs to activate caspase-1. There are five confirmed members of inflammasomes: nucleotide-binding oligomerization domain (NOD), leucine-rich repeat (LRR)-containing proteins (NLR) family representative NLRP1, NLRP3, NLRC4, absent-in-melanoma 2 (AIM2) and pyrin [71,72]. Additionally, other members of PRRs known for forming inflammasomes are NLRP2, NLRP6, NLRP7, NLRP12 and IFI16 [73-77]. Among various inflammasomes, the NLRP3 inflammasome is a well-characterized and most studied molecular platform that responds to RNA viruses and gets activated against microbial infection (PAMPs) and host cell damage or aggregates (DAMPs). NLRP3 inflammasome is composed of the NLRP3 receptor, the adaptor molecule apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC), and caspase-1. NLRP3 is a tripartite protein having an amino-terminal pyrin domain (PYD), a central NOD or NACHT, and an LRR domain [78]. To initiate inflammasome assembly PYD of NLRP3 interacts with PYD of ASC [79]. The NOD of NLRP3 possesses ATPase activity and is crucial for oligomerization of NLRP3 following the activation [80]. A commonly used NLRP3 inhibitor MCC950 targets ATPase activity of NOD [81,82]. Following NLRP3 inflammasome assembly, caspase-1 becomes activated by autoproteolytic cleavage and promotes maturation of proinflammatory cytokines, IL-1β, IL-18, IL-6, TNFα and gasdermin-D (GSDMD), a pore-forming protein that incites pyroptosis (a type of inflammatory programmed cell death) [52,83]. The hallmark of the active inflammasome is the presence of spec (puncta), these puncta or spec are micron-size structures formed by polymerization of ASC as a consequence of the activation by PAMPs, and DAMPs [84,85].
Inflammasome activation and regulation
Inflammasomes are key signaling platforms responsible for detecting pathogenic microorganisms and sterile stressors resulting in the activation and secretion of proinflammatory cytokines. The mechanisms of activation and regulation of inflammasomes are complex and ensure an effective but balanced inflammasome-mediated immune response. Owing to the well-characterized and most studied inflammasome NLRP3, we will further contemplate activation and regulation mechanism using this. Despite the controversy, a two-step signaling model for the activation of NLRP3 inflammasome is widely accepted, consisting of primming (first signal) and activation steps (second signal) [86-88]. Primming signals are generally ligands for toll-like receptors (TLRs), NLRs or cytokine receptors which in turn activate the NF-kB. Primming signals regulate the NLRP3 inflammasome by upregulating transcription of key components (NLRP3 and IL-1β), as a basal expression of these components is insufficient for activation of inflammasome in resting cells [78,89]. In response to TLR ligands, NF-kB induces signaling components MyD88 and TRIF, which regulate the induction of NLRP3 as well as pro-IL-1β [89]. However, the primming signal does not alter the expression profile of ACS, pro-caspase-1, and pro-IL-18 [78]. Further, it was reported that apoptotic signaling molecules caspase-8 and FADD prime NLRP3 induction [90,91]. Interaction of IKK complex with caspase-8 promotes NFkB transcription and translocation for downstream signaling [92]. In contrast, the transcription-independent role of priming was reported using rapid priming with lipopolysaccharide (LPS), where NLRP3 activation occurs in the absence of its induction [93]. This rapid transcription-independent priming is believed to be mediated by a signaling molecule downstream of TLRs and MyD88 known as IL-1 receptor-associated kinase 1 (IRAK-1), [94]. Phosphorylation of IRAK-1 induced by LPS primes activation of inflammasome in IKK complex independent manner [95]. Nevertheless, beyond the transcriptional regulation, the primming step does more to license NLRP3 inflammasome activation.
Following the primming step, activation of NLRP3 inflammasome can be induced by a plethora of stimuli which includes, ATP, K+ ionophores [96], particulate matter [97,98], heme [99], pathogen-associated RNA [100,101], microbial toxins and integrant [102-104]. These biochemically diverse agonists induce multiple cellular and molecular signals including ionic flux, dysfunction of mitochondria, generation of reactive oxygen species (ROS), and lysosomal damage for the NLRP3 inflammasome activation. Activated inflammasome plays a critical role in host defense in anticipation of infectious agents mounting immune responses. Stringent inflammasome activation regulation is required as dysregulated NLRP3 inflammasome has been involved in the pathogenesis of numerous inflammatory diseases. Precise regulation of activation of NLRP3 inflammasome is critical for adequate immune protection to the host by subverting the tissue damage. The mechanisms for the NLRP3 inflammasome regulation include post-translational alterations of NLRP3 as well as its interacting partners.
NLRP3 inflammasome in COVID-19
The involvement of excessive inflammation and resultant cytokine storm owing to uncontrolled release of cytokines is accountable for the unfavorable clinical outcomes of the COVID-19 [105,106]. The mechanism of inflammasome activation in SARS-CoV-2 is poorly explored, despite confirmed participation of NLRP3 inflammasome in SARS-Co-V and MERS-CoV [107,108]. However, the engagement of pyroptosis and cytokine storm, where inflammasome-associated products such as IL-1β, IL-18, and LDH were detected in COVID-19 patient sera, suggests the association of inflammasome in the COVID-19 [105,106,109-111]. Further, as an inflammasome-independent pathway can also produce inflammatory substances, it demands a definitive confirmation of the involvement of inflammasome in the SARS-CoV-2 infection [112-114]. In an elegant article by Rodrigue and coworkers [85] on COVID-19, the involvement of NLRP3 inflammasome was confirmed in patients with moderate to severe infection of SARS-CoV-2. Other contemporary studies have shown the involvement of SARS-CoV-2-induced activation of NLRP3 inflammasome in the COVID-19 [45,115-119].
COVID-19 induced NLRP3 inflammasome activation and cytokine storm
The immune system defends our body against invaders such as SARS-CoV-2. This defense system includes 2-arms, the innate and adaptive immune systems. The innate-immunity-mediated antiviral responses are triggered by recognizing PAMPs, leading to the induction of adaptive immunity which is critical to protecting the host [120,121]. The host cells in response to viral infection release cytokines, chemokines, leukotrienes, proteases, and ROS to enable viral clearance [122]. A stringent equilibrium between antagonistic signals and cellular response maintains the immune response evoked by pathogens and is responsible for preventing host tissue damage, by preventing continuous activation of the immune system [122]. Usually, acute viral infections evoke systemic inflammatory responses leading to excessive synthesis and release of proinflammatory cytokines as a defense measure [122]. This systemic inflammatory response triggered by infections and drugs causes cytokine storm or CRS. Previously, influenza viral infection causing respiratory illness has been characterized as a stimulus for CRS [123]. Influenza patients undergo robust cytokine-mediated responses which are associated with fever, hypoxemia and hypotension [10,123]. The syndrome may be mild or can develop into persistent high-grade fevers, vasodilatory shock with hemodynamic instability demanding mechanical ventilation [10,123]. Similarly, the existence of elevated inflammatory cytokine levels in COVID-19 patients leading to CRS or cytokine storm culminate in lung collapse, respiratory failure and may lead to multiorgan failures [105]. Enhanced induction in IL-1β, TNFα, IL-6, IL-18, IL-10, IL-1RA, and C-X-C motif chemokine ligand 10 (CXCL10) was reported in severe COVID-19 patients exhibiting cytokine storm phenotype [106,124]. These excessively released cytokines may produce eosinopenia (low count of eosinophil) and lymphocytopenia (low count of CD4+T, CD8+T, and NK cells) and can induce naïve B-cell activation, Th17 differentiation, neutrophil recruitment and monocyte stimulation [125-128]. Recently, it was demonstrated that SARS-CoV-2 infection could lead to NLRP3 overactivation resulting in cytokine storms in the hematopoietic stem cells [116]. The NLRP3 is one of the most critical innate immune components that accelerate inflammation by releasing IL-1β, IL-18 and provokes pyroptosis. There have been reports of a positive correlation of IL-18 and caspase-1 with other inflammatory markers, including C-reactive protein (CRP), LDH, and IL-6 in the activation of inflammasome in COVID-19 patients [85]. Further investigation to find out the cytokine storm in other organs and tissues causing the disease severity in COVID-19 is imperative. Thus, to control cytokine storm in SARS-CoV-2 infected patient NLRP3 inflammasome inhibitors can be harnessed alongside other inflammatory inhibitors.
NLRP3 inflammasome activation and COVID 19-associated neurological symptoms
The activation of NLRP3 inflammasome and associated pathophysiology of various neurological disorders have been reported in numerous neurological disorders, such as in Alzheimer’s disease (AD) [129-133], Parkinson’s disease (PD) [134-136], multiple sclerosis (MS) [137,138], and traumatic brain injury [139-141]. Viral infection in the brain (neuroinvasion) leads to inflammatory response resulting in neuroinflammation [142]. At the beginning of the COVID-19 pandemic, a general belief was that SARS-CoV-2 infection solely affects the respiratory system of humans [143], but the appearance of newer symptoms such as olfactory and taste indicates an involvement of the CNS [144]. There is evidence that SARS-CoV-2 can infect neuronal cells, the BBB endothelial cells, microglia, and astrocyte via ACE2 and another receptor known as cluster differentiation 147 (CD147) [145,146] and cause a neurological deficit in a substantial proportion of COVID-19 patients [147,148]. Figure 2 is the graphical representation of COVID-19 induced neurological syndromes.
In the CNS, the critical player of neuroinflammation is microglial cells, a major source of proinflammatory cytokines [149]. As resident macrophages in the brain, microglia are accountable for phagocytosis and removal of pathogens and neurotoxic agents [150]. They are also the target cells in SARS-CoV-2 CNS infection. Studies using postmortem samples of COVID-19 patients have demonstrated that SARS-CoV-2 was present in the brain compartments [151,152]. SARS-CoV-2 infection to the CNS was further confirmed by the observation that this virus can infect neural progenitors and brain organoids [153]. The entry of SARS-CoV-2 into the brain may take a similar approach as other viruses to invade brain tissues, the hematogenous route, the retrograde route [154], or the direct access to the brain via the olfactory canal [105,155]. Following entry, the virus and viral proteins can cause direct neural cell injury or promote microglial pro-inflammatory responses causing an indirect neural injury [156]. Inflammasome activation through SARS-CoV-2 infection or entrance of viral proteins into the brain and other CNS compartments was reported. [157,158]. It has been shown that incubation of spike glycoprotein S1 of SARS-CoV-2 with BV2 microglial cells elevated levels of NLRP3, IL-1β, TNFα, IL-6, nitric oxide (NO) and enhanced activity of NFkB and caspase-1 [157]. The S1 protein also triggered the production of interferon-beta (IFNβ), TNFα, and NFkB in the human microglia [159].
Extensive activation of cascades of neuroinflammation is provoked by excessive release of cytokines and chemokines, which drives neuronal hyperexcitability through activation of glutamate receptors accompanied by induction of seizures [160,161]. COVID-19 patients may give rise to inflammatory injury and brain edema owing to the immune system over-exuberance response may lead to defective consciousness [162]. Such immunologic response resulting in hyperinflammation may further amplify the cytokine storm [163]. It was presumed that intracranial cytokine storm further potentiates the breakdown of BBB and the consequent leukocytes migration. COVID-19-induced defects in gustation and olfaction could be a result of olfactory bulb injury due to, nasal cavity inflammation. Thus, the binding of odorants to the olfactory receptors is blocked due to nasal inflammation resulting in dysfunction in the olfactory response [164]. Additionally, loss of taste (ageusia) occurs due to dysfunction in the taste buds [165]. In COVID-19 patients, it usually takes a longer time for the regeneration and recovery of damaged neurons in the olfactory lobes [164]. Depending on the extent of viral insult, the loss of smell and taste may persist for months to years [164]. It is widely recognized that senior individuals with compromised health conditions are more vulnerable to COVID-19-associated fatal and long-term consequences. The profile of NLRP3 inflammasome involvement in neurological consequences induced by COVID-19 is an unmet need for research. Considering age-related neurological deficits, COVID-19 brain infection could further exacerbate neurological symptoms in senior patients.
Mechanisms of NLRP3 inflammasome activation in COVID-19
Inflammasomes are large cytosolic multiprotein oligomers of the innate immune system which assemble in response to PAMPs and DAMPs leading to proinflammatory cytokine release and lytic cell death termed pyroptosis [52,53]. The NLRP3 is a well-characterized sensor of the NLR family, detects a broad range of microbial motifs in addition to environmental irritants and endogenous danger signals. The exact molecular mechanism for activation of NLRP3 inflammasome is largely unknown. It occurs in a two-step process: primming, the first signal and subsequent activation is the second signal. For SARS-CoV-2 primming triggers are not well established, however, a recent study has shown that spike glycoprotein can initiate inflammasome activation [166]. Generally, NLRP3 inflammasome responds to an agglomeration of affront to the cells, which causes Ca2+ influx, K+ efflux, ROS production by mitochondria, mitochondrial dysfunction, and lysosomal rupture [52,167-169]. In addition, the NLRP3 inflammasome can be activated by pore-forming toxins, extracellular ATP and large extracellular aggregates including cholesterol and uric acid crystals, and amyloid [53]. Viral infection evokes the NLRP3 inflammasome activation, which is also admissible with coronaviruses [170,171].
Previous studies using SARS-CoV have demonstrated that activation of the inflammasome occurs by E and ORF3a proteins via altering the permeability of K+ ions across the plasma membrane and ROS production by mitochondria [50,172]. In a recent study, inflammasome-derived products including Casp1p20, IL-1β, IL-6 and IL-18 were also detected at higher levels in SARS-CoV-2 infected patients and were correlated with the severity of disease [85,117]. It is conventionally agreed that activation of NLRP3 occurs in viral infection by assembling NLRP3 with ASC and recruiting caspase-1, resulting in proinflammatory cytokine production and GSDMD-mediated pyroptosis [45,85,170,173]. Further, SARS-CoV-2 N-protein was shown to directly interact with the NLRP3 and promote the ASC assembly. This sequel of N-protein and NLRP3 interaction enhances proteolytic activity of caspase-1 and enhanced release of IL-1β and IL-6, resulting in hyperinflammation and cytokine storm in the lungs [118]. Using the NLRP3 double knockout mice, Pan and colleagues have demonstrated that the lung NLRP3 inflammasomes activation was the cause of ARDS and resulting fetal death [118].
Other contemporary studies have revealed that SARS-CoV-2 S, E, ORF3a and ORF8 can induce inflammasome activation and hyperinflammation, leading to severe clinical outcomes [45,119,174-176]. Nevertheless, investigations on the mechanisms of inflammasome activation induced by SARS-CoV-2 could be of immense interest in the battle against the pandemic of COVID-19. In the following section, we report current advances in NLRP3 inflammasome activation induced by proteins of SARS-CoV-2.
SARS-CoV-2 proteins-mediated NLRP3 activation
In addition to SARS-CoV-2 infection-associated activation of the NLRP3 inflammasome, SARS-CoV-2 proteins were also found to induce NLRP3 activation. Among them are S, N, E, Viroprin (ORF3a), and ORF8 proteins [45,118,119,157,174-176]. How these proteins cause NLRP3 inflammasome activation and resultant biological consequences are discussed tentatively in the following subsections.
SARS-CoV-2 Spike (S)
The SARS-CoV-2 S-protein is a glycoprotein and a crucial player in infectivity as it binds to host cell receptors ACE2, which facilitates virus entry into the cells [177,178]. It has been consistently, demonstrated that the SARS-CoV-2 S-protein interacts with ACE2 and causes the release of various cytokines including IL-1β, IL-6, IL-8, and IL-18 via the NLRP3 inflammasome-mediated activation of caspase-1 [171,174,179]. The recombinant nucleic acid-based and subunit vaccines are against this antigenic protein [17,18,180]. The S-protein can trigger NLRP3 inflammasome activation, resulting in hyperinflammation and related cytokine storms [157,166,174]. It has been reported that incubation of recombinant spike glycoprotein S1 of SARS-CoV-2 with peripheral blood mononuclear cells (PBMCs) of humans evoked excessive cytokine production, which was abolished by dexamethasone [174]. The spike glycoprotein S1 evoked cytokine release through mechanisms involving activation of NFkB, p38MAPK, and NLRP3 inflammasome as demonstrated by harnessing different specific inhibitors [174]. Further studies on BV2 microglial cells demonstrated that spike glycoprotein S1 activated the NLRP3 inflammasome resulting in enhancement of pro-inflammatory cytokine production and nitric oxide [157]. These results indicate that viral spike glycoprotein S1, perhaps other viral proteins as well, plays a crucial role in the activation of the NLRP3 inflammasome. This exemplifies the involvement of NLRP3 inflammasome in alveolar cells in COVID-19. Using primary rat microglial cells and spike glycoprotein S1 of SARS-CoV-2, we observed activation of the NLRP3 inflammasome by measuring enhanced production of IL-1β and other cytokines, as well as the expression of NO, and iNOS (Dutta and Xiong, unpublished data). Our results further demonstrated the neuroinflammatory properties of spike glycoprotein S1 via activation of the NLRP3 inflammasome. The spike glycoprotein S1-mediated inflammasome activation in rat microglial cultures was blocked by an NLRP3 specific inhibitor MCC950 (Dutta and Xiong, unpublished data). In addition, the S-protein of SARS-CoV-2 was found to damage hematopoietic stem/progenitor cells through pyroptosis mediated by NLRP3 inflammasome-dependent mechanism [115]. Nevertheless, these experimental results, including ours, demonstrated that SARS-CoV-2 spike glycoprotein induces NLRP3 inflammasome activation.
SARS-CoV-2 nucleocapsid
It was shown that SARS-CoV-2 nucleocapsid (N-protein) interacts with NLRP3 and resulted in NLRP3 inflammasome activation in both cell cultures in vitro and a mouse model of viral infection in vivo [118]. The study demonstrated that N-protein interacted in a dose-dependent manner with NLRP3 and promoted the IL-1β maturation and release. Application of a specific NLRP3 inflammasome inhibitor MCC950 and an inhibitor for Caspase-1 Ac-YVAD-cmk blocked the NLRP3 inflammasome activation [118]. This is a classic finding with an elucidation of a mechanistic view of NLRP3 inflammasome activation by N-protein, where N-protein not only interacted with NLRP3 but also facilitated its assembly with ASC, a process of polymerization and activation. This was detected by increased spec or puncta formation after N-protein treatment. This study further identified the exact domain of N-protein (CTD, 260-340aa), which interacts with the NLRP3 inflammasome using deletion mutation constructs and Co-IP. Deletion of this particular domain of N-protein resulted in a significant reduction in p17, p20, and ASC oligomerization, demonstrating that the physical interaction between NLRP3 and N-protein was indispensable for the activation of the NLRP3 inflammasome [118]. Hence, treatments targeting N-protein might suppress cytokine storms and reduce lung injury and complications in other organs mediated by SARS-CoV-2-associated NLRP3 overactivation.
The N-protein of SARS-CoV-2 possesses dual actions on innate immune responses. At lower doses, it suppresses type I interferon (IFN-I) signaling as well as inflammatory cytokine expression. In contrast, it promotes IFN-I signaling and expression of inflammatory cytokine at higher doses [181]. Such dual functions were also observed in regulating the phosphorylation status of IRF3, STAT1, and STAT2 and their nuclear translocation [181]. N-protein combined with TRIM25-protein was found to suppress the ubiquitination as well as activation of the retinoic acid-inducible gene (RIG-I) [181]. In addition, N-protein bounds to GSDMD linker region and hindered GSDMD cleavage by caspase-1 [182]. These findings indicate that the N-protein of SARS-CoV-2 plays a pivotal role in triggering inflammatory responses in COVID-19.
SARS-CoV-2 envelope
The envelope protein (E-protein) of SARS-CoV-2 is a small structural protein and plays a crucial role in the activation of the NLRP3 inflammasome [176]. The E-protein was shown to have dual roles in the modulation of NLRP3 inflammasome in human and murine macrophages. It suppressed activation of NLRP3 inflammasome in the stage of early infection but activated NLRP3 inflammasome at the advanced stage of the infection [176]. The mechanisms underlying its dual effects on NLRP3 inflammasome are not well characterized at present and further investigations are definitely desirable.
SARS-CoV-2 ORF3a (viroporin)
Open reading frame 3a (ORF3a) is an accessory protein conserved in both SARS-CoV and SARS-CoV-2 [183,184]. It is a viroporin, a transmembrane protein, that works as an ion channel and helps in the viral release [185,186]. In SARS-CoV, ORF3a protein activates NLRP3 inflammasomes leading to dysregulated immune response and elevated levels of proinflammatory cytokines [48,187]. It was shown that SARS-CoV ORF3a protein can activate the NLRP3 inflammasome in lipopolysaccharide primed macrophages [172]. Owing to conservation in ORF3a protein in SARS-CoV and SARS-CoV-2 [183,184], the role of ORF3a in NLRP3 inflammasome activation is apparent. A recent study has shown that ectopically expressed SARS-CoV-2 ORF3a activated the NLRP3 inflammasome by triggering IL-1β expression via the NFkB pathway [175]. Furthermore, the study revealed that this viroporin primed and activated inflammasome through both ASC-dependent and independent pathways. It was also shown that ORF3a-mediated activation of inflammasome was mediated via potassium ion efflux and by NEK7 and NLRP3 oligomerization. Application of MCC950 blocked ORF3a-mediated inflammasome activation.
