Abstract
Recognition of invading microbes as non-self is the first step of immune responses. In insects, peptidoglycan recognition proteins (PGRPs) detect peptidoglycans (PGs) of bacterial cell wall, leading to the activation of defense responses. Twelve PGRPs have been identified in the silkworm, Bombyx mori, through bioinformatics analysis. However, their biochemical functions are mostly uncharacterized. In this study, we found PGRP-S5 transcript levels were up-regulated in fat body and midgut after bacterial infection. Using recombinant protein isolated from Escherichia coli, we showed that PGRP-S5 binds to PGs from certain bacterial strains and induces bacteria agglutination. Enzyme activity assay confirmed PGRP-S5 is an amidase; we also showed it is an antibacterial protein effective against both Gram-positive and -negative bacteria. Additionally, we demonstrated that specific recognition of PGs by PGRP-S5 is involved in the prophenoloxidase activation pathway. Together, these data suggest the silkworm PGRP-S5 functions as a pattern recognition receptor for the prophenoloxidase pathway initiation and as an effecter to inhibit bacterial growth as well. We finally discussed possible roles of PGRP-S5 as a receptor for antimicrobial peptide gene induction and as an immune modulator in the midgut.
Keywords: Pattern recognition, Melanization, Insect immunity, Bombyx mori
1. Introduction
Insects and other arthropods rely solely on innate immunity to protect themselves against harmful microbes (Lavine and Strand, 2002; Lemaitre and Hoffmann, 2007). Insect immune system consists of antimicrobial peptide production through IMD and Toll pathways (Hetru and Hoffmann, 2009), phenoloxidase (PO) cascade (Cerenius et al., 2008), and cellular responses such as phagocytosis (Strand, 2008). These pathways and responses are provoked by recognition of the intruders as non-self through interaction between host pattern recognition receptors (PRRs) and pathogen-associated molecular patterns (PAMPs) (Medzhitov and Janeway, 2002). PAMPs are conserved molecules on the microbial surface, such as lipopolysaccharide (LPS) and peptidoglycans (PGs) in bacteria and β-1,3-glucan in fungi. Peptidoglycans are β-1,4-linked N-acetylglucosamine and N-acetylmuramic acid polymers crosslinked by short peptides. The crosslinking peptides vary in amino acid residues and in the linkage of stem peptides in different bacterial species. In Gram-negative bacteria and the Gram-positive genera Bacillus and Clostridium, the glycan chains are directly crosslinked by meso-diaminopimelic acid (DAP) containing peptides. In most Gram-positive bacteria, PGs have an interpeptide bridge and a lysine instead of DAP in the stem peptide (Kurata et al., 2006).
Peptidoglycan recognition proteins (PGRPs) are a family of PRRs in innate immune system, which was first purified from the silkworm Bombyx mori (Yoshida et al., 1996). The silkworm PGRP binds to Lys-type PG from Micrococcus luteus and is involved in prophenoloxidase (proPO) activation (Yoshida et al., 1996). Its mRNA is expressed in fat body, hemocytes and epidermal cells and is up-regulated in fat body by injected bacteria (Ochiai and Ashida, 1999). Using ‘PGRP’ to search the NCBI Unigene database, we retrieved over a hundred entries, over half of which are from insects. All PGRPs have a highly conserved carboxyl-terminal PGRP domain (~160 amino acid residues in length) homologous to bacteriophage T7 lysozyme (Ochiai and Ashida, 1999; Kang et al., 1998), while the amino-terminal region is unique for a given PGRP with no or low similarity to other PGPR members. Drosophila genome contains thirteen PGRP genes encoding nineteen proteins through alternative splicing (Werner et al., 2000). A recent review summarized the structures and functions of these PGRPs in Drosophila immune system (Kurata, 2014). Drosophila PGRP-SA and -SD circulate in hemolymph, detect Lys-type PGs of most Gram-positive bacteria, and activate a serine protease cascade to induce antimicrobial peptide gene transcription through the Toll pathway (Michel et al., 2001; Bischoff et al., 2004; Charroux et al., 2009). PGRP-LC and -LE are receptors in the IMD signaling pathway (Choe et al., 2002; Gottar et al., 2002; Ramet et al., 2002; Takehana et al., 2002, 2004). Membrane-bound PGRP-LCx plays a key role in sensing DAP-type PGs of Gram-negative bacteria and Gram-positive Bacilli, while PGRP-LCa/x heterodimers recognize tracheal cytotoxin (TCT) of Gram-negative bacteria (Neyen et al., 2012). Cytosolic PGRP-LE detects TCT and forms homodimers to trigger the IMD pathway (Lim et al., 2006). In an intracellular bacteria Listeria monocytogenes-Drosophila infection model, PGPR-LE plays a crucial role as the receptor for autophagy and is responsible for induced synthesis of an antibacterial peptide listericin (Yano and Kurata, 2008; Goto et al., 2010). PGRP-LC and -LE cooperatively contribute to gut immune response, with a master role of PGRP-LE that induces limited responses to pathogenic bacteria and tolerance to microbiota in the intestine (Neyen et al., 2012; Bosco-Drayon et al., 2012). PGRP-LC and PGRP-SC1 are involved in phagocytosis as well, though the mechanisms are still unknown (Ramet et al., 2002; Garver et al., 2006). Besides the roles as a receptor to detect PG from bacteria, PGRPs also function as regulators to modulate immune responses. PGRP-LA is primarily expressed in barrier epithelia and positively regulates the IMD pathway (Gendrin et al., 2013). PGRP-LF acts as a specific negative regulator of IMD pathway, probably through competition with PGRP-LCa for the binding to PGRP-LCx (Maillet et al., 2008; Basbous et al., 2011). Owing to the N-acetylmuramoyl-L-alanine amidase activity that hydrolyzes the amide bond between MurNAc and L-alanine in PG, PGRP-SC1/2 and -LB may function as scavengers to break down PGs and thus down-regulate inflammatory responses via IMD pathway (Mellroth et al., 2003; Bischoff et al., 2006; Zaidman-Rémy et al., 2006). PGRP-SB1 shows bactericidal effect stemmed from its amidase activity, while its role in immune system needs further study (Mellroth and Steiner, 2006; Zaidman-Rémy et al., 2011).
There are twelve PGRP genes in the B. mori genome (Tanaka et al., 2008). PGRP-S1, the first PGRP discovered by Yoshida et al. (1996), is functionally characterized. In this study, we produced the recombinant PGRP-S5 and investigated its PG-binding specificity, amidase activity, antibacterial activity, and function in immune response. Our results provided evidence that PGRP-S5 plays dual roles in the silkworm, serving as a receptor for the proPO activation pathway and as an antibacterial protein.
2. Materials and methods
2.1. Insect rearing, bacterial challenge, and tissue collection
Silkworm (Nistari) eggs were kindly provided by Dr. Erjun Ling in Shanghai Life Science Institute. Newly hatched larvae were reared to 5th instar on fresh mulberry leaves at 27 ± 1 °C, photoperiod 14L:10D, and 65 ± 10% relative humidity. The indigenous bacteria were eliminated by 5 μl (10 μg/μl) tetracycline in diet block (4 mm × 4 mm × 2 mm) for each 5th instar larva (day 3) before oral infection. The bacteria Pseudomonas aeruginosa and Staphylococcus aureus were cultured overnight in Luria-Bertani (LB) medium with constant shaking at 37 °C and then harvested by centrifugation at 8000g for 10 min. Infection was conducted by feeding each silkworm on artificial diet containing sterilized 0.85% NaCl (negative control), 2 × 109 P. aeruginosa or 1.4 × 108 of S. aureus in the sterilized saline. Each treatment group included 15 larvae. At 8 and 24 h after the larvae consumed the diet blocks, all treated insects from each group were dissected with midgut tissue collected and stored in Trizol reagent (Invitrogen) at −80°C. For collecting fat body and hemocytes from challenged larvae, 1 × 107 cells of bacteria in 50 μl sterilized saline were injected into hemocoel of each larva. The controls were injected with 50 μl sterilized saline. At different time points after injection, 15 larvae from each group were dissected to collect hemolymph and fat body. Cell-free hemolymph was removed after centrifugation at 500g for 10 min at 4 °C. The hemocyte pellets and fat body samples were treated with Trizol reagent and stored at −80 °C.
