Summary
Targeting immune checkpoints, such as Programmed cell Death 1 (PD1), has improved survival in cancer patients by restoring antitumor immune responses. Most patients, however, relapse or are refractory to immune checkpoint blocking therapies. Neuropilin-1 (NRP1) is a transmembrane glycoprotein required for nervous system and angiogenesis embryonic development, also expressed in immune cells. We hypothesized that NRP1 could be an immune checkpoint co-receptor modulating CD8+ T cells activity in the context of the antitumor immune response. Here, we show that NRP1 is recruited in the cytolytic synapse of PD1+CD8+ T cells, cooperates and enhances PD-1 activity. In mice, CD8+ T cells specific deletion of Nrp1 improves anti-PD1 antibody antitumor immune responses. Likewise, in human metastatic melanoma, the expression of NRP1 in tumor infiltrating CD8+ T cells predicts poor outcome of patients treated with anti-PD1. NRP1 is a promising target to overcome resistance to anti-PD1 therapies.
Subject areas: Immunology, Cancer
Graphical abstract
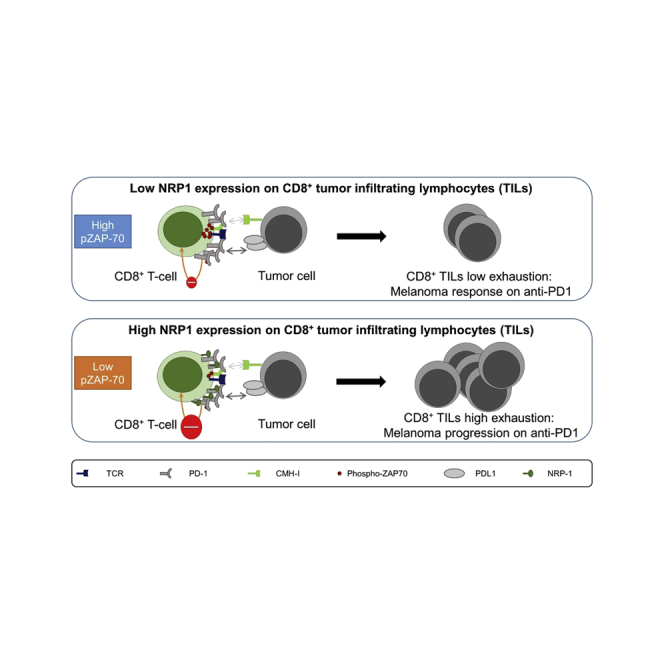
Highlights
-
•
NRP1 modulates PD1 activity secondary to complexes formation on CD8+ T cells
-
•
Anti-PD1 therapy is synergistic with NRP1 specific deletion on CD8+ T cells in mouse
-
•
NRP1 expression on CD8+ TILs predicts poor outcome in patients treated with anti-PD1
Immunology ; Cancer;
Introduction
For decades, cancer therapy has mainly relied on surgery, chemotherapy and radiotherapy. More recently targeted therapies of pathways involved in metabolism, proliferation, survival of tumor cells, and in angiogenesis have emerged as new paradigms of cancer treatment, with significant successes. Better knowledge of the physiology of the immune system have led to the discovery of key regulatory pathways of T cell activation like immune checkpoints that control activation of the immune system after antigen challenges. These checkpoints are involved in immune escape of cancer cells and are responsible for T cell exhaustion in the tumor environment (Zitvogel et al., 2006). T cell exhaustion is a mechanism of immune tolerance induced by chronic antigen stimulation (Schietinger and Greenberg, 2014). Initially described in chronic viral infection, exhaustion is also involved in tumor progression and is particularly driven by programmed cell death 1 (PD1) on CD8+ T cell and its ligand PD-L1 expressed by cancer cells and the tumor microenvironment (Baitsch et al., 2011; Blackburn et al., 2009; Butte et al., 2007). The development of immune checkpoint therapies in cancer, particularly against PD1/PD-L1 axis has led to successful therapies of cancer (Reck et al., 2016; Robert et al., 2015). However, most patients relapse or are refractory to these therapies. Several mechanisms have been shown to be involved in this resistance (Blackburn et al., 2008; Ngiow et al., 2015; Zaretsky et al., 2016), including loss of class I HLA expression, JAK1/2 mutation in the tumor cells, and high PD1 expression by tumor-infiltrating CD8+ T-cells. Deciphering mechanisms involved in PD1 regulation is critically needed to improve patients’ survival.
Neuropilin-1 (NRP1) is a transmembrane glycoprotein required for vasculogenesis and axonal guidance during embryonic development (Kawakami et al., 1996; Nakamura and Goshima, 2002). Beyond its role in development, we and others have demonstrated that NRP1 is expressed in several types of immune cells and is involved in immunological synapse formation and immune cell activation and migration (Kumanogoh and Kikutani, 2013; Takamatsu et al., 2010; Tordjman et al., 2002). Recent works have shown that NRP1 facilitates immune subversion because of its suppressive effect on tumor-associated macrophage induction, regulatory T cell activity (Casazza et al., 2013; Delgoffe et al., 2013; Hansen et al., 2012). Based on these data, we hypothesized that NRP1 could be an immune checkpoint co-receptor in CD8+ tumor infiltrating lymphocytes.
Results
NRP1 is expressed in activated CD8+ T cells and controls their antitumoral function in mice
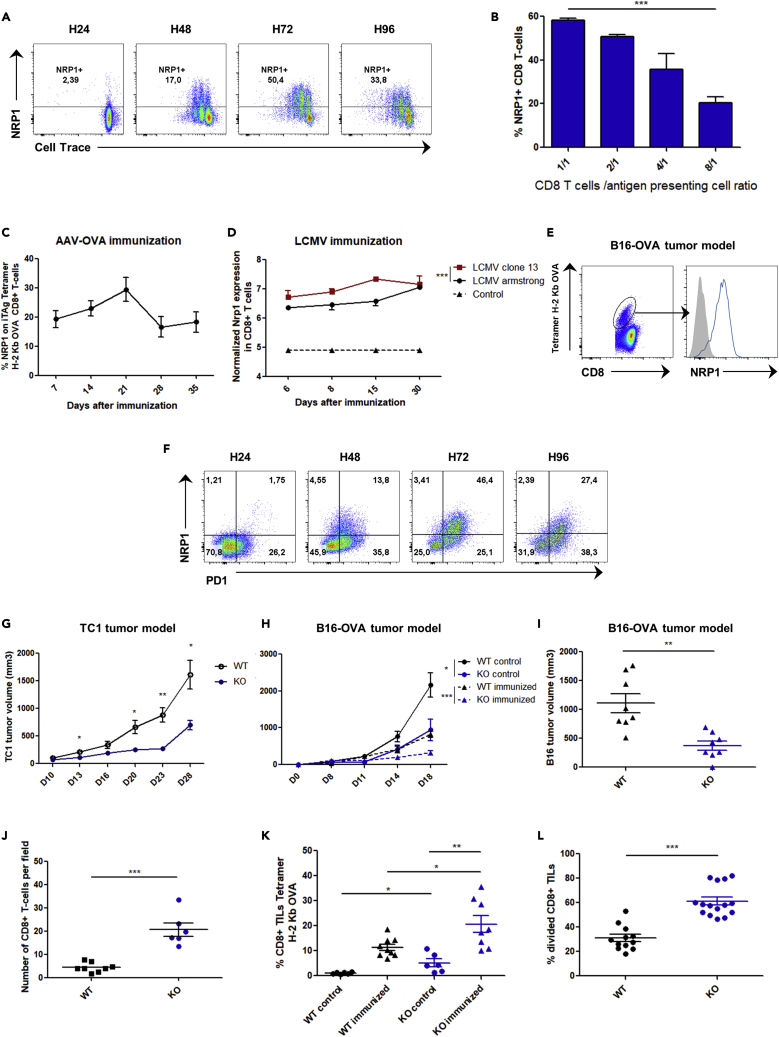
Although PD1 is a key factor of exhaustion, its expression is not sufficient to induce an exhaustion profile in CD8+ T cells. For example, in the mice LCMV clone 13 infection model, most antigen-specific CD8+ T cells have been induced; however, while maintaining PD1 expression after antigen withdrawal, a fraction of these CD8+ T cells retain their ability to produce cytokines upon new LCMV antigen challenge (Utzschneider et al., 2013). This observation suggests the involvement of a potential additional partner. Because NRP1 is unable to signal autonomously (Pellet-Many et al., 2008) and is also expressed in activated T cells at the synapse level, we hypothesized that NRP1 may be involved in PD1 inhibitory activity. In vitro NRP1 was expressed on murine CD8+ T cells after activation driven by OVA peptide (SIINFEKL), and the intensity of its expression correlated positively with antigen availability (Figures 1A and 1B). To investigate in vivo the expression of NRP1 on CD8+ T cells, we studied 3 models of acute or persistent antigen-specific immunization. As previously reported (Hwang et al., 2019; Jackson et al., 2014), NRP1 was not expressed on naive CD8+ T cells. In contrast, activated specific CD8+ T cells expressed NRP1 after intramuscular adeno-associated virus - OVA immunization (AAV-OVA), with a peak of expression at day 21 post-immunization (Figure 1C). NRP1 was also highly expressed in mice by specific CD8+ T cells in the exhaustion model of LCMV clone 13 viral infection when compared with LCMV Armstrong infection and in specific anti-OVA257 CD8+ tumor infiltrating lymphocytes (TILs) in a model of B16-OVA tumor progression (Figures 1D and 1E). In the latter model, NRP1+CD8+ TILs were PD1+. Likewise, in vitro upon activation, mice CD8+ Tcells expressed NRP1 on a subset expressing high levels of PD1 (PD1highCD8+ T-cells, Figure 1F).
Figure 1.
NRP1 is expressed in activated CD8+ T cells and controls their antitumoral function in mice
(A) NRP1 expression and cell trace intensity analyzed by flow cytometry in OT1 murine CD8+ T cells activated during 24, 48, 72, and 96 h with OVA257 peptide pulsed on dendritic cells (SIINFEKL, 10−9 mg/mL). Data are representative of 5 independent experiments.
(B) Flow cytometry analysis of NRP1 expression in OT1 murine CD8+ T cells, 72 h after activation with OVA peptide (SIINFEKL, 10−9 mg/mL). Expression is shown according to different effector CD8+ T cell / antigen-presenting cell (DC) ratios (E/A ratio: 1/1, 2/1, 4/1, and 8/1). p value (p = 0.0006) was determined by one-way ANOVA. Data are representative of 2 independent experiments. Expression was assessed by flow cytometry and data are presented as the mean ± SEM of percentage of NRP1 on CD8+ T cells (c) Expression of NRP1 in CD8+ T cells from B6 mice, after intramuscular immunization with AAV-OVA vector. Expression was assessed by flow cytometry and data are presented as the mean ± SEM of percentage of NRP1 on iTAg Tetramer/PE - H-2 Kb OVA, at day 7, 14, 21, 28, and 35 after immunization. Data are representative of 5 independent experiments.
(D) NRP1 expression profiles in H2-Db GP33-specific CD8+ T cells according to in vivo infection in mice with LCMV Armstrong (n = 16), LCMV clone 13 (n = 16), or naïve CD44low CD8+ T cells from controls (n = 4) at days 6, 8, 15, and 30. Raw transcriptomic data were from Doering et al. (2012) microarray experiments [32]. Data are presented as the mean ± SEM p value was determined by two-way ANOVA(p = 0.0008).
(E) Flow cytometry analysis of NRP1 expression (blue line curve) on iTAg Tetramer/PE - H-2 Kb OVA CD8+ TILs collected from C57BL/6 mice bearing a B16-OVA tumor at day 14 post-immunization with ovalbumin and poly-IC. Data are representative of 3 independent experiments.
(F) Flow cytometry analysis of NRP1 and PD1 expression in OT1 CD8+ T cells activated with OVA257 peptide (SIINFEKL, 10−9M) at 24, 48, 72, and 96 h post-activation. Data are representative of 3 independent experiments.
(H) Mice were pre-immunized (immunized) or not pre-immunized (control) with ovalbumin and poly-IC. B16-OVA tumor volume was assessed at day 0, 8, 11, 14, and 18 post-immunization in CD8Nrp1KO (KO) and control C8Cre (WT) mice. Data are presented as mean ± SEM. p values were determined by using student t-test ∗∗∗p < 0.001, ∗p < 0.05. Data are representative of 3 independent experiments.
(G) CD8Nrp1KO (KO) and control (WT) mice were injected in the right flank with 1 × 105 TC1 lung tumor cells subcutaneously. Data are presented as mean ± SEM p values was determined by using student t-test ∗∗p < 0.01, ∗p < 0.05. Data are representative of 3 independent experiments (I) CD8Nrp1KO (KO) and control (WT) mice were pre-immunized with ovalbumin and poly-IC. Tumor volume was assessed 21 days after immunization. Data are presented as mean ± SEM p values was determined by using student t-test (p = 0.0012). Data are representative of 3 independent experiments.
(J) Number of CD8+ TILs per fields with highest CD8+ T cells infiltration from CD8Nrp1KO mice (KO) or controls (WT) assessed by confocal microscopy at day 21 post immunization with ovalbumin and poly-IC. Data are presented as mean ± SEM. p values (p < 0.0001) was determined by using student t-test. Data are representative of 3 tumors per group.
(K) Percentages of Tetramer/PE - H-2 Kb OVA CD8+ TILs in B16-OVA tumors of four different mice group assessed at day 14 post-immunization by flow cytometry from CD8Nrp1KO (KO) and control (WT) mice immunized or not immunized (control) with ovalbumin and poly-IC. Data are presented as the mean percentage of CD8+ TILs Tetramer positive ± SEM. p values were determined by using student T test ∗∗p < 0.01, ∗p < 0.05. Data are representative of 2 independent experiments.
(L) Ex vivo TILs proliferation was analyzed by flow cytometry 72 h post-activation with anti-CD3 and anti-CD28. TILs were collected from B16-OVA tumors at day 21 post-immunization from 3 mice. Data are presented as the mean percentage of divided CD8+ T cells ± SEM. p values was determined by using student t-test ∗∗∗p < 0.001. Data are representative of 2 independent experiments.
To further study the role of NRP1 expression in CD8+ T cells in vivo, we generated a mouse model in which CD8+ T cells were specifically invalidated for Nrp1 (CD8Nrp1KO) by breeding Nrp1flox/flox mice with CD8CreTg mice. At steady state, the CD8Nrp1KO mouse harbored no immunological phenotype, and as expected, CD8+ T cells did not express NRP1 upon activation (Figures S1 and S2). In both antigen-specific and not specific antitumor immune response, tumor growth was significantly decreased in CD8Nrp1KO mice as compared to the control (Figures 1G–1I). Accordingly, analysis of the tumor immune microenvironment in CD8Nrp1KO and control mice showed an increase in CD8+ TILs frequency in CD8Nrp1KO mice (Figures 1J–1K). After ex vivo activation, CD8Nrp1KO TILs harbored an increased proliferation capacity (Figure 1L). These results suggest that NRP1 expression on CD8+ TILs might be involved in the negative regulation of antitumor immune responses.
NRP1 modulates PD1 activity at the synapse between CD8+ T cells and tumor cells
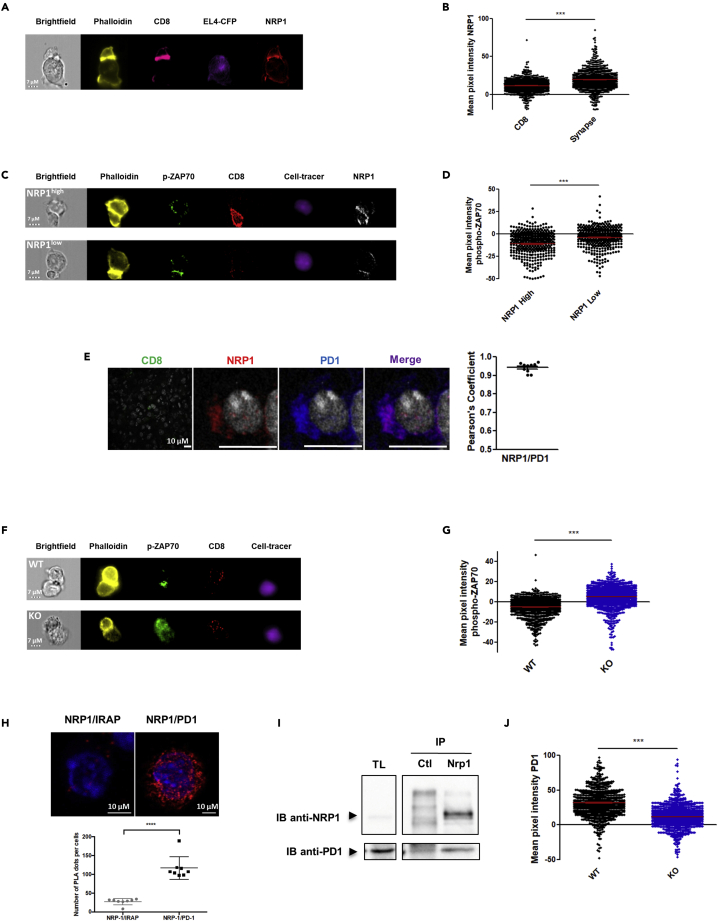
Because we previously reported that NRP1 was involved in the immunological synapse between T cells and dendritic cells (Tordjman et al., 2002), we then investigated whether NRP1 could be localized in the synapse between T cells and tumor cells and could be involved in the effector function of CD8+ TILs in this specific context. To address this question, we developed a synapse model between transgenic TCR OT1 T cells and tumor cells (EL4-CFP cells) bearing the cognate SIINFEKL antigen (OVA257) and between activated CD8+ T cells from CD8Nrp1KO mice or littermate and allogeneic tumor cells (A20 cells). In these models, imaging flow cytometry analysis of cell conjugates showed that NRP1 and PD1 were recruited together to the synapse between activated CD8+ T cells and tumor cells (Figures 2A, 2B, and S3); similarly, as demonstrated for PD1highCD8+ T cells, NRP1+CD8+ T cells were characterized by low levels of ZAP70 phosphorylation expression (Figures 2C and 2D), indicating a low TCR signaling activation. Because it has been previously reported that the clustering and colocalization of PD1 and TCR is critical in inducing low level of phospho-ZAP70 in the synapse junction in response to the binding of PD-L1 to PD-1 (Yokosuka et al., 2012; Zinselmeyer et al., 2013), which characterize the exhaustion synapse, we then investigated whether NRP1 was involved in PD1 recruitment and function at the synapse. First, by immunofluorescence, in vivo we showed that PD1 and NRP1 were co-localized in CD8+ TILs from mice (Figure 2E), and phospho-ZAP70 expression was significantly increased in synapses established with CD8+ T cells from CD8Nrp1KO compared with controls, strongly suggesting that NRP1 could be mechanistically involved in T cell exhaustion at the molecular level (Figures 2F and 2G). In accordance with this hypothesis, the proximity between NRP1 and PD1 was demonstrated on activated mice CD8+ T cells by a proximity ligation assay (Duolink) in vitro (Figure 2H). By performing co-immunoprecipitation experiments, we provided additional evidence for this interaction in a protein complex (Figure 2I). In contrast, in CD8Nrp1KO, although PD1 was expressed, its localization within the synapse was significantly reduced as compared with NRP1+CD8+ T cells from WT mice (Figure 2J). Taken together, our data suggest that NRP1 is a partner of PD1 enhancing its recruitment and activity at the synapse between CD8+ T cells and tumor cells.
Figure 2.
NRP1 modulates PD1 activity at the synapse between CD8+ T cells and tumor cells
(A) Illustrative image of phalloidin (yellow), CD8 (pink), CFP from EL4 (purple), and NRP1 (red) labeling in the synapse model between activated OT1 CD8+ T cells and EL4-CFP tumor cells bearing OVA257 (SIINFEKL), observed by ImageStream. The synapse is the high phalloidin labeling zone. Bright field image is in white (scale bar = 7μm). Data are representative of 4 independent experiments from 2 synapse models.
(B) Quantification by ImageStream of NRP1 expression (mean pixel intensity/MPI) in an allogeneic synapse model between activated CD8+ T cells and cell tracer violet labeled A20 cells. NRP1 expression was analyzed in activated CD8+ T cells at the synapse junction (high phalloidin labeling zone). Data are presented as the mean MPI ± SEM. p value (p < 0.0001) was determined by Wilcoxon matched pairs test. Data are representative of 4 independent experiments from 2 synapse models.
(C) Illustrative image of phalloidin (yellow), phospho-ZAP70 (green), CD8 (red), cell tracer violet labeled A20 tumor cells (purple), and NRP1 (white) between activated NRP1high or NRP1low CD8+ T cells and cell tracer violet labeled A20 tumor cells observed by ImageStream. The synapse is the high phalloidin labeling zone. Bright field image is in white (scale bar = 7μm). Data are representative of 2 independent experiments.
(D) Quantification by Imagestream of phospho-ZAP70 amounts (mean pixel intensity/MPI) in the synapse junction (high phalloidin labeling zone) between activated NRP1high or NRP1low CD8+ T cells and cell tracer violet labeled A20 tumor cells. Data are presented as mean MPI ± SEM. p value (p < 0.0001) was determined by Mann Whitney test. Data are representative of 2 independent experiments.
(E) Left panel: CD8 (green) NRP1 (red), PD1 (blue), and NRP1/PD1 merge (purple) expression observed by confocal microscopy in CD8+ TILs from control mice (WT) at day 21 post-activation (x63 oil objective, scale bar = 10 μm). Data are representative of 3 tumors. Right panel: Colocalization of NRP1 and PD1 was assessed by the calculation of Pearson coefficient. Data from 10 CD8+ TILs analyzed are presented as mean ± SEM.
(F) Phospho-ZAP70 signal according to its localization within the synapse between the CD8+ T cells and the tumor cells. Illustrative image of phalloidin (yellow), phospho-ZAP70 (green), CD8 (red), and Cell Tracer (A20 cells, purple) labeling in the synapse model between activated CD8+ T cells from CD8Nrp1KO mice (KO) or controls (WT) and allogeneic A20 tumor cells, by ImageStream. Bright field image is in white (scale bar = 7μm). Data are representative of 2 independent experiments.
(G) Phospho-ZAP70 signal according to its localization within the synapse between the CD8+ T cells and the tumor cells. Quantification by Image stream of phospho-ZAP70 amounts (mean pixel intensity/MPI) in the synapse junction (high phalloidin labeling zone) between activated CD8+ T cells from CD8Nrp1KO mice (KO) or control mice (WT), and cell tracer violet labeled A20 tumor cells. Data are presented as mean MPI ± SEM. p value (p < 0.0001) was determined by Mann Whitney test. Data are representative of 2 independent experiments.
(H) Proximity of NRP1 and PD1 proteins demonstrated by Duolink assay on in vitro activated CD8+ T cells from C57BL/6J mice. Upper panel: Left: Negative control experiments performed using anti-IRAP and anti-NRP1 antibodies (PLA-Duolink). Right: NRP1/PD1 complexes (anti-NRP1 and anti-PD1 antibodies with PLA-Duolink). The red spots indicate less than 40nm proximity between cellular-bound antibodies. Nuclei are stained with DAPI (blue). Images have been observed by confocal microscopy (x63 oil objective, scale bar = 10μm). Data are representative of 5 independent experiments. Lower panel: Comparison of number of PLA plots per cell. Data are presented as mean ± SEM.
(I) NRP1 and PD1 interaction was demonstrated by CoIP experiments performed in splenocytes from C57BL/6J mice activated with anti-CD3 and anti-CD28 antibodies. NRP1 and PD1 immunoblot (IB) detection is shown in total lysate (TL) as control, in eluate from IgG Control (ctl) IP, and from NRP1 IP (N = 1 experiment). NRP1/PD1 Co-IP was also observed after PD1 IP (N = 2 experiment). Data are representative of 3 independent experiments.
(J) Quantification by Imagestream of PD1 expression (MPI) in the synapse junction (high phalloidin labeling zone) between activated CD8+ T cells from CD8Nrp1KO mice (KO) or controls (WT), and allogeneic A20 tumor cells. Data are presented as mean MPI ± SEM. p value (p < 0.0001) was determined by Mann Whitney test. Data are representative of 2 independent experiments.
Targeting both NRP1 and PD1 has a synergistic effect in human and mouse CD8+ T cells immune response
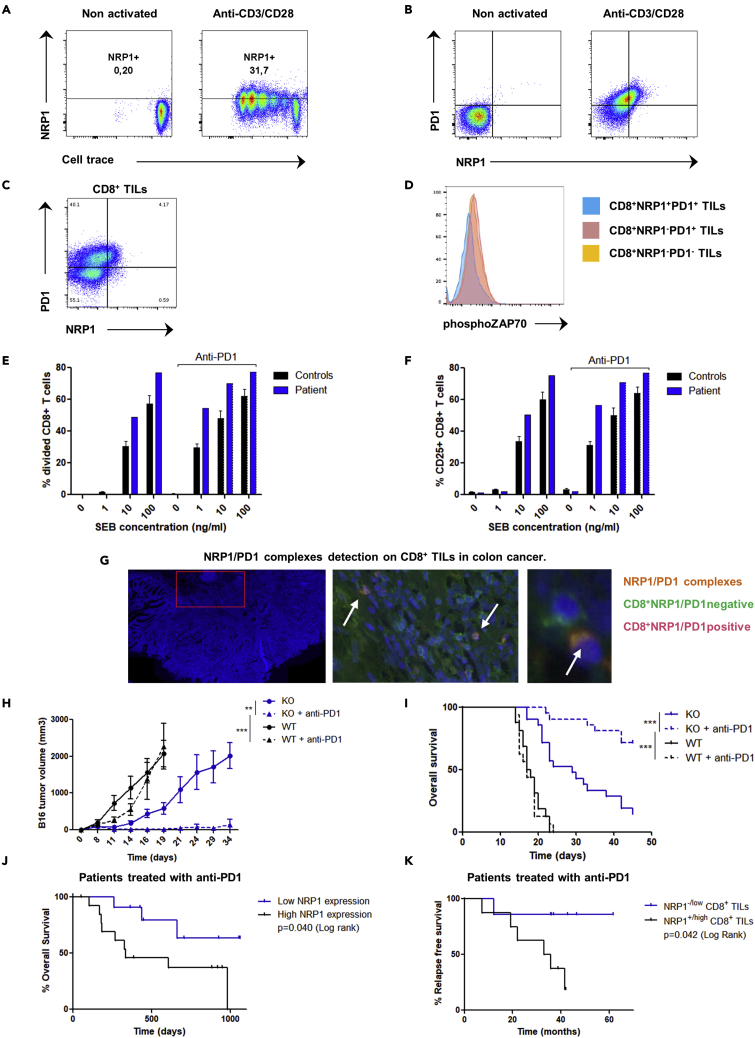
We next investigated whether the role in exhaustion of NRP1 in mice held true in human CD8+ T cells. First, we found that similarly to mice, NRP1 was also expressed after TCR activation in human CD8+ T cells (Figures 3A and 3B) and coexpressed within the PD1highCD8+ T cells population. Within the human tumor microenvironment, NRP1 expression was found on CD8+ TILs, specifically on PD1+CD8+ T cells and identified a subset of PD1+CD8+ TILs with low phospho-ZAP70 expression (phospho-ZAP70lowNRP1+PD1+CD8+ TILs) (Figures 3C, 3D, and S4). No patient bearing a homozygous NRP1 mutation had been described so far, potentially because of the lethality of homozygous NRP1 deletion in utero (Gu et al., 2003). However, we identified a unique patient with NRP1 haploinsufficiency caused by a heterozygous deletion of the chromosomal region (10p11.22) including the NRP1 gene (Heide et al., 2017). CD8+ T cells from the NRP1+/− patient have an increased ability to proliferate and to express CD25 after in vitro activation by the staphylococcal enterotoxin b (SEB) superantigen (Figures 3E, 3F, and S5). In addition, the increase of patient’s CD8+ T cells activation was synergistic in combination with an anti-PD1 antibody. Finally, we confirmed the presence of the NRP1/PD1 complexes on human CD8+ T cells in vitro and in vivo in the colon cancer microenvironment (Figure 3G).
Figure 3.
Targeting both NRP1 and PD1 has a synergistic effect in human and mouse CD8+ T cells immune response
(A) Flow cytometry analysis of NRP1 expression according to cell trace on human CD8+ T cells 96 h after activation with anti-CD3 and anti-CD28, or on nonactivated cells. Data are representative of 5 independent experiments.
(B) Flow cytometry analysis of NRP1 and PD1 expression in human CD8+ T cells 96 h after in vitro activation with anti-CD3 and anti-CD28 or non-activated cells. Data are representative of 3 independent experiments.
(C) Flow cytometry analysis of NRP1 and PD1 expression in CD8+ TILs. Data are representative of 3 independent experiments in human endometrial, kidney, and ovarian cancer.
(D) Flow cytometry analysis of phospho-ZAP70 in PD1+CD8+ TILs according to NRP1 expression. Data from one experiment in human endometrial cancer.
(E) Flow cytometry analysis of percentage of divided CD8+ T cells from a patient bearing an NRP1 haploinsufficiency (patient) or from controls (N = 5), respective to SEB superantigen concentration (0, 1, 10, or 100 ng/mL), in the presence or not of anti-PD1 antibody. Activation was performed during 72 h. Data are presented as the mean ± SEM. Data representative of 1 experiment.
(F) Flow cytometry analysis of CD25 expression in CD8+ T cells from a patient bearing an NRP1 haploinsufficiency (patient) or from controls (N = 5), respective to SEB superantigen concentration (0, 1, 10, or 100 ng/mL), in the presence or not of anti-PD1 antibody. Activation was performed during 72 h. Data are presented as the mean % of CD25 expression ± SEM. Data representative of 1 experiment.
(G) NRP1/PD-1 complexes detection by Proximity-Ligation-Assay (PLA) technology on CD8+ TILs in human colon cancer. Left panel: Representative area of tumor tissue observed. Acquisition with NDPI view software. Middle panel: Tumor infiltrating NRP1+PD1+CD8+ T cells (pink): Merge of green CD8 staining (FITC) and orange NRP1/PD-1 spots (TRITC). Acquisition with NDPI view software: zoom in x20. Right panel: CD8+ NRP1/PD1 positive cell (Pink) and CD8+ NRP1/PD1 negative cells (green). Acquisition with NDPI view software: zoom in x40. Nuclei are stained with DAPI (blue). Images have been observed by confocal microscopy ×20 oil objective. Data are representative of 10 independent experiments.
(H) CD8Nrp1KO (KO) and control (WT) mice were pre-immunized with ovalbumin and poly-IC and treated or not with anti-PD1 antibody in vivo. B16-OVA tumor volume was assessed until 35 days after immunization. Data are presented as the mean ± SEM and as Kaplan Meyer curve. p values were determined by two-way ANOVA test ∗∗∗p < 0.001 ∗∗p < 0.01. Data are representative of 5 experiments.
(I) CD8Nrp1KO (KO) and control (WT) mice were pre-immunized with ovalbumin and poly-IC and treated or not with anti-PD1 antibody in vivo. Overall survival was assessed until 50 days after immunization. Data are presented as the mean ± SEM and as Kaplan Meyer curve. p values were determined by Log rank test ∗∗∗p < 0.001. Data are representative of 5 experiments.
(J) Analysis of overall survival of patients with metastatic melanoma treated with anti-PD1, according to RNA NRP1 expression (low or high expression: groups have been determined according to ROC curve analysis) assessed in the tumor before anti-PD1 treatment. Data from transcriptomics analysis of metastatic melanoma tumors were available from Hugo et al. (Hugo et al., 2016) Data are presented as Kaplan Meyer curve. p value (p = 0.040) was determined by Log-rank test (n = 25 patients).
(K) Analysis of relapse free survival of patients with metastatic melanoma treated with anti-PD1 and reached at least a partial response, according to NRP1 expression (NRP1-/low compared with NRP1+/high) in CD8+ TILs assessed by immunohistochemistry before starting therapy. Blind analysis has been performed to assess NRP1 expression. Data are presented as the Kaplan Meyer curve. p value (p = 0.042) was determined by Log-rank test (n = 15 patients).
To address this synergistic effect between PD1 and NRP1, we evaluated the in vivo efficacy of anti-PD1 antibody in the B16-OVA tumor growth mouse model. As previously reported in this model, anti-PD1 treatment had no effect on overall survival in WT mice. In contrast, a significant increase in mouse survival was observed in the CD8Nrp1KO, which was more pronounced upon anti-PD1 treatment indicating a strong synergistic effect (Figures 3H and 3I). To assess the role of NRP1 in human cancer, we next performed an in silico study analyzing microarray data from metastatic melanoma cancer treated in clinical trials with anti-PD1 therapy (Hugo et al., 2016) (Figure 3J). In accordance with our hypothesis, a low expression of NRP1 in tumor before therapy was associated with improved patients’ overall survival (p = 0.040). Because NRP1 might be expressed in other cells than CD8+ T cells, we then investigated the outcome of 28 patients with metastatic melanoma treated with anti-PD1 therapy, depending on the expression of NRP1 on CD8+ TILs before starting therapy. Following our hypothesis, we found a trend for highest complete response rate and a significant increase of relapse-free survival in patients with NRP1-/lowCD8+ TILs compared with NRP1+/highCD8+ TILs (p = 0.042, Figure 3K). Taken together, our data demonstrate that NRP1 should be considered as an actor of exhaustion by enhancing PD1 activity on CD8+ TILs.