NLRP3 inflammasome as a therapeutic target for COVID-19
Because of poor immune fitness, dysregulated NLRP3 inflammasome and resultant hyperinflammation contribute to the COVID-19 severity [10,188]. Inhibitors targeting the pathways of NLRP3 inflammasome activation can be contemplated as a prospective therapy. The specific inhibitor against NLRP3, MCC950, may have therapeutic potential for patients at the early stage of the disease to prevent cytokine storms, ameliorate complications, and reduce fatal outcomes [118]. Other inhibitors, like Ac-YVAD-cmk which is a specific inhibitor for caspase-1, can also be considered for the treatment, either alone or in combination with MCC950. The therapeutic efficacies of these inhibitors need to be evaluated by in vitro and in vivo models of disease and ultimately in clinical trials. Indeed, in the treatment of cardiovascular disease, the use of NLRP3 inhibitors has been reviewed in the literature [189], paving the path for using NLRP3 inhibitors in COVID-19. It is worth noting that inflammasome and pyroptosis are proposed as potential therapeutic targets for the COVID-19 treatment [190]. Thus, inhibitors targeting critical components of inflammasome activation, cellular pyroptosis, and downstream cytokine can be implicated in the treatment of COVID-19 [43]. Indeed, it has been shown that targeting NLRP3 inflammasome could be a promising immune intervention against the severe COVID-19 [191]. Thus, we tentatively discuss a few NLRP3 inhibitors, both specific (selectively blocks the NLRP3 inflammasome) and non-specific (indirect inhibition of NLRP3-mediated signaling), on their therapeutic potential for COVID-19. Treatment with these inhibitors can block caspase-1 activity, resulting in an inhibition of IL-1β and other cytokine production, and consequent reduction in inflammation (Figure 3).
Figure 3.
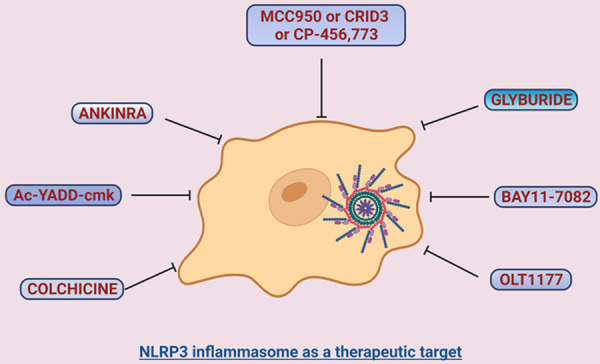
Inhibition of NLRP3 inflammasome may have therapeutic potential for COVID-19 and related neurological syndromes. Graphical representation of inflammasome inhibition by various inhibitors. The NLRP3 inflammasome is a prospective target for therapeutic approaches in the treatment of COVID-19. Inhibitors against NLRP3 inflammasome may ameliorate SARS-CoV-2-induced hyperinflammation and related cytokine storms. These inhibitors, in addition to their beneficial effects in lung and other organs, could also have therapeutic potential for COVID-19 neurological syndromes associated with microglial NLRP3 inflammasome activation.
MCC950 (CRID3 or CP-456,773)
MCC950 is a specific inhibitor for NLRP3 inflammasome with a small molecular weight. MCC950 blocks the ATPase activity by binding non-covalently near the Walker B motif of NLRP3 resulting in inhibition of NLRP3 [81,192-197]. Pigs and mice treated with MCC950 exhibited a decreased infiltration of neutrophils, reduced expression of myocardial IL-1β and diminished infarct size and cardiac dysfunction [198-200]. In lung ischemia-reperfusion (IR), the NLRP3 inflammasome was found overactive in the murine lung IR model [201]. Animals pretreated with MCC950 significantly alleviated IR-induced lung injury by restricting the proinflammatory cytokine release and inhibiting infiltration of neutrophils [201]. As neutrophil infiltration has been reported in severe COVID-19 patients along with cytokine storms [202], the use of MCC950 may attenuate neutrophil infiltrations and cytokine storms. In a recent study using monocytes from healthy donors, it was shown that MCC950 can inhibit SARS-CoV-2 infection-induced caspase-1 activation leading to IL-1β production [85]. Also, SARS-CoV-2 N-protein- and S-protein-induced NLRP3 inflammasome induction in lung cells, PBMCs, BV2 neural cells, and mice were inhibited when treated with MCC950 [118,157,174]. In addition to its treatment in lung injury and cardiovascular complications associated with NLRP3 inflammasome activation, MCC950 was used to treat neuroinflammation-related injury as well. A study on spinal cord injury mice model showed that MCC950 restricted the inflammatory response and improved neurological sequel [203]. Such therapeutic effects were achieved by blocking the assembly of the NLRP3 inflammasome, including NLRP3-ACS and NLRP3-caspase-1 complex formation, and inhibiting the release of cytokines TNFα, IL-18 and IL-1β [203]. Nevertheless, NLRP3 inflammasome inhibition by MCC950 has been applied for the treatment of multiple sclerosis (MS)-associated central neuropathic pain in patients with RR-MS [204]. Inspired by this study, MCC950 could be considered for treatment of neuroinflammatory disease conditions due to SARS-CoV-2 infection.
Glyburide
Glyburide is the first NLRP3 inhibitor that works in vitro but at a high dose [205]. It was shown that glyburide can inhibit NLRP3 inflammasome and lung tumorigenesis in mice [206], which implicates for the treatment of lung hyperinflammation in COVID-19 patients.
BAY11-7082
As a synthetic NFkB inhibitor, BAY11-7082 acts by alkylating the cysteine residue of the ATPase region of NLRP3 resulting in inhibition of the NF-kB pathway [207]. To explore its anti-inflammatory properties, BAY11-7-82 was tested in PBMCs treated with spike glycoprotein S1 of SARS-CoV-2. Treatment with S1 increased phosphorylation of NFkB p65, IkBα, and IkBα degradation, resulting in NFkB activation [174]. Such effects were blocked by either dexamethasone or BAY11-7082, leading to an inhibition of inflammasome activation [174]. In another study aiming to recapitulate neuroinflammation by SARS-CoV-2 in BV2 cells, it was shown that BAY11-7082 inhibited the SARS-CoV-2 S1-induced activation of NLRP3 inflammasome as assayed by a reduction in IL-1β, TNFα and IL-6 production [157]. These observations suggest that BAY11-7082 might have therapeutic potential for COVID-19 cytokine storm and neurological consequences associated with neuroinflammation.
OLT1177
OLT1177 is a specific small-molecule inhibitor for the NLRP3 functioning by blocking the ATPase activity [133,208,209]. It acts against mutants of NLRP3 in the cryopyrin-associated periodic syndrome patients [210] and may have therapeutic potential for COVID-19. In a phase 2A clinical trial, OLT1177 was found safe and effective in reducing targeted joint pain in gout patients [211]. As the compound is already in a clinical trial, testing its efficacy in COVID-19 patients may open another avenue for the treatment of COVID-19.
Colchicine
This is a tricyclic alkaloid compound that is currently in use to treat familial Mediterranean fever, gout and acute as well as chronic pericarditis [212,213]. Colchicine impedes NLRP3 and ASC interaction via disruption of microtubule and inhibits the inflammasome activation [214]. In a recent review, colchicine was suggested to repurpose for COVID-19 treatment owing to its anti-inflammatory and immunomodulatory nature [215]. Based on its current use in the CNS disease treatment, neuro-behcet’s syndrome, a severe chronic inflammatory vascular disease [216], colchicine may have the potential in treating COVID-19-induced neuroinflammation.
Ac-YVAD-cmk
This is an efficacious inhibitor of caspase-1 [217]. Caspase-1 plays a terminal effector role in inflammasome by converting proinflammatory pro-IL-18 and pro-IL-1β into respective mature forms by cleavage [218]. Inhibition of caspase 1 blocks inflammatory processes associated with inflammasome activation. Ac-YVAD-cmk possesses activity to effectively block activation of inflammasomes and display anti-inflammatory and anti-pyroptotic effects [219,220]. Recently, it was shown that SARS-CoV-2 N-protein mediated activation of NLRP3 inflammasome was inhibited using Ac-YVAD-cmk [118]. In a mouse model of depression, its inhibitory effect on caspase-1 was also demonstrated where immune activation and NLRP3-mediated depression were ameliorated by Ac-YVAD-cmk [220]. In view of its ability to selectively block IL-1β converting enzyme, Ac-YVAD-cmk could be considered as a potential treatment option for neuroinflammation and multiorgan damage in COVID-19 patients.
Anakinra
As an interleukin-1 type I receptor (IL-1RI) antagonist, anakinra is in use for the treatment against multiple autoinflammatory diseases including familial Mediterranean fever, MS, and rheumatoid arthritis [221-223]. These clinical disorders, which benefited from anakinra, share the same pathophysiological hallmarks of COVID-19, including macrophage activation syndrome (MAS), hemophagocytic lymphohystiocytosis, and septic shock [224-226], raising the hope for its application for COVID-19 patients. The use of anakinra was indeed found to have clinical benefits in saving the lives of hospitalized COVID-19 patients with moderate to severe manifestations [227].
Nevertheless, suitable NLRP3 inflammasome inhibitors with high efficacy and safety in clinical trials may be considered for immediate COVID-19 treatment emergency use authorization. While dexamethasone and other immunosuppressive drugs block type I IFN signaling, this may leave viral replication unchecked. The advantage of NLRP3 inflammasome inhibitors in COVID-19 leaves type I IFN signaling intact, which increases virus clearance by limiting the viral replication [228]. Also, it was shown that the NLRP3 inflammasome was repressed by type I IFN via reducing the pro-IL-1β level and its cleavage into mature IL-1β [229]. Thus, treatment of COVID-19 patients with inflammasome inhibitors will encounter dual effects: leaving type I IFN levels high promoting antiviral state and restraining the hyperinflammation induced by the NLRP3 inflammasome.
