Abstract
The Coronavirus disease 2019 (COVID-19) has posed a serious threat to global health and the world economy. Antiviral therapies targeting coronavirus are urgently required. The Cepharanthine (CEP) is a traditional Chinese herbal extract. Our previous research revealed that CEP has a very potent anti-coronavirus effect, but its mechanism of action was not fully understood. To investigate the effect of novel coronavirus on protein glycosylation in infected cells and to further investigate the mechanism of action of CEP against coronavirus, a cellular model using coronavirus GX_P2V infection of Vero E6 cells was established. The effect of coronavirus GX_P2V on host cell protein glycosylation was investigated by N-glycoproteomic analysis, and the antagonistic effect of CEP on the abnormal protein glycosylation caused by coronavirus was analyzed. The results showed that GX_P2V could cause abnormal changes in protein glycosylation levels in host cells, while CEP could partially antagonize the abnormal protein glycosylation caused by GX_P2V. In addition, we also found that CEP could regulate the glycosylation level of coronavirus S protein. In conclusion, this article provides important ideas about the infection mechanism of novel coronaviruses, providing evidence for CEP as a promising therapeutic option for coronavirus infection.
Keywords: Cepharanthine, Coronavirus, N-glycosylation, SARS-CoV-2, Viral pathogenesis
1. Introduction
The global pandemic of COVID-19 caused by the highly infectious and mutating novel coronavirus SARS-CoV-2 has had a tremendous impact on the international community. For nearly three years since the outbreak of COVID-19 pandemic in late 2019, there have been ongoing efforts to study the mechanism of action of the novel coronavirus and to develop potent anti-SARS-CoV-2 drugs. Although considerable progress has been made in the study of molecular mechanisms and drug development of against coronaviruses, SARS-CoV-2 drug targets as well as anti-SARS-CoV-2 drugs are still being validated [1].
Cepharanthine (CEP) is a traditional Chinese medicine extract from Stephania cepharantha Hayata first discovered by Japanese pharmacologists in 1934 [2]. In January 2020, after the novel coronavirus outbreak, we systematically screened more than 2400 compounds already clinically approved for use and identified CEP as the most effective drug for the treatment of novel coronavirus infections [3]. We patented our discovery, and conducted a study on the antiviral mechanism of action of CEP. In November 2021, we published a paper in BRIEFINGS IN BIOINFORMATICS revealing the CEP's anti-novel coronavirus action at the transcriptional level: it acts by effectively antagonizing most dysregulated genes and pathways in infected cells, including the ER stress/unfolded protein response and the HSF1-mediated heat shock response [2]. Further studies revealed that the anti-novel coronavirus effect of CEP may also be reflected in the inhibition of viral genome replication and viral protein translation [4]. CEP also prevents SARS-CoV-2 virus from attaching and entering cells. The CEP molecule binds to the S protein of the virus, preventing it from binding to the viral receptor-ACE2 [5].
It was found that the S proteins of the novel coronavirus have extensive glycosylation [6]. Glycosylation is the most abundant and complex post-translational modification which can have profound structural and functional effects on the conjugate [7]. The oligosaccharide fraction is considered to be related with multiple biological processes and affects the physical properties of proteins. It has been known that protein glycosylation is stable among healthy people, although it is different from person to person. Only when homeostasis changes due to lifestyle or pathological conditions, will glycosylation changes significantly [8]. The glycosylation has been regarded as an important quality attribute of biopharmaceuticals [9,10]. Coronavirus S glycans can mask the protein surface and consequently limit access to neutralizing antibodies and thwart the humoral immune response [[11], [12], [13], [14]]. Therefore, we hypothesize that glycosylation may potentially help the treatment of coronavirus infestation.
GX_P2V is a SARS-CoV-2 related coronavirus isolated from pangolin. Its spike protein shares 92.2% amino acid identity with the spike protein of SARS-CoV-2 [15]. Besides, GX_P2V also has the shared receptor with SARS-CoV-2 but no pathogenicity to human. Based on these characteristics, it has been reported that GX_P2V can be used as an accessible in vitro model for developing therapies or for mechanism investigation against SARS-CoV-2 [3]. Here we adopted this well-established coronavirus SARS-CoV-2 cellular model by using GX_P2V infestation with Vero E6 to investigate the mechanisms of coronavirus SARS-CoV-2 infestation with respect to the dysregulation of glycoprotein and screening for potential targets for therapeutic intervention.
And recent studies have proven that CEP had the same performance in the cell culture model of SARS-CoV-2 [5]. However, the mechanism of CEP on glycosylation targets remains unclear. In this paper, we carried out an N-glycosylation modification proteomics analysis to study the mechanisms of SARS-CoV-2 infection and the anti-coronavirus effects of CEP. The 4D label-free quantitative proteomic approach, which is highly sensitive and capable of accurate quantification, has been applied in proteomic analysis for detecting more low-abundance proteins, and it also has excellent performance in identifying modifications of proteins [[16], [17], [18]]. Using the advanced technique 4D label-free quantification technology, we constructed the N-glycoproteomic profiling, revealing new therapeutic targets against coronavirus infestation and CEP's intervention. Our results provide important insights into the mechanism of coronaviruses infection and the identification of drug targets for anti-coronavirus of multi-target drug combination usage.
2. Materials and methods
2.1. Cell culture and coronavirus
SARS-CoV-2 related coronavirus GX_P2V/pangolin/2017/Guangxi was isolated from frozen tissue samples of pangolins (M. javanica), which were collected between August 2017–January 2018. The pangolins had died during the smuggling operation, and these dead pangolins tissue samples were seized by the Guangxi Customs during their routine anti-smuggling operations. Using the intestine-lung mixed sample we were able to isolate GX_P2V using the Vero-E6 cell line. And its complete genome has been submitted to GenBank with accession number MT072864.1 [15]. The compound CEP was purchased from TOPscience (Shanghai, China). Vero E6 cells (American Type Culture Collection, Manassas, VA, USA) (ATCC, No. 1586) were cultured in high-glucose containing DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS) in 37 °C incubator with 5% CO2.
2.2. CEP and GX_P2V treatment
Vero E6 cells were cultured in T175 flask and treated with or without GX_P2V (MOI of 0.01) in the presence or absence of CEP in triplicates. Group 1 (Vero): normal control; group 2 (Vero_C): CEP only treatment for 72 h; group 3 (Vero_P): GX_P2V treatment for 72 h; group 4 (Vero_C_P): GX_P2V and CEP GX_P2V treatment for 72 h. The cytotoxicity of CEP to Vero E6 cells was measured by CellTiter blue according to the manufacturer's protocol (Promega, Catalog Number: PR-G8081; Madison, WI, USA). Cell samples were harvested according to the protocol for Glycoproteomics analysis from PTM-Biolab (Hangzhou, China), and stored in −80 °C refrigerator for further analysis.
2.3. Protein extraction and trypsin digestion
Samples were stored in −80 °C refrigerator before thawed on ice and centrifuged at 12,000 g at 4 °C for 10 min to remove cell debris. The supernatant was transferred to a new centrifuge tube for protein concentration determination using BCA kit.
Equal amount of each protein sample was enzymatically lysed with the same volume of lysis buffer containing the appropriate amount of reference protein. The protein samples were mixed with the lysis buffer by five times of the sample volume and precipitated with pre-cooled acetone at −20 °C for 2 h, followed by centrifugation at 4500g for 5 min. After that, the precipitate was collected and washed twice with pre-cooled acetone. The protein precipitate from each sample was dried, and then sonicated in buffer containing 200 mM Triethylammonium bicarbonate (TEAB). Digestion was performed by incubation with trypsin at a ratio of 1: 50 (enzyme: protein, m/m) with protein samples for overnight. Finally, the samples were desalted according to the C18 ZipTips instructions, and vacuum freeze-dried for HPLC analysis.
2.4. The enrichment of glycosylation modification of peptides
The peptide fragments were dissolved in 40 μL of enrichment buffer (80% acetonitrile/1% trifluoroacetic acid) and transferred to a hydrophilic micro-column. The enrichment was completed by centrifugation at 4000g for approximately 15 min in HILIC. The hydrophilic micro-column was washed for 3 times with enrichment buffer. The glycopeptides were eluted with 10% acetonitrile, then the eluate was collected and vacuum dried. After drying, the eluate was reconstituted in 50 μL of 50 mM ammonium bicarbonate buffer dissolved in 50 μL of hydrogen peroxide with 2 μL of water. The glycopeptides were incubated with PNGase F glycosidase overnight at 37 °C. Finally, salt was removed according to the C18 ZipTips instructions and vacuum freeze-dried for liquid-liquid analysis.
2.5. Liquid chromatography-mass spectrometry (LC-MS)
The peptides were dissolved in liquid chromatography mobile phase A (0.1% (v/v) formic acid aqueous solution) and then separated using NanoElute ultra-efficient liquid phase system. Mobile phase A is an aqueous solution containing 0.1% formic acid; mobile phase B is an acetonitrile solution containing 0.1% formic acid. Liquid phase gradient setting: 0–50 min, 2%∼22%B; 50–52 min, 22%∼35%B; 52–55 min, 35%∼90%B; 55–60 min, 90%B, the flow rate was maintained at 450 nL/min.
The peptides were separated by an ultra-high-performance liquid phase system and injected into a capillary ion source. Then the peptide segments are analyzed by a TIMS-TOF Pro mass spectrometer. The ion source voltage was set to 1.6 kV and both the peptide parent ion and its secondary fragments were detected and analyzed using TOF. The secondary mass spectrometry scan range was set to 100–1700 m/z. The data acquisition mode used parallel cumulative serial fragmentation (PASEF) mode. One primary mass spectrometry acquisition followed by 10 PASEF mode acquisitions of secondary spectra with parent ion charge numbers in the range 0–5. The dynamic exclusion time of the tandem mass spectrometry scan was set to 30 s to avoid repeated scans of the parent ion.
2.6. Database search
Retrieval parameter settings: the database was FA105LPNg_ Chlorocebus_sabaeus_60711_ Ensembl_GX_P2V_protein_TX_combine_20200413 (19267 sequences), an anti-library was added to calculate the false positive rate (FDR) caused by random matching, and a common pollution library was added to the database to eliminate the contamination protein in the identification results Impact; the digestion method was set to Trypsin/P; the number of missed cleavage sites was set to 2; the minimum length of the peptide was set to 7 amino acid residues; the maximum modification number of the peptide was set to 5.
The mass error tolerance of the primary precursor ions in First search and Main search was both set to 20 ppm, and the mass error tolerance of the secondary fragment ions was 0.02 Da. The cysteine alkylation was set as a fixed modification, and the variable modification was the oxidation of methionine, the acetylation of the N-terminus of the protein, and the deamination of asparagine (18O). The FDR for protein identification and PSM identification was set to 1%.
2.7. Bioinformatics analyses
Glycoproteomic data of GX_P2V infection group and 15 glycosylation sites in 12 glycoprotein targets of CEP treatment in presence of GX_P2V l was analyzed by Metascape web-based platform [19]. Pathway and process enrichment analysis were carried out with the following ontology sources: KEGG Pathway, GO Biological Processes, Reactome Gene Sets, Canonical Pathways and CORUM. PPI networks were constructed by string (STRING; http://string-db.org. version 11.0) [20] and Cytoscape (version 3.7.1) [21], the most relevant modules in the PPI networks were identified using MCODE [22], and the network adjusted by the Files Layout algorithm. Reactome analysis tool (http://reactome.org) was used to identify the enriched pathways of glycosylation sites on glycoproteins remains at normal state in response to GX_P2V treatment in presence of CEP. (p ≤ 0.05) (https://reactome.org/PathwayBrowser/#TOOL=AT) [23]. Uniprot was used to find the subcellular localization of proteins (https://www.uniprot.org/) [24]. One-way ANOVA followed by Dunnett's multiple comparisons test was performed using GraphPad Prism version 8.0.0 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com.
3. Results and discussion
3.1. N-glycoproteomic profiling highlights the differentially expressed glycoproteins in SARS-CoV-2-related coronavirus GX-P2V infection cellular model and CEP intervention
N-Glycosylation is one of the most important post-translational modifications of proteins [25]. Although there are only a few hundreds of glycoproteins in total, their highly complex glycosylation increases the number of theoretical protein morphologies by several orders of magnitude [26]. Many cell surface and extracellular matrix-related proteins become glycosylated through secretory pathways, thereby regulating their adhesion interactions, physical and chemical properties, and diversifying their functions [27].
To systematically study the differentially expressed glycoproteins during SARS-CoV-2 infestation, we adopted our well-established SARS-CoV-2 cellular model [3] with GX_P2V-treated Vero E6 cells, and performed the N-glycoproteomic profiling via the 4D label-free LC-MS/MS analysis in the presence or absence of GX_P2V and/or CEP. The workflow for the experiment was outlined in supplementary Figure S1. First, the cell samples were harvested and enzymatically digested and then analyzed by 4D label-free LC-MS/MS and bioinformatics. A total of 737,202 secondary spectrograms were obtained by mass spectrometry (supplementary Figure S1B). A total of 1770.0 N-glycosylation modification sites on 828.0 proteins were identified with database searching, where 1298.0 sites on 624.0 proteins have quantitative information. The detected N-glycosylation tends to obey N-X-T, N-C-S, N-G-S, N-X-S and N-X-C rules (supplementary Figure S1C).
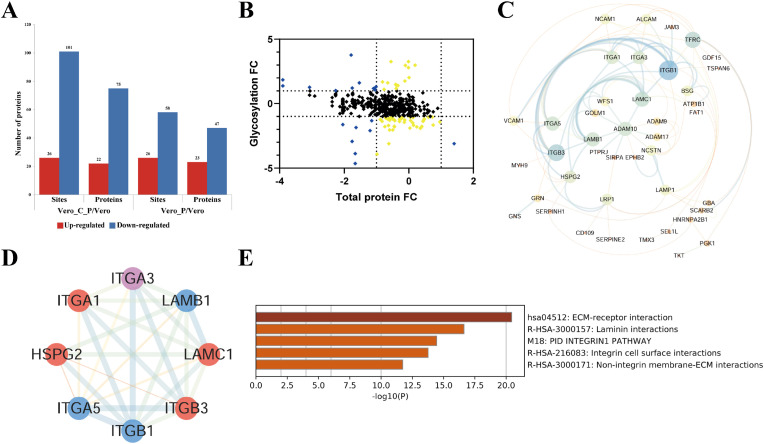
We took the fold changes (Fc = expression levels of glycosylated peptide in Group 2 (or 3 or 4)/Group 1 ratios) at Fc ≥ 2 (up-regulated) or Fc ≤ 0.5 (down-regulated) as differentially expressed protein, where group 1 was the mock control (Vero), group 2 was treated with CEP only (Vero_C), group 3 was treated by coronavirus GX_P2V only (Vero_P) and group 4 with both CEP and GX_P2V (Vero_C_P). In comparison with the mock control group, GX_P2V treatment can up-regulate 26 glycosylation sites in 23 proteins and down-regulate 58 glycosylation sites in 47 proteins, whereas in the presence of CEP, GX_P2V treatment induced 26 glycosylation sites up-regulated in 22 proteins and 101 glycosylation sites down-regulated in 75 proteins in host Vero E6 cells (Fig. 1 A). We then performed the analyses of global proteomics and respective of glycosylation occupancy based on the effects of GX_P2V infection (Fig. 1B).
Fig. 1.
Effects of GX_P2V infection on the aberrant glycosylation of proteins in Vero E6 cells. A. Effects of GX_P2V treatment on glycosylation (sites or proteins) in presence or absence of CEP. Fold change (Vero_C_P/Vero or Vero_P/Vero) ≥2 or ≤0.5 was considered as up-regulation or down-regulation, respectively. B. Global presentation of proteomic and respective glycosylation occupancy based on the effects of GX_P2V infection. Yellow dots represent proteins which have not changed significantly in global proteomics data but the peptides with glycosylation have differentially expressed over 2 folds, and blue dots represent proteins which are differentially expressed both in the levels of global protein and glycosylation. C. Protein-protein interaction (PPI) network and the interaction modules of key proteins in response to GX_P2V treatment. The PPI network of 66 differentially expressed glycoproteins (Fc ≥ 2 or Fc ≤ 0.5) in GX_P2V treatment was produced using STRING and Cytoscape. D. The key interaction module was obtained from the PPI network by Cytoscape with MCODE plugin using differentially expressed glycosproteins in GX_P2V-treated Vero E6 cells. Proteins marked in red color represent those proteins containing up-regulated glycosylation sites, the blue ones indicate proteins containing down-regulated glycosylation sites, and the purple color labelled protein contains both up- and down-regulated glycosylation sites. E. Functional enrichment analysis of the key modules induced by GX_P2V treatment was obtained from Metascape. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The protein-protein interaction (PPI) networks and the interaction modules of these differentially expressed proteins in Vero E6 cells in response to GX_P2V treatment analysis was conducted (Fig. 1C). We found that the key interaction module obtained from PPI network with MCODE plugin involved in 8 aberrant glycoproteins named 4 up-regulated glycoproteins namely Integrin Subunit Alpha 1(ITGA1), Integrin Subunit Beta 3(ITGB3), Laminin Subunit Gamma 1 (LAMC1) and Heparan Sulfate Proteoglycan 2(HSPG2), and 3 down-regulated glycoproteins named Integrin Subunit Beta 1 (ITGB1), Laminin Subunit Beta 1 (LAMB1), Integrin Subunit Alpha 5(ITGA5). The protein Integrin subunit alpha 3(ITGA3) contains both up- and down-regulated glycosylation sites (Fig. 1D). LAMC1 was reported to be involved in cell proliferation, angiogenesis, growth, migration and invasion [28]. Remarkably, the majority of these 8 aberrant glycoproteins in the key interaction module were enriched in ECM-receptor interaction pathways (Fig. 1D and E). Specifically, KEGG pathway enrichment showing GX_P2V infection affected glycoproteins in ECM-receptor interaction (Fig. 2 A). ECM components including collagens, laminins, and fibronectins are the major ligands that bind and activate integrin receptors. It helps in mediating cell-cell interactions by binding additional cell receptors or other soluble molecules. ECM plays an important role in cell adhesion, and viral infection may cause significant changes in cell adhesion. Functional enrichment analysis of the key modules revealed that these modules fall into the functional interaction categories of ECM-receptor interaction, laminin interactions, integrin cell surface interactions, endoderm development and receptor-mediated endocytosis. The heavily affected ECM and cell surface interactions might give a reasonable explanation on the gross glass-like changes in the clinical lung image.
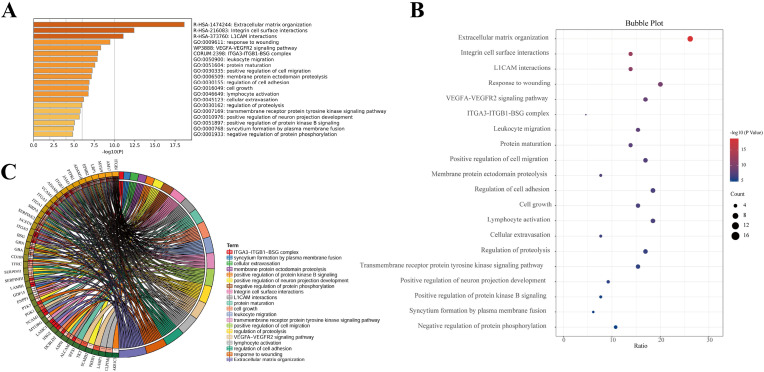
Fig. 2.
Functional enrichment clustering analysis of deferentially expressed glycoproteins in GX_P2V -treated Vero E6 cells. A. Functional enrichment analysis of proteins with aberrant glycosylation induced by GX_P2V infection was constructed by Metascape. B. Enriched bubble diagram of proteins with aberrant glycosylation after GX_P2V treatment was drawn by SangerBox. C. Circle diagram of protein functional enrichment analysis showing the relationship of aberrant glycosylation sites and the involved functions. The inner circle on the left side represents the significance p value of the gene corresponding pathway, and the right represents the corresponding term. The denseness of the left semicircular line represents the number of pathways associated with the protein. The density of the right semicircle line indicates how many proteins are enriched in that pathway. The figure shows that ITGB1 is involved in the most relevant pathways and the extracellular matrix is enriched for the most genes.
3.2. Coronavirus GX_P2V-induced alteration in protein glycosylation affects various functions of infected cells
N-linked glycosylation was involved in multiple biological processes, such as receptor interactions, immune responses, protein secretion and transport, cell adhesion, signal transduction, etc. As described above, we detected 84 sites of aberrant glