Abstract
The diversity of sulfate-reducing bacteria (SRB) in brackish sediment was investigated using small-subunit rRNA and dissimilatory sulfite reductase (DSR) gene clone libraries and cultivation. The phylogenetic affiliation of the most commonly retrieved clones for both genes was strikingly similar and produced Desulfosarcina variabilis-like sequences from the inoculum but Desulfomicrobium baculatum-like sequences from a high dilution in natural media. Related organisms were subsequently cultivated from the site. PCR bias appear to be limited (or very similar) for the two primersets and target genes. However, the DSR primers showed a much higher phylogenetic specificity. DSR gene analysis is thus a promising and specific approach for investigating SRB diversity in complex habitats.
Sulfate-reducing bacteria (SRB) constitute a paraphyletic group of physiologically diverse anaerobes, which all share the ability to obtain energy from dissimilatory reduction of inorganic sulfate. SRB are widespread in nature and, besides being key organisms in the sulfur cycle, also play an important role in the degradation of organic matter in many anoxic environments (12, 13). SRB are also known to play a role in the biodegradation of a number of environmental pollutants (7). Their activity in oil reservoirs results in biocorrosion and other problems of great economic impact in the offshore oil and gas industries (9). Due to their great ecological and economic importance, SRB populations have been intensively studied during the last few decades. Although important for ecological studies, it is generally acknowledged that traditional culturing only recovers a very limited fraction of the natural microbial diversity (1, 8, 32), indicating that new cultivation procedures need to be developed. Recently, a new most-probable-number method was described which greatly improved the growth and recovery of SRB from sludge and sediments (3, 30).
Most of the molecular studies on the diversity of SRB in complex communities, e.g., in granular sludge (23), the water column of a stratified Fjord (28), biofilm (2), microbial mats (27), and sediments (6, 21, 22), have been based on small-subunit (SSU) rRNA gene analysis. However, retrieved SSU rRNA sequences frequently are not related to any cultivated organism, and it thus becomes impossible to infer a likely ecophysiology for the organism containing the gene. SSU rRNA-based phylogeny has revealed that SRB constitute a paraphyletic group with members among the δ-Proteobacteria, gram-positive bacteria, and even Archaea (4). Unlike monophyletic traits such as oxygenic photosynthesis, some lineages of sulfate reducers are closely related to organisms that are unable to carry out dissimilatory sulfate reduction. This reduces the ability to identify SRB solely based on their SSU rRNA sequence.
An alternative approach to infer physiology from environmental sequences is to retrieve functional gene sequences coding for enzymes that are essential to the target organisms. Dissimilatory sulfite reductase (DSR) is a key enzyme in sulfate respiration, which so far has been found only in dissimilatory SRB. Recently, partial sequences of the gene encoding for DSR were used to evaluate the phylogeny of SRB, and the derived phylogeny was found to be consistent with the SSU rRNA-based phylogeny (31). The PCR primerset used by Wagner et al. (31) amplifies most of the α- and β-subunits of the DSR gene and allows the detection of members of all known groups of SRB (31). Studies of natural diversity of SRB based on PCR amplification of the DSR gene have recently revealed environmental DSR sequences unrelated to any known SRB gene in a microbial mat (18) and in sediments (29).
In this study, we combined both selective SSU rRNA and DSR primers to investigate the diversity of SRB in brackish-water sediments (Kysing Fjord, Denmark). We compared the phylogeny derived from partial SSU rRNA and DSR sequences retrieved from clone libraries. Subsequently, SRB were cultivated from the site, and their phylogeny was established and compared to that obtained by molecular analysis of the same sample. The aim of this study was to (i) evaluate the diversity in the sample, (ii) determine the ability and/or failure of cultivation methods to retrieve common species, and (iii) evaluate PCR bias by comparing two different primersets.