Summary
As SARS-CoV-2 is the etiologic agent for global COVID-19 pandemic fatalities, research into the underlying mechanisms is an unmet need. While specific molecular mechanisms influencing disease severity remain to be determined, a number of studies have demonstrated that activation of inflammasomes and inflammatory mediators including IL-1β, IL-18, IL-6 and LDH are intimately associated with the severity of COVID-19. While it is not restricted to infect the respiratory system, SARS-CoV-2 can attack other organ systems including the brain. COVID-19 gives rise to a group of disease manifestations, and one of the dreadful consequences is the massive inflammatory response mediated by the NLRP3 inflammasome. Inflammasome signaling on one hand provides defense against microbial invasion, and on the other hand, as a countermeasure, produces hyperinflammatory responses. The NLRP3 is one of the well-characterized inflammasomes which plays a salient role in hyperinflammatory responses in SARS-CoV and MERS-CoV as well as in SARS-CoV-2 infection. NLRP3 inflammasome activation induced by infection of SARS-CoV-2 not only causes severe respiratory complications but provokes neurological syndromes. This review has assembled current advances on the SARS-CoV-2-mediated activation of NLRP3 inflammasome that may help us better understand the disease in a broader way and find ways to contain the COVID-19 severity and death.
Acknowledgements
Figures are created with BioRender.com. Supported by NIH grant R01DA050540 (H.X.) from the National Institute on Drug Abuse.
Disclosure of conflict of interest
None.
References
- 1.Gao GF. From “A”IV to “Z”IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, Dong L, Cui X, Miao Y, Wang D, Dong J, Xiao C, Chen W, Wang H. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in wuhan, china, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand R, Thayer R, Srinivasan A, Nayyar S, Gardner M, Luciw P, Dandekar S. Biological and molecular characterization of human immunodeficiency virus (HIV-1BR) from the brain of a patient with progressive dementia. Virology. 1989;168:79–89. doi: 10.1016/0042-6822(89)90406-6. [DOI] [PubMed] [Google Scholar]
- 7.Ding Q, Lu P, Fan Y, Xia Y, Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol. 2020;92:1549–1555. doi: 10.1002/jmv.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machhi J, Herskovitz J, Senan AM, Dutta D, Nath B, Oleynikov MD, Blomberg WR, Meigs DD, Hasan M, Patel M, Kline P, Chang RC, Chang L, Gendelman HE, Kevadiya BD. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman TL, Swartz TH. Targeting the NLRP3 inflammasome in severe COVID-19. Front Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seitz BM, Aktipis A, Buss DM, Alcock J, Bloom P, Gelfand M, Harris S, Lieberman D, Horowitz BN, Pinker S, Wilson DS, Haselton MG. The pandemic exposes human nature: 10 evolutionary insights. Proc Natl Acad Sci U S A. 2020;117:27767–27776. doi: 10.1073/pnas.2009787117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Wei L, Niu P. The novel coronavirus outbreak in Wuhan, China. Glob Health Res Policy. 2020;5:6. doi: 10.1186/s41256-020-00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bambra C, Riordan R, Ford J, Matthews F. The COVID-19 pandemic and health inequalities. J Epidemiol Community Health. 2020;74:964–968. doi: 10.1136/jech-2020-214401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han E, Tan MMJ, Turk E, Sridhar D, Leung GM, Shibuya K, Asgari N, Oh J, Garcia-Basteiro AL, Hanefeld J, Cook AR, Hsu LY, Teo YY, Heymann D, Clark H, McKee M, Legido-Quigley H. Lessons learnt from easing COVID-19 restrictions: an analysis of countries and regions in Asia Pacific and Europe. Lancet. 2020;396:1525–1534. doi: 10.1016/S0140-6736(20)32007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 16.Shyr ZA, Gorshkov K, Chen CZ, Zheng W. Drug discovery strategies for SARS-CoV-2. J Pharmacol Exp Ther. 2020;375:127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T COVE Study Group. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Perez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW Jr, Hammitt LL, Tureci O, Nell H, Schaefer A, Unal S, Tresnan DB, Mather S, Dormitzer PR, Sahin U, Jansen KU, Gruber WC C4591001 Clinical Trial Group. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach JF, Berche P, Chatenoud L, Costagliola D, Valleron AJ. COVID-19: individual and herd immunity. C R Biol. 2021;344:7–18. doi: 10.5802/crbiol.41. [DOI] [PubMed] [Google Scholar]
- 20.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ CMMID COVID-19 Working Group; COVID-19 Genomics UK (COG-UK) Consortium. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL COVID-19 Genomics UK (COG-UK) Consortium. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, Conlon EG, Schaefer-Babajew DJ, DaSilva J, Muecksch F, Gaebler C, Lifton R, Nussenzweig MC, Hatziioannou T, Bieniasz PD, Darnell RB. Vaccine breakthrough infections with SARS-CoV-2 variants. N Engl J Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan W, Zhao X, Ma X, Wang W, Niu P, Xu W, Gao GF, Wu G. A novel coronavirus genome identified in a cluster of pneumonia cases - Wuhan, China 2019-2020. China CDC Wkly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- 24.Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jockusch S, Tao C, Li X, Anderson TK, Chien M, Kumar S, Russo JJ, Kirchdoerfer RN, Ju J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antiviral Res. 2020;180:104857. doi: 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hidalgo P, Valdes M, Gonzalez RA. Molecular biology of coronaviruses: an overview of virus-host interactions and pathogenesis. Bol Med Hosp Infant Mex. 2021;78:41–58. doi: 10.24875/BMHIM.20000249. [DOI] [PubMed] [Google Scholar]
- 29.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik YA. Properties of coronavirus and SARS-CoV-2. Malays J Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 31.Djomkam ALZ, Olwal CO, Sala TB, Paemka L. Commentary: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Front Oncol. 2020;10:1448. doi: 10.3389/fonc.2020.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanclemente-Alaman I, Moreno-Jimenez L, Benito-Martin MS, Canales-Aguirre A, Matias-Guiu JA, Matias-Guiu J, Gomez-Pinedo U. Experimental models for the study of central nervous system infection by SARS-CoV-2. Front Immunol. 2020;11:2163. doi: 10.3389/fimmu.2020.02163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baig AM, Khaleeq A, Ali U, Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 37.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci U S A. 2020;117:15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alenina N, Bader M. ACE2 in brain physiology and pathophysiology: evidence from transgenic animal models. Neurochem Res. 2019;44:1323–1329. doi: 10.1007/s11064-018-2679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukiw WJ, Pogue A, Hill JM. SARS-CoV-2 infectivity and neurological targets in the brain. Cell Mol Neurobiol. 2022;42:217–224. doi: 10.1007/s10571-020-00947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paniz-Mondolfi A, Bryce C, Grimes Z, Gordon RE, Reidy J, Lednicky J, Sordillo EM, Fowkes M. Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) J Med Virol. 2020;92:699–702. doi: 10.1002/jmv.25915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdin SM, Elgendy SM, Alyammahi SK, Alhamad DW, Omar HA. Tackling the cytokine storm in COVID-19, challenges and hopes. Life Sci. 2020;257:118054. doi: 10.1016/j.lfs.2020.118054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polidoro RB, Hagan RS, de Santis Santiago R, Schmidt NW. Overview: systemic inflammatory response derived from lung injury caused by SARS-CoV-2 infection explains severe outcomes in COVID-19. Front Immunol. 2020;11:1626. doi: 10.3389/fimmu.2020.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toldo S, Bussani R, Nuzzi V, Bonaventura A, Mauro AG, Cannata A, Pillappa R, Sinagra G, Nana-Sinkam P, Sime P, Abbate A. Inflammasome formation in the lungs of patients with fatal COVID-19. Inflamm Res. 2021;70:7–10. doi: 10.1007/s00011-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y, Xu J, Zhou C, Wu Z, Zhong S, Liu J, Luo W, Chen T, Qin Q, Deng P. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 48.Reghunathan R, Jayapal M, Hsu LY, Chng HH, Tai D, Leung BP, Melendez AJ. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nieto-Torres JL, Verdia-Baguena C, Jimenez-Guardeno JM, Regla-Nava JA, Castano-Rodriguez C, Fernandez-Delgado R, Torres J, Aguilella VM, Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the Cytokine Storm’ in COVID-19. J Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 53.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karnik M, Beeraka NM, Uthaiah CA, Nataraj SM, Bettadapura ADS, Aliev G, Madhunapantula SV. A review on SARS-CoV-2-induced neuroinflammation, neurodevelopmental complications, and recent updates on the vaccine development. Mol Neurobiol. 2021;58:4535–4563. doi: 10.1007/s12035-021-02399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yachou Y, El Idrissi A, Belapasov V, Ait Benali S. Neuroinvasion, neurotropic, and neuroinflammatory events of SARS-CoV-2: understanding the neurological manifestations in COVID-19 patients. Neurol Sci. 2020;41:2657–2669. doi: 10.1007/s10072-020-04575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Awogbindin IO, Ben-Azu B, Olusola BA, Akinluyi ET, Adeniyi PA, Di Paolo T, Tremblay ME. Microglial implications in SARS-CoV-2 infection and COVID-19: lessons from viral rna neurotropism and possible relevance to parkinson’s disease. Front Cell Neurosci. 2021;15:670298. doi: 10.3389/fncel.2021.670298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abboud H, Abboud FZ, Kharbouch H, Arkha Y, El Abbadi N, El Ouahabi A. COVID-19 and SARS-Cov-2 infection: pathophysiology and clinical effects on the nervous system. World Neurosurg. 2020;140:49–53. doi: 10.1016/j.wneu.2020.05.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pennisi M, Lanza G, Falzone L, Fisicaro F, Ferri R, Bella R. SARS-CoV-2 and the nervous system: from clinical features to molecular mechanisms. Int J Mol Sci. 2020;21:5475. doi: 10.3390/ijms21155475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glass WG, Subbarao K, Murphy B, Murphy PM. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 60.Li K, Wohlford-Lenane C, Perlman S, Zhao J, Jewell AK, Reznikov LR, Gibson-Corley KN, Meyerholz DK, McCray PB Jr. Middle east respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis. 2016;213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pleasure SJ, Green AJ, Josephson SA. The spectrum of neurologic disease in the severe acute respiratory syndrome Coronavirus 2 pandemic infection: neurologists move to the frontlines. JAMA Neurol. 2020;77:679–680. doi: 10.1001/jamaneurol.2020.1065. [DOI] [PubMed] [Google Scholar]