2.2. RNA isolation and cDNA preparation
The midgut, fat body and hemocyte samples were homogenized by a pellet pestle (Kontes) and total RNA of each tissue was extracted using Trizol reagent according to the protocol. The total RNA was purified by RNeasy MinElute Cleanup Kit (Qiagen) with genomic DNA removed by DNase treatment (Promega). First-strand cDNA was synthesized from purified RNA (1 μg) using superscript™ III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. A260 and A280 of each sample were determined on a Biophotometer (Eppendorf) to calculate concentrations and A260/A280 values. The cDNA samples were diluted to 50 ng/μl with deionized water and employed as templates in quantitative real-time (QRT)-PCR analysis.
2.3. QRT-PCR analysis
Two BmPGRP-S5 gene-specific primers 5′-TGACTTCTGCCGACCTGACAC-3′ and 5′-TTTCCATCCATTGCCACACACC-3′ were used to amplify a product of 172 bp. A 192 bp cDNA fragment of the house-keeping gene, IF4A (DQ443290.1), was amplified as an internal control to normalize the transcript level of PGRP-S5 using 5′-TCTGGCATCATACCTTCTACAA-3′ and 5′-TCTGTGTCATCTTTTCCCTGTT-3′ (Wu et al., 2010). QRT-PCR was performed in a total volume of 20 μl containing 10 μl of 2× FastStart Essential DNA Green Master (Roche), 1 μl of cDNA, 1 μl of each primer (10 μM) and 7 μl of double distilled water. QRT-PCR was conducted under the following conditions: initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 20 s, and extension at 72 °C for 30 s. Melt curve (55–95 °C) was determined to confirm the amplification of specific PCR product (Bio-Rad IQ5). Standard curves of the two genes (BmPGRP-S5 and IF4A) were measured to find out the amplification efficiency and r2 value. The relative quantitative method (2−ΔΔCt) was used to determine the expression level changes (Schmittgen and Livak, 2008). The data were plotted using Prism 5.0 (GraphPad Software, Inc) and analyzed by Student’s t-test.
2.4. Cloning, expression and purification of BmPGRP-S5
The entire PGRP-S5 coding region was amplified by PCR under the same conditions (Section 2.3) using the following primers (forward: 5′-GCATATGCATCCTCGGCTTATCGAAAA-3′ with NdeI site; reverse: 5′-GCTCGAGTGTCTGTTTATTTAGTTCTC with XhoI site). The PCR product was isolated from gel and purified by Gel Extraction (Omega A). The fragment was inserted into pMD19-T vector (TaKaRa) and then transformed into Escherichia coli JM109. The plasmid was extracted by Plasmid Mini Kit (Omega A) and confirmed by sequencing (Shanghai Sunny Biotechnology Co).
A modified pET28b was used to express, purify, and detect PGRP-S5, which encodes a small ubiquitin-like modifier (SUMO) led by an amino-terminal hexahistidine affinity tag. The 106-residue peptide domain, encoded by an NcoI-NdeI fragment, was employed to increase solubility of recombinant protein and can be removed by a protease that specifically recognizes the tertiary structure of SUMO. Two oligonucleotides (5′-TATCGATTACAAGGATGACGACGATAAG CATATG-3′ and 3’-AGCTAATGCTTCTACTGCTGCTATTCGTATACCTAG-5′, encoding the FLAG tag DYKDDDDK) were phosphorylated, annealed, and inserted between the NdeI and BamHI sites. The original NdeI site was eliminated by the mutation (CATATC) and a new NdeI site was added in front of the BamHI site. Similarly, a c-Myc tag was introduced between HindIII and XhoI sites of the vector using 5′-AGCTTCTCGAGCAGAAGCTCATCTCTGAAGAGGATCTGTAGT-3′ and 3′-AGAGCTCGTCTTCGAGTAGAGACTTCTCCTAGACATCAAGCT-5′ (encoding EQKLISEEDL*). The original XhoI site was eliminated by the mutation (TTCGAG) and a new XhoI site is added next to the HindIII site. In-frame cloning of the coding sequence into the NdeI and XhoI sites of pSFM (standing for SUMO, FLAG, c-Myc) would render recombinant fusion proteins detected by commercially available monoclonal antibodies against either tag. After sequencing verification, pSFM was double digested with NdeI and XhoI, ligated with the PGRP-S5 cDNA insert retrieved from pMD19-T, and then transformed into E. coli BL21.
The E. coli BL21 cells were inoculated into LB medium containing kanamycin (50 μg/ml), grown to the logarithmic phase at 37 °C with shaking at 240 rpm, and then induced by IPTG at a final concentration of 0.5 mM for 10 h at 25 °C with shaking at 150 rpm. The bacteria were harvested by centrifugation at 8000g for 10 min at 4°C, suspended in phosphate buffered saline (PBS, pH 7.4) at 5 ml per gram of wet bacteria, incubated with lysozyme at a final concentration of 0.1 μg/ml for 30 min at room temperature, and finally lysed by ultrasonication (130 Watt, 20 kHz, 10 × 10 s). The BmPGRP-S5 was first purified by affinity chromatography on a Ni2+-NTA agarose column (Qiagen). The purified protein was digested by SUMO protease (GeneCopoeia) at 4°C overnight and purified on the Ni2+-NTA column again to remove cleaved SUMO and uncleaved fusion protein. To detect induced expression of PGRP-S5 and monitor its purification, proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie Brilliant Blue R250 and immunoblot analysis using anti-c-Myc (1:20,000 dilution, Cwbio) as primary antibody and goat-anti-rabbit IgG (1:20,000 dilution, Santa Cruze) as secondary antibody, and then mass spectrometry was used to verify the purified recombinant PGRP-S5.
2.5. Binding of BmPGRP-S5 to PGs
Insoluble PGs from M. luteus, S. aureus, Bacillus megaterium, and Bacillus subtilis were prepared as described previously (Sumathipala and Jiang, 2010). E. coli PG was purchased from InvivoGen. PGs (10 μl, 10 μg/μl) or 10 μl PBS was incubated with the purified PGRP-S5 (20 μl, 0.2 μg/μl) at room temperature for 2 h. The mixture was centrifuged at 4 °C, 10,000g for 15 min. The supernatant was saved as unbound sample; PBS mixed with PGRP-S5 was regarded as control of total proteins. Then the pellet was spun down after being washed with 30 μl of 50 mM NaCl, 150 mM NaCl, and 500 mM NaCl in PBS sequentially, and the 150 mM NaCl supernatant was taken as the wash sample. Finally, the pellet was suspended with 30 μl PBS as the bound sample. The four samples were treated with 10 μl 4 × SDS sample buffer at 95 °C for 5 min. After centrifugation at 13,000g for 5 min, the samples (10 μl) were separated by 12% SDS-PAGE followed by immunoblot analysis using anti-c-Myc and goat-anti-rabbit IgG as described in Section 2.4.
2.6. Bacterial agglutination assay
M. luteus, P. aeruginosa, S. aureus, E. coli and Serratia marcescens were used to test possible role of PGRP-S5 in bacterial agglutination. The bacteria in mid-logarithmic phase were harvested and diluted with PBS to 2 × 108 cells/ml (M. luteus, E. coli, P. aeruginosa) and 1 × 107 cells/ml (S. aureus, S. marcescens). The diluted bacteria suspensions (10 μl) were separately mixed with 10 μl PGRP-S5 (200 μg/ml) or BSA (200 μg/ml), incubated in a 96-well plate at room temperature for 2 h, and observed under an inverted fluorescence microscope (AMG, EVOS FL).
2.7. Amidase activity determination
Insoluble PGs from E. coli, M. luteus, B. subtilis, S. aureus and B. megaterium were suspended in PBS and briefly sonicated. Each PG sample (10 μl, 1 mg/ml) was incubated with 50 μl PGRP-S5 (40 μg/ml) or 50 μl lysozyme (40 μg/ml) in PBS at room temperature on a Tube Tumbler (Select Bioproducts). The decay of light scattering at 405 nm was measured after 0, 1, 2, 4, and 8 h on a Biophotometer (Eppendorf).