Sediment samples were obtained from shallow-water sediments in Kysing Fjord (Norsminde, Denmark), previously shown to support high rates of sulfate reduction (11). Surface sediment, used as an inoculum for diversity studies, consisted of a mixture of oxidized (brown) and reduced (black) sediment collected from the top 0 to 4 cm. The organic-rich (∼10%, dry weight) sediment mixture was initially stored at 4°C (in situ temperature) in the dark under anaerobic conditions in order to establish a large population of SRB. During storage, a further blackening of the sediment mixture was observed. Prior to making dilutions of the sediment sample, aliquots were frozen at −20°C for subsequent molecular analysis.
Kysing Fjord sediments (hereafter referred to as the inoculum) were 10-fold serially diluted in anoxic natural medium consisting of sterilized top layer sediment (0 to 2 cm) from the site (30). The inoculum was also 10-fold diluted in anoxic synthetic medium (33) containing a mixture of carbon sources (lactate, methanol, pyruvate, acetate, formate, and glucose, each at a 5 mM final concentration) and 0.1 g of yeast extract per liter. The final pH of the medium was ca. 7. Dilution tubes were sampled (0.5 ml) for molecular analysis after 3 weeks of incubation at 25°C. SRB from the highest positive dilution in natural media were reinoculated into different synthetic media, each containing only one of the carbon sources mentioned above (20 mM final concentration). Growth in culture tubes was monitored by measuring H2S production (5). Purification of the SRB strains was performed by repeated application of the roll tube method (10). Anaerobic Hungate techniques (10, 15) were used throughout this work. Pure cultures and enrichments were characterized by molecular methods.
Genomic DNA was extracted by bead beating using the Bio 101 FastPrep Instrument, the FastDNA Spin Kit for Soil (inoculum and serial dilutions), and the FastDNA Kit MH (enrichments and pure cultures), according to the protocol of the manufacturer (Bio 101, Vista, Calif.). DNA extracts were stored at −20°C. PCR amplification was performed on extracted DNA or directly using a Thermocycler (PTC-200 Peltier Thermal Cycler; M. J. Research). SSU rRNA gene was amplified with the primerset 385F and 907R (Table 1). Reaction conditions were as follows: 93°C for 1 min, 25 cycles of 30 s at 92°C, 1 min at 57°C, and 45 s at 72°C, and a final extension of 5 min at 72°C. For denaturing gradient gel electrophoresis (DGGE) analysis, 35 PCR cycles and the primer 385F with a GC clamp (Table 1) were used. DSR gene was amplified with the primerset DSR1F and DSR4R (Table 1). Reaction conditions were as follows: 94°C for 1 min, 30 or 35 cycles of 20 s at 94°C, 1 min at 54°C, and 3 min at 72°C, and a final extension of 10 min at 72°C.
TABLE 1.
Primers used for PCR amplification and sequencing of SSU rRNA and DSR genes
| Expt type and primer | Sequence (5′-3′)c | Specificity | Reference |
|---|---|---|---|
| SSU rRNA gene amplification | |||
| 385Fab | CGG CGT CGC TGC GTC AGG | SRB belonging to the δ-Proteobacteria, other δ-Proteobacteria, and some gram-positive bacteria | 2 |
| 907Ra | CCG TCA ATT CCT TTR AGT TT | All organisms | 14 |
| DSR gene amplification | |||
| DSR1F | ACS CAC TGG AAG CAG CAC G | All groups of SRB | 31 |
| DSR4R | GTG TAG CAG TTA CCG CA | All groups of SRB | 31 |
| Sequencing | |||
| VectorF | TTG GGC CCT CTA GAT G | TOPO XL cloning vector | 29 |
| VectorR | CTA TGC ATC AAG CTT GG | TOPO XL cloning vector | 29 |
Primers used also for SSU rRNA sequencing.
Primer used with a 5′-end GC clamp for DGGE analysis: 5′-CGC CCG CCG CGC GGC GGG CGG GGC GGG GGC ACG GGC.
R, G or A; S, C or G.
Partial SSU ribosomal DNA (rDNA) amplificates were separated by DGGE according to their melting behavior (19). We used the D-GENE DGGE system from Bio-Rad (Munich, Germany), and 8% acrylamide gels with a denaturating gradient ranging from 25 to 75% (100% denaturant was 7 M urea and 40% [vol/vol] formamide). Electrophoresis was conducted at a constant voltage of 200 V at 60°C for 7 h. Bands of interest were excised from the gel, eluted in 20 μl of sterile distilled water. Each band was subsequently reamplified, and its position was confirmed by DGGE before sequencing. For sequencing, bands were reamplified using the primerset without GC clamp.