- 62.Marshall M. How COVID-19 can damage the brain. Nature. 2020;585:342–343. doi: 10.1038/d41586-020-02599-5. [DOI] [PubMed] [Google Scholar]
- 63.Helms J, Kremer S, Merdji H, Clere-Jehl R, Schenck M, Kummerlen C, Collange O, Boulay C, Fafi-Kremer S, Ohana M, Anheim M, Meziani F. Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helms J, Kremer S, Meziani F. More on neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382:e110. doi: 10.1056/NEJMc2015132. [DOI] [PubMed] [Google Scholar]
- 65.Chen X, Laurent S, Onur OA, Kleineberg NN, Fink GR, Schweitzer F, Warnke C. A systematic review of neurological symptoms and complications of COVID-19. J Neurol. 2021;268:392–402. doi: 10.1007/s00415-020-10067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McAbee GN, Brosgol Y, Pavlakis S, Agha R, Gaffoor M. Encephalitis Associated with COVID-19 Infection in an 11-Year-Old Child. Pediatr Neurol. 2020;109:94. doi: 10.1016/j.pediatrneurol.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moriguchi T, Harii N, Goto J, Harada D, Sugawara H, Takamino J, Ueno M, Sakata H, Kondo K, Myose N, Nakao A, Takeda M, Haro H, Inoue O, Suzuki-Inoue K, Kubokawa K, Ogihara S, Sasaki T, Kinouchi H, Kojin H, Ito M, Onishi H, Shimizu T, Sasaki Y, Enomoto N, Ishihara H, Furuya S, Yamamoto T, Shimada S. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int J Infect Dis. 2020;94:55–58. doi: 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Remmelink M, De Mendonca R, D’Haene N, De Clercq S, Verocq C, Lebrun L, Lavis P, Racu ML, Trepant AL, Maris C, Rorive S, Goffard JC, De Witte O, Peluso L, Vincent JL, Decaestecker C, Taccone FS, Salmon I. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Crit Care. 2020;24:495. doi: 10.1186/s13054-020-03218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020;92:552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaivola J, Nyman TA, Matikainen S. Inflammasomes and SARS-CoV-2 infection. Viruses. 2021;13:2513. doi: 10.3390/v13122513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma D, Kanneganti TD. The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J Cell Biol. 2016;213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, Stehlik C. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36:464–476. doi: 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minkiewicz J, de Rivero Vaccari JP, Keane RW. Human astrocytes express a novel NLRP2 inflammasome. Glia. 2013;61:1113–1121. doi: 10.1002/glia.22499. [DOI] [PubMed] [Google Scholar]
- 77.Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, Conlon JE, Burbage JJ, Proulx MK, Liu Q, Reed G, Mecsas JC, Iwakura Y, Bertin J, Goguen JD, Fitzgerald KA, Lien E. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vajjhala PR, Mirams RE, Hill JM. Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein. J Biol Chem. 2012;287:41732–41743. doi: 10.1074/jbc.M112.381228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, Zimmermann AG, Ting JP. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proceedings of the National Academy of Sciences. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coll RC, Hill JR, Day CJ, Zamoshnikova A, Boucher D, Massey NL, Chitty JL, Fraser JA, Jennings MP, Robertson AAB, Schroder K. MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat Chem Biol. 2019;15:556–559. doi: 10.1038/s41589-019-0277-7. [DOI] [PubMed] [Google Scholar]
- 82.Tapia-Abellan A, Angosto-Bazarra D, Martinez-Banaclocha H, de Torre-Minguela C, Ceron-Carrasco JP, Perez-Sanchez H, Arostegui JI, Pelegrin P. MCC950 closes the active conformation of NLRP3 to an inactive state. Nat Chem Biol. 2019;15:560–564. doi: 10.1038/s41589-019-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hauenstein AV, Zhang L, Wu H. The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr Opin Struct Biol. 2015;31:75–83. doi: 10.1016/j.sbi.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodrigues TS, de Sa KSG, Ishimoto AY, Becerra A, Oliveira S, Almeida L, Goncalves AV, Perucello DB, Andrade WA, Castro R, Veras FP, Toller-Kawahisa JE, Nascimento DC, de Lima MHF, Silva CMS, Caetite DB, Martins RB, Castro IA, Pontelli MC, de Barros FC, do Amaral NB, Giannini MC, Bonjorno LP, Lopes MIF, Santana RC, Vilar FC, Auxiliadora-Martins M, Luppino-Assad R, de Almeida SCL, de Oliveira FR, Batah SS, Siyuan L, Benatti MN, Cunha TM, Alves-Filho JC, Cunha FQ, Cunha LD, Frantz FG, Kohlsdorf T, Fabro AT, Arruda E, de Oliveira RDR, Louzada-Junior P, Zamboni DS. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. 2019;20:3328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell. 2016;165:792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, Hornung V, Latz E. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gurung P, Anand PK, Malireddi RK, Vande Walle L, Van Opdenbosch N, Dillon CP, Weinlich R, Green DR, Lamkanfi M, Kanneganti TD. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol. 2014;192:1835–1846. doi: 10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allam R, Lawlor KE, Yu EC, Mildenhall AL, Moujalled DM, Lewis RS, Ke F, Mason KD, White MJ, Stacey KJ, Strasser A, O’Reilly LA, Alexander W, Kile BT, Vaux DL, Vince JE. Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non-apoptotic caspase-8 is required for inflammasome priming. EMBO Rep. 2014;15:982–990. doi: 10.15252/embr.201438463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lemmers B, Salmena L, Bidere N, Su H, Matysiak-Zablocki E, Murakami K, Ohashi PS, Jurisicova A, Lenardo M, Hakem R, Hakem A. Essential role for caspase-8 in Toll-like receptors and NFkappaB signaling. J Biol Chem. 2007;282:7416–7423. doi: 10.1074/jbc.M606721200. [DOI] [PubMed] [Google Scholar]
- 93.Schroder K, Sagulenko V, Zamoshnikova A, Richards AA, Cridland JA, Irvine KM, Stacey KJ, Sweet MJ. Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology. 2012;217:1325–1329. doi: 10.1016/j.imbio.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 94.Lin KM, Hu W, Troutman TD, Jennings M, Brewer T, Li X, Nanda S, Cohen P, Thomas JA, Pasare C. IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proc Natl Acad Sci U S A. 2014;111:775–780. doi: 10.1073/pnas.1320294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim SJ, Cha JY, Kang HS, Lee JH, Lee JY, Park JH, Bae JH, Song DK, Im SS. Corosolic acid ameliorates acute inflammation through inhibition of IRAK-1 phosphorylation in macrophages. BMB Rep. 2016;49:276–281. doi: 10.5483/BMBRep.2016.49.5.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 97.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 98.Dostert C, Pétrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Silveira AA, Cunningham C, Corr E, Ferreira WA Jr, Costa FF, Almeida CB, Conran N, Dunne A. Heme induces NLRP3 inflammasome formation in primary human macrophages and may propagate hemolytic inflammatory processes by inducing S100A8 expression. Blood. 2016;128:1256–1256. [Google Scholar]
- 100.Sha W, Mitoma H, Hanabuchi S, Bao M, Weng L, Sugimoto N, Liu Y, Zhang Z, Zhong J, Sun B, Liu YJ. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc Natl Acad Sci U S A. 2014;111:16059–16064. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kanneganti TD, Ozören N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Núñez G. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 102.Greaney AJ, Leppla SH, Moayeri M. Bacterial exotoxins and the inflammasome. Front Immunol. 2015;6:570. doi: 10.3389/fimmu.2015.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mathur A, Feng S, Hayward JA, Ngo C, Fox D, Atmosukarto II, Price JD, Schauer K, Märtlbauer E, Robertson AAB, Burgio G, Fox EM, Leppla SH, Kaakoush NO, Man SM. A multicomponent toxin from Bacillus cereus incites inflammation and shapes host outcome via the NLRP3 inflammasome. Nat Microbiol. 2019;4:362–374. doi: 10.1038/s41564-018-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kasper L, König A, Koenig PA, Gresnigt MS, Westman J, Drummond RA, Lionakis MS, Groß O, Ruland J, Naglik JR, Hube B. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nature Communications. 2018;9:4260. doi: 10.1038/s41467-018-06607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He L, Ding Y, Zhang Q, Che X, He Y, Shen H, Wang H, Li Z, Zhao L, Geng J, Deng Y, Yang L, Li J, Cai J, Qiu L, Wen K, Xu X, Jiang S. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, Chan JFW, Woo PCY, Yuen KY. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 109.Chi Y, Ge Y, Wu B, Zhang W, Wu T, Wen T, Liu J, Guo X, Huang C, Jiao Y, Zhu F, Zhu B, Cui L. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020;222:746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han Y, Zhang H, Mu S, Wei W, Jin C, Tong C, Song Z, Zha Y, Xue Y, Gu G. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: a retrospective and observational study. Aging (Albany NY) 2020;12:11245–11258. doi: 10.18632/aging.103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Yale IT, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alfaidi M, Wilson H, Daigneault M, Burnett A, Ridger V, Chamberlain J, Francis S. Neutrophil elastase promotes interleukin-1beta secretion from human coronary endothelium. J Biol Chem. 2015;290:24067–24078. doi: 10.1074/jbc.M115.659029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guma M, Ronacher L, Liu-Bryan R, Takai S, Karin M, Corr M. Caspase 1-independent activation of interleukin-1beta in neutrophil-predominant inflammation. Arthritis Rheum. 2009;60:3642–3650. doi: 10.1002/art.24959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joosten LA, Netea MG, Fantuzzi G, Koenders MI, Helsen MM, Sparrer H, Pham CT, van der Meer JW, Dinarello CA, van den Berg WB. Inflammatory arthritis in caspase 1 gene-deficient mice: contribution of proteinase 3 to caspase 1-independent production of bioactive interleukin-1beta. Arthritis Rheum. 2009;60:3651–3662. doi: 10.1002/art.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kucia M, Ratajczak J, Bujko K, Adamiak M, Ciechanowicz A, Chumak V, Brzezniakiewicz-Janus K, Ratajczak MZ. An evidence that SARS-Cov-2/COVID-19 spike protein (SP) damages hematopoietic stem/progenitor cells in the mechanism of pyroptosis in Nlrp3 inflammasome-dependent manner. Leukemia. 2021;35:3026–3029. doi: 10.1038/s41375-021-01332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ratajczak MZ, Kucia M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells. Leukemia. 2020;34:1726–1729. doi: 10.1038/s41375-020-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferreira AC, Soares VC, de Azevedo-Quintanilha IG, Dias S, Fintelman-Rodrigues N, Sacramento CQ, Mattos M, de Freitas CS, Temerozo JR, Teixeira L, Damaceno Hottz E, Barreto EA, Pao CRR, Palhinha L, Miranda M, Bou-Habib DC, Bozza FA, Bozza PT, Souza TML. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7:43. doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pan P, Shen M, Yu Z, Ge W, Chen K, Tian M, Xiao F, Wang Z, Wang J, Jia Y, Wang W, Wan P, Zhang J, Chen W, Lei Z, Chen X, Luo Z, Zhang Q, Xu M, Li G, Li Y, Wu J. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat Commun. 2021;12:4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shah A. Novel Coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Betakova T, Kostrabova A, Lachova V, Turianova L. Cytokines induced during influenza virus infection. Curr Pharm Des. 2017;23:2616–2622. doi: 10.2174/1381612823666170316123736. [DOI] [PubMed] [Google Scholar]
- 121.Freeman CM, Martinez FJ, Han MK, Ames TM, Chensue SW, Todt JC, Arenberg DA, Meldrum CA, Getty C, McCloskey L, Curtis JL. Lung dendritic cell expression of maturation molecules increases with worsening chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;180:1179–1188. doi: 10.1164/rccm.200904-0552OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gu Y, Hsu AC, Pang Z, Pan H, Zuo X, Wang G, Zheng J, Wang F. Role of the innate cytokine storm induced by the influenza a virus. Viral Immunol. 2019;32:244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- 123.Shimabukuro-Vornhagen A, Godel P, Subklewe M, Stemmler HJ, Schlosser HA, Schlaak M, Kochanek M, Boll B, von Bergwelt-Baildon MS. Cytokine release syndrome. J Immunother Cancer. 2018;6:56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soy M, Keser G, Atagunduz P, Tabak F, Atagunduz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Root-Bernstein R. Innate receptor activation patterns involving TLR and NLR synergisms in COVID-19, ALI/ARDS and sepsis cytokine storms: a review and model making novel predictions and therapeutic suggestions. Int J Mol Sci. 2021;22:2108. doi: 10.3390/ijms22042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Severini C, Barbato C, Di Certo MG, Gabanella F, Petrella C, Di Stadio A, de Vincentiis M, Polimeni A, Ralli M, Greco A. Alzheimer’s disease: new concepts on the role of autoimmunity and NLRP3 inflammasome in the pathogenesis of the disease. Curr Neuropharmacol. 2021;19:498–512. doi: 10.2174/1570159X18666200621204546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Y, Dong Z, Song W. NLRP3 inflammasome as a novel therapeutic target for Alzheimer’s disease. Signal Transduct Target Ther. 2020;5:37. doi: 10.1038/s41392-020-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Scott XO, Stephens ME, Desir MC, Dietrich WD, Keane RW, de Rivero Vaccari JP. The inflammasome adaptor protein asc in mild cognitive impairment and Alzheimer’s disease. Int J Mol Sci. 2020;21:4674. doi: 10.3390/ijms21134674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lonnemann N, Hosseini S, Marchetti C, Skouras DB, Stefanoni D, D’Alessandro A, Dinarello CA, Korte M. The NLRP3 inflammasome inhibitor OLT1177 rescues cognitive impairment in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2020;117:32145–32154. doi: 10.1073/pnas.2009680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Araujo FM, Cuenca-Bermejo L, Fernandez-Villalba E, Costa SL, Silva VDA, Herrero MT. Role of microgliosis and NLRP3 inflammasome in Parkinson’s disease pathogenesis and therapy. Cell Mol Neurobiol. 2021;42:1283–1300. doi: 10.1007/s10571-020-01027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang W, Nguyen LT, Burlak C, Chegini F, Guo F, Chataway T, Ju S, Fisher OS, Miller DW, Datta D, Wu F, Wu CX, Landeru A, Wells JA, Cookson MR, Boxer MB, Thomas CJ, Gai WP, Ringe D, Petsko GA, Hoang QQ. Caspase-1 causes truncation and aggregation of the Parkinson’s disease-associated protein alpha-synuclein. Proc Natl Acad Sci U S A. 2016;113:9587–9592. doi: 10.1073/pnas.1610099113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou Y, Lu M, Du RH, Qiao C, Jiang CY, Zhang KZ, Ding JH, Hu G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener. 2016;11:28. doi: 10.1186/s13024-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jahanbazi Jahan-Abad A, Karima S, Sahab Negah S, Noorbakhsh F, Borhani-Haghighi M, Gorji A. Therapeutic potential of conditioned medium derived from oligodendrocytes cultured in a self-assembling peptide nanoscaffold in experimental autoimmune encephalomyelitis. Brain Res. 2019;1711:226–235. doi: 10.1016/j.brainres.2019.01.035. [DOI] [PubMed] [Google Scholar]
- 138.Malhotra S, Costa C, Eixarch H, Keller CW, Amman L, Martinez-Banaclocha H, Midaglia L, Sarro E, Machin-Diaz I, Villar LM, Trivino JC, Oliver-Martos B, Parlade LN, Calvo-Barreiro L, Matesanz F, Vandenbroeck K, Urcelay E, Martinez-Gines ML, Tejeda-Velarde A, Fissolo N, Castillo J, Sanchez A, Robertson AAB, Clemente D, Prinz M, Pelegrin P, Lunemann JD, Espejo C, Montalban X, Comabella M. NLRP3 inflammasome as prognostic factor and therapeutic target in primary progressive multiple sclerosis patients. Brain. 2020;143:1414–1430. doi: 10.1093/brain/awaa084. [DOI] [PubMed] [Google Scholar]
- 139.Irrera N, Russo M, Pallio G, Bitto A, Mannino F, Minutoli L, Altavilla D, Squadrito F. The role of NLRP3 inflammasome in the pathogenesis of traumatic brain injury. Int J Mol Sci. 2020;21:104. doi: 10.3390/ijms21176204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ismael S, Ahmed HA, Adris T, Parveen K, Thakor P, Ishrat T. The NLRP3 inflammasome: a potential therapeutic target for traumatic brain injury. Neural Regen Res. 2021;16:49–57. doi: 10.4103/1673-5374.286951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.O’Brien WT, Pham L, Symons GF, Monif M, Shultz SR, McDonald SJ. The NLRP3 inflammasome in traumatic brain injury: potential as a biomarker and therapeutic target. J Neuroinflammation. 2020;17:104. doi: 10.1186/s12974-020-01778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dahm T, Rudolph H, Schwerk C, Schroten H, Tenenbaum T. Neuroinvasion and inflammation in viral central nervous system infections. Mediators Inflamm. 2016;2016:8562805. doi: 10.1155/2016/8562805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients With 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Agyeman AA, Chin KL, Landersdorfer CB, Liew D, Ofori-Asenso R. Smell and taste dysfunction in patients with COVID-19: a systematic review and meta-analysis. Mayo Clin Proc. 2020;95:1621–1631. doi: 10.1016/j.mayocp.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ribeiro DE, Oliveira-Giacomelli A, Glaser T, Arnaud-Sampaio VF, Andrejew R, Dieckmann L, Baranova J, Lameu C, Ratajczak MZ, Ulrich H. Hyperactivation of P2X7 receptors as a culprit of COVID-19 neuropathology. Mol Psychiatry. 2021;26:1044–1059. doi: 10.1038/s41380-020-00965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen R, Wang K, Yu J, Howard D, French L, Chen Z, Wen C, Xu Z. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in the human and mouse brains. Front Neurol. 2020;11:573095. doi: 10.3389/fneur.2020.573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heneka MT, Golenbock D, Latz E, Morgan D, Brown R. Immediate and long-term consequences of COVID-19 infections for the development of neurological disease. Alzheimers Res Ther. 2020;12:69. doi: 10.1186/s13195-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Asadi-Pooya AA, Simani L. Central nervous system manifestations of COVID-19: a systematic review. J Neurol Sci. 2020;413:116832. doi: 10.1016/j.jns.2020.116832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kwon HS, Koh SH. Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl Neurodegener. 2020;9:42. doi: 10.1186/s40035-020-00221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Andoh M, Koyama R. Comparative review of microglia and monocytes in CNS phagocytosis. Cells. 2021;10:2555. doi: 10.3390/cells10102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cama VF, Marin-Prida J, Acosta-Rivero N, Acosta EF, Diaz LO, Casadesus AV, Fernandez-Marrero B, Gilva-Rodriguez N, Cremata-Garcia D, Cervantes-Llanos M, Piniella-Matamoros B, Sanchez D, Del Rosario-Cruz L, Borrajero I, Diaz A, Gonzalez Y, Penton-Arias E, Montero-Gonzalez T, Guillen-Nieto G, Penton-Rol G. The microglial NLRP3 inflammasome is involved in human SARS-CoV-2 cerebral pathogenicity: a report of three post-mortem cases. J Neuroimmunol. 2021;361:577728. doi: 10.1016/j.jneuroim.2021.577728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman OE, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Levavasseur E, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Kazmi SAJ, Zhang K, Wilen CB, Horvath TL, Plu I, Haik S, Thomas JL, Louvi A, Farhadian SF, Huttner A, Seilhean D, Renier N, Bilguvar K, Iwasaki A. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021;218:e20202135. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang BZ, Chu H, Han S, Shuai H, Deng J, Hu YF, Gong HR, Lee AC, Zou Z, Yau T, Wu W, Hung IF, Chan JF, Yuen KY, Huang JD. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zubair AS, McAlpine LS, Gardin T, Farhadian S, Kuruvilla DE, Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 2020;77:1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kumar A, Pareek V, Prasoon P, Faiq MA, Kumar P, Kumari C, Narayan RK. Possible routes of SARS-CoV-2 invasion in brain: in context of neurological symptoms in COVID-19 patients. J Neurosci Res. 2020;98:2376–2383. doi: 10.1002/jnr.24717. [DOI] [PubMed] [Google Scholar]