2.8. Antibacterial activity assay
M. luteus, S. aureus, S. marcescens and E. coli cultures were grown to OD600 0.6 in LB medium at 37 °C with shaking. Bacteria in 200 μl cultures were harvested by centrifugation at 8000g for 10 min at 4 °C and resuspended in 200 μl of PBS. After 1:100 dilution, the bacterial suspensions (10 μl) were mixed with 25 μl, 40 μM ZnCl2 in PBS or 200 μg/ml PGRP-S5 in the same buffer in 1.5 ml tubes. After shaking on a Tube Tumbler for 5 h at room temperature, the mixtures were individually mixed with 1 ml LB and cultured at 37 °C overnight prior to absorbance measurement at 600 nm.
2.9. Prophenoloxidase activation
Day 1, 5th instar larvae were placed on the ice and surface disinfected with 70% ethyl alcohol and hemolymph was collected from cut caudal horn, and the hemolymph was centrifuged at 16,000g for 30 s to remove hemocytes. Four reactions were prepared: (1) 15 μl PBS and 5 μl plasma; (2) 2 μl PGRP-S5 (200 μg/ml), 5 μl plasma and 13 μl PBS; (3) 5 μl plasma and 13 μl PBS; (4) 2 μl PGRP-S5 (200 μg/ml), 5 μl plasma and 11 μl PBS. After incubation on ice for five min, the reaction mixtures (3) and (4) were reacted with 2 μl PG (1 mg/ml). After incubation on ice for two more minutes, the reaction mixtures were centrifuged at 1000g for 1 min. The supernatants (5 μl) were individually transferred to microplate wells and mixed with 100 μl of 2 mM dopamine in 50 mM sodium phosphate, pH 6.5 for PO activity determination at 490 nm on a plate reader (Jiang et al., 2003).
3. Results
3.1. PGRP-S5 sequence analysis, expression profiling upon bacterial infection, and recombinant protein production
From the silkworm cDNA database (http://sgp.dna.affrc.go.jp/FLcDNA), we identified a cDNA sequence (accession number: NM_001043393) for PGRP-S5 (Tanaka et al., 2008; Suetsugu et al., 2013). The first part of the cDNA (nucleotides 1–102) represent exon 1 including a 5′ UTR and MFLSFCIFIVFCAYTSSHPRLIEK coding region. The underlined sequence represents a predicted 17-residue signal peptide. The remaining cDNA sequence (nucleotides 103–673) corresponds to exons 2–4 coding for DHLSVDFPVCSRDCWGA… GALLEEVSTWDNYHPGHVNFRELNKQTKF*, where the omitted part is a 191-residue PGRP domain. The open reading frame is directly followed by a poly(A) tail since there is a polyad-enylation signal AATAAA encoding NK near the carboxyl terminus. In the structure of Drosophila PGRP-LB (an active amidase), His42, Tyr78, His152 and Cys160 interact with zinc and Thr158 is a surface residue at the active site for PG binding (Kim et al., 2003). In Drosophila PGRP-SA (non-catalytic), the replacement of His152 and Cys160 (PGRP-LB numbering) by Gly150 and Ser158 result in the loss of zinc binding activity (Reiser et al., 2004). Sequence alignment shows that the five conserved residues critical for zinc binding and PG interaction in Drosophila PGRP-LB are present in silkworm PGRP-S5 (Fig. 1A). Phylogenetic analysis revealed that silkworm PGRP-S5 was grouped with Drosophila PGRP-LB and Manduca sexta PGRP-2 with the T7 lysozyme as the root (Fig. 1B) suggesting B. mori PGRP-S5 may have amidase activity.
Fig. 1.
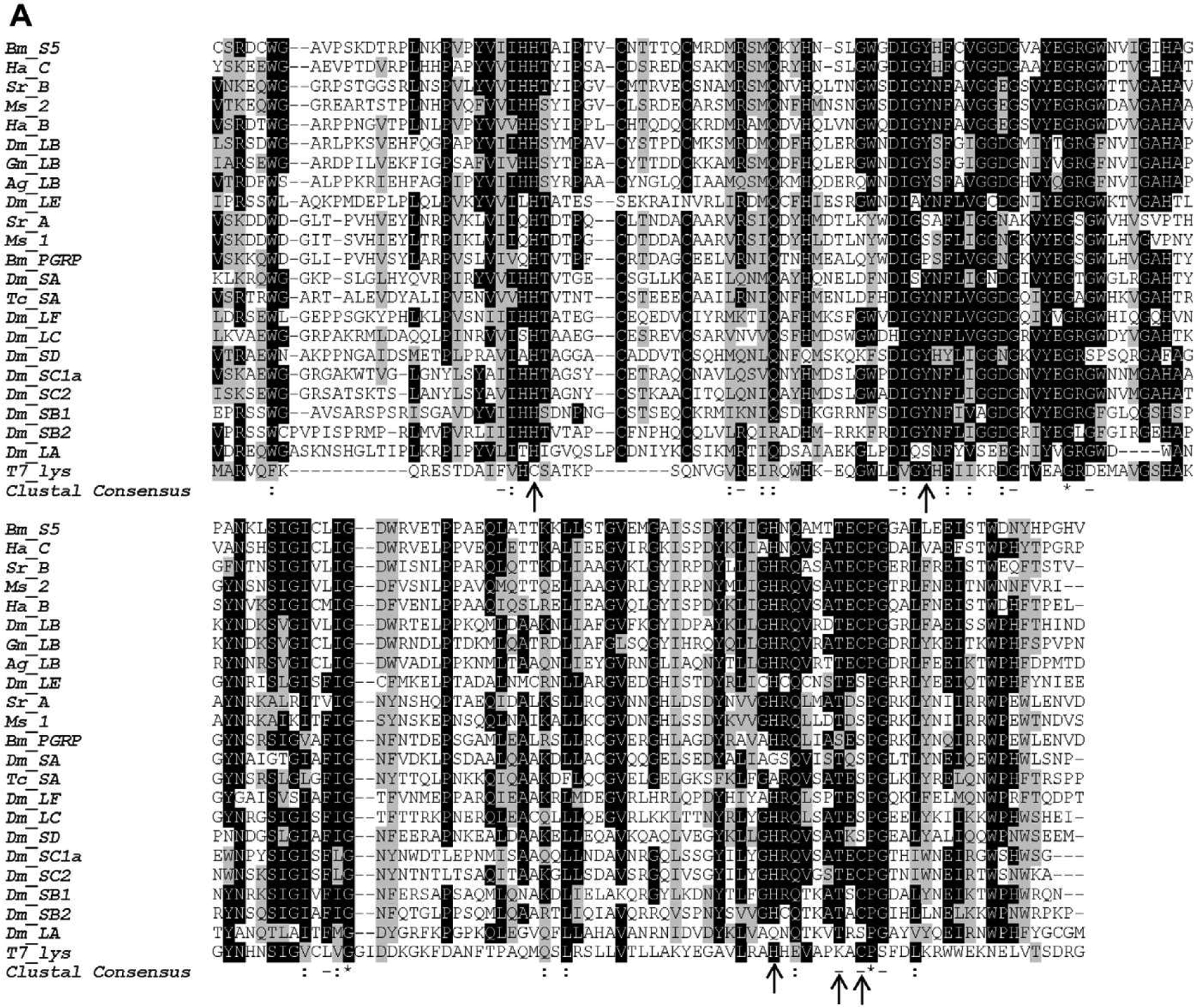

Multiple PGRP sequence alignment using ClustalW (A) and construction of phylogenetic tree (B). Identical and similar amino acid residues are shaded in black and gray, respectively. The residues responsible for zinc binding and amidase are indicated with arrows. The scale bar on the bottom of the tree indicates the distance. Numbers on the branches represent the distances. Bm, B. mori (PGRP-S5: NP_001036858.1; PGRP: BAA77209); Ha, Helicoverpa armigera (PGRP-B: AFP23116; PGRP-C AFP23117); Sr, Samia Cynthia ricini (PGRP-A: BAF03522; PGRP-B: BAF03520); Ms, M. sexta (PGRP-1: AF413068; PGRP-2: ACX49764); Dm, D. melanogaster (PGRP-LA: CG32042; PGRP-LB: CG14704; PGRP-LC: CG4432; PGRP-LE: CG8995; PGRP-LF: CG4437; PGRP-SA: CG11709; PGRP-SB1: CG9681; PGRP-SB2: CG9697; PGRP-SC1a: CG14746; PGRP-SC2: CG14745; PGRP-SD: CG7496); Gm, Glossina morsitans (PGRP-LB: ABC25064); Ag, Anopheles gambiae (PGRP-LB: XP_003435776); Tc, Tribolium castaneum (PGRP-SA: XP_969883).