Discussion
NRP1 has already been implicated in the immune response against tumors (Casazza et al., 2013; Delgoffe et al., 2013; Hansen et al., 2012) by acting as a break on both innate and adaptive immunity. Immune checkpoint therapies have led to multiple successes in patients with cancer (Reck et al., 2016; Robert et al., 2015). Unfortunately, most patients relapse or are refractory even with a combination of immune checkpoints inhibitors (Postow et al., 2015). Data from our observations in humans suggest that NRP1 inhibition could be a potential therapeutic strategy to improve anti-PD1 efficacy. With respect to safety, no side effects were reported in the experiments in mice evaluating the association with anti-PD1 therapy. Furthermore, CD8Nrp1KO mice that were cured of B16-OVA tumor cells with anti-PD1 did not exhibit any autoimmune or inflammatory phenotype. This observation argues for the potential safety of using either a drug able to reduce NRP1 expression or an antibody blocking both NRP1 and PD1 on CD8+ T cells. In line with this strategy, recently, Leclerc et al. have reported that NRP1+CD8+ TILs migration and lytic function were inhibited by the NRP1 ligand, semaphorin-3A. They showed that combining anti-NRP1 and anti-PD1 antibodies was synergistic in a mouse tumor model, without proving, however, that this effect was CD8+ T cell-mediated (Leclerc et al., 2019). Interestingly, in a recent paper by Liu et al. (2020), it has been shown that NRP1 expression in CD8+ T cells may impair durability of CD8+ T cell-mediated tumor immunosurveillance. Here, we report that specific deletion of Nrp1 on CD8+ T cells dramatically enhances survival of mice bearing B16-OVA tumors, with potential cure with the addition of anti-PD1 therapy. Our data suggest that strategies using NRP1-deleted CD8+ chimeric antigen receptor (CAR) T cells alone or combined with an anti-PD1 antibody could be a way to improve efficacy of CAR-T cells by decreasing exhaustion and increasing durability of the response.
In conclusion, we confirm recent findings that identify NRP1 as an immune checkpoint and provide here an additional mechanism of its action by enhancing PD-1 inhibitory effect at the synapse level, thus being a potential therapeutic target to overcome immune therapies resistance.
Limitations of the study
Mechanisms of recruitment of NRP1 to the synapse and of the complex formation allowing cooperation between NRP1 and PD1 remain to be understood.
The heterogeneity of NRP1 expression in CD8+ T cells remains to be understood.
A larger number of patients treated by anti PD-1 antibodies in an independent study would be necessary to confirm the clinical role of NRP1 expression.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse PD-1 (CD279) (J43) | BioXcell | Cat# BE0033-2, RRID:AB_1107747 |
| polyclonal Armenian hamster IgG antibody | BioXcell | Cat# BE0091, RRID:AB_1107773 |
| anti-mouse CD3ε (145-2C11) | BioXcell | Cat# BE0001-1, RRID:AB_1107634 |
| anti-mouse CD28 (37.51) | BioXcell | Cat# BE0015-1, RRID:AB_1107624 |
| anti-human CD3e (OKT3) | BioXcell | Cat# BE0001-2, RRID:AB_1107632 |
| anti-human/monkey CD28 (CD28.2) | BioXcell | Cat# BE0291, RRID:AB_2687814 |
| iTAg Tetramer/PE – H-2 Kb OVA (SIINFEKL) | MBL int corporation | Code: TB-5001-1 |
| CD8 MicroBeads, human | Miltenyi Biotec | Order no 130-045-201 |
| CD8a (Ly-2) MicroBeads, mouse | Miltenyi Biotec | Order no 130-117-044 |
| anti-human CD3 Antibody (OKT3) | eBioscience | Cat# 48-0037-42; RRID:AB_1272055 |
| anti-human CD8a Antibody (RPA-T8) | eBioscience | Cat# 12-0088-42; RRID:AB_1724104 |
| anti-human CD25 Antibody (CD25-4E3) | eBioscience | Cat# 17-0257-42; RRID:AB_11218671 |
| anti-human CD279 (PD-1) Antibody (MIH4) | eBioscience | Cat# 11-9969-42; RRID:AB_10548937 |
| anti-mouse CD3e Antibody (145-2C11) | eBioscience | Cat# 48-0031-82; RRID:AB_10735092 |
| anti-mouse CD8a Antibody (53-6.7) | eBioscience | Cat# 25-0081-82; RRID:AB_469584 |
| anti-mouse CD279 (PD-1) Antibody (J43) | eBioscience | Cat# 11-9985-82; RRID:AB_465472 |
| anti-mouse CD25 Antibody (PC61.5) | eBioscience | Cat# 45-0251-82; RRID:AB_914324 |
| anti-mouse/human Phospho-ZAP70/Syk (Tyr319, Tyr352) Antibody (n3kobu5) | eBioscience | Cat# 17-9006-42; RRID:AB_2573268 |
| anti-mouse/human IRAP (D7C5) XP Rabbit mAb antibody | Cell Signaling Technology | Cat# 6918; RRID:AB_10860248 |
| anti-mouse/human Phospho-Zap-70 (Tyr319)/Syk (Tyr352) (65E4) Rabbit mAb antibody | Cell Signaling Technology | Cat# 73382; RRID:AB_2799838 |
| anti-human CD45 (HI30) 100Tst antibody | BD Biosciences | Cat# 612891; RRID:AB_2870179 |
| anti-human CD3e (HIT3a) antibody | BD Biosciences | Cat# 740283; RRID:AB_2740022 |
| anti-human CD4 (M-T477) antibody | BD Biosciences | Cat# 750175; RRID:AB_2874380 |
| anti-human CD8 (HIT8A) 25Tst antibody | BD Biosciences | Cat# 566856; RRID:AB_2869910 |
| anti-human CD279 (MIH4) antibody | BD Biosciences | Cat# 564323; RRID:AB_2738745 |
| anti-mouse Neuropilin-1 Affinity Purified PAb antibody | R and D Systems | Cat# FAB566A; RRID:AB_2267476 |
| anti-human Neuropilin-1 MAb (Cl 446921) antibody | R and D Systems | Cat# FAB3870A; RRID:AB_1241850 |
| donkey anti-rat IgG (H + L) Highly Cross-Adsorbed Secondary Antibody | Thermo Fisher Scientific | Cat# A48269TR; RRID:AB_2896335 |
| goat anti-hamster IgG (H + L) Cross-Adsorbed Secondary Antibody | Thermo Fisher Scientific | Cat# A-21451; RRID:AB_2535868 |
| donkey anti-goat IgG (H + L) Cross-Adsorbed Secondary Antibody | Thermo Fisher Scientific | Cat# A-11057; RRID:AB_2534104 |
| duolink in situ PLA probe anti-rabbit MINUS antibody | Sigma-Aldrich | Cat# DUO92005; RRID:AB_2810942 |
| duolink in situ PLA probe anti-mouse PLUS antibody | Sigma-Aldrich | Cat# DUO92001, RRID:AB_2810939 |
| duolink in situ PLA probe anti-goat PLUS antibody | Sigma-Aldrich | Cat# DUO92003 |
| polyclonal rabbit anti-human NRP1 antibody (Asp525) | LifeSpan Biosciences | Cat# LS-C177530 |
| anti-mouse NRP1 [EPR3113] antibody | Abcam | Cat# ab81321; RRID:AB_1640739 |
| anti-mouse PD-1 (ab1) antibody | Sigma-Aldrich | Cat# PRS4065; RRID:AB_1855098 |
| anti-mouse PD-1 antibody | Abcam | Cat# ab117420; RRID:AB_10902420 |
| anti-mouse Neuropilin-1 Affinity Purified PAb antibody | R and D Systems | Cat# AF566; RRID:AB_355445 |
| anti-CD8b (H35–17.2) unconjugated monoclonal antibody | BD Biosciences | Cat# 550797; RRID:AB_393886 |
| Bacterial and virus strains | ||
| rAAV1/SPc5-12-mOVA | Bartolo et al. (2019) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Albumin from chicken egg white | Sigma-Aldrich | CAS number: 9006-59-1 |
| Poly(I:C) HMW | Invivogen | CAS number: 31852-29-6 |
| Staphylococcal enterotoxin B from Staphylococcus aureus (SEB) | Sigma-Aldrich | CAS number: 11100-45-1 |
| Zombie Aqua Fixable Viability dye | Biolegend | Cat No 423101 |
| CellTrace Violet | Thermo fisher | Cat No C34557 |
| Fixable Viability Dye eFluor 450 | Thermo fisher | Cat No 65-0863-14 |
| Phalloidin AF555 | Thermo fisher | Cat No A34055 |
| Deposited data - Data accession numbers | ||
| Rossignol, Julien (2022), “Neuropilin-1 cooperates with PD-1 in CD8+ T-cells predicting outcomes in melanoma patients treated with anti-PD1”, | Mendeley Data, V1 | https://doi.org/10.17632/5xgbf6fdty.1 |
| Experimental models: Cell lines | ||
| EL4-CFP cell line (EL4 cell line transfected with membrane-bound CFP) | ATCC | Cat# TIB-39, RRID:CVCL_0255 |
| A20 cell line | ATCC | Cat# TIB-208, RRID:CVCL_1940) |
| B16-OVA cell line (B16-F10 transfected with ovalbumin) | ATCC | Cat# CRL-6475, RRID:CVCL_0159) |
| TC1 cell line | ATCC | Cat# JHU-1, RRID:CVCL_4699) |
| Experimental models: Organisms/strains | ||
| Mice: B6.129(SJL)-Nrp1tm2Ddg/J | Jackson Laboratory, California, USA | Cat# JAX:005247; RRID:IMSR_JAX:005247 |
| Mice: C57BL/6-Tg 1Itan/J | Jackson Laboratory, California, USA | Cat# JAX:008766; RRID:IMSR_JAX:008766 |
| Mice: C57BL/6J | Janvier Labs, France | Cat# 5752053; RRID:MGI:5656904 |
| Mice: B6.129S6-Rag2tm1Fwa Tg(TcraTcrb)1100Mjb | Gift from Jean Davoust, INSERM U1151 | catalog # TAC:2334; RRID:IMSR_TAC:2334 |
| Experimental models and subject details | ||
| Patient with NRP1 haploinsufficiency | Gender: Male | Age: 16 |
| Healthy donors | Gender: Female 3 Male 2 | Median age: 22 [18:32] |
| Melanoma patients | Gender: Female 15 Male 13 | Median age: 69 [24;89] |
| Software and algorithms | ||
| GraphPadPrism, version 5.0 | GraphPad Software | GraphPad Prism; RRID:SCR_002798 |
| FlowJo v10 software | Tresstar Inc | FlowJo; RRID:SCR_008520 |
| INSPIRE software | Amnis Corp, Millipore | RRID:SCR_018589 |
| IDEAS software | Amnis Corp, Millipore | RRID:SCR_018589 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Julien Rossignol (julien.rossignol@aphp.fr).