PCR products were purified with the QIAquick PCR Purification kit (Qiagen GmbH, Hilden, Germany) and cloned using the TOPO XL Cloning Kit (Invitrogen, San Diego, Calif.). The clone library was screened by direct PCR amplification from a colony. Plasmids containing the insert of the right length were isolated using a QIAprep Miniprep Kit (Qiagen) and kept at −20°C.
Purified PCR products and plasmids were sequenced directly with an ALFexpress automated DNA sequencer, using a Thermosequenase Fluorescent Sequencing Kit (Pharmacia Biotech, Uppsala, Sweden). The primers used for sequencing are listed in Table 1. Partial SSU rRNA sequences were aligned with the sequence editor SeqPup (D. G. Gilbert, SeqPup version 0.6). Only unambiguously aligned sequence positions (485 nucleotides) were exported to the PAUP∗ version 4.0 program (D. L. Swofford, PAUP∗ 4th ed.; Sinauer Associates) for phylogenetic analysis. Different phylogenetic algorithms, i.e., maximum parsimony, distance matrix, and maximum-likelihood analysis, were applied to the dataset using default parameters. Partial α- and β-subunit sequences of the DSR gene were aligned according to the amino acid sequences with the sequence editor GDE implemented in the ARB package (26). Dendrograms were constructed for unambiguous amino acids (301 amino acids) using maximum parsimony, distance matrix, and maximum-likelihood analysis as implemented in the ARB package (26). The robustness of DSR and SSU rRNA trees was tested by bootstrap analysis with 100 resamplings.
Sequence accession numbers.
All sequences have been deposited in GenBank under accession numbers AF360632 to AF360642 for SSU rRNA sequences and AF360643 to AF360694 for DSR sequences.
SRB growth.
When Kysing Fjord sediment was serially diluted in natural media (sterilized sediments from the site), the growth of SRB was detected after 3 weeks of incubation down to the 10−10 dilution, using both the SSU rRNA and the DSR primerset. No amplification was obtained using these selective primers in dilutions higher than the 10−10 dilution or in samples from uninoculated natural media even after 12 weeks of incubation. When serial dilutions were made in synthetic media containing a mixture of common carbon sources for SRB, growth occurred only down to the 10−6 dilution. These results, confirmed by subsequent chemical analysis of H2S production, agree with recent studies which show that viable counts of SRB were up to 4 orders of magnitude higher when sediment media were used for enumeration (3, 30). Recently, Sass et al. (25) reported that only a minor fraction of the total community of SRB in a lake sediment was recovered by cultivation in a synthetic medium. Natural media seem to improve the recovery of both cultured and uncultured SRB from complex environments. However, the growth of SRB down to the dilution 10−10 was unexpected since recovery of SRB in marine sediment is usually much lower (reference 30 and references therein). There could be several explanations for the high recovery we observed: (i) enrichment of SRB in the organic-rich sediment during the preincubation period, (ii) the presence of “hot spots” of SRB, and (iii) an event of low probability so that a SRB was diluted out to the 10−10 dilution despite a small population size. High rates of sulfate reduction (up to 4 μmol cm−3 day−1) have been reported for this organic-rich sediment (11). Using literature values of specific sulfate reduction rates of 10−14 to 10−15 mol of sulfate cell−1 day−1 (30) would yield population sizes of 4 × 108 to 4 × 109 cells cm−3 operating at their Vmax, indicating that the number of active cells may have been significantly higher.
DGGE analysis of serial dilutions.