To investigate the role of BmPGRP-S5 in silkworm immunity, we first examined the change of its mRNA level by quantitative real-time PCR after injection of live bacteria into hemocoel or through natural oral infection. In fat body, injection of P. aeruginosa caused BmPGRP-S5 transcription level increased significantly until 48 hpi; S. aureus injection caused the increase until 24 hpi (Fig. 2A). In hemocytes, injection of P. aeruginosa led to an increase from 4 to 24 hpi, whereas S. aureus injection induced BmPGRP-S5 expression from 4 to 8 hpi (Fig. 2B). After we challenged silkworm larvae through oral route, the PGRP-S5 transcript level in midgut increased remarkably in response to S. aureus from 8 to 16 hpi; P. aeruginosa ingestion did not induced a significant change in the PGRP-S5 mRNA levels on the contrary (Fig. 2C).
Fig. 2.
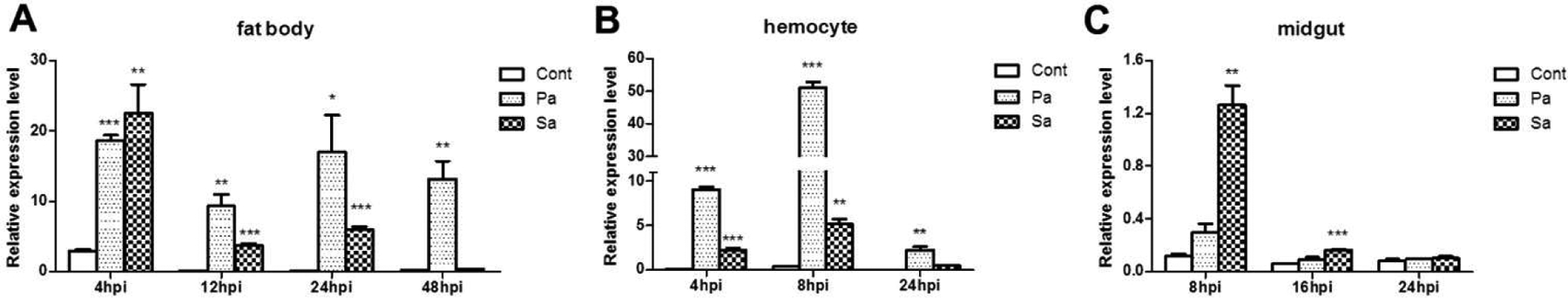
Changes in BmPGRP-S5 mRNA levels in response to sterile 0.85% NaCl (control), P. aeruginosa (Pa), or S. aureus (Sa) in fat body after injection (A), in hemocytes after injection (B), and in midgut after oral feeding (C). PCR amplification efficiency and correlation coefficient (r2) are 99.8% and 0.994% for BmPGRP-S5; 100.7% and 0.995 for IF4A. Asterisks indicate significant differences: *p < 0.05; **p < 0.01 ; ***p < 0.001 for pairwise comparisons by Student’s t-tests between control (Cont) and Pa or Sa infected for the same hours post infection (hpi).
To produce sufficient protein for functional study, we cloned the entire open reading frame, expressed the mature PGRP-S5 (HPR…NKQT) in E. coli, and purified the recombinant protein. The cDNA was inserted into the pSFM expression vector, which has a 6 × His tag followed by a SUMO cleavable tag at the amino-terminus and FLAG and c-Myc tags flanking the NdeI and XhoI sites. The soluble fusion protein was produced at a high level in E. coli. After we isolated the fusion protein using Ni2+-NTA column, SUMO protease was used to cleave the 6 × His tagged SUMO and release PGRP-S5 with the FLAG and c-Myc tags at its amino and carboxyl termini, respectively. Since amino-terminal fusion partner has a 6 × His tag, we used Ni2+-NTA column again to capture the released SUMO, and the PGRP-S5 (HIDYKDDDDKHMHPR-LIEK… ELNKOTLEOKLISEEDL* was present in the flow-through fraction. The carboxyl terminal c-Myc tag was used to detect recombinant PGRP-S5 with c-Myc antibody by immunoblotting. The purified fusion protein and PGRP-S5 were shown in Fig. 3A and B. Mass spectrometry analysis confirmed 67% of the recombinant PGRP-S5 sequence (Fig. 3C).
Fig. 3.
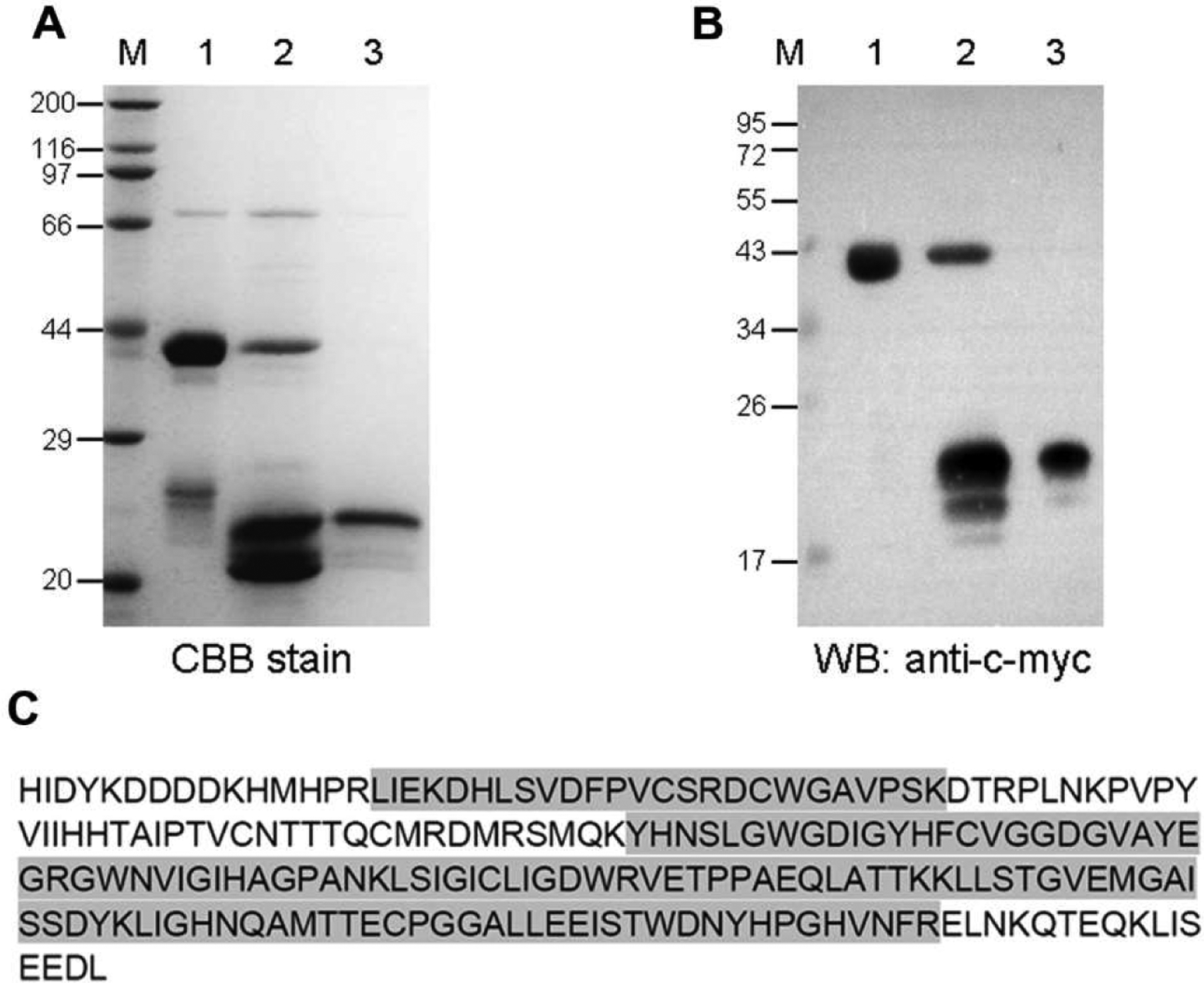
Purification and identification of the recombinant BmPGRP-S5. (A) Coomassie Brilliant Blue (CBB) stained gel; (B) Western blot (WB) probed with anti-c-Myc antibodies. M, protein size markers with their molecular masses (kDa) labeled on the left. Lane 1, purified fusion protein; lane 2, fusion protein digested with 6 × His-tagged SUMO protease (20 kDa); lane 3, purified PGRP-S5 fused with FLAG and c-Myc tags. (C) Mass spectrometry analysis of the purified protein with the matched regions shaded.