Materials availability
This study did not generate new unique reagents.
CD8Nrp1KO mice were generated by breeding B6.129(SJL)-Nrp1tm2Ddg/J (Nrp1lox/lox) and C57BL/6-Tg 1Itan/J (CD8cre) mice were obtained from Jackson Laboratories.
Experimental model and subject details
Mice
B6.129(SJL)-Nrp1tm2Ddg/J (Nrp1lox/lox) and C57BL/6-Tg 1Itan/J (CD8cre) mice were obtained from Jackson Laboratories. Wild Type (WT) mice were littermate C57BL/6-Tg 1Itan/J. All in vivo experiments were performed in 8–10 weeks old male mice and in vitro with 8–10 weeks old female mice. C57BL/6J mice were purchased from Janvier Labs (CS 4105 LE GENEST ST ISLE, SAINT BERTHEVIN Cedex France). Rag1−/−OT-I (CD45.2) mice, carrying a TCR specific for OVA257 bound to H-2Kb, were bred in Imagine Institute animal facility. For all experiments, littermates of the same sex were randomly assigned to experimental groups. Mice were given ad libitum access to food and water with a 12 h light/12 h dark cycle. Experiments were performed in the Imagine Institute, specific-pathogen-free facilities in Necker hospital. Animal protocols were approved by local ethic comitee according to the recommendations of the French Institutional Committee (A75-15-34).
Patients
Peripheral blood (PB) was collected from a patient with NRP1 haploinsufficiency caused by a heterozygous deletion of the chromosomal region including NRP1 gene (Age: 16, Gender: Male). Informed consent was obtained from the patient, according to the recommendations of the appropriate ethics committee (Comité de Protection des Personnes Ile-de-France). PB from 5 healthy donors has been collected for control CD8+ T-cells activation (Median age was 22 [18:32] and 3 healthy donors were female). They gave their informed consent according to the recommendations of the institutional ethics committee (Comité de Protection des Personnes Ile-de-France X). Peripheral blood mononuclear cells (PBMC) were obtained from PB samples after performing a Ficoll-Hypaque gradient. For melanoma patients study, all patients received appropriate information and signed an informed consent form authorizing tumor biopsies and molecular studies in the context of an institutional review board and Center of Biological Resources–approved protocol (MSN-08–027 Comité de Protection des Personnes Ile-de-France). Median age was 69 [24;89], 15 patients were female and 13 patients were male.
Method details
Tumor models
Nrp1lox/loxCD8cre (CD8Nrp1KO) and CD8cre mice were injected in the right flank with 5 × 105 B16-OVA melanoma cells or 1 × 105 TC1 lung tumor cells subcutaneously. Tumors were measured every 3 days with digital caliper and tumor size was calculated according to the following formula: (length x width2)/2. In the immunization model, ovalbumin (400μg, Sigma-Aldrich) combined with poly-IC (40μg, Invivogen) were injected in the opposite flank at day 7 and 14. In the anti-PD1 therapy model, mice were injected 5 times with 200μg of anti-PD1 antibody (J43, BioXcell) or isotype (Armenian Hamster IgG, BioXcell) every 48 h starting at day 7. Tumors were collected at day 21. TILs were analyzed by FACS after tumor mechanical disruption. Tumor fragments were embedded in the optimum cutting temperature (O.C.T.) compound and snap-frozen until analysis.
In vivo immunization model with AAV-OVA
Plasmid constructions and rAAV1 vector productions have already been published (Bartolo et al., 2019). Briefly, cDNA were inserted by PCR in pSMD2 rAAV1 plasmid between the SPc5-12 muscle-specific promoter and a polyA signal to create rAAV1/SPc5-12-mOVA (rAAV1/mOVA) targeting the muscle. C57BL/6JRj mice were immunized against OVA257 with rAAV1 vector diluted in 25 μL PBS injected into the left tibialis anterior using a 30-gauge RN Hamilton syringe.
In vitro mice assays and synapse conjugates
For in vitro CD8+ T-cells activation, splenocytes from CD8Nrp1KO and WT were cultured in 96 U-bottom plates, which had been coated with 5 μg/mL anti-CD3e (145-2C11, Bio X cell). Culture medium consisted of 5 μg/mL anti-CD28 (37.51, BioXcell) in RPMI 1640 supplemented with 10% FCS, 2 mML-glutamine, 5μM betamercaptoethanol, 100U/mL penicillin and 100 mg/mL streptomycin.
For in vitro antigen specific activation, CD8+ T-cells from C57BL/6-Tg (TcraTcrb) 1100Mjb/J (OT1) mice were activated with SIINFEKL presented by bone marrow derived dendritic cells. CD8+ T-cells were labeled with 5 μM CellTrace Violet (Termo fisher).
For synapses conjugates, CD8+ T-cells from CD8Nrp1KO and CD8cre or mice were activated with plate bound anti-CD3 (5 μg/mL, 145-2C11) and soluble anti-CD28 (5 μg/mL, 37.51). At 72h, activated CD8+ T-cells were mixed with allogeneic cell tracer violet labeled A20 tumor cells (ratio effector/target: 4/1) and incubated at 37 °C for 1 h. In OVA257 specific synapse between CD8+ T-cells and OVA257 pulsed EL4-CFP tumor cells, spleen CD8+ T-cells from Rag1−/−OT-I mice were activated in vitro for 96 h with OVA257 peptide (10−7 mg/mL). Synapses between CD8+ T-cells and tumor cells were then fixed with PFA 1.5% for 30 min at 4 °C and then stained for ImageStream analysis.
In vitro human assays
For in vitro CD8+ T-cells activation in human, CD8+ T-cells were sorted using magnetic positive selection (Miltenyi Biotec) from donor PBMCs and were cultures in 96 U-bottom plates which had been coated with 5 μg/mL anti-CD3e (OKT3, BioXcell) with 5 μg/mL anti-CD28 (CD28.2, BioXcell). Culture medium consisted of RPMI 1640 supplemented with 10% FCS, 2 mML-glutamine, 5μM betamercaptoethanol, 100 U/mL penicillin and 100 mg/mL streptomycin.
For in vitro superantigen Staphyolococcus enterotoxin B (SEB, Sigma-Aldrich) activation, CD8+ T-cells were sorted using magnetic positive selection (Miltenyi Biotec) from patient bearing a NRP1 haploinsufficiency and controls (n = 5) and were cultures in 96 U-bottom plates.
Preparation of bone-marrow-derived dendritic cells (BMDCs)
BMDCs were generated from C57BL/6-Tg 1Itan/J mice by a 7-day culture of bone marrow precursors in IMDM supplemented with 10% FCS, 2 mM L-glutamine, 50 μM β-mercaptoethanol and 20 ng/mL GM-CSF.
Antibodies and flow cytometry
Fluorescently conjugated antibodies were purchased from Ebioscience (anti-human CD3, anti-human CD8, anti-human PD1, anti-human CD25, anti-mouse CD3, anti-mouse CD8, anti-mouse PD1, anti-mouse CD25). Anti-human neuropilin-1 (FAB3870R) and anti-mouse neuropilin-1 (FAB566A) were purchased from R&D Systems. ITAg Tetramer - H-2 Kb OVA (SIINFEKL) conjugated to PE was purchased from MBL International Corporation, live/dead Fixable Viability Dye eFluor 450 and near infrared (Thermo Fisher). Celltrace violet cell proliferation kit was purchased from Thermo Fisher. Flow cytometric cell acquisition was done on a Canto II flow cytometer (BD Biosciences). Cell sorting was performed on a FACSAria II instrument (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (TreeStar). Cells were gated on live cells (live/dead Fixable Viability Dye eFluor 450 negative).
Imaging flow cytometry (ImageStream)
Samples were run on an ImageStream ISX mkII (Amnis Corp, Millipore) and a 60× magnification was used for all acquisitions. Data were acquired using the INSPIRE software (Amnis Corp) and analyzed using the IDEAS™ software (version 6.2 Amnis Corp). On average, 30,000 events were collected in all experiments.
Phalloidine was obtained from Thermoficher and anti-Phospho-ZAP70/Syk (Tyr319, Tyr352) APC-conjugated antibody (clone n3kobu5) from Ebioscience. Permeabilization and intra-cellular staining were performed using phalloidin and anti-phospho-ZAP70 diluted in PBS, Triton ×-100 at 0.1%. Single stain controls were run for each fluorochrome used and spectral compensation was performed. Cells were gated for single cell using the area and aspect ratio of the brightfield image, gated for focused cells using the gradient RMS feature and synapses were selected according to the coexpression of cell tracer (tumor cells) and CD8. Synapses were gated using phalloidine high area between cells. Results were expressed as mean pixel intensity value, which is the intensity normalized to surface area.
Confocal analysis
For tumor microenvironment analysis, frozen tumors onto slides were fixed with acetone. They were then incubated with the primary antibodies overnight at 4 °C: anti-NRP1 (goat), anti-PD1 (Hamster) and anti-CD8 (rat) and then incubated with secondary antibodies for 2 h at room temperature (Donkey anti-rat AF488, goat anti-hamster AF647 and donkey anti-goat AF568: Thermo Fisher). Nuclei were stained with DAPI, and the slides were examined with a confocal microscope (LSM 700 Carl Zeiss). Image analysis and figures were performed using ImageJ software (v1.52p) and Figure J, Cell counter and JACoP plugin. For each independent experiment, 6-8 fields were collected per slide. For each field, CD8 and DAPI-positive staining were used to define the nuclear region of interest (ROI). For quantification analysis, fields with highest CD8+ T-cells infiltration have been selectionned.
Duolink assay
To analyze the NRP1/PD1 interaction both in mice and in human, we used the Duolink II technology (Olink Bioscience), which is an in-situ proximity ligation assay technology. Slides were incubated with primary antibodies (anti-PD1 and anti-NRP1) and with secondary antibodies conjugated with oligonucleotides (PLA probe Minus anti-rabbit and PLA probe PLUS anti-mouse or goat). Ligation and amplification reactions were performed according to manufacturer’s instructions. Two types of negative controls were used: secondary antibodies alone and rabbit anti-IRAP antibody clone D7C5 (Cell signaling Technology).
Immunoprecipitation and immunoblot
Ten million of activated Murine splenocytes where washed with PBS at 4°c, then lysed in buffer: EDTA 1mM, NaCl150mM, Tris HCl 25mM, glycerol 10%. Antiproteases, antiphosphatases and detergent (final concentration 0.1% NP40) were added extemporaneously. Lysate was centrifuged at 20,000 G for 15 min at 4°c to remove debris. Preclearing was performed: 15μL of Protein G Sepharose 4 Fast Flow beads (Sigma-Aldrich) were added to the lysate and incubated for 1h at 4°c under rotating wheel. Supernatant was divided in two conditions for incubation with 2μG of antibodies: either rabbit anti-mouse NRP1 (ab81321, AbCam) or rabbit IgG control, for 1h at 4°c under rotating wheel. Then, lysates were respectively incubated with 15μL of Protein G Sepharose for 90 min at 4°c under rotating wheel. Beads were washed with PBS at 4°c. Protein complexes were eluted by adding 4X Laemmli buffer followed by heating at 95°c for 5 min. Beta-mercaptoethanol was added and the whole eluate was reheated for 1min and centrifugated. Proteins were resolved on 10% PAGE and analyzed by immunoblotting. Antibodies used included rabbit anti-mouse NRP1 (ab81321, AbCam) and rabbit Anti mouse PD1 (PRS4065, Sigma-Aldrich), diluted in TTBS BSA 5%.
Immunohistochemical technique and interpretation
After antigen unmasking, a polyclonal antibody directed to NRP1 (LifeSpan Biosciences) was applied at 1/100 dilution, during 1 h, to deparaffinized 4 μm-thick sections of formalin-fixed tumor tissue samples, using an automated slide stainer (Benchmark Ultra, Ventana, Tucson, AZ). Detection was performed through the Ultraview Universal Alkaline phosphatase Red detection kit (Ventana); the red chromogen was chosen to avoid confusion with the brown melanin deposits often observed in melanoma tissues. Positive and negative controls were included. Acquisitions were performed with a Leica DM2500 microscope (Wetzlar, Germany). The interpretation of immunostained tissue sections was performed by a trained pathologist (blind analysis). The density of NRP1+ cells per surface unit and their distribution within the tumor tissue (intra-tumoral, stromal, interface with peritumoral tissue) were evaluated.
Human TILs phenotyping
Fresh tumor tissue was dissociated at 37 °C for 1 h in a mixture of RPMI, DNaseI, Collagenase Type IV and Dispase using “gentle MACS dissociator” (Miltenyi Biotec). Cells were filtered and washed in 1X PBS −/− buffer and resuspended in 1X ACK lysis buffer (BioLegend, CA) in the dark for 10 min at Room Temperature before dilution in 1X PBS−/− and washed in FACS buffer (1XPBS, 2% FCS) at 4 °C. Flow cytometry staining was performed on 500,000 cells for membrane phenotype using the following monoclonal antibodies: anti-CD45, CD3, CD4, CD8, PD1 (Beckton Dickinson); anti-NRP1 (R&D Systems) anti-phosphoZAP70 (Cell Signaling Technology). All antibodies were used at concentrations recommended by the manufacturer. Cells were stained for viability using the Zombie Aqua Fixable Viability dye (BioLegend) according to the manufacturer’s protocol. The samples were acquired on the BD LSR Fortessa X-20 (Beckton Dickinson) and analysed using FlowJo v10 software (Tresstar Inc.).
Quantification and Statistical analysis
Statistical analyses were performed using GraphPadPrism (version 5.0; GraphPad Software). The data are expressed as the mean ± SEM unless noted otherwise. Two-tailed Student’s unpaired t-tests, two-way ANOVA(column factor), Wilcoxon matched pairs test, Mann-Whitney U-tests and Pearson linear regression were used as appropriate. p values 0.05 were considered significant, values smaller than this are indicated in figure legends ∗p < 0.05 ∗∗p < 0.01 ∗∗∗p < 0.001. Overall survival and relapse free survival were calculated with the Kaplan-Meier method and curves were compared by log rank test.
Acknowledgments
We would like to thank the Plateforme PETRA (Institut Gustave Roussy, Villejuif) and the Cell Imaging Plat-form of Imagine Institute (Necker, Paris) for providing assistance for the imaging experiment.
This work was supported (Research Grants) by “fondation BMS” and “Sauvez la vie, appel à projet Université Paris Descartes” (16FND998UPDE).
Patent: EP19305003.6 METHODS AND PHARMACEUTICAL COMPOSITIONS FOR ENHANCING CD8+ T CELL-DEPENDENT IMMUNE RESPONSES IN SUBJECTS SUFFERING FROM CANCER.
None of the material reported here has been published or is under consideration elsewhere, including the Internet.