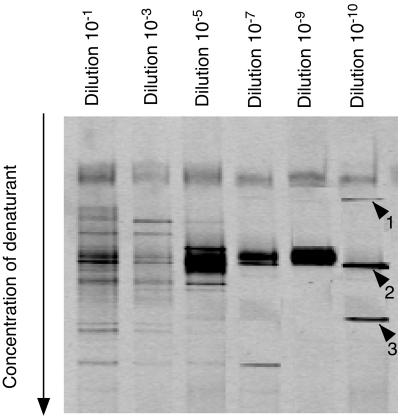
DGGE analysis of partial SSU rRNA gene amplificates showed only three bands in the last positive dilution (10−10) (Fig. 1). Phylogenetic analysis showed that only one sequence (band 3) was related to the SRB within the δ-Proteobacteria. The sequence was highly similar (>99% similarity) to the SSU rDNA sequence of Desulfomicrobium baculatum (Fig. 2B). The sequences of bands 1 and 2 were affiliated with the gram-positive bacteria but were not related to any Desulfotomaculum sp. They are thus most likely not SRB, i.e., false positives. The DGGE pattern showed a general reduction in diversity for increasing dilutions; however, band 1 and band 3 were detected in the 10−10 dilution but not in the lower dilutions (Fig. 1). In particular, band 3 corresponding to the D. baculatum sequence did not appear in lower dilutions. It is unlikely to be due to a preferential PCR amplification of SSU rDNA of organisms corresponding to the prominent bands in lower dilutions, since mixing DNA extracts from the dilutions 10−10 and 10−7 or 10−9 did not inhibit the amplification of SSU rDNA of D. baculatum (data not shown). D. baculatum was probably overgrown by other organisms in the lower dilutions and therefore was present in too low numbers to be detected by DGGE analysis of SSU rDNA amplificates. These fast-growing organisms could have been less abundant in the inoculum than D. baculatum and therefore absent and unable to inhibit its growth in the 10−10 dilution.
FIG. 1.
DGGE gel containing partial SSU rRNA gene amplificates obtained after serial dilutions of inoculum in natural media. Lanes show PCR amplificates of DNA extract from serial dilutions of the inoculum after 3 weeks of incubation. Denaturant concentration varied from 25% at the top to 75% at the bottom of the gel. The numbers indicate bands that were excised from the gel, reamplified, and sequenced.
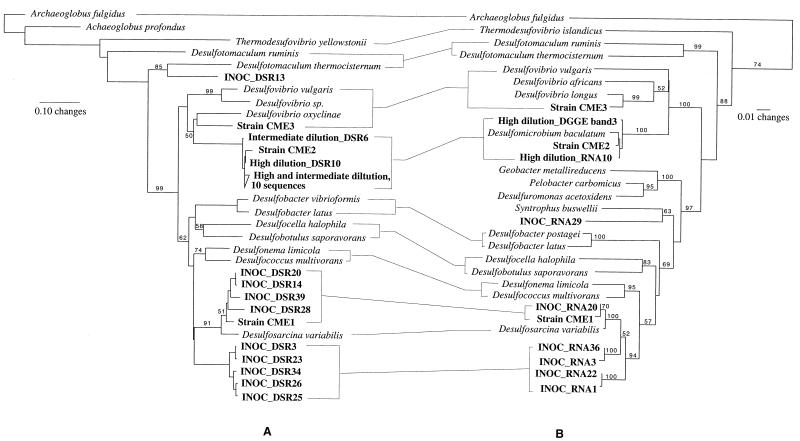
FIG. 2.
Comparison of the phylogenetic trees derived from retrieved DSR sequences (A) and SSU rRNA sequences (B). Species present in both trees are connected by horizontal lines. Environmental sequences and sequences from newly isolated strains are shown in boldface. The numbers on branching points are bootstrap values with 100 replicates (values of <50 were not included).
SRB diversity.
The phylogeny of retrieved amino acid sequences (DSR gene) and nucleotide sequences (SSU rRNA gene) was estimated by distance matrix, parsimony, and maximum-likelihood analysis. Similar tree topologies were obtained with all phylogenetic methods. Figure 2 shows a comparison of DSR and SSU rRNA sequence-based trees obtained by distance matrix analysis. From the inoculum, 10 DSR clones and 23 SSU rRNA clones were sequenced. We retrieved a cluster of nine DSR sequences closely related to Desulfosarcina variabilis (Fig. 2A). One DSR sequence (INOC-DSR13) was deeply branching and was only distantly affiliated with two Desulfotomaculum species (Fig. 2A). However, it was closely related to a cluster of 14 DSR sequences previously retrieved in another brackish sediment in Denmark (Mariager Fjord) (data not shown). Only 6 of the 23 partial SSU rRNA sequences retrieved were branching within the δ-Proteobacteria (Fig. 2B). Five of these six sequences were related to known SRB, the closest known relative being Desulfosarcina variabilis (1 to 4.4% sequence similarity for partial rDNA sequence). The last δ-Proteobacteria sequence (INOC_RNA29) was only distantly related to Syntrophus buswelii (<90% sequence similarity for partial rDNA sequence).
From the highest positive dilution in natural media (10−10), 7 DSR clones and 10 SSU rRNA clones were sequenced. Five DSR clones from an intermediate dilution (10−7) were also sequenced. All retrieved DSR sequences were clustering together and were closely related to strain CME2 subsequently identified as D. baculatum (Fig. 2A). As already observed from cloning from the inoculum, most of the retrieved SSU rRNA sequences were probably not SRB sequences; 9 sequences out of 10 clones sequenced were related to the genus Clostridium (data not shown). Only 1 of the 10 retrieved SSU rRNA sequences was affiliated with known SRB. This clone was closely related to D. baculatum and strain CME2 (>99% sequence similarity for partial rDNA sequence) (Fig. 2B), as was the excised band from the high-dilution DGGE gel (>98.8% sequence similarity for partial rDNA sequence) (band 3, Fig. 1).
Culture studies conducted with Kysing Fjord sediment generally agreed with results obtained by the molecular approach. D. baculatum, detected in dilutions 10−7 and 10−10 by cloning of the SSU rRNA and DSR genes, was cultivated from the dilution 10−10 of the inoculum in natural media. It also appeared in several enrichments and was isolated (strain CME2) from dilution 10−5 of a previous dilution series in synthetic media using lactate as carbon source. Desulfosarcina variabilis, evidenced in the inoculum by both cloning of the SSU rRNA and DSR genes, has previously been cultivated from a high-dilution tube (10−6) inoculated with fresh sediment from the same site (strain CME1). In addition, a strain belonging to the genus Desulfovibrio, which was not retrieved by molecular analysis, was also cultivated from the site from dilution 10−2 of a previous dilution series in synthetic media using methanol as the carbon source (strain CME3). Enrichment of Desulfovibrio sp. on methanol was unexpected. To our knowledge, only one species of this genus, Desulfovibrio carbinolicus, has so far been isolated on methanol (20), and most members of this genus are not reported to grow on this substrate. Based on partial SSU rRNA sequences, strain CME3 closest relative is Desulfovibrio longus (>97% sequence similarity for partial rDNA sequence). However, Desulfovibrio longus is not able to grow on methanol (16). For all three isolates, the phylogeny based on DSR sequences agreed with the phylogeny based on SSU rRNA sequences (Fig. 2).
Our results from cultivation and clone libraries indicated that D. baculatum was the dominant SRB species in high serial dilutions of the inoculum grown in natural media. In contrast, Desulfosarcina variabilis was detected by direct cloning of the inoculum but was never observed in the highest dilution. However, previous cultivation of strain CME1, closely related to Desulfosarcina variabilis (>99% sequence similarity for partial rDNA sequence) from a high dilution indicated that Desulfosarcina variabilis is probably also abundant in this environment. It is known that Desulfosarcina variabilis is able to form strong aggregates that are difficult to separate into individual cells, which can explain why it was not detected in high dilutions. Nevertheless, a limited number of clones were sequenced in this study, and it is likely that other sequences, in particular sequences related to the genera Desulfovibrio and Desulfomicrobium, could be retrieved from the inoculum.
The phylogenetic comparison of retrieved SSU rRNA and DSR sequences showed that similar SRB populations were detected by cloning either gene, D. baculatum-like sequences from high dilutions and Desulfosarcina variabilis-like sequences from the inoculum. This is in accordance with recent studies on diversity and abundance of SRB in a microbial mat where similar populations were detected and described based on retrieved DSR sequences and SSU rRNA hybridization (17, 18). It is remarkable, however, that two different primersets, amplifying two different genes with presumably different PCR bias, made highly similar predictions about the retrieved populations in two different samples. Consequently, the PCR bias must have been very low or at least very similar for both of the two primersets.
However, our results demonstrated that the phylogenetic specificity of the DSR primerset was superior to the specificity of the SSU rRNA primerset. All sequences retrieved with the DSR primers were DSR sequences (22 of 22 clones sequenced), but only 6 of the SSU rRNA sequences of 33 clones sequenced were clearly related to SRB. The primer 385F does amplify SSU rDNA from many SRB, but there is ample evidence that distantly related taxons, which are not currently thought to contain SRB, are also targeted by this primer (2, 24, 25). The lack of specificity greatly limits the usefulness of selective SSU rRNA primers for accessing the diversity of SRB in complex environments. Besides being highly specific, the DSR primerset targets all known groups of SRB (31), whereas the primer 385F was only designed to amplify SSU rDNA from SRB belonging to the δ-Proteobacteria (2). Finally, the DSR gene approach is directly linked to the process of dissimilatory sulfate reduction, whereas the ecophysiology function of an organism characterized only by an environmental SSU rRNA sequence remains largely unknown, especially if it belongs to a novel phylogenetic lineage. For example, it is not certain whether the organism with the SSU rRNA sequence INOC-RNA29 is a dissimilatory SRB, although it clearly falls within the δ-Proteobacteria because some members of the δ-Proteobacteria do not reduce sulfate. In contrast, we can infer from the retrieved DSR sequence INOC_DSR13, which was not related to any known SRB sequence, that a new, hitherto-uncultured, SRB was present in Kysing Fjord sediment. A related cluster of DSR sequences has been retrieved from another coastal environment (Mariager Fjord), indicating that this uncultured organism may be common in brackish sediments. In other studies, DSR sequences unrelated to any cultivated and sequenced organisms have also been retrieved from a microbial mat (18) and from sediment (29).
The similarity of the phylogenetic trees derived from SSU rRNA and DSR sequences and the higher specificity and the broad target range of the DSR primers makes DSR gene analysis a promising approach for studying the diversity of SRB in complex habitats.
Acknowledgments
This work was supported by Statens Naturvidenskabelige Forskningsråd, Statens Tekniske Videnskabelige Forskningsråd, and the Carlsberg Foundation.
We thank Dorte T. Ganzhorn, Pernille V. Thykjær, Tove Wiegers, and Jane Frydenberg for excellent technical assistance. We also thank Michael Wagner (Technische Universität München) and Dave Stahl (Northwestern University) for helpful advice and access to their DSR sequences of pure cultures.
REFERENCES
- 1.Amann R, Ludwig W, Schleifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Stromley J, Devereux R, Key R, Stahl D. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandt K K, Vester F, Jensen A N, Ingvorsen K K. Sulfate reduction dynamics and enumeration of sulfate-reducing bacteria in hypersaline sediments of the Great Salt Lake (Utah, USA) Microb Ecol. 2001;41:1–11. doi: 10.1007/s002480000059. [DOI] [PubMed] [Google Scholar]
- 4.Castro H F, Williams N H, Ogram A. Phylogeny of sulfate-reducing bacteria. FEMS Microbiol Ecol. 2000;31:1–9. doi: 10.1111/j.1574-6941.2000.tb00665.x. [DOI] [PubMed] [Google Scholar]
- 5.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 6.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ensley B D, Suflita J M. Metabolism of environmental contaminants by mixed and pure cultures of sulfate-reducing bacteria. In: Barton L L, editor. Sulfate-reducing bacteria. Vol. 8. New York, N.Y: Peplum Press; 1995. pp. 293–332. [Google Scholar]
- 8.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton W A, Lee W. Biocorrosion. In: Barton L L, editor. Sulfate-reducing bacteria. Vol. 8. New York, N.Y: Peplum Press; 1995. pp. 243–264. [Google Scholar]
- 10.Hungate R E. A roll tube method for cultivation of strict anaerobes. In: Norris J R, Ribbons D W, editors. Methods in microbiology. 3B. London, England: Academic Press; 1969. pp. 117–132. [Google Scholar]