3.2. BmPGRP-S5 binds to PGs and causes bacteria agglutination
Using purified PGs from the bacteria, we tested binding of the recombinant PGRP-S5 to these insoluble polysaccharides (Fig. 4A). PGRP-S5 showed strong affinity to DAP-PGs from E. coli, B. megaterium and B. subtilis, Lys-PG from M. luteus, and weak binding to Lys-PG from S. aureus. Furthermore, we examined if the PGRP-S5 can agglutinate bacteria. Compared to bovine serum albumin, the recombinant PGRP-S5 caused pronounced agglutination of E. coli and P. aeruginosa (Fig. 4B), but the agglutination of M. luteus, S. marcescens or S. aureus was negligible (data not shown).
Fig. 4.
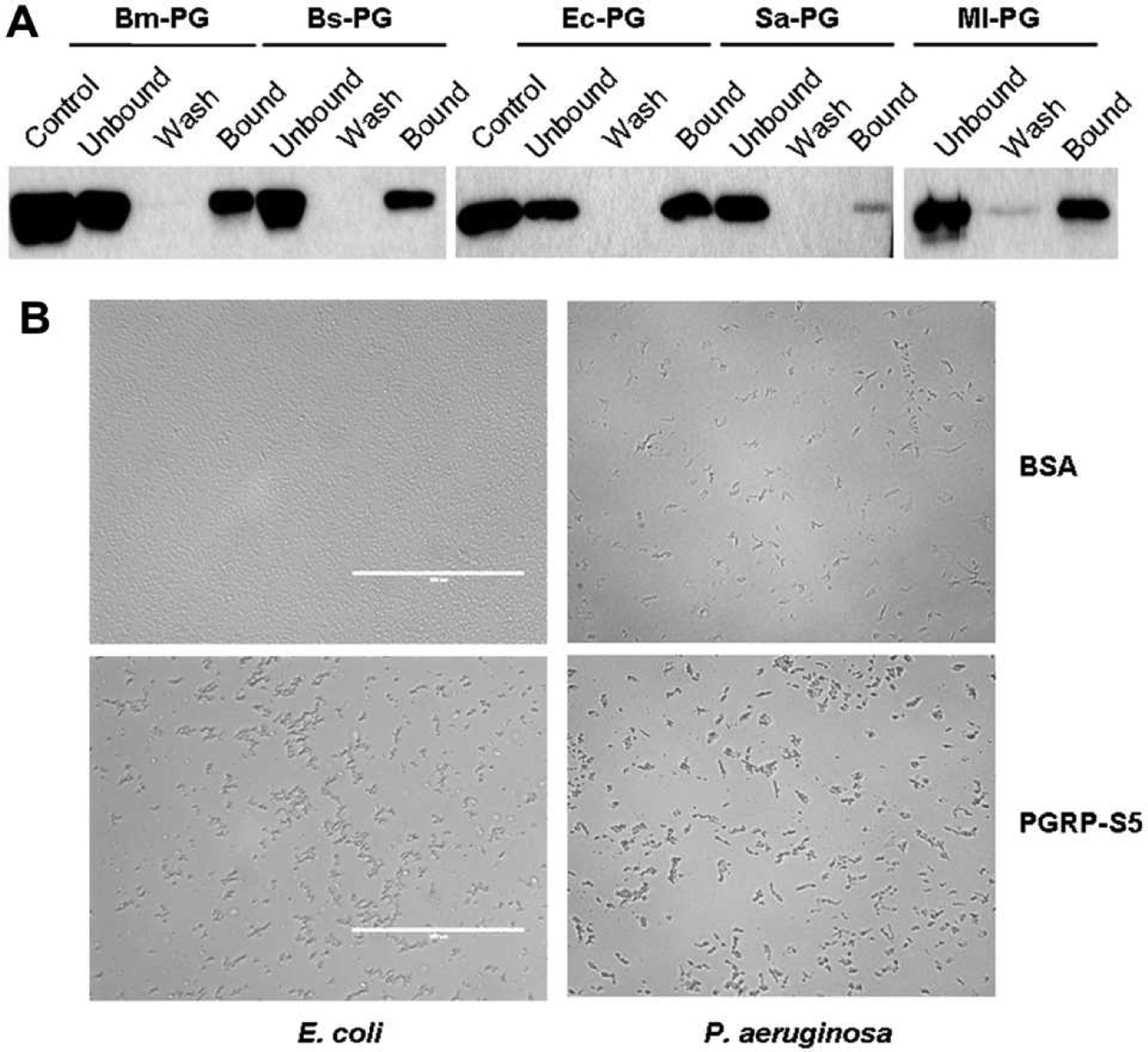
Binding of BmPGRP-S5 to PGs (A) and bacterial agglutination caused by BmPGRP-S5 (B). As described in Section 2.4, the total (i.e., control), unbound, wash, and bound fractions were separated by SDS-PAGE and visualized by immunoblotting using anti-c-Myc as the primary antibody. Bm-PG, Bs-PG, Sa-PG, MI-PG and Ec-PG: PGs from B. megaterium, B. subtilis, S. aureus, M. luteus and E. coli, respectively. Bacterial suspension (10 μl) was incubated with the purified PGRGP-S5 (10 μl, 200 μg/ml) for 2 h and then photographed under microscope (40×).
3.3. BmPGRP-S5 shows amidase and antibacterial activity
Sequence analysis implied the PGRP-S5 has an amidase activity (Fig. 1A). We found that the purified PGRP-S5 displayed equivalent activity of egg lysozyme towards PGs from B. megaterium, S. aureus, B. subtilis, E. coli, and M. luteus. We also observed that Zn2+ may enhance PGRP-S5 amidase activity to PGs from B. megaterium, B. subtilis and E. coli, but not to PGs from M. luteus and S. aureus (Fig. 5).
Fig. 5.