Author contributions
J.R. designed the study, performed the experiments, analyzed the data and wrote the manuscript. M.D. performed Imagestream experiments, analyzed the data, and helped to write the manuscript. Z.B., M.W., and E.C. performed duolink experiments, analyzed the data, and helped to write the manuscript. F.G. performed Co-IP experiments, analyzed the data, and helped to write the manuscript. G.F., R.R., J.B., and T.C. performed in vivo experiments, in vitro human experiments, and helped to write the manuscript. P.M, A.R., M. D’A., J.B., L.P., F.C., Y.L., I.M., and M.T.R. helped to analyze data and to write the manuscript. C.C. managed mice breeding and genotyping. N.G. and T.M. helped to perform confocal microscopy acquisitions and analyses and helped to write the manuscript. N.C. helped to perform in silico expirement and analyzed data. K.B. helped to perform Co-IP experiments. A.E., H.H., A.M., and S.M. performed CD8+ TILs phenotyping. D.H. and J.L. identified and studied the patient with NRP1 haploinsufficiency. C.R., I.G., D.L., J.Y., and L.C. provided tumor samples of patients with metastatic melanoma treated with anti-PD1 therapy, performed microscopy acquisitions, and analyzed data. F.P. provided tumor samples of patients with colon cancer. J.D. and D.G. provided AAV-OVA vector, analyzed the data, and helped to write the manuscript. O.H. designed the study, supervised the overall project, analyzed the data, edited, and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104353.
Supplemental information
Data and code availability
Section 1: Data
Standardized datatype data have been deposited at [datatype-specific repository] and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
All data reported in this paper will be shared by the lead contact upon request.
Section 2: Code
This paper does not report original code.
Section 3:
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Baitsch L., Baumgaertner P., Devevre E., Raghav S.K., Legat A., Barba L., Wieckowski S., Bouzourene H., Deplancke B., Romero P., et al. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J. Clin. Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolo L., Li Chung Tong S., Chappert P., Urbain D., Collaud F., Colella P., Richard I., Ronzitti G., Demengeot J., Gross D.A., et al. Dual muscle-liver transduction imposes immune tolerance for muscle transgene engraftment despite preexisting immunity. JCI Insight. 2019;4:e127008. doi: 10.1172/jci.insight.127008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn S.D., Shin H., Freeman G.J., Wherry E.J. Selective expansion of a subset of exhausted CD8 T cells by αPD-L1 blockade. Proc. Natl. Acad. Sci. U S A. 2008;105:15016–15021. doi: 10.1073/pnas.0801497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn S.D., Shin H., Haining W.N., Zou T., Workman C.J., Polley A., Betts M.R., Freeman G.J., Vignali D.A.A., Wherry E.J. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte M.J., Keir M.E., Phamduy T.B., Sharpe A.H., Freeman G.J. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casazza A., Laoui D., Wenes M., Rizzolio S., Bassani N., Mambretti M., Deschoemaeker S., Van Ginderachter J.A., Tamagnone L., Mazzone M. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Delgoffe G.M., Woo S.R., Turnis M.E., Gravano D.M., Guy C., Overacre A.E., Bettini M.L., Vogel P., Finkelstein D., Bonnevier J., et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501:252–256. doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering T.A., Crawford A., Angelosanto J.M., Paley M.A., Ziegler C.G., Wherry E.J. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Rodriguez E.R., Reimert D.V., Shu T., Fritzsch B., Richards L.J., Kolodkin A.L., Ginty D.D. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev. Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen W., Hutzler M., Abel S., Alter C., Stockmann C., Kliche S., Albert J., Sparwasser T., Sakaguchi S., Westendorf A.M., et al. Neuropilin 1 deficiency on CD4+Foxp3+ regulatory T cells impairs mouse melanoma growth. J. Exp. Med. 2012;209:2001–2016. doi: 10.1084/jem.20111497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide S., Keren B., Billette de Villemeur T., Chantot-Bastaraud S., Depienne C., Nava C., Mignot C., Jacquette A., Fonteneau E., Lejeune E., et al. Copy number variations found in patients with a corpus callosum abnormality and intellectual disability. J. Pediatr. 2017;185:160–166.e1. doi: 10.1016/j.jpeds.2017.02.023. [DOI] [PubMed] [Google Scholar]
- Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.Y., Sun Y., Carroll C.R., Usherwood E.J. Neuropilin-1 regulates the secondary CD8 T cell response to virus infection. mSphere. 2019;4:1–12. doi: 10.1128/msphere.00221-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.R., Berrien-Elliott M., Yuan J., Hsueh E.C., Teague R.M. Neuropilin-1 expression is induced on tolerant self- Reactive cd8+ t cells but is dispensable for the tolerant phenotype. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110707. e110707-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A., Kitsukawa T., Takagi S., Fujisawa H. Developmentally regulated expression of a cell surface protein, neuropilin, in the mouse nervous system. J. Neurobiol. 1996;29:1–17. doi: 10.1002/(SICI)1097-4695(199601)29:1<1::AID-NEU1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Kumanogoh A., Kikutani H. Immunological functions of the neuropilins and plexins as receptors for semaphorins. Nat. Rev. Immunol. 2013;13:802–814. doi: 10.1038/nri3545. [DOI] [PubMed] [Google Scholar]
- Leclerc M., Voilin E., Gros G., Corgnac S., de Montpréville V., Validire P., Bismuth G., Mami-Chouaib F. Regulation of antitumour CD8 T-cell immunity and checkpoint blockade immunotherapy by Neuropilin-1. Nat. Commun. 2019;10:3345–3414. doi: 10.1038/s41467-019-11280-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Somasundaram A., Manne S., Gocher A.M., Szymczak-Workman A.L., Vignali K.M., Scott E.N., Normolle D.P., John Wherry E., Lipson E.J., et al. Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. Nat. Immunol. 2020;21:1010–1021. doi: 10.1038/s41590-020-0733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Goshima Y. Structural and functional relation of neuropilins. Adv. Exp. Med. Biol. 2002;515:55–69. doi: 10.1007/978-1-4615-0119-0_5. [DOI] [PubMed] [Google Scholar]
- Ngiow S.F., Young A., Jacquelot N., Yamazaki T., Enot D., Zitvogel L., Smyth M.J. A threshold level of intratumor CD8+ T-cell PD1 expression dictates therapeutic response to anti-PD1. Cancer Res. 2015;75:3800–3811. doi: 10.1158/0008-5472.CAN-15-1082. [DOI] [PubMed] [Google Scholar]
- Pellet-Many C., Frankel P., Jia H., Zachary I. Neuropilins: structure, function and role in disease. Biochem. J. 2008;411:211–226. doi: 10.1042/BJ20071639. [DOI] [PubMed] [Google Scholar]
- Postow M.A., Chesney J., Pavlick A.C., Robert C., Grossmann K., McDermott D., Linette G.P., Meyer N., Giguere J.K., Agarwala S.S., et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 2015;372:2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M., Rodriguez-Abreu D., Robinson A.G., Hui R., Csoszi T., Fulop A., Gottfried M., Peled N., Tafreshi A., Cuffe S., et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- Robert C., Long G.V., Brady B., Dutriaux C., Maio M., Mortier L., Hassel J.C., Rutkowski P., McNeil C., Kalinka-Warzocha E., et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- Schietinger A., Greenberg P.D. Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol. 2014;35:51–60. doi: 10.1016/j.it.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu H., Takegahara N., Nakagawa Y., Tomura M., Taniguchi M., Friedel R.H., Rayburn H., Tessier-Lavigne M., Yoshida Y., Okuno T., et al. Semaphorins guide the entry of dendritic cells into the lymphatics by activating myosin II. Nat. Immunol. 2010;11:594–600. doi: 10.1038/ni.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordjman R., Lepelletier Y., Lemarchandel V., Cambot M., Gaulard P., Hermine O., Romeo P.H. A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat. Immunol. 2002;3:477–482. doi: 10.1038/ni789. [DOI] [PubMed] [Google Scholar]
- Utzschneider D.T., Legat A., Fuertes Marraco S.A., Carrie L., Luescher I., Speiser D.E., Zehn D. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat. Immunol. 2013;14:603–610. doi: 10.1038/ni.2606. [DOI] [PubMed] [Google Scholar]
- Yokosuka T., Takamatsu M., Kobayashi-Imanishi W., Hashimoto-Tane A., Azuma M., Saito T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 2012;209:1201–1217. doi: 10.1084/jem.20112741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeyer B.H., Heydari S., Sacristan C., Nayak D., Cammer M., Herz J., Cheng X., Davis S.J., Dustin M.L., McGavern D.B. PD-1 promotes immune exhaustion by inducing antiviral T cell motility paralysis. J. Exp. Med. 2013;210:757–774. doi: 10.1084/jem.20121416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Tesniere A., Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat. Rev. Immunol. 2006;6:715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Section 1: Data
Standardized datatype data have been deposited at [datatype-specific repository] and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
All data reported in this paper will be shared by the lead contact upon request.
Section 2: Code
This paper does not report original code.
Section 3:
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.