- 11.Ingvorsen K, Jørgensen B. Seasonal variation in H2S emission to the atmosphere from intertidal sediments in Denmark. Atmos Environ. 1982;16:855–865. [Google Scholar]
- 12.Jørgensen B B. Mineralization of organic metter in the sea bed-the role of sulfate reduction. Nature. 1982;296:643–645. [Google Scholar]
- 13.Jørgensen B B, Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark) Appl Environ Microbiol. 1991;57:847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane D J. 16/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: Wiley; 1991. pp. 113–175. [Google Scholar]
- 15.Macy J M, Snellen J E, Hungate R E. Use of syringe methods for anaerobiosis. Am J Clin Nutr. 1972;25:1318–1323. doi: 10.1093/ajcn/25.12.1318. [DOI] [PubMed] [Google Scholar]
- 16.Magot M, Caumette P, Desperrier J M, Matheron R, Dauga C, Grimont F, Carreau L. Desulfovibrio longus sp. nov., a sulfate-reducing bacterium isolated from an oil-producing well. Int J Syst Bacteriol. 1992;42:398–403. doi: 10.1099/00207713-42-3-398. [DOI] [PubMed] [Google Scholar]
- 17.Minz D, Fishbain S, Green S J, Muyzer G, Cohen Y, Rittmann B E, Stahl D A. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl Environ Microbiol. 1999;65:4659–4665. doi: 10.1128/aem.65.10.4659-4665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minz D, Flax J L, Green S J, Muyzer G, Cohen Y, Wagner M, Rittmann R, Stahl D A. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl Environ Microbiol. 1999;65:4666–4671. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muyzer G, de Waal E, Uitterlinden A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanninga H J, Gottschal J C. Properties of Desulfovibrio carbinolicus sp. nov. and other sulfate-reducing bacteria isolated from an anaerobic-purification plant. Appl Environ Microbiol. 1987;53:802–809. doi: 10.1128/aem.53.4.802-809.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rooney-Varga J, Devereux R, Evans R, Hines M. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santegoeds C M, Damgaard L R, Hesselink G, Zopfi J, Lens P, Muyzer G, de Beer D. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl Environ Microbiol. 1999;65:4618–4629. doi: 10.1128/aem.65.10.4618-4629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santegoeds C M, Ferdelman T G, Muyzer G, de Beer D. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl Environ Microbiol. 1998;64:3731–3739. doi: 10.1128/aem.64.10.3731-3739.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sass H, Wieringa E, Cypionka H, Babenzien H, Overmann J. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch Microbiol. 1998;170:243–521. doi: 10.1007/s002030050639. [DOI] [PubMed] [Google Scholar]
- 26.Strunk O, Ludwig W, Gross O, Reichel B, Stuckmann N, May M, Nunhoff B, Lenke M, Ginhart T, Vilbig A, Westran R. ARB—a software environment for sequence data. Munich, Germany: Technishe Universitet München; 1998. [Google Scholar]
- 27.Teske A, Ramsing N, Habicht K, Fukui M, Küver J, Jr/gensen B, Cohen Y. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt) Appl Environ Microbiol. 1998;64:2943–2951. doi: 10.1128/aem.64.8.2943-2951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teske A, Wawer C, Muyzer G, Ramsing N. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomsen T R, Finster K, Ramsing N B. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl Environ Microbiol. 2001;67:1646–1656. doi: 10.1128/AEM.67.4.1646-1656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vester F, Ingvorsen K. Improved most-probable-number method to detect sulfate-reducing bacteria with natural media and a radiotracer. Appl Environ Microbiol. 1998;64:1700–1707. doi: 10.1128/aem.64.5.1700-1707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner M, Roger A, Flax J, Brusseau G, Stahl D. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward D W, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 33.Widdel F, Bak F. Gram-negative sulfate-reducing bacteria. In: Balows T H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Vol. 4. New York, N.Y: Springer-Verlag; 1992. p. 3351. [Google Scholar]