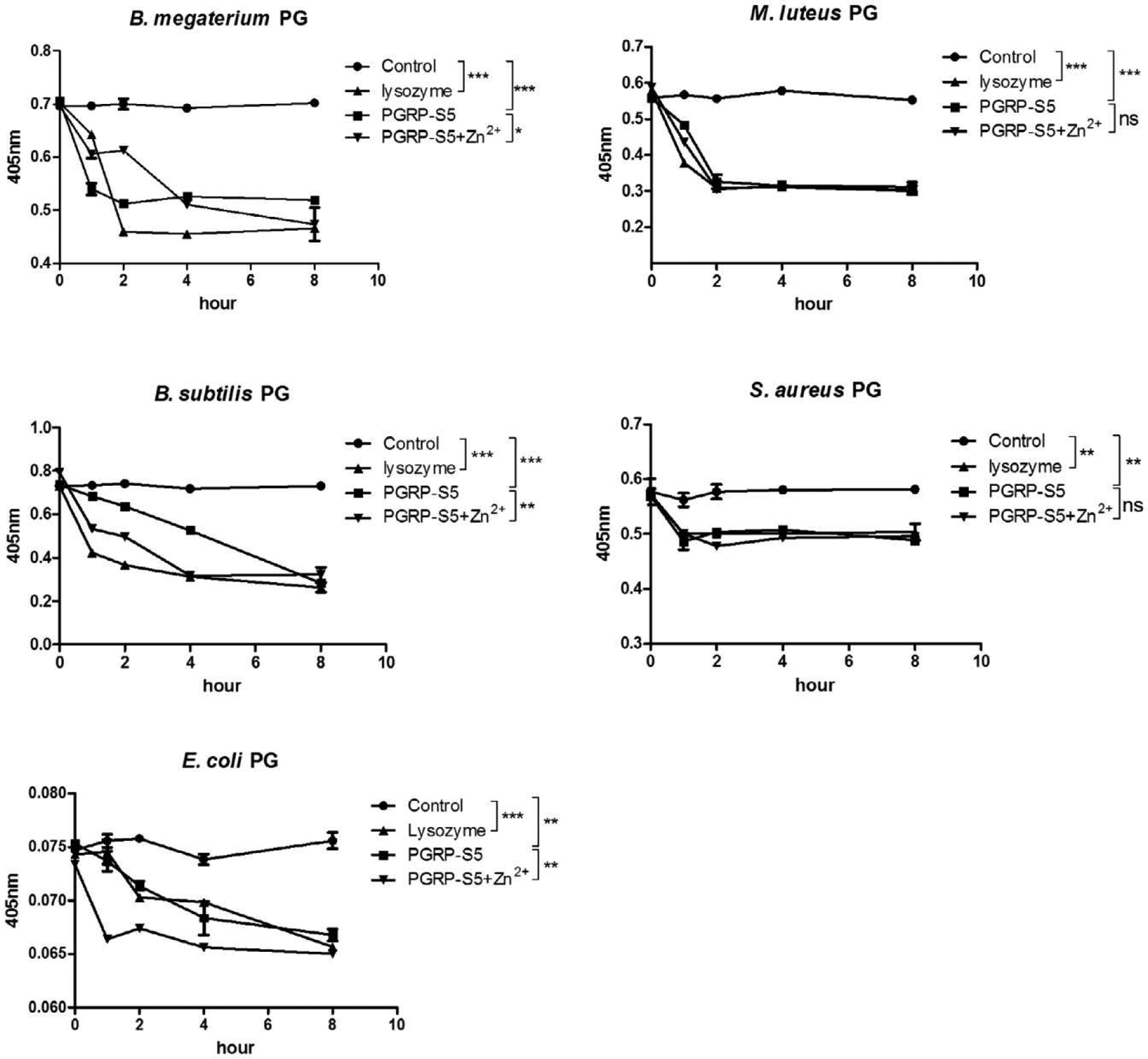
Hydrolysis of PGs by BmPGRP-S5 or lysozyme. Insoluble PGs from B. megaterium, B. subtilis, E. coli, S. aureus, and M. luteus were separately incubated with buffer (i.e., control), BmPGRP-S5 (with or without Zn2+), or lysozyme. Light scattering at 405 nm was monitored for 8 h. Asterisks indicate significant differences: *p < 0.05; **p < 0.01; ***p < 0.001 for pairwise comparisons by Student’s t-tests. ns, not significant.
We also demonstrated antibacterial activity of BmPGRP-S5 (Fig. 6). The recombinant protein alone exhibited strong antibacterial activity to Gram-positive bacteria M. luteus and S. aureus, Gram-negative bacteria S. marcescens and E. coli. When we supplemented zinc ions in the reaction systems, we found Zn2+ itself had no or negligible antibacterial effect. However, zinc ions significantly enhanced PGRP-S5 antibacterial activity. This observation suggested that PGRP-S5 antibacterial activity comes from zinc dependent and independent mechanisms.
Fig. 6.
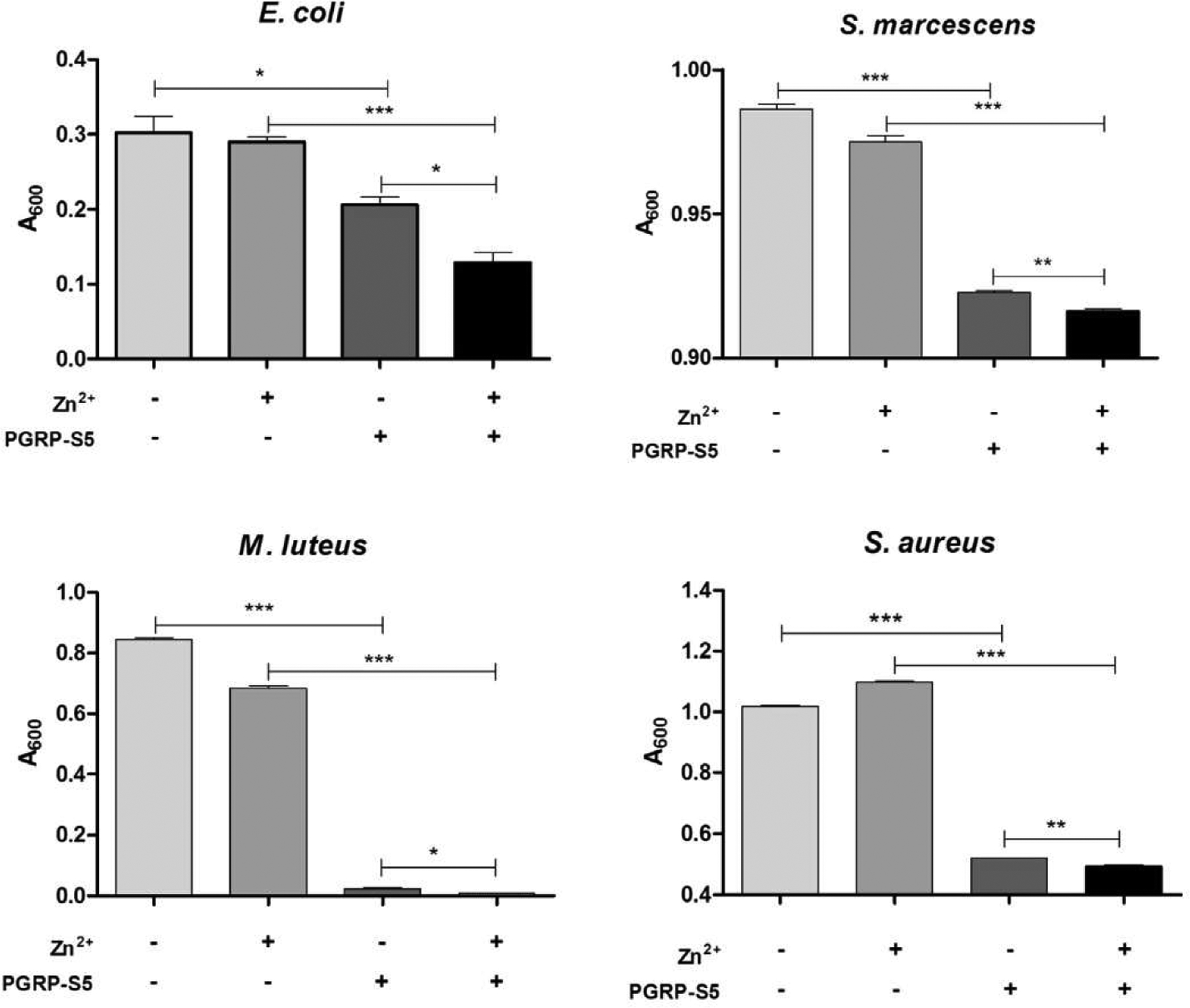
Antibacterial activity of BmPGRP-S5. Bacteria were separately incubated with the purified PGRP-S5, Zn2+, or both for 5 h at room temperature then cultured in LB overnight. Turbidity of the cultures was measured at 600 nm. Asterisks indicate significant differences: *p < 0.05; **p < 0.01; ***p < 0.001 for pairwise comparisons by Student’s t-tests.
