Summary
Background
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with high phenotypic and genetic heterogeneity. The common variants of specific oxytocin-related genes (OTRGs), particularly OXTR, are associated with the aetiology of ASD. The contribution of rare genetic variations in OTRGs to ASD aetiology remains unclear.
Methods
We catalogued publicly available de novo mutations (DNMs) [from 6,511 patients with ASD and 3,391 controls], rare inherited variants (RIVs) [from 1,786 patients with ASD and 1,786 controls], and both de novo copy number variations (dnCNVs) and inherited CNVs (ihCNVs) [from 15,581 patients with ASD and 6,017 controls] in 963 curated OTRGs to explore their contribution to ASD pathology, respectively. Finally, a combined model was designed to prioritise the contribution of each gene to ASD aetiology by integrating DNMs and CNVs.
Findings
The rare genetic variations of OTRGs were significantly associated with ASD aetiology, in the order of dnCNVs > ihCNVs > DNMs. Furthermore, 172 OTRGs and their connected 286 ASD core genes were prioritised to positively contribute to ASD aetiology, including top-ranked MAPK3. Probands carrying rare disruptive variations in these genes were estimated to account for 10∼11% of all ASD probands.
Interpretation
Our findings suggest that rare disruptive variations in 172 OTRGs and their connected 286 ASD core genes are associated with ASD aetiology and may be potential biomarkers predicting the effects of oxytocin treatment.
Funding
Guangdong Key Project, National Natural Science Foundation of China, Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province.
Keywords: Autism spectrum disorder, Oxytocin-related biomarkers, Rare genetic variations, Combined model, MAPK3
Research in context.
Evidence before this study
Multiple clinical studies have investigated the beneficial effects of oxytocin intervention in social functioning and within non-social domains (repetitive behaviour) in patients with ASD; however, their findings are controversial. Although genome-wide association studies have attempted to link common variants in certain OTRGs, especially OXTR, to the efficacy of oxytocin intervention in ASD, findings remain paradoxical.
Added value of this study
Publicly available rare genetic variations from large cohort studies showed that the rare genetic variations of OTRGs are significant contributors to ASD aetiology (in the order of dnCNVs > ihCNVs > DNMs). Leveraging our combination model, 458 potential oxytocin-related molecular biomarkers based on rare disruptive variations were prioritised as contributors to ASD pathology.
Implications of all the available evidence
This study established the correlation between rare genetic variations of OTRGs and ASD aetiology. In addition, the potential oxytocin-related biomarkers identified herein could be valuable in both understanding the underlying mechanism of ASD pathophysiology and clinical investigation of ASD treatment with oxytocin.
Alt-text: Unlabelled box
Introduction
Autism spectrum disorder (ASD) is a highly prevalent and highly heritable neurodevelopmental disorder (NDD); however, no effective therapeutic options are available for curing core symptoms, including impaired social interactions and repetitive behaviours.1,2 Oxytocin is a nine-amino acid neuropeptide produced in the paraventricular nucleus of the hypothalamus and released from the posterior pituitary gland.3 Owing to its indispensable role in the intervention of a variety of social behaviours4, 5, 6 and its potential to treat ASD,7 oxytocin has attracted considerable attention in ASD research. Several studies have mentioned or inferred that dysregulated oxytocin signalling is associated with ASD aetiology.8, 9, 10, 11 Studies of murine models carrying ASD-associated rare genetic mutations have shown connections to the oxytocin system, and the social behavioural deficits in these mice could be ameliorated by intranasal oxytocin administration.12, 13, 14 By profiling plasma microRNAs of 30 patients with ASD as well as 30 gender- and age- matched healthy controls, the downregulated miR-6126, which targets genes enriched in the oxytocin signalling pathway, was identified.15 Moreover, the overall plasma oxytocin level was reportedly lower in patients with ASD than in controls.16, 17, 18 ASD patients with the lowest pre-treatment oxytocin level exhibited the most remarkable social improvement, and this enhanced social functioning was consistent with their increased blood oxytocin levels post-treatment.19 These findings support the hypothesis that ASD patients with deficient oxytocin system may benefit from oxytocin intervention.15,19
Compared to intravenous administration, intranasal administration of oxytocin in patients with ASD is a promising approach as it effectively reaches the central nervous system (CNS) directly through the nose-to-brain route.4 Although an increasing number of clinical trials have investigated the efficacy of intranasal oxytocin intervention, the conclusions vary and are controversial.19, 20, 21, 22, 23, 24, 25, 26, 27, 28 Certain studies supported the beneficial effects of oxytocin intervention in social functioning and within non-social domains (repetitive behaviour) in patients with ASD,20 whereas other studies support only one of these effects19, 20, 21, 22, 23, 24, 25,28, 29, 30, 31, 32, 33 or neither.26,27,34,35 Given that these clinical trials employed different experimental designs, inclusion and exclusion criteria for recruiting participants, intervention methods, and approaches for evaluating intervention efficiencies, inconsistencies in the outcomes are inevitable.19, 20, 21, 22, 23, 24, 25, 26, 27, 28
Genome-wide association studies (GWASs) have revealed that several common variants of OXTR, including rs7632287, rs237887, rs2268491, and rs2254298, might be associated with ASD,36 and their accumulation might be positively associated with the severity of ASD symptoms.37 In addition, there have been attempts to link the common OXTR variants, rs2254298 and rs53576, to the efficacy of intranasal oxytocin in alleviating ASD symptoms, but the findings are controversial.38,39 Rare variants, including de novo mutations (DNMs) and rare inherited variants (RIVs), contribute to ASD aetiology, with DNMs showing relatively strong functional effects.40 Several ASD studies, employing whole-exome sequencing and whole-genome sequencing (WGS), have detected functional DNMs, leading to the prioritisation of some candidate genes in ASD.41, 42, 43, 44 DNMs in coding regions reportedly contribute to ∼30% of all cases of simplex autism families and ∼45% of cases diagnosed in females.44 We have identified several ASD genes45,46 and demonstrated the contribution of DNMs to ASD aetiology in brain-size-related genes47 and vitamin-related genes,48 as well as revealed the convergence and divergence in DNMs among three ASD subcategories.49 Furthermore, copy number variations (CNVs) contributed to the risk of developing ASD50, 51, 52, 53 and might account for ∼4% of ASD cases.53 However, whether the rare genetic variants in OTRGs are associated with ASD pathology remains unclear.
Herein, this study systematically compared the rare SNV variants and CNVs in OTRGs between ASD and control groups, in terms of sex and non-verbal intelligence quotient (NVIQ), to investigate their potential roles in ASD pathology. Furthermore, by performing a comprehensive integrative analysis, we prioritised potential oxytocin-related molecular biomarkers that may contribute to the aetiology of ASD.
Methods
Curation of OTRGs
A total of 963 OTRGs (Supplemental Table 1) were manually curated based on the following selection criteria: (i) directly or indirectly related to oxytocin [genes directly related to oxytocin had the following features: 1) encode a precursor protein that is processed to produce oxytocin and neurophysin I; 2) encode oxytocin receptors; 3) alter plasma oxytocin levels or oxytocin expression following knockout; 4) exhibit differential expression following oxytocin administration; 5) their mRNA expression level can be used as a marker for oxytocin signalling. Genes indirectly related to oxytocin were defined as additional genes that co-occurred with “oxytocin” in the title of PubMed studies]; (ii) interacted with genes directly related to oxytocin in the inBio Map database54 and with a confidence score equal to 1; (iii) involved in the oxytocin signalling pathway (hsa04921) in the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database55; and (iv) interacted with oxytocin in manually curated chemical–gene/protein interactions sourced from the Comparative Toxicogenomics Database (CTD).56 In addition, 208 core OTRGs were defined as OTRGs solely sourced from the PubMed, KEGG, and CTD databases. The OTRGs, as well as the core OTRGs, were used for burden analysis.
Data collection and processing
DNMs and clinical information (sex and NVIQ) from 6,511 probands with ASD and from 3,391 controls were sourced from the Gene4Denovo database57 and, if available, from the supplementary data of corresponding published papers, respectively (Supplemental Table 2). Inherited variants and clinical information (sex and NVIQ) were gathered from 1,786 quad families in the Simons Simplex Collection (SSC).58 De novo CNVs (dnCNVs), inherited CNVs (ihCNVs), and clinical information (sex and NVIQ) from 15,581 probands with ASD and 6,107 controls were collated from the AutDB database59 (Supplemental Table 3). All the diagnostic and selection criteria for probands and controls, matching criteria for controls have been described in the original papers.57, 58, 59 Similar to our previous studies,60,61 detailed variant annotation was performed using ANNOVAR62 and VarCards.63 RIVs were defined as inherited variants with a minor-allele frequency below 0·1%. ReVe64 was used to predict deleterious missense (Dmis) variants with scores > 0·7. Loss-of-function (LoF) and Dmis variants were considered functional mutations. Gene annotation and pathogenicity estimation of CNVs were performed using ClassifyCNV.65 CNVs covering at least one of the OTRGs were defined as OTRG-associated CNVs and used for burden analysis.