- 156.de Rivero Vaccari JC, Dietrich WD, Keane RW, de Rivero Vaccari JP. The inflammasome in times of COVID-19. Front Immunol. 2020;11:583373. doi: 10.3389/fimmu.2020.583373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Olajide OA, Iwuanyanwu VU, Adegbola OD, Al-Hindawi AA. SARS-CoV-2 spike glycoprotein S1 induces neuroinflammation in BV-2 microglia. Mol Neurobiol. 2022;59:445–458. doi: 10.1007/s12035-021-02593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Albornoz E, Amarilla AA, Modhiran N, Parker S, Li XX, Wijesundara DK, Zamora AP, McMillan CL, Liang B, Peng NYG, Sng JDJ, Saima FT, Paramitha D, Parry R, Avumegah MS, Isaacs A, Lo M, Miranda-Chacon Z, Bradshaw D, Salinas-Rebolledo C, Rajapakse NW, Munro T, Rojas-Fernandez A, Young PR, Stacey KJ, Khromykh AA, Chappell KJ, Watterson D, Woodruff TM. SARS-CoV-2 drives NLRP3 inflammasome activation in human microglia through spike-ACE2 receptor interaction. bioRxiv. 2022 doi: 10.1038/s41380-022-01831-0. 2022.2001.2011.475947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mishra R, Banerjea AC. SARS-CoV-2 spike targets USP33-IRF9 axis via exosomal miR-148a to activate human microglia. Front Immunol. 2021;12:656700. doi: 10.3389/fimmu.2021.656700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Libbey JE, Kennett NJ, Wilcox KS, White HS, Fujinami RS. Interleukin-6, produced by resident cells of the central nervous system and infiltrating cells, contributes to the development of seizures following viral infection. J Virol. 2011;85:6913–6922. doi: 10.1128/JVI.00458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Singhi P. Infectious causes of seizures and epilepsy in the developing world. Dev Med Child Neurol. 2011;53:600–609. doi: 10.1111/j.1469-8749.2011.03928.x. [DOI] [PubMed] [Google Scholar]
- 162.Durrani M, Kucharski K, Smith Z, Fien S. Acute transverse myelitis secondary to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a case report. Clin Pract Cases Emerg Med. 2020;4:344–348. doi: 10.5811/cpcem.2020.6.48462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ Hlh Across Speciality Collaboration UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Soler ZM, Patel ZM, Turner JH, Holbrook EH. A primer on viral-associated olfactory loss in the era of COVID-19. Int Forum Allergy Rhinol. 2020;10:814–820. doi: 10.1002/alr.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vaira LA, Salzano G, Fois AG, Piombino P, De Riu G. Potential pathogenesis of ageusia and anosmia in COVID-19 patients. Int Forum Allergy Rhinol. 2020;10:1103–1104. doi: 10.1002/alr.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Theobald SJ, Simonis A, Georgomanolis T, Kreer C, Zehner M, Eisfeld HS, Albert MC, Chhen J, Motameny S, Erger F, Fischer J, Malin JJ, Grab J, Winter S, Pouikli A, David F, Boll B, Koehler P, Vanshylla K, Gruell H, Suarez I, Hallek M, Fatkenheuer G, Jung N, Cornely OA, Lehmann C, Tessarz P, Altmuller J, Nurnberg P, Kashkar H, Klein F, Koch M, Rybniker J. Long-lived macrophage reprogramming drives spike protein-mediated inflammasome activation in COVID-19. EMBO Mol Med. 2021;13:e14150. doi: 10.15252/emmm.202114150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tseng WA, Thein T, Kinnunen K, Lashkari K, Gregory MS, D’Amore PA, Ksander BR. NLRP3 inflammasome activation in retinal pigment epithelial cells by lysosomal destabilization: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2013;54:110–120. doi: 10.1167/iovs.12-10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Okada M, Matsuzawa A, Yoshimura A, Ichijo H. The lysosome rupture-activated TAK1-JNK pathway regulates NLRP3 inflammasome activation. J Biol Chem. 2014;289:32926–32936. doi: 10.1074/jbc.M114.579961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zheng M, Williams EP, Malireddi RKS, Karki R, Banoth B, Burton A, Webby R, Channappanavar R, Jonsson CB, Kanneganti TD. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J Biol Chem. 2020;295:14040–14052. doi: 10.1074/jbc.RA120.015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Campbell GR, To RK, Hanna J, Spector SA. SARS-CoV-2, SARS-CoV-1, and HIV-1 derived ssRNA sequences activate the NLRP3 inflammasome in human macrophages through a non-classical pathway. iScience. 2021;24:102295. doi: 10.1016/j.isci.2021.102295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Courjon J, Dufies O, Robert A, Bailly L, Torre C, Chirio D, Contenti J, Vitale S, Loubatier C, Doye A, Pomares-Estran C, Gonfrier G, Lotte R, Munro P, Visvikis O, Dellamonica J, Giordanengo V, Carles M, Yvan-Charvet L, Ivanov S, Auberger P, Jacquel A, Boyer L. Heterogeneous NLRP3 inflammasome signature in circulating myeloid cells as a biomarker of COVID-19 severity. Blood Adv. 2021;5:1523–1534. doi: 10.1182/bloodadvances.2020003918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Olajide OA, Iwuanyanwu VU, Lepiarz-Raba I, Al-Hindawi AA. Induction of exaggerated cytokine production in human peripheral blood mononuclear cells by a recombinant SARS-CoV-2 spike glycoprotein S1 and its inhibition by dexamethasone. Inflammation. 2021;44:1865–1877. doi: 10.1007/s10753-021-01464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu H, Chitre SA, Akinyemi IA, Loeb JC, Lednicky JA, McIntosh MT, Bhaduri-McIntosh S. SARS-CoV-2 viroporin triggers the NLRP3 inflammatory pathway. bioRxiv. 2020 doi: 10.1016/j.virol.2022.01.003. 2020.2010.2027.357731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yalcinkaya M, Liu W, Islam MN, Kotini AG, Gusarova GA, Fidler TP, Papapetrou EP, Bhattacharya J, Wang N, Tall AR. Modulation of the NLRP3 inflammasome by Sars-CoV-2 Envelope protein. Sci Rep. 2021;11:24432. doi: 10.1038/s41598-021-04133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020;11:576622. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Potere N, Candeloro M, Porreca E, Marinari S, Federici C, Auciello R, Di Nisio M. Direct oral anticoagulant plasma levels in hospitalized COVID-19 patients treated with dexamethasone. J Thromb Thrombolysis. 2021;53:346–351. doi: 10.1007/s11239-021-02561-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pino M, Abid T, Pereira Ribeiro S, Edara VV, Floyd K, Smith JC, Latif MB, Pacheco-Sanchez G, Dutta D, Wang S, Gumber S, Kirejczyk S, Cohen J, Stammen RL, Jean SM, Wood JS, Connor-Stroud F, Pollet J, Chen WH, Wei J, Zhan B, Lee J, Liu Z, Strych U, Shenvi N, Easley K, Weiskopf D, Sette A, Pollara J, Mielke D, Gao H, Eisel N, LaBranche CC, Shen X, Ferrari G, Tomaras GD, Montefiori DC, Sekaly RP, Vanderford TH, Tomai MA, Fox CB, Suthar MS, Kozlowski PA, Hotez PJ, Paiardini M, Bottazzi ME, Kasturi SP. A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Sci Immunol. 2021;6:eabh3634. doi: 10.1126/sciimmunol.abh3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhao Y, Sui L, Wu P, Wang W, Wang Z, Yu Y, Hou Z, Tan G, Liu Q, Wang G. A dual-role of SARS-CoV-2 nucleocapsid protein in regulating innate immune response. Signal Transduct Target Ther. 2021;6:331. doi: 10.1038/s41392-021-00742-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ma J, Zhu F, Zhao M, Shao F, Yu D, Ma J, Zhang X, Li W, Qian Y, Zhang Y, Jiang D, Wang S, Xia P. SARS-CoV-2 nucleocapsid suppresses host pyroptosis by blocking Gasdermin D cleavage. EMBO J. 2021;40:e108249. doi: 10.15252/embj.2021108249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ren Y, Shu T, Wu D, Mu J, Wang C, Huang M, Han Y, Zhang XY, Zhou W, Qiu Y, Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bianchi M, Borsetti A, Ciccozzi M, Pascarella S. SARS-Cov-2 ORF3a: mutability and function. Int J Biol Macromol. 2021;170:820–826. doi: 10.1016/j.ijbiomac.2020.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5:e00266-20. doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Scott C, Griffin S. Viroporins: structure, function and potential as antiviral targets. J Gen Virol. 2015;96:2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- 187.Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JS, Lau YL. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.van den Berg DF, Te Velde AA. Severe COVID-19: NLRP3 inflammasome dysregulated. Front Immunol. 2020;11:1580. doi: 10.3389/fimmu.2020.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Toldo S, Mezzaroma E, Buckley LF, Potere N, Di Nisio M, Biondi-Zoccai G, Van Tassell BW, Abbate A. Targeting the NLRP3 inflammasome in cardiovascular diseases. Pharmacol Ther. 2021;236:108053. doi: 10.1016/j.pharmthera.2021.108053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yap JKY, Moriyama M, Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID-19. J Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zeng J, Xie X, Feng XL, Xu L, Han JB, Yu D, Zou QC, Liu Q, Li X, Ma G, Li MH, Yao YG. Specific inhibition of the NLRP3 inflammasome suppresses immune overactivation and alleviates COVID-19 like pathology in mice. EBioMedicine. 2022;75:103803. doi: 10.1016/j.ebiom.2021.103803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, Butler MS, Haneklaus M, Sutton CE, Nunez G, Latz E, Kastner DL, Mills KH, Masters SL, Schroder K, Cooper MA, O’Neill LA. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Daniels MJ, Rivers-Auty J, Schilling T, Spencer NG, Watremez W, Fasolino V, Booth SJ, White CS, Baldwin AG, Freeman S, Wong R, Latta C, Yu S, Jackson J, Fischer N, Koziel V, Pillot T, Bagnall J, Allan SM, Paszek P, Galea J, Harte MK, Eder C, Lawrence CB, Brough D. Fenamate NSAIDs inhibit the NLRP3 inflammasome and protect against Alzheimer’s disease in rodent models. Nat Commun. 2016;7:12504. doi: 10.1038/ncomms12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dempsey C, Rubio Araiz A, Bryson KJ, Finucane O, Larkin C, Mills EL, Robertson AAB, Cooper MA, O’Neill LAJ, Lynch MA. Inhibiting the NLRP3 inflammasome with MCC950 promotes non-phlogistic clearance of amyloid-beta and cognitive function in APP/PS1 mice. Brain Behav Immun. 2017;61:306–316. doi: 10.1016/j.bbi.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 195.Fan Y, Du L, Fu Q, Zhou Z, Zhang J, Li G, Wu J. Inhibiting the NLRP3 inflammasome with MCC950 ameliorates isoflurane-induced pyroptosis and cognitive impairment in aged mice. Front Cell Neurosci. 2018;12:426. doi: 10.3389/fncel.2018.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fu Q, Li J, Qiu L, Ruan J, Mao M, Li S, Mao Q. Inhibiting NLRP3 inflammasome with MCC950 ameliorates perioperative neurocognitive disorders, suppressing neuroinflammation in the hippocampus in aged mice. Int Immunopharmacol. 2020;82:106317. doi: 10.1016/j.intimp.2020.106317. [DOI] [PubMed] [Google Scholar]
- 197.Vande Walle L, Stowe IB, Sacha P, Lee BL, Demon D, Fossoul A, Van Hauwermeiren F, Saavedra PHV, Simon P, Subrt V, Kostka L, Stivala CE, Pham VC, Staben ST, Yamazoe S, Konvalinka J, Kayagaki N, Lamkanfi M. MCC950/CRID3 potently targets the NACHT domain of wild-type NLRP3 but not disease-associated mutants for inflammasome inhibition. PLoS Biol. 2019;17:e3000354. doi: 10.1371/journal.pbio.3000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.van Hout GP, Bosch L, Ellenbroek GH, de Haan JJ, van Solinge WW, Cooper MA, Arslan F, de Jager SC, Robertson AA, Pasterkamp G, Hoefer IE. The selective NLRP3-inflammasome inhibitor MCC950 reduces infarct size and preserves cardiac function in a pig model of myocardial infarction. Eur Heart J. 2017;38:828–836. doi: 10.1093/eurheartj/ehw247. [DOI] [PubMed] [Google Scholar]
- 199.Cheng P, Yang G, Zhao X, Zeng W, Sun D, Zeng L, Li G, Guan G, Tan J, Zhu C. Precisely and efficiently enzyme response microspheres with immune removal escape loaded with MCC950 ameliorate cardiac dysfunction in acute myocardial infarction. J Biomed Nanotechnol. 2020;16:153–165. doi: 10.1166/jbn.2020.2885. [DOI] [PubMed] [Google Scholar]
- 200.Gao R, Shi H, Chang S, Gao Y, Li X, Lv C, Yang H, Xiang H, Yang J, Xu L, Tang Y. The selective NLRP3-inflammasome inhibitor MCC950 reduces myocardial fibrosis and improves cardiac remodeling in a mouse model of myocardial infarction. Int Immunopharmacol. 2019;74:105575. doi: 10.1016/j.intimp.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 201.Xu KY, Wu CY, Tong S, Xiong P, Wang SH. The selective Nlrp3 inflammasome inhibitor Mcc950 attenuates lung ischemia-reperfusion injury. Biochem Biophys Res Commun. 2018;503:3031–3037. doi: 10.1016/j.bbrc.2018.08.089. [DOI] [PubMed] [Google Scholar]
- 202.Tomar B, Anders HJ, Desai J, Mulay SR. Neutrophils and neutrophil extracellular traps drive necroinflammation in COVID-19. Cells. 2020;9:1383. doi: 10.3390/cells9061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jiao J, Zhao G, Wang Y, Ren P, Wu M. MCC950, a selective inhibitor of NLRP3 inflammasome, reduces the inflammatory response and improves neurological outcomes in mice model of spinal cord injury. Front Mol Biosci. 2020;7:37. doi: 10.3389/fmolb.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Khan N, Kuo A, Brockman DA, Cooper MA, Smith MT. Pharmacological inhibition of the NLRP3 inflammasome as a potential target for multiple sclerosis induced central neuropathic pain. Inflammopharmacology. 2018;26:77–86. doi: 10.1007/s10787-017-0401-9. [DOI] [PubMed] [Google Scholar]
- 205.Lamkanfi M, Mueller JL, Vitari AC, Misaghi S, Fedorova A, Deshayes K, Lee WP, Hoffman HM, Dixit VM. Glyburide inhibits the Cryopyrin/Nalp3 inflammasome. J Cell Biol. 2009;187:61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Li M, Liu H, Shao H, Zhang P, Gao M, Huang L, Shang P, Zhang Q, Wang W, Feng F. Glyburide attenuates B(a)p and LPS-induced inflammation-related lung tumorigenesis in mice. Environ Toxicol. 2021;36:1713–1722. doi: 10.1002/tox.23293. [DOI] [PubMed] [Google Scholar]
- 207.Juliana C, Fernandes-Alnemri T, Wu J, Datta P, Solorzano L, Yu JW, Meng R, Quong AA, Latz E, Scott CP, Alnemri ES. Anti-inflammatory compounds parthenolide and Bay 11-7082 are direct inhibitors of the inflammasome. J Biol Chem. 2010;285:9792–9802. doi: 10.1074/jbc.M109.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Marchetti C, Swartzwelter B, Koenders MI, Azam T, Tengesdal IW, Powers N, de Graaf DM, Dinarello CA, Joosten LAB. NLRP3 inflammasome inhibitor OLT1177 suppresses joint inflammation in murine models of acute arthritis. Arthritis Res Ther. 2018;20:169. doi: 10.1186/s13075-018-1664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sanchez-Fernandez A, Skouras DB, Dinarello CA, Lopez-Vales R. OLT1177 (Dapansutrile), a selective NLRP3 inflammasome inhibitor, ameliorates experimental autoimmune encephalomyelitis pathogenesis. Front Immunol. 2019;10:2578. doi: 10.3389/fimmu.2019.02578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Marchetti C, Swartzwelter B, Gamboni F, Neff CP, Richter K, Azam T, Carta S, Tengesdal I, Nemkov T, D’Alessandro A, Henry C, Jones GS, Goodrich SA, St Laurent JP, Jones TM, Scribner CL, Barrow RB, Altman RD, Skouras DB, Gattorno M, Grau V, Janciauskiene S, Rubartelli A, Joosten LAB, Dinarello CA. OLT1177, a beta-sulfonyl nitrile compound, safe in humans, inhibits the NLRP3 inflammasome and reverses the metabolic cost of inflammation. Proc Natl Acad Sci U S A. 2018;115:E1530–E1539. doi: 10.1073/pnas.1716095115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kluck V, Jansen T, Janssen M, Comarniceanu A, Efde M, Tengesdal IW, Schraa K, Cleophas MCP, Scribner CL, Skouras DB, Marchetti C, Dinarello CA, Joosten LAB. Dapansutrile, an oral selective NLRP3 inflammasome inhibitor, for treatment of gout flares: an open-label, dose-adaptive, proof-of-concept, phase 2a trial. Lancet Rheumatol. 2020;2:e270–e280. doi: 10.1016/s2665-9913(20)30065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Leung YY, Yao Hui LL, Kraus VB. Colchicine-update on mechanisms of action and therapeutic uses. Semin Arthritis Rheum. 2015;45:341–350. doi: 10.1016/j.semarthrit.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mauro AG, Bonaventura A, Vecchie A, Mezzaroma E, Carbone S, Narayan P, Potere N, Cannata A, Paolini JF, Bussani R, Montecucco F, Sinagra G, Van Tassel BW, Abbate A, Toldo S. The role of nlrp3 inflammasome in pericarditis: potential for therapeutic approaches. JACC Basic Transl Sci. 2021;6:137–150. doi: 10.1016/j.jacbts.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 215.Ghaith HS, Gabra MD, Nafady MH, Elshawah HE, Negida A, Kamal MA. A review of the rational and current evidence on colchicine for COVID-19. Curr Pharm Des. 2021 doi: 10.2174/1381612827666211210142352. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 216.Nava F, Ghilotti F, Maggi L, Hatemi G, Del Bianco A, Merlo C, Filippini G, Tramacere I. Biologics, colchicine, corticosteroids, immunosuppressants and interferon-alpha for Neuro-Behcet’s Syndrome. Cochrane Database Syst Rev. 2014:CD010729. doi: 10.1002/14651858.CD010729.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 218.Man SM, Kanneganti TD. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat Rev Immunol. 2016;16:7–21. doi: 10.1038/nri.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Van Opdenbosch N, Gurung P, Vande Walle L, Fossoul A, Kanneganti TD, Lamkanfi M. Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat Commun. 2014;5:3209. doi: 10.1038/ncomms4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang Y, Liu L, Peng YL, Liu YZ, Wu TY, Shen XL, Zhou JR, Sun DY, Huang AJ, Wang X, Wang YX, Jiang CL. Involvement of inflammasome activation in lipopolysaccharide-induced mice depressive-like behaviors. CNS Neurosci Ther. 2014;20:119–124. doi: 10.1111/cns.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dzhidoian Z. The role of interleukin-1 and interleukin-1-receptor antagonist in development of familial mediterranean fever and other autoinflammatory diseases. Georgian Med News. 2011:53–56. [PubMed] [Google Scholar]
- 222.Schrijver HM, Crusius JB, Uitdehaag BM, Garcia Gonzalez MA, Kostense PJ, Polman CH, Pena AS. Association of interleukin-1beta and interleukin-1 receptor antagonist genes with disease severity in MS. Neurology. 1999;52:595–599. doi: 10.1212/wnl.52.3.595. [DOI] [PubMed] [Google Scholar]
- 223.Dinarello CA. The role of the interleukin-1-receptor antagonist in blocking inflammation mediated by interleukin-1. N Engl J Med. 2000;343:732–734. doi: 10.1056/NEJM200009073431011. [DOI] [PubMed] [Google Scholar]
- 224.Lind-Holst M, Hartling UB, Christensen AE. High-dose anakinra as treatment for macrophage activation syndrome caused by refractory Kawasaki disease in an infant. BMJ Case Rep. 2019;12:e229708. doi: 10.1136/bcr-2019-229708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rajasekaran S, Kruse K, Kovey K, Davis AT, Hassan NE, Ndika AN, Zuiderveen S, Birmingham J. Therapeutic role of anakinra, an interleukin-1 receptor antagonist, in the management of secondary hemophagocytic lymphohistiocytosis/sepsis/multiple organ dysfunction/macrophage activating syndrome in critically ill children*. Pediatr Crit Care Med. 2014;15:401–408. doi: 10.1097/PCC.0000000000000078. [DOI] [PubMed] [Google Scholar]
- 226.Shakoory B, Carcillo JA, Chatham WW, Amdur RL, Zhao H, Dinarello CA, Cron RQ, Opal SM. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: reanalysis of a prior phase III trial. Crit Care Med. 2016;44:275–281. doi: 10.1097/CCM.0000000000001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kyriazopoulou E, Huet T, Cavalli G, Gori A, Kyprianou M, Pickkers P, Eugen-Olsen J, Clerici M, Veas F, Chatellier G, Kaplanski G, Netea MG, Pontali E, Gattorno M, Cauchois R, Kooistra E, Kox M, Bandera A, Beaussier H, Mangioni D, Dagna L, van der Meer JWM, Giamarellos-Bourboulis EJ, Hayem G International Collaborative Group for Anakinra in COVID-19. Effect of anakinra on mortality in patients with COVID-19: a systematic review and patient-level meta-analysis. Lancet Rheumatol. 2021;3:e690–e697. doi: 10.1016/S2665-9913(21)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Park A, Iwasaki A. Type I and Type III Interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]