3.4. BmPGRP-S5 is involved in proPO activation
Next we asked the question what immune responses might be activated by the interaction between PGRP-S5 and PG. By in vitro reconstituting the proPO activation system, we found that recombinant PGRP-S5 or tested PG from different bacterial strains alone increased PO activity. Incubation of silkworm plasma (without hemocytes) with both PGRP-S5 and PG from E. coli, S. aureus, B. megaterium, M. luteus, or B. subtilis boosted proPO activation considerably (Fig. 7).
Fig. 7.
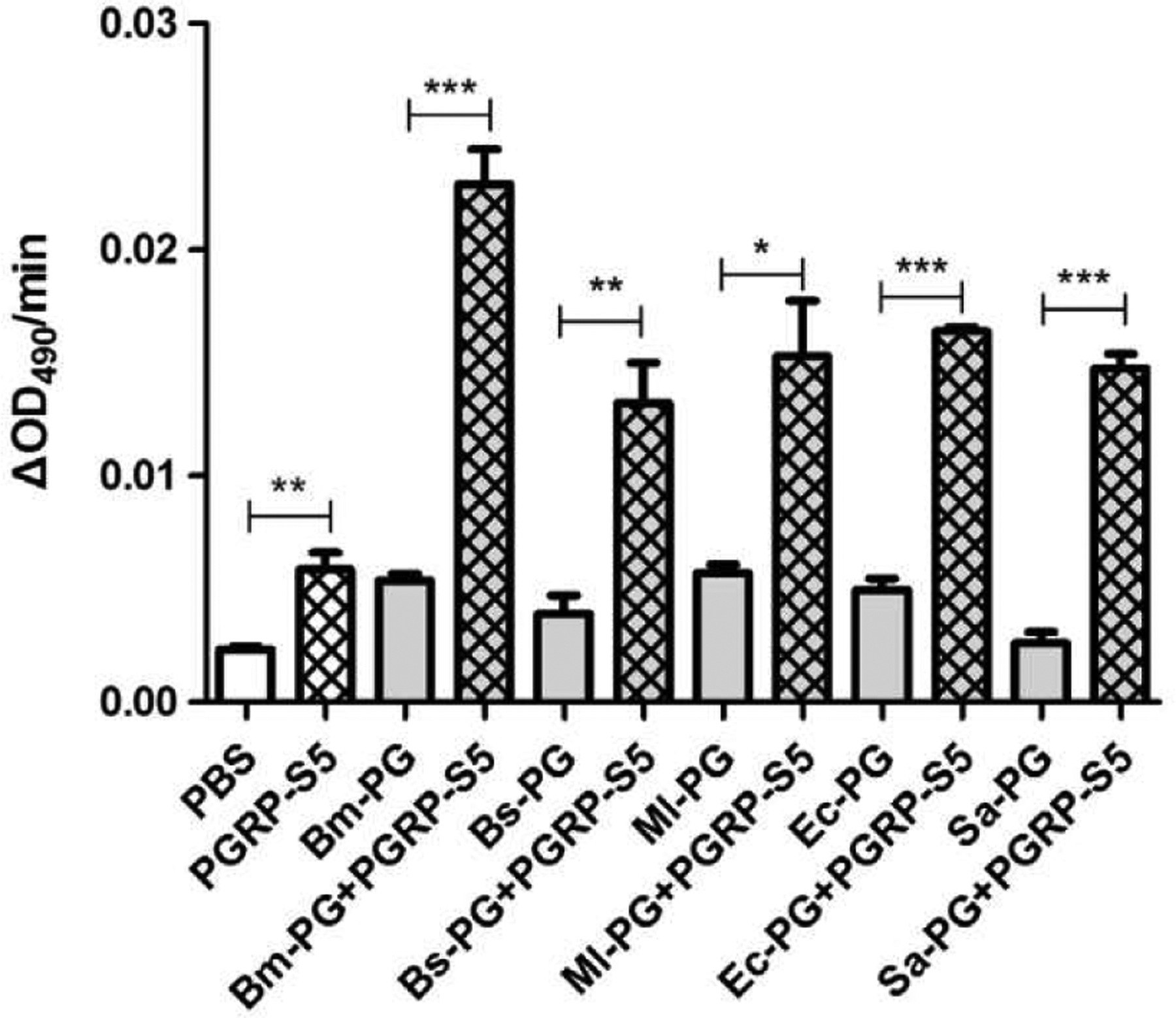
Role of BmPGRP-S5 in the silkworm proPO activation pathway. Silkworm plasma was incubated with the recombinant PGRP-S5, PG, or both. PGs were added after plasma had been incubated with PGRP-S5 for 5 min at room temperature. Two minutes after PG addition, PO activity of the reactions was measured using dopamine as substrate. Asterisks indicate significant differences: *p < 0.05; **p < 0.01; ***p < 0.001 for pairwise comparisons by Student’s t-tests.
4. Discussion
In the present study, we found PGRP-S5 transcription was up-regulated significantly in the silkworm fat body and midgut after bacterial infection. Using the recombinant PGRP-S5, we further demonstrated that BmPGRP-S5 was an active amidase with anti-bacterial activity. More importantly, we revealed that PGRP-S5 was involved in proPO activation pathway through interaction with strain specific PG. However, there are two questions remained to be answered in the future.
4.1. PGRP-S5 serves as a receptor or an effecter?
As a family of pattern recognition proteins, PGRPs serve as receptors to detect DAP-type or Lys-type PGs of different bacteria and the recognition activates immune pathways, including IMD, Toll and autophagy in the fruit fly (Charroux et al., 2009) and proPO pathways in the silkworm, beetle and tobacco hornworm (Yoshida et al., 1996; Park et al., 2007; Sumathipala and Jiang, 2010). Some PGRPs exhibit antibacterial activity. These antibacterial PGRPs include Drosophila PGRP-SB1 (Mellroth and Steiner, 2006; Zaidman-Rémy et al, 2011), bumblebee PGRP-S (You et al., 2010), cotton bollworm PGRP-B and -C (Yang et al., 2013), rockfish PGRP-L2 (Kim et al., 2010), zebra fish PGRPs (Li et al., 2007), and mammalian PGRPs (Dziarski and Gupta, 2010). Drosophila PGRP-SB1/2 null mutation did not alter antimicrobial peptide gene expression after immune challenges (Zaidman-Rémy et al., 2011). Similarly, PGRP-S knock-down did not affect antimicrobial peptide genes expression in bumblebee (You et al., 2010). This suggests these PGRPs only function as antimicrobial proteins but not as receptors for antimicrobial peptide genes induction pathway. Our biochemical evidence suggested silkworm PGRP-S5 serves as a receptor for PG leading to the proPO activation pathway, it may act as an antimicrobial protein as well. Whether it is the receptor for antimicrobial peptide genes induction pathway remains unknown and needs further investigation.
4.2. Its role in the gut immune response: a negative regulator?
When we tried to detect the expression of twelve PGRP genes in silkworm midgut tissue after bacteria feeding infection, we found PGRP-S5 and PGRP-L6 expression up-regulated dramatically (Fig. 2, PGRP-L6 data not shown). Phylogenetic analysis revealed silkworm PGRP-S5 is close to Drosophila PGRP-LB in the phylogenetic tree (Tanaka et al., 2008). Sequence analysis implied silkworm PGRP-S3, -S4, -S5, and -S6 might have amidase activity. We speculate that PGRP-S5 may play a negative regulatory role in the silkworm midgut as Drosophila PGRP-LB. This hypothesis requires further experimental evidence to support.
4.3. Concluding remarks
PGRP-S5 was up-regulated in the fat body and midgut of silkworm upon bacterial infection. To elucidate the function of this molecule, we generated and purified recombinant protein from E. coli. Biochemical studies demonstrated PGRP-S5 binds to PGs specifically and causes bacteria agglutination. It is also an active amidase and antibacterial protein. Its interaction with specific PG triggers proPO activation pathway. Collectively, our results confirmed PGRP-S5 plays dual roles as a receptor for the pathway and an antibacterial protein in silkworm. Whether it serves as a receptor for antimicrobial peptide genes transcription signaling and as a negative regulator in gut immune response are two open questions and need further investigations.