Burden analysis of OTRGs
The mutation burden analysis was performed between the following: ASD and control subjects; ASD and control subjects of the same sex; male and female probands; and ASD probands within each of the different classes of NVIQs (IQ ≤ 50, 50 < IQ ≤ 80, and IQ > 80) and control subjects in the DNM, RIV, and CNV datasets.
CNVs of different lengths may carry different numbers of OTRGs in ASD probands and controls. Therefore, the following normalised equation was designed to calculate the CNV abundance in OTRGs as follows:
where, i is the ith affected individual (ASD proband or control); n is the total number of affected individuals (ASD probands or controls); j is the jth affected OTRG in the ith affected individual, and m is the number of affected OTRGs in an affected individual.
For comparison analysis, CNV count and normalised abundance of OTRG-associated dnCNVs were both used for CNV burden analysis. A similar burden analysis between ASD probands and controls was performed with the normalised abundance of OTRG-associated ihCNV.
Combined model for gene prioritisation
Inspired by the methods used by Krumm et al.58,66 we designed a comprehensive model by integrating DNMs and CNVs to prioritise the contribution of each protein-coding gene to ASD aetiology, which can be summarised as follows:
where t stands for the three types of genetic variations (t = 1 for DNM, t = 2 for dnCNV, and t = 3 for ihCNV), bi,t is the burden ratio of gene i in the tth type of rare genetic variations between ASD probands and controls, calculated as follows:
Where wt represents the weight of the tth type of rare genetic variations contributing to ASD aetiology, quantified using the odds ratio between ASD and control subjects with all reference genes from our collected data. Estimation of gene scores used disruptive genetic variations consisting of functional DNMs, pathogenic dnCNVs, and pathogenic ihCNVs. In addition, the odds ratios (weights) estimated by Krumm et al.66 were included for comparative analysis. Therefore, by combining this model with two sources of weights, two contribution scores were produced for each gene: the O-Gene_score (using weights estimated by our study) and the K-Gene_score (using weights estimated by Krumm et al.66). Finally, the contribution scores were used for OTRG prioritisation, and genes for which the O-Gene_score and K-Gene_score were both > 0 were considered to positively contribute to ASD aetiology.
Metascape67 was used for biological pathway enrichment with genes of interest. Significantly enriched biological pathways were defined by a Q value < 0·05.
Data collection for comprehensive integration analysis
ASD genes identified by genetic variations were collected from the following three sources: (1) ASD candidates defined in our previous study on NDDs;68 (2) genes retrieved from the Simons Foundation Autism Research Initiative (SFARI) database (https://gene.sfari.org/); and (3) genes documented in the AutDB database.59 Gene-level LoF intolerance (pLI) scores were downloaded from the Genome Aggregation Database (gnomAD)69 (v2.2.1). Publicly available ASD brain transcriptome data were retrieved from related studies70,71 and our previous study.68 Genes with a false discovery rate (FDR) below 0·05 were deemed significantly differentially expressed, as in our previous study.68 Brain expression data of humans were downloaded from the BrainSpan database (http://www.brainspan.org/). Human protein-protein interaction (PPI) data were retrieved from the STRING (https://string-db.org/) and InWeb_IM50 (http://www.intomics.com/inbio/map) databases.
Permutation test and network construction
The permutation test was performed on the positively contributed OTRGs (PC-OTRGs) and the collected known ASD genes based on BrainSpain co-expression and PPI data to evaluate their functional connections. Similar to previous studies,47,48,72 gene pairs co-expressed with |R| > 0·8 in the human brain73 or PPI with a score > 400 in STRING or a score > 0·8 in InWeb_IM50,74 were considered as connected. In brief, the numbers of connected genes within the 172 PC-OTRGs and the collected known ASD genes and their connections were compared with corresponding numbers produced by 172 random genes of 1,000,000 iterations, as in our previous study.73
PC-OTRGs and their connected ASD core genes were used to construct a weighted network based on co-expressed and PPI gene pairs used in the permutation test. The node in the weighted network was quantified by adjusted weight (NW) expressed as follows:
Network figures were generated using Cytoscape v.3.8.0 (https://cytoscape.org).
Statistical analysis
A one-sided rate ratio (RR) test was used to estimate whether the burden of OTRGs is significantly higher in the ASD probands than in corresponding controls using the ‘rateratio.test’ function in the ‘rateratio.test’ R package (v1.0-2; https://cran.r-project.org/web/packages/rateratio.test/index.html). A chi-square test was used to validate associations between the OTRG-associated CNVs and ASD aetiology. Participants without sex or NVIQ information were excluded in corresponding subgroup comparisons. For all comparisons, differences were considered statistically significant at p < 0·05.
Role of funding source
The funders had no role in the study design, data collection, data analyses, interpretation, and writing of the manuscript.
Results
OTRG summary and mutation burden of OTRGs in patients with ASD
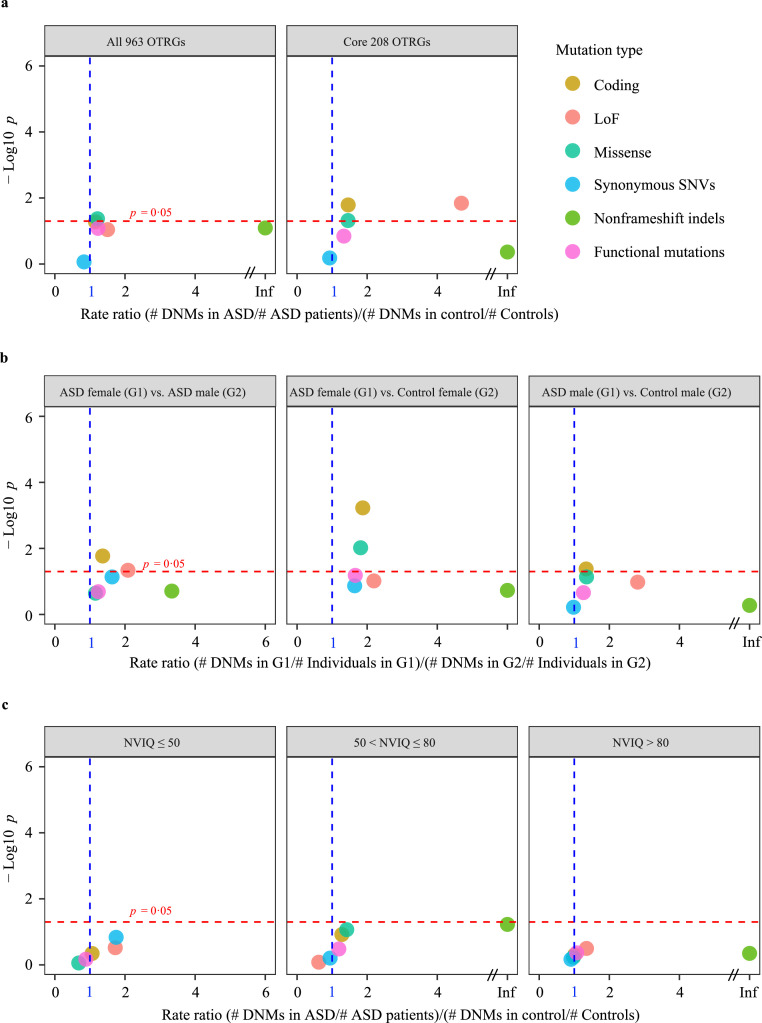
Of the 963 OTRGs curated from four sources, 208 were defined as core OTRGs (Methods section). In addition, we catalogued 8,175 coding DNMs in 6,511 ASD probands and 3,629 coding DNMs in 3,391 controls from 16 studies (Supplemental Figure 1). The coding DNMs in the 963 OTRGs were used for mutation burden analysis. In the ASD probands, there was a slightly higher non-significant number of both coding DNMs (RR = 1·16, p = 5·27 × 10−2; RR test) and LoF DNMs (RR = 1·50, p = 9·08 × 10−2; RR test) than those in the controls; this difference was significantly higher in missense DNMs (RR = 1·21, p = 4·26 × 10−2; RR test, Figure 1a and Supplemental Tables 4–5). When focusing on the 208 core OTRGs, significant differences in mutation burdens between patients with ASD and the controls were observed in both coding DNMs (RR = 1·46, p = 1·62 × 10−2; RR test) and LoF DNMs (RR = 4·69, p = 1·44 × 10−2; RR test, Figure 1a). These results indicate that the ASD probands carry a higher coding DNM burden in core OTRGs than the controls, especially LoF DNMs.
Figure 1.
DNM burden of OTRGs in patients with ASD compared with that in controls. (a) Differences in DNM burden between patients with ASD (n = 6,511) and control subjects (n = 3,391) for each mutation type in all OTRGs (n = 963) and core OTRGs (n = 208). DNM, de novo mutation; OTRGs, oxytocin-related genes; loss-of-function (LoF) including stop-gain, stop-loss, and splicing sites SNVs and frameshift indels; Deleterious missense, variants with ReVe score > 0.7; Functional mutations, combination of LoF mutations and deleterious missense mutations. (b) Differences in DNM burden between sex-stratified patients with ASD (nfemale = 769; nmale = 3,846) and control subjects (nfemale = 1,011; nmale = 900) for each mutation type in all curated OTRGs. (C) Differences in DNM burden between NVIQ-stratified patients with ASD (nNVIQ ≤ 50 = 333, n50 < NVIQ ≤ 80 = 617, and nNVIQ > 80 = 1,558) and control subjects (n = 1,911) for each mutation type in all the OTRGs. All the p values were calculated by the rate ratio test. NVIQ, non-verbal intelligence quotient.
Notably, ASD exhibits a strong male bias, with a male: female ratio of 4: 1.75 Therefore, mutation burden analysis was employed to investigate whether there was a sex-based difference in the DNM burden. The mutation rates of coding DNMs were significantly higher in female than in male probands with ASD (RR = 1·36, p = 0·02; RR test) as well as in female probands with ASD than in female controls (RR = 1·87, p = 5·91 × 10−4; RR test, Figure 1b). However, only a slightly significant difference in coding DNM burden was observed when comparing male probands with ASD and male controls (RR = 1·34, p = 0·04; RR test, Figure 1b). Patients with ASD have diverse intelligence levels, a crucial phenotypic marker of ASD;76 therefore, proband stratification was attempted to evaluate the difference in DNM burden in OTRGs between NVIQ-stratified probands and the controls, but no significant difference was observed (Figure 1c).
To identify whether there were significant differences in the burden of RIVs of OTRGs between probands with ASD and controls, we catalogued 4,244 and 4,223 coding RIVs of OTRGs from 1,786 patients with ASD and 1,786 controls, respectively (Supplemental Tables 6–7). The results exhibited no significant differences in mutation burdens between ASD probands and controls, even when the RIVs were stratified by different inheritance patterns (Supplemental Figure 2a). Furthermore, no significant differences in the mutation burden were observed between a subset of ASD probands and controls stratified by sex or NVIQ for any type of RIVs (Supplemental Figure 2b–c).
OTRG-associated CNVs significantly associated with ASD aetiology
CNVs, including dnCNVs and ihCNVs, significantly contribute to ASD aetiology.50 However, whether the burden of CNVs of OTRGs significantly differs between ASD probands and control subjects remains unexplored. Therefore, 16,902 CNVs (697 dnCNVs and 16,205 ihCNVs) from 15,581 ASD probands and 11,921 CNVs (212 dnCNVs and 11,709 ihCNVs) from 6,107 controls of 12 studies were collected (Supplemental Figure 1; Supplemental Tables 3 and 8). Of the dnCNVs in ASD probands, ∼28·98% (202/697) were associated with OTRGs, which was significantly higher than that in the controls (∼9·91%, 21/212; Chi-square test, p = 1⋅58 × 10−8; Supplemental Table 9). In addition, there were significantly more OTRG-associated dnCNVs in ASD probands than in the controls in “All CNVs” (RR = 3·71, p = 1·03 × 10−11; RR test), “Deletion CNVs” (RR = 5·41, p = 5·20 × 10−10; RR test), and “Pathogenic CNVs” (RR = 9·65, p = 4·22 × 10−8; RR test, Figure 2a). When focusing on the 208 core OTRGs with dnCNVs, similar trends were observed (Figure 2a).
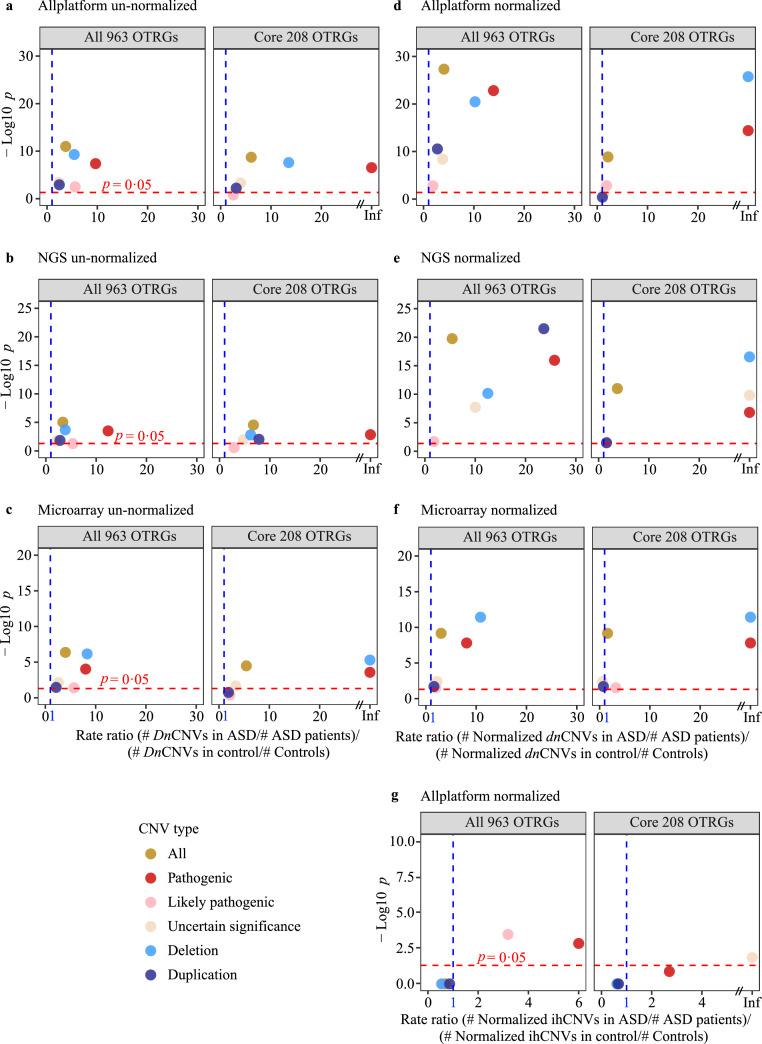
Figure 2.
CNV burden of OTRGs in patients with ASD compared with that in controls. (a) Differences in dnCNV burden between patients with ASD (n = 15,581) and control subjects (n = 6,017) for each CNV type in all OTRGs and core OTRGs based on raw dnCNV counts. dnCNV, de novo CNV. (b) Differences in dnCNV burden of OTRGs between patients with ASD (nNGS = 8,338) and control subjects (nNGS = 3,437) based on raw dnCNV counts from NGS platform. (c) Differences in dnCNV burden of OTRGs between patients with ASD (nmicroarray = 7,243) and control subjects (nmicroarray = 2,580) using raw dnCNV counts from microarray analysis. (d) Differences in dnCNV burden of OTRGs between patients with ASD (n = 15,581) and control subjects (n = 6,017) with normalised dnCNV counts. (e) Differences in dnCNV burden of OTRGs between patients with ASD (nNGS = 8,338) and control subjects (nNGS = 3,437) using normalised dnCNV counts from the NGS platform. (f) Differences in dnCNV burden of OTRGs between patients with ASD (nmicroarray = 7,243) and control subjects (nmicroarray = 2,580) using normalised dnCNV counts from microarray analysis. (g) Differences among the ihCNV burden between patients with ASD (n = 15,581) and control subjects (n = 6,017) for each CNV type in all the OTRGs and the core OTRGs based on normalised ihCNV counts. All the p values were generated by the rate ratio test. ihCNV, inherited CNV.
As CNV data were collected mainly from two platforms, namely next-generation sequencing (NGS) and microarrays, all ASD probands and control subjects were separated by platforms to estimate whether significant differences in the dnCNV burden were affected by specific platforms. Consequently, irrespective of whether the dataset was used with NGS or microarrays alone, significantly higher burdens of OTRG-associated dnCNVs were consistently observed for several CNV types in ASD probands than in controls, including “All CNVs” (RR = 3·37, p = 9·37 × 10−6 for NGS; RR = 3·99, p = 4·29 × 10−7 for microarrays; RR test), “Deletion CNVs” (RR = 3·85, p = 2·02 × 10−4 for NGS; RR = 8·31, p = 6·94 × 10−7 for microarrays; RR test), and “Pathogenic CNVs” (RR = 12·37, p = 3·11 × 10−4 for NGS; RR = 8·01, p = 9·35 × 10−5 for microarrays; RR test, Figure 2b–c).
Potential bias may result from directly using the number of OTRG-associated CNVs for burden analysis, as each specific CNV may cover different numbers of OTRGs. To curtail this bias, CNV abundance in OTRGs was normalised based on the gene length and used for burden analysis. Significantly higher dnCNV burdens in OTRGs were found in ASD probands than in controls using normalised data from both platforms, NGS alone, or microarray alone in most CNV types (Figure 2d–f). In addition, as more accurate findings can be achieved with larger sample sizes, the normalised CNV data from all platforms were used for downstream analysis.
As ihCNVs also contribute significantly to ASD aetiology, we evaluated the burden of OTRG-associated ihCNVs. There was a significantly higher burden in ASD probands than in controls for several types of CNVs, including “Pathogenic CNVs” (RR = Inf, p = 1·46 × 10−3; RR test) and “Likely pathogenic CNVs” (RR = 3·19, p = 3·38 × 10−4; RR test, Figure 2g and Supplemental Table 10). However, this contribution was much lower than that estimated for the corresponding types of normalised dnCNVs (Figure 2d, g). These results indicate that the genetic contribution of OTRGs is significantly associated with ASD aetiology in the order of dnCNVs > ihCNVs, and that “Pathogenic CNVs” play a prominent role in both, which was confirmed by association analysis (Supplemental Table 11 and Supplemental Figure 3a).
To further explore whether the sex bias observed in the DNM burden (Figure 1b) existed for the CNV burden, we used the normalised dnCNV data from all platforms for burden analysis. Consistent with those from the DNM burden analysis, OTRG-associated dnCNV burdens were significantly higher in female ASD probands than in male ASD probands and female controls (Supplemental Table 9 and Supplemental Figure 3b). Moreover, significantly higher OTRG-associated dnCNV burdens were found in ASD probands with NVIQ ≤ 50 than in the controls for several types of CNVs, especially “Pathogenic CNVs” (RR = 99·29, p = 1·10 × 10−14; RR test, Supplemental Figure 3c).
Combined model for OTRG prioritisation
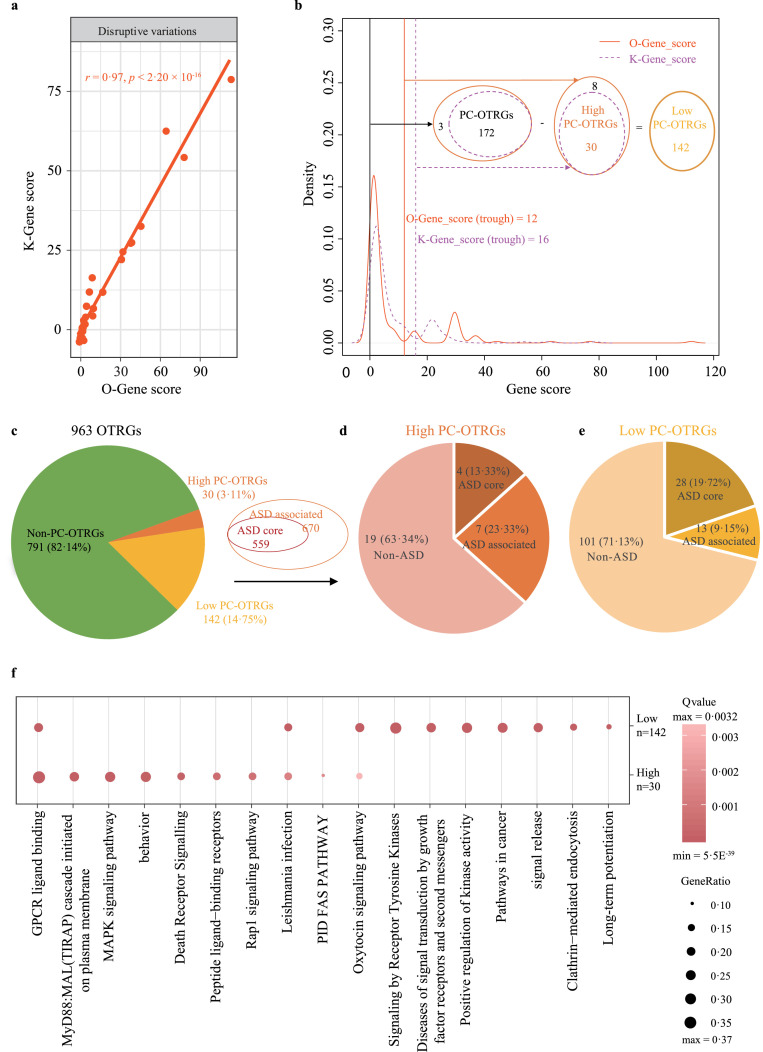
By integrating disruptive DNMs, dnCNVs, and ihCNVs, we designed a combined model to comprehensively evaluate the contribution of each OTRG to ASD aetiology, with weights estimated in our study (O-Gene_score) (Supplemental Tables 12–13) and those determined by Krumm et al.66 (K-Gene_score). We found a significant correlation (r = 0·97, p < 2·20 × 10−16, Pearson correlation) between gene scores generated using the two sources of weights, indicating the reliability of weights estimated in our study using collected data (Figure 3a). In addition, based on gene score density distribution (Figure 3b), 172 OTRGs were identified to positively contribute to ASD aetiology (PC-OTRGs) with both O-Gene_score > 0 and K-Gene_score > 0. There was an evident trough adjacent to the highest peak in the density distributions for both O-Gene_score and K-Gene_score (Figure 3b), indicating that two distinct gene clusters contribute to ASD aetiology in these PC-OTRGs. Therefore, the bottom values of 12 for O-Gene_score and 16 for K-Gene_score were used for PC-OTRG stratification. From 172 PC-OTRGs, we obtained 30 high PC-OTRGs with both O-Gene_score > 12 and K-Gene_score >16, whereas the remaining 142 PC-OTRGs were considered low PC-OTRGs.
Figure 3.
Summary of OTRG prioritisation using a combined model. (a) Correlation of K-Gene score and O-Gene score for OTRGs. K-Gene and O-Gene scores were produced using a combined model with rare disruptive variations and weights estimated by either Krumm et al. or the present study, respectively. Rare disruptive variations consisted of functional DNMs from patients with ASD (n = 6,511) and control subjects (n = 3,391), as well as pathogenic dnCNVs and pathogenic ihCNVs from patients with ASD (n = 15,581) and control subjects (n = 6,017). The p value was calculated by the Pearson correlation test. (b) A total of 172 PC-OTRGs were identified, consisting of 30 high PC-OTRGs and 142 low PC-OTRGs based on the OTRG density plot of K-Gene scores and O-Gene scores. PC-OTRGs, positively contributed OTRGs. (c) Composition of high and low PC-OTRGs in all 963 OTRGs. (d) Composition of ASD core and ASD-associated genes in the 30 high PC-OTRGs based on the annotation of collected 559 ASD core and 670 ASD-associated genes. (e) Composition of ASD core and ASD-associated genes in 142 low PC-OTRGs based on the annotation of collected 559 ASD core and 670 ASD-associated genes. (f) Top 10 enriched terms based on 30 high PC-OTRGs and 142 low PC-OTRGs.
To further explore the relationship between these high/low PC-OTRGs and ASD risk, 1,229 ASD genes consisting of 559 ASD core genes and the remaining 670 ASD-associated genes were collected from our previous NDD study,68 SFARI, and AutDB databases (Figure 3c). We found that 36·66% of the high PC-OTRGs and 28·87% of the low PC-OTRGs (Figure 3d-e) were shared with ASD genes (32 ASD core and 20 ASD-associated genes; Supplemental Figure 4a, p = 1·16 × 10−20, hypergeometric test). The top three genes among the 30 high PC-OTRGs were MAPK3 (ASD-associated gene), SHANK3 (ASD core gene), and PRKAB2 (Supplemental Figure 5). In addition, 78 of the 172 PC-OTRGs were significantly differentially expressed between ASD and control cortices (Supplemental Figure 4b, p = 9·67 × 10−8, hypergeometric test).
The Metascape tool67 was used to further annotate the 30 high and 142 low PC-OTRGs. As expected, “GPCR ligand binding” and “oxytocin signalling pathway” were significantly enriched in both high and low PC-OTRGs (Figure 3f and Supplemental Table 14). Moreover, several terms were specifically enriched in either high PC-OTRGs (e.g., “MAPK signalling pathway”) or low PC-OTRGs (e.g., “signalling by receptor tyrosine kinase”) (Figure 3f).
PC-OTRGs are functionally associated with ASD genes
A permutation test with 1,000,000 iterations was employed to determine the relationship between the 172 PC-OTRGs and each of the 1,177 unshared ASD genes and 527 ASD core genes (Supplemental Figure 4a) based on the co-expression data from the human brain and PPI data. PC-OTRGs were significantly more connected (co-expressed and/or PPI) with ASD core genes than random expectations. Overall, 169 of the 172 PC-OTRGs (p = 0; permutation test, Supplemental Figure 6a) were connected with 427/527 ASD core genes (p = 3·0 × 10−5; permutation test, Supplemental Figure 6b), with 3,052 connections (p = 0; permutation test, Supplemental Figure 6c). Specifically, based on human brain expression data, 60 of the 172 PC-OTRGs (p = 7·27 × 10−4; permutation test, Supplemental Figure 6d) were co-expressed with 194/527 ASD core genes (p = 1·5 × 10−5; permutation test, Supplemental Figure 6e), with 618 connections, which is significantly higher than that of random expectations (p = 1·58 × 10−2; permutation test, Supplemental Figure 6f). In the PPI data, 168/172 PC-OTRGs (p = 0; permutation test, Supplemental Figure 6g) interacted with 387/527 ASD core genes (p = 3·56 × 10−4; permutation test, Supplemental Figure 6h) through 2,507 connections (p = 0; permutation test, Supplemental Figure 6i), which also significantly exceeded that of random expectations. Similarly, a significantly higher connection was observed for PC-OTRGs and all ASD genes than for random expectations (Supplemental Figure 7).
The 172 PC-OTRGs accounted for 0·67% (104/15,581) of patients with ASD with pathogenic CNVs and 2·26% (147/6,511) of patients with functional DNMs in the CNV and DNM cohorts (Supplemental Figure 8a). In addition, 66·98% (286/427) of ASD core genes (Supplemental Figure 6b), connected to 172 PC-OTRGs, positively contributed to ASD aetiology. Considering the 172 PC-OTRGs and the 286 ASD core genes connected to them (458 potential molecular biomarkers), 1·09% (170/15,581) of patients with ASD in the CNV cohort were estimated to carry pathogenic CNVs (Supplemental Figure 8b). Due to the unavailability of sex and NVIQ information for most patients with ASD in the CNV cohorts, the stratification based on sex and NVIQ was not pursued further. In the DNM cohort, 9·72% (633/6,511) of patients with ASD carried functional DNMs in 458 molecular biomarkers (Supplemental Figure 8b) where information on either sex, NVIQ, or both was available for 74·09% (469/633), 39·18% (248/633), and 35·55% (225/633) of patients with ASD, respectively (Supplemental Table 15). Therefore, sex and NVIQ differences were estimated between patients affected by the functional DNMs of these molecular biomarkers and those unaffected. These affected patients showed a significantly higher female: male ratio (p = 5·21 × 10−6; chi-square test) and a low (NVIQ ≤ 80): high (NVIQ > 80) NVIQ ratio, which were 1·69 and 1·97 times higher than those in the unaffected group, respectively (p = 3·21 × 10−7; chi-square test; Supplemental Table 15). In addition, the low: high NVIQ ratio was significantly higher (2·26 times) in female than in male patients (p = 1·92 × 10−2; chi-square test; Supplemental Table 15).
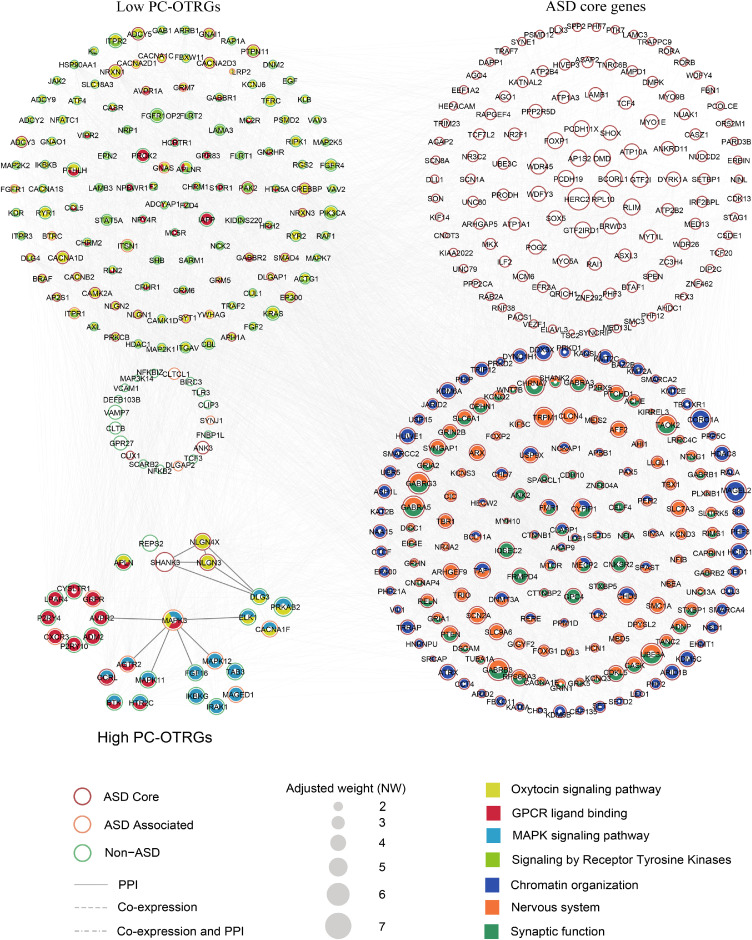
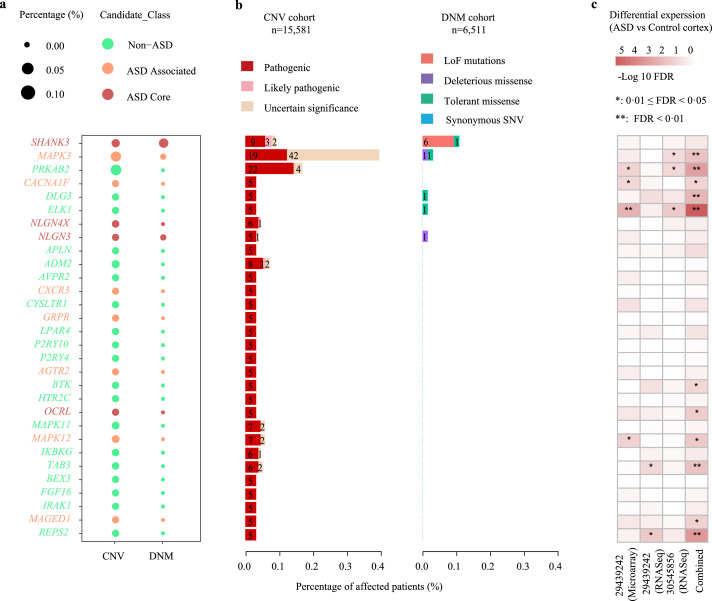
Next, the 172 PC-OTRGs and their connected 286 ASD core genes, which positively contributed to ASD aetiology, were used for network construction, respectively, based on co-expression and PPI. Our results showed that these genes were functionally convergent (Figure 4a). Specifically, 93·33% (28/30) of high- and 85·92% (122/142) of low-PC-OTRGs were involved in at least one of the four enriched pathways, namely “oxytocin signalling pathway”, “GPCR ligand binding”, “MAPK signalling pathway”, and “signalling by receptor tyrosine kinases” (Figure 4a and Supplemental Table 14). Furthermore, ∼59·44% (170/286) of their connected ASD core genes are involved in chromatin organisation, nervous system development, and synaptic function (Figure 4a and Supplemental Table 15), which are known networks contributing to ASD aetiology.77 Among these high PC-OTRGs (Figure 5), MAPK3 was the most prominent gene, participating in three of the enriched pathways, followed by SHANK3, NLGN4X, and NLGN3 (Figure 4b and Supplemental Table 16). In addition, MAPK3 seemed to be affected more frequently by CNVs rather than DNMs (Figure 5a–b) and was significantly differentially expressed in the ASD cortex as compared to that in the normal control cortex (Figure 5c). These results imply oxytocin-related complex networks that may be associated with the aetiology of ASD.
Figure 4.
Co-expression and PPI of the potential biomarkers. Co-expression and PPI networks for the potential molecular biomarkers identified, consisting of 172 PC-OTRGs and their connected 286 ASD core genes. Only potential molecular biomarkers with connections are displayed. Biomarkers involved in different oxytocin or ASD-related pathways are filled with corresponding colours. Circle size represents the adjusted contribution of each potential molecular biomarker to ASD aetiology. Interconnections of interested genes within high PC-OTRGs are highlighted with a darker colour.
Figure 5.
Summary of 30 potential molecular biomarkers (high PC-OTRGs) for ASD. (a) Percentage and number of patients affected by each type of rare genetic variations in potential molecular biomarkers in the corresponding CNV and DNM cohorts. (b) Percentage of patients affected by rare disruptive variations of each potential molecular biomarker in each respective cohort. Rare disruptive variations include pathogenic CNVs and functional DNMs. The size of the dot represents the percentage of affected patients. (c) Differential expression results based on publicly available ASD cortex transcriptome analysis data.
Discussion
It is well documented that ASD is primarily attributed to rare mutations, which may cause distinct pathophysiological characteristics that may yield heterogeneous effects on treatment response.14,40 Nonetheless, the potential intricate correlations between rare genetic variations in OTRGs and ASD aetiology have not been systematically investigated. Therefore, in this study, we compared the rare genetic variations in OTRGs between ASD probands and controls in terms of sex, NVIQ, and inheritance. Our results revealed that: (1) OTRGs significantly contributed to ASD aetiology in the order of dnCNVs > ihCNVs > DNMs at the individual level; (2) significantly higher mutation burdens of OTRGs exist in female patients than in male patients and female controls, supporting the female-protective effect in ASD;78 and (3) significantly higher dnCNV burdens in OTRGs are observed in stratified ASD patients with NVIQ ≤ 50 than in controls. In addition, rare disruptive variations of OTRGs exhibited a significantly dominant association compared with other types of rare variations. Furthermore, we prioritised 458 potential oxytocin-related molecular biomarkers, based on rare disruptive variations, comprising 172 PC-OTRGs and 286 ASD core genes connected to them, with functional DNMs and pathogenic CNVs covering ∼10% (633/6 511) and ∼1% (170/15 581) of patients with ASD, respectively.
By integrating rare disruptive variations, our combination model identified 172 PC-OTRGs that were significantly enriched in several ASD-related terms, especially “MAPK signalling pathway” contributing to ASD pathogenesis by affecting brain development.79 Although, the oxytocin-regulated MAPK pathway, via MEK1/2 (MAPK1/MAPK3), has been demonstrated to be essential for the anxiolytic effect of oxytocin in the paraventricular nucleus,5 the mechanism underlying how oxytocin regulates the MAPK pathway, thereby affecting ASD core symptoms, warrants further exploration. With comprehensive integration of publicly available genetic and transcriptomic data, two PC-OTRGs, MAPK3 and SHANK3, were highlighted as top two genes for their association with ASD aetiology. Notably, we observed that an ∼1Mb MAPK3-carrying CNV hotspot in the 16p11.2 region presents in 0·39% (61/15 581) of patients with ASD (Supplemental Figure 9). In addition, the 16p11.2 region contained two additional ASD genes, TAOK2 and KCTD13 (Supplemental Figure 9), which reportedly affect autism-related neurodevelopment and cognition as well as synaptic transmission, via RhoA signalling,80,81 a downstream target of oxytocin signalling (https://www.kegg.jp/pathway/hsa04921). These previous studies show that MAPK3 could be a promising oxytocin-related risk gene for ASD even though the intricate regulatory network needs further exploration. In addition, we observed that DNMs most frequently occurred in SHANK3, with six functional DNMs and nine pathogenic CNVs (Figure 5 and Supplemental Table 17). Given that loss of oxytocin-positive neurons has been observed in Shank3b-knockout mice,82 oxytocin administration could improve social behaviour and alleviate synaptic plasticity deficits in Shank3-deficient rats,83 we speculate that patients with SHANK3 mutations could potentially benefit from oxytocin intervention. Although OXTR is not known to carry any functional coding DNMs or to be covered by any pathogenic CNVs, this receptor may be related to ASD behaviour, clinical phenotypes, and oxytocin response via common variants36,37,39 and epigenetic modifications.84 The difference between the burden of pathogenic CNVs of OTRGs in ASDs and controls was found to be significantly higher than that of functional DNMs, whereas the occurrence of pathogenic CNVs of these molecular biomarkers in all patients with ASD was 10 times lower than that of functional DNMs. These findings are consistent with the notion that rare CNV variants exert a major effect on the ASD risk of individual patients, implying that the overall effect of functional DNMs of these molecular biomarkers at the population level may be a major factor associated with ASD aetiology.
Several limitations should be considered when interpreting these results in this study. A meta-analysis examining oxytocin trials in ASD reported no significant heterogeneity in the oxytocin responses,85 which may be due to the small number of clinical trials and sample size used in the study, as well as approaches for evaluating intervention efficiencies in the collected cohorts.14 Considering that 10∼11% of ASD probands carried oxytocin-related rare variants, it is possible that only a small portion of ASD probands might respond to oxytocin intervention. All these factors may contribute to the statistically non-significant difference in oxytocin response among different patients with ASD. Moreover, it should be acknowledged that the therapeutic responses to oxytocin treatment for ASD core symptoms may be associated with various treatment factors, such as dosage or dose frequency of oxytocin intervention in different developmental stages,14,30,86,87 OXTR genetics,39 and the interactions of dose frequency with OXTR genetics.33,38 Notably, ∼17.47% (80 of 458) of potential oxytocin-related molecular biomarkers and ∼19.72% (156 of 791) of non-PC-OTRGs (Supplemental Table 19) were significantly differentially expressed through modification of histone and/or DNA methylation in ASD cortex compared with that in control.88,89 For example, epigenetic modifications in OXTR,84,90,91 ASH1L,92 and RELN93 have showed associations to social deficits in ASD, raising the possibility in which epigenetic modifications could also play a role in oxytocin response. Furthermore, this study only focused on the genetic contribution of coding DNMs and RIVs, as well as dnCNV and ihCNVs from independent cohorts of ASD genetic studies. Each type of genetic variant was evaluated independently; more accurate findings may be obtained by considering their interactions using WGS, which could detect different types of genetic variations simultaneously. In addition, we did not include variants that could regulate gene expression at both the transcriptional and post-transcriptional levels.
In conclusion, based on rare disruptive variations, 458 potential oxytocin-related molecular biomarkers were prioritised as contributors to ASD aetiology, which may be valuable in both understanding the underlying mechanism of ASD pathophysiology and clinical investigation of oxytocin-based treatment of ASD. Therefore, future studies on oxytocin intervention in patients with ASD should consider the dosage or dose frequency of oxytocin in different developmental stages, genetic background, clinical phenotypes (sex and NVIQ), approaches for evaluating intervention efficiencies, and same-patient plasma oxytocin levels before and after the intervention to maximise clinical benefit.
Contributors
Zhongsheng Sun, Kun Xia, and Jinchen Li conceived, designed, and supervised the study. Tao Wang, Liqiu Liu and Tingting Zhao acquired and analysed the data. Tao Wang and Liqiu Liu verified the underlying data. Tianda Fan and Yi Li organized the article tables. Tao Wang, Huajing Teng and Yan Wang interpreted the data. Zhongsheng Sun, Tao Wang, and Tingting Zhao drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
The study was based on the data available at Gene4Denovo database (http://www.genemed.tech/gene4denovo/download), Simons Simplex Collection (SSC) (DOI: 10.1038/ng.3303), and AutDB database (http://autism.mindspec.org/autdb). The code of this study is available from the corresponding author upon reasonable request.
Declaration of interests
A patent application has been submitted by Tao Wang, Liqiu Liu, Zhongsheng Sun and the Institute of Zoology, Chinese Academy of Sciences based on these results.
Acknowledgements
This work was supported by the Guangdong Key Project “Development of new tools for diagnosis and treatment of Autism” (#2018B030335001 to ZSS), “Early diagnosis and treatment of autism spectrum disorders” (#202007030002 to ZSS), Strategic Priority Research Program of Chinese Academy of Sciences (XDPB16 to ZSS), National Natural Science Foundation of China (#31871191 to ZSS), Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, National Natural Science Foundation of China (#82130043 and #81730036 to Kun Xia), Science and Technology Major Project of Hunan Provincial Science and Technology Department (#2021SK1010 to Kun Xia), National Natural Science Foundation of China (#81801133 to JCL), Young Elite Scientist Sponsorship Program by CAST (#2018QNRC001 to JCL), Innovation-Driven Project of Central South University (#20180033040004 to JCL), and Natural Science Foundation of Hunan Province for outstanding Young Scholars (#2020JJ3059 to JCL).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104091.
Contributor Information
Jinchen Li, Email: lijinchen@csu.edu.cn.
Kun Xia, Email: xiakun@sklmg.edu.cn.
Zhongsheng Sun, Email: sunzs@biols.ac.cn.
Appendix. Supplementary materials
References
- 1.Levy SE, Mandell DS, Schultz RT. Autism. Lancet. 2009;374(9701):1627–1638. doi: 10.1016/S0140-6736(09)61376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LJ, Barrett CE. Neuroscience. Can oxytocin treat autism? Science. 2015;347(6224):825–826. doi: 10.1126/science.aaa8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viero C, Shibuya I, Kitamura N, et al. REVIEW: Oxytocin: Crossing the bridge between basic science and pharmacotherapy. CNS Neurosci Ther. 2010;16(5):e138–ee56. doi: 10.1111/j.1755-5949.2010.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintana DS, Lischke A, Grace S, Scheele D, Ma Y, Becker B. Advances in the field of intranasal oxytocin research: lessons learned and future directions for clinical research. Mol Psychiatry. 2021;26(1):80–91. doi: 10.1038/s41380-020-00864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grinevich V, Neumann ID. Brain oxytocin: how puzzle stones from animal studies translate into psychiatry. Mol Psychiatry. 2021;26(1):265–279. doi: 10.1038/s41380-020-0802-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Froemke RC, Young LJ. Oxytocin, neural plasticity, and social behavior. Annu Rev Neurosci. 2021;44:359–381. doi: 10.1146/annurev-neuro-102320-102847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen H. Neuroscience: The hard science of oxytocin. Nature. 2015;522(7557):410–412. doi: 10.1038/522410a. [DOI] [PubMed] [Google Scholar]
- 8.Jurek B, Neumann ID. The oxytocin receptor: From intracellular signaling to behavior. Physiol Rev. 2018;98(3):1805–1908. doi: 10.1152/physrev.00031.2017. [DOI] [PubMed] [Google Scholar]
- 9.Voinsky I, Bennuri SC, Svigals J, Frye RE, Rose S, Gurwitz D. Peripheral blood mononuclear cell oxytocin and vasopressin receptor expression positively correlates with social and behavioral function in children with autism. Sci Rep. 2019;9(1):13443. doi: 10.1038/s41598-019-49617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakos J, Srancikova A, Havranek T, Bacova Z. Molecular mechanisms of oxytocin signaling at the synaptic connection. Neural Plast. 2018;2018 doi: 10.1155/2018/4864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebstein RP, Knafo A, Mankuta D, Chew SH, Lai PS. The contributions of oxytocin and vasopressin pathway genes to human behavior. Horm Behav. 2012;61(3):359–379. doi: 10.1016/j.yhbeh.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa K, Matsumura K, Baba M, et al. Intranasal oxytocin administration ameliorates social behavioral deficits in a POGZ(WT/Q1038R) mouse model of autism spectrum disorder. Mol Brain. 2021;14(1):56. doi: 10.1186/s13041-021-00769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penagarikano O, Lazaro MT, Lu XH, et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci Transl Med. 2015;7(271):271ra8. doi: 10.1126/scitranslmed.3010257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geschwind DH. Oxytocin for autism spectrum disorder - down, but not out. N Engl J Med. 2021;385(16):1524–1525. doi: 10.1056/NEJMe2110158. [DOI] [PubMed] [Google Scholar]
- 15.Nakata M, Kimura R, Funabiki Y, Awaya T, Murai T, Hagiwara M. MicroRNA profiling in adults with high-functioning autism spectrum disorder. Mol Brain. 2019;12(1):82. doi: 10.1186/s13041-019-0508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modahl C, Green L, Fein D, et al. Plasma oxytocin levels in autistic children. Biol Psychiatry. 1998;43(4):270–277. doi: 10.1016/s0006-3223(97)00439-3. [DOI] [PubMed] [Google Scholar]
- 17.Al-Ayadhi LY. Altered oxytocin and vasopressin levels in autistic children in Central Saudi Arabia. Neurosciences. 2005;10(1):47–50. [PubMed] [Google Scholar]
- 18.Elissar A, Jean-René D, Tiziana Z, Evelyn H, Marion L, Angela S. Promoting social behavior with oxytocin in high-functioning autism spectrum disorders. PNAS USA. 2010;107(9):4389–4394. doi: 10.1073/pnas.0910249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker KJ, Oztan O, Libove RA, et al. Intranasal oxytocin treatment for social deficits and biomarkers of response in children with autism. PNAS USA. 2017;114(30):8119. doi: 10.1073/pnas.1705521114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernaerts S, Boets B, Steyaert J, Wenderoth N, Alaerts K. Oxytocin treatment attenuates amygdala activity in autism: a treatment-mechanism study with long-term follow-up. Transl Psychiatry. 2020;10(1):383. doi: 10.1038/s41398-020-01069-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe T, Abe O, Kuwabara H, et al. Mitigation of sociocommunicational deficits of autism through oxytocin-induced recovery of medial prefrontal activity: a randomized trial. JAMA Psychiatry. 2014;71(2):166–175. doi: 10.1001/jamapsychiatry.2013.3181. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T, Kuroda M, Kuwabara H, et al. Clinical and neural effects of six-week administration of oxytocin on core symptoms of autism. Brain. 2015;138(Pt 11):3400–3412. doi: 10.1093/brain/awv249. [DOI] [PubMed] [Google Scholar]
- 23.Yatawara CJ, Einfeld SL, Hickie IB, Davenport TA, Guastella AJ. The effect of oxytocin nasal spray on social interaction deficits observed in young children with autism: a randomized clinical crossover trial. Mol Psychiatry. 2016;21(9):1225–1231. doi: 10.1038/mp.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamasue H, Okada T, Munesue T, et al. Effect of intranasal oxytocin on the core social symptoms of autism spectrum disorder: a randomized clinical trial. Mol Psychiatry. 2020;25(8):1849–1858. doi: 10.1038/s41380-018-0097-2. [DOI] [PubMed] [Google Scholar]
- 25.Bernaerts S, Boets B, Bosmans G, Steyaert J, Alaerts K. Behavioral effects of multiple-dose oxytocin treatment in autism: a randomized, placebo-controlled trial with long-term follow-up. Mol Autism. 2020;11(1):6. doi: 10.1186/s13229-020-0313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guastella AJ, Gray KM, Rinehart NJ, et al. The effects of a course of intranasal oxytocin on social behaviors in youth diagnosed with autism spectrum disorders: a randomized controlled trial. J Child Psychol Psychiatry. 2015;56(4):444–452. doi: 10.1111/jcpp.12305. [DOI] [PubMed] [Google Scholar]
- 27.Dadds MR, Macdonald E, Cauchi A, Williams K, Levy F, Brennan J. Nasal oxytocin for social deficits in childhood autism: A randomized controlled trial. J Autism Dev Disord. 2014;44(3):521–531. doi: 10.1007/s10803-013-1899-3. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Huang X, Ebstein RP, Yu R. Intranasal oxytocin in the treatment of autism spectrum disorders: A multilevel meta-analysis. Neurosci Biobehav Rev. 2021;122:18–27. doi: 10.1016/j.neubiorev.2020.12.028. [DOI] [PubMed] [Google Scholar]
- 29.Kato Y, Kuwabara H, Okada T, et al. Oxytocin-induced increase in N,N-dimethylglycine and time course of changes in oxytocin efficacy for autism social core symptoms. Mol Autism. 2021;12(1):15. doi: 10.1186/s13229-021-00423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamasue H, Kojima M, Kuwabara H, et al. Effect of a novel nasal oxytocin spray with enhanced bioavailability on autism: A randomized trial. Brain. 2022 doi: 10.1093/brain/awab291. [DOI] [PubMed] [Google Scholar]
- 31.Alaerts K, Bernaerts S, Prinsen J, Dillen C, Steyaert J, Wenderoth N. Oxytocin induces long-lasting adaptations within amygdala circuitry in autism: a treatment-mechanism study with randomized placebo-controlled design. NPP. 2020;45(7):1141–1149. doi: 10.1038/s41386-020-0653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anagnostou E, Soorya L, Chaplin W, et al. Intranasal oxytocin versus placebo in the treatment of adults with autism spectrum disorders: A randomized controlled trial. Mol Autism. 2012;3(1):16. doi: 10.1186/2040-2392-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosaka H, Okamoto Y, Munesue T, et al. Oxytocin efficacy is modulated by dosage and oxytocin receptor genotype in young adults with high-functioning autism: a 24-week randomized clinical trial. Transl Psychiatry. 2016;6(8):e872. doi: 10.1038/tp.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikich L, Kolevzon A, King BH, et al. Intranasal oxytocin in children and adolescents with autism spectrum disorder. N Engl J Med. 2021;385(16):1462–1473. doi: 10.1056/NEJMoa2103583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munesue T, Nakamura H, Kikuchi M, et al. Oxytocin for male subjects with autism spectrum disorder and comorbid intellectual disabilities: A randomized pilot study. Front Psychiatry. 2016;7:2. doi: 10.3389/fpsyt.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loparo D, Waldman ID. The oxytocin receptor gene (OXTR) is associated with autism spectrum disorder: a meta-analysis. Mol Psychiatry. 2015;20(5):640. doi: 10.1038/mp.2014.77. [DOI] [PubMed] [Google Scholar]
- 37.Hernandez LM, Krasileva K, Green SA, et al. Additive effects of oxytocin receptor gene polymorphisms on reward circuitry in youth with autism. Mol Psychiatry. 2016;22(8):1134. doi: 10.1038/mp.2016.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kou J, Zhang Y, Zhou F, et al. A randomized trial shows dose-frequency and genotype may determine the therapeutic efficacy of intranasal oxytocin. Psychol Med. 2020:1–10. doi: 10.1017/S0033291720003803. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Otowa T, Abe O, et al. Oxytocin receptor gene variations predict neural and behavioral response to oxytocin in autism. Soc Cogn Affect Neurosci. 2017;12(3):496–506. doi: 10.1093/scan/nsw150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iakoucheva LM, Muotri AR, Sebat J. Getting to the cores of autism. Cell. 2019;178(6):1287–1298. doi: 10.1016/j.cell.2019.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi C, Wu J, Jiang T, et al. Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum Mutat. 2012;33(12):1635–1638. doi: 10.1002/humu.22174. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, Yu P, Jin X, et al. Genomic landscapes of Chinese sporadic autism spectrum disorders revealed by whole-genome sequencing. J Genet Genomics. 2018;45(10):527–538. doi: 10.1016/j.jgg.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Yuen RKC, Merico D, Bookman M, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20(4):602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Zeng C, Li J, et al. PAK2 Haploinsufficiency Results in Synaptic Cytoskeleton Impairment and Autism-Related Behavior. Cell Rep. 2018;24(8):2029–2041. doi: 10.1016/j.celrep.2018.07.061. [DOI] [PubMed] [Google Scholar]