Acknowledgements
This study was supported by the National Basic Research Program of China 2012CB114600 (to Z.Q.L.) and National Institutes of Health Grant GM58634 (to H.J.).
Abbreviations:
- PG and PGRP
peptidoglycan and its recognition protein
- LPS
lipopolysaccharide
- DAP
meso-diaminopimelic acid
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PO and proPO
phenoloxidase and its precursor
- QRT-PCR
quantitative real-time PCR
- PBS
phosphate buffered saline
- BSA
bovine serum albumin
References
- Basbous N, Coste F, Leone P, Vincentelli R, Royet J, Kellenberger C, Roussel A, 2011. The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to down-regulate the Imd pathway. EMBO Rep. 12, 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J, 2004. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol 5, 1175–1180. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J, 2006. Down-regulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco-Drayon V, Poidevin M, Boneca, Ivo G, Narbonne-Reveau K, Royet J, Charroux B, 2012. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12, 153–165. [DOI] [PubMed] [Google Scholar]
- Cerenius L, Lee BL, Söderhäll K, 2008. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. [DOI] [PubMed] [Google Scholar]
- Charroux B, Rival T, Narbonne-Reveau K, Royet J, 2009. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 11, 631–636. [DOI] [PubMed] [Google Scholar]
- Choe K-M, Werner T, Stöven S, Hultmark D, Anderson KV, 2002. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296, 359–362. [DOI] [PubMed] [Google Scholar]
- Dziarski R, Gupta D, 2010. Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun. 16, 168–174. [DOI] [PubMed] [Google Scholar]
- Garver LS, Wu J, Wu LP, 2006. The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proc. Natl. Acad. Sci. USA 103, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrin M, Zaidman-Rémy A, Broderick NA, Paredes J, Poidevin M, Roussel A, Lemaitre B, 2013. Functional analysis of PGRP-LA in Drosophila immunity. PLoS One 8. e69742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Yano T, Terashima J, Iwashita S, Oshima Y, Kurata S, 2010. Cooperative regulation of the induction of the novel antibacterial listericin by peptidoglycan recognition protein LE and the JAK-STAT pathway. J. Biol. Chem 285, 15731–15738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J, 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644. [DOI] [PubMed] [Google Scholar]
- Hetru C, Hoffmann JA, 2009. NF-kB in the immune response of Drosophila. Cold Spring Harb. Perspect. Biol 1, a000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Kanost MR, 2003. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J. Biol. Chem 278,3552–3561. [DOI] [PubMed] [Google Scholar]
- Kang D, Liu G, Lundström A, Gelius E, Steiner H, 1998. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 95, 10078–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Byun M, Oh BH, 2003. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol 4, 787–793. [DOI] [PubMed] [Google Scholar]
- Kim MY, Jang JH, Lee J-W, Cho JH, 2010. Molecular cloning and characterization of peptidoglycan recognition proteins from the rockfish, Sebastes schlegeli. Fish Shellfish Immunol. 28, 632–639. [DOI] [PubMed] [Google Scholar]
- Kurata S, 2014. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol 42, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata S, Ariki S, Kawabata S-I, 2006. Recognition of pathogens and activation of immune responses in Drosophila and horseshoe crab innate immunity. Immunobiology 211, 237–249. [DOI] [PubMed] [Google Scholar]
- Lavine MD, Strand MR, 2002. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol 32, 1295–1309. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J, 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol 25, 697–743. [DOI] [PubMed] [Google Scholar]
- Li X, Wang S, Qi J, Echtenkamp SF, Chatterjee R, Wang M, Boons G-J, Dziarski R, Gupta D, 2007. Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immunity 27, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH, 2006. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem 281, 8286–8295. [DOI] [PubMed] [Google Scholar]
- Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J, 2008. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe 3, 293–303. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA, 2002. Decoding the patterns of self and nonself by the innate immune system. Science 296, 298–300. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Steiner H, 2006. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem. Biophys. Res. Commun 350, 994–999. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Steiner H, 2003. A scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem 278, 7059–7064. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart J-M, Hoffmann JA, Royet J, 2001. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414, 756–759. [DOI] [PubMed] [Google Scholar]
- Neyen C, Poidevin M, Roussel A, Lemaitre B, 2012. Tissue- and ligand-specific sensing of Gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J. Immunol 189, 1886–1897. [DOI] [PubMed] [Google Scholar]
- Ochiai M, Ashida M, 1999. A pattern recognition protein for peptidoglycan: cloning the cDNA and the gene of the silkworm, Bombyx mori. J. Biol. Chem 274, 11854–11858. [DOI] [PubMed] [Google Scholar]
- Park JW, Kim CH, Kim JH, Je BR, Roh KB, Kim SJ, Lee HH, Ryu JH, Lim JH, Oh BH, Lee WJ, Ha NC, Lee BL, 2007. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. USA 104, 6602–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB, 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648. [DOI] [PubMed] [Google Scholar]
- Reiser J-B, Teyton L, Wilson LA, 2004. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 Å resolution. J. Mol. Biol 340, 909–917. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ, 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Strand MR, 2008. The insect cellular immune response. Insect Sci. 15, 1–14. [Google Scholar]
- Suetsugu Y, Futahashi R, Kanamori H, Kadono-Okuda K, Sasanuma S-I, Narukawa J, Ajimura M, Jouraku A, Namiki N, Shimomura M, Sezutsu H, Osanai-Futahashi M, Suzuki MG, Daimon T, Shinoda T, Taniai K, Asaoka K, Niwa R, Kawaoka S, Katsuma S, Tamura T, Noda H, Kasahara M, Sugano S, Suzuki Y, Fujiwara H, Kataoka H, Arunkumar KP, Tomar A, Nagaraju J, Goldsmith MR, Feng Q, Xia Q, Yamamoto K, Shimada T, Mita K, 2013. Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect Bombyx mori. G3 (Bethesda) 3 (1), 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumathipala N, Jiang H, 2010. Involvement of Manduca sexta peptidoglycan recognition protein-1 in the recognition of bacteria and activation of prophenoloxidase system. Insect Biochem. Mol. Biol 40, 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S, 2002. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates Imd/Relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. USA 99, 13705–13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S, 2004. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 23, 4690–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M, 2008. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol 38, 1087–1110. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D, 2000. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97, 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang X, Chen X, Cao P, Beerntsen BT, Ling E, 2010. BmToll9, an arthropod conservative Toll, is likely involved in the local gut immune response in the silkworm. Bombyx mori. Dev. Comp. Immunol 34, 93–96. [DOI] [PubMed] [Google Scholar]
- Yang DQ, Su ZL, Qiao C, Zhang Z, Wang JL, Li F, Liu XS, 2013. Identification and characterization of two peptidoglycan recognition proteins with zinc-dependent antibacterial activity from the cotton bollworm, Helicoverpa armigera. Dev. Comp. Immunol 39, 343–351. [DOI] [PubMed] [Google Scholar]
- Yano T, Kurata S, 2008. Induction of autophagy via innate bacterial recognition. Autophagy 4, 958–960. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M, 1996. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm. Bombyx mori. J. Biol. Chem 271, 13854–13860. [DOI] [PubMed] [Google Scholar]
- You H, Wan H, Li J, Jin BR, 2010. Molecular cloning and characterization of a short peptidoglycan recognition protein (PGRP-S) with antibacterial activity from the bumblebee Bombus ignitus. Dev. Comp. Immunol 34, 977–985. [DOI] [PubMed] [Google Scholar]
- Zaidman-Rémy A, Hervé M, Poidevin M, Pili-Floury S, Kim M-S, Blanot D, Oh B-H, Ueda R, Mengin-Lecreulx D, Lemaitre B, 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473. [DOI] [PubMed] [Google Scholar]
- Zaidman-Rémy A, Poidevin M, Hervé M, Welchman DP, Paredes JC, Fahlander C, Steiner H, Mengin-Lecreulx D, Lemaitre B, 2011. Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS One 6, e17231. [DOI] [PMC free article] [PubMed] [Google Scholar]