- 46.Song W, Li Q, Wang T, et al. Putative complement control protein CSMD3 dysfunction impairs synaptogenesis and induces neurodevelopmental disorders. Brain Behav Immun. 2022;102:237–250. doi: 10.1016/j.bbi.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Wang L, Guo H, et al. Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders. Mol Psychiatry. 2017;22(9):1282–1290. doi: 10.1038/mp.2017.140. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Wang L, Yu P, et al. Vitamin D-related genes are subjected to significant de novo mutation burdens in autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2017;174(5):568–577. doi: 10.1002/ajmg.b.32543. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Hu S, Zhang K, et al. A comparative study of the genetic components of three subcategories of autism spectrum disorder. Mol Psychiatry. 2019;24(11):1720–1731. doi: 10.1038/s41380-018-0081-x. [DOI] [PubMed] [Google Scholar]
- 50.Leppa VMM, Kravitz SNN, Martin CLL, et al. Rare inherited and de novo CNVs reveal complex contributions to ASD risk in multiplex families. Am J Hum Genet. 2016;99(3):540–554. doi: 10.1016/j.ajhg.2016.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo H, Peng Y, Hu Z, et al. Genome-wide copy number variation analysis in a Chinese autism spectrum disorder cohort. Sci Rep. 2017;7:44155. doi: 10.1038/srep44155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bacchelli E, Cameli C, Viggiano M, et al. An integrated analysis of rare CNV and exome variation in autism spectrum disorder using the infinium PsychArray. Sci Rep. 2020;10(1):3198. doi: 10.1038/s41598-020-59922-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanders SJ, He X, Willsey AJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87(6):1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li T, Wernersson R, Hansen RB, et al. A scored human protein–protein interaction network to catalyze genomic interpretation. Nat Methods. 2016;14(1):61. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–DD61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis AP, Murphy CG, Johnson R, et al. The Comparative Toxicogenomics Database: Update 2011. Nucleic Acids Res. 2013;41:D1104–D1D14. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao G, Li K, Li B, et al. Gene4Denovo: an integrated database and analytic platform for de novo mutations in humans. Nucleic Acids Res. 2020;48(D1):D913–DD26. doi: 10.1093/nar/gkz923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krumm N, Turner TN, Baker C, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Wang T, Zhao X, et al. AI-Driver: an ensemble method for identifying driver mutations in personal cancer genomes. NAR Genom Bioinform. 2020;2(4):lqaa084. doi: 10.1093/nargab/lqaa084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang T, Ruan S, Zhao X, et al. OncoVar: an integrated database and analysis platform for oncogenic driver variants in cancers. Nucleic Acids Res. 2021;49(D1):D1289–DD301. doi: 10.1093/nar/gkaa1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kai W, Mingyao L, Hakon H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Shi L, Zhang K, et al. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2017;46:D1039–D1D48. doi: 10.1093/nar/gkx1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J, Zhao T, Zhang Y, et al. Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 2018;46(15):7793–7804. doi: 10.1093/nar/gky678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gurbich TA, Ilinsky VV. ClassifyCNV: a tool for clinical annotation of copy-number variants. Sci Rep. 2020;10(1):20375. doi: 10.1038/s41598-020-76425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krumm N, O'Roak BJ, Karakoc E, et al. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet. 2013;93(4):595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang T, Zhang Y, Liu L, et al. Targeted sequencing and integrative analysis of 3195 Chinese patients with neurodevelopmental disorders prioritized 26 novel candidate genes. J Genet Genomics. 2021;48(4):312–323. doi: 10.1016/j.jgg.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 69.Karczewski KJ, Francioli LC, Tiao G, et al. Author Correction: The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2021;590(7846):E53. doi: 10.1038/s41586-020-03174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gandal MJ, Zhang P, Hadjimichael E, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420) doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gandal MJ, Haney JR, Parikshak NN, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Focus (Am Psychiatr Publ) 2019;17(1):66–72. doi: 10.1176/appi.focus.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang T, Zhang Y, Liu L, et al. Targeted sequencing and integrative analysis of 3,195 Chinese patients with neurodevelopmental disorders prioritized 26 novel candidate genes. J Genet Genomics. 2021;48(4):312–323. doi: 10.1016/j.jgg.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y, Wang T, Wang Y, Xia K, Li J, Sun Z. Targeted sequencing and integrative analysis to prioritize candidate genes in neurodevelopmental disorders. Mol Neurobiol. 2021;58(8):3863–3873. doi: 10.1007/s12035-021-02377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li T, Wernersson R, Hansen RB, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods. 2017;14(1):61–64. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morales-Hidalgo P, Roigé-Castellví J, Hernández-Martínez C, Voltas N, Canals J. Prevalence and characteristics of autism spectrum disorder among spanish school-age children. J Autism Dev Disord. 2018;48(105):1–15. doi: 10.1007/s10803-018-3581-2. [DOI] [PubMed] [Google Scholar]
- 76.Black DO, Wallace GL, Sokoloff JL, Kenworthy L. Brief report: IQ split predicts social symptoms and communication abilities in high-functioning children with autism spectrum disorders. J Autism Dev Disord. 2009;39(11):1613–1619. doi: 10.1007/s10803-009-0795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Li N, Li C, et al. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl Psychiatry. 2020;10(1):4. doi: 10.1038/s41398-020-0699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiao J, Zhang M, Yang P, et al. Identification of de novo JAK2 and MAPK7 Mutations Related to Autism Spectrum Disorder Using Whole-Exome Sequencing in a Chinese child and adolescent trio-based sample. J Mol Neurosci. 2020;70(2):219–229. doi: 10.1007/s12031-019-01456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richter M, Murtaza N, Scharrenberg R, et al. Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling. Mol Psychiatry. 2019;24(9):1329–1350. doi: 10.1038/s41380-018-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Escamilla CO, Filonova I, Walker AK, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551(7679):227–231. doi: 10.1038/nature24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sgritta M, Dooling SW, Buffington SA, et al. Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron. 2019;101(2) doi: 10.1016/j.neuron.2018.11.018. 246–59 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harony-Nicolas H, Kay M, du Hoffmann J, et al. Oxytocin improves behavioral and electrophysiological deficits in a novel Shank3-deficient rat. eLife. 2017;6:e18904. doi: 10.7554/eLife.18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andari E, Nishitani S, Kaundinya G, et al. Epigenetic modification of the oxytocin receptor gene: implications for autism symptom severity and brain functional connectivity. NPP. 2020;45(7):1150–1158. doi: 10.1038/s41386-020-0610-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martins D, Paduraru M, Paloyelis Y. Heterogeneity in response to repeated intranasal oxytocin in schizophrenia and autism spectrum disorders: A meta-analysis of variance. Br J Pharmacol. 2022;179(8):1525–1543. doi: 10.1111/bph.15451. [DOI] [PubMed] [Google Scholar]
- 86.Kou J, Zhang Y, Zhou F, et al. Anxiolytic effects of chronic intranasal oxytocin on neural responses to threat are dose-frequency dependent. Psychother Psychosom. 2022:1–12. doi: 10.1159/000521348. [DOI] [PubMed] [Google Scholar]
- 87.Martins D, Brodmann K, Veronese M, et al. “Less is more”: A dose-response account of intranasal oxytocin pharmacodynamics in the human brain. Prog Neurobiol. 2022;211 doi: 10.1016/j.pneurobio.2022.102239. [DOI] [PubMed] [Google Scholar]
- 88.Sun W, Poschmann J, Cruz-Herrera Del Rosario R, et al. Histone Acetylome-wide Association Study of Autism Spectrum Disorder. Cell. 2016;167(5):1385–1397. doi: 10.1016/j.cell.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 89.Ramaswami G, Won H, Gandal MJ, et al. Integrative genomics identifies a convergent molecular subtype that links epigenomic with transcriptomic differences in autism. Nat Commun. 2020;11(1):4873. doi: 10.1038/s41467-020-18526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Liang Y, Jiang X, et al. Maternal Diabetes-Induced Suppression of Oxytocin Receptor Contributes to Social Deficits in Offspring. Front Neurosci. 2021;15 doi: 10.3389/fnins.2021.634781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andari E, Rilling JK. Genetic and epigenetic modulation of the oxytocin receptor and implications for autism. Neuropsychopharmacology. 2021;46(1):241–242. doi: 10.1038/s41386-020-00832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qin L, Williams JB, Tan T, et al. Deficiency of autism risk factor ASH1L in prefrontal cortex induces epigenetic aberrations and seizures. Nat Commun. 2021;12(1):6589. doi: 10.1038/s41467-021-26972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhubi A, Chen Y, Guidotti A, Grayson DR. Epigenetic regulation of RELN and GAD1 in the frontal cortex (FC) of autism spectrum disorder (ASD) subjects. Int J Dev Neurosci. 2017;62:63–72. doi: 10.1016/j.ijdevneu.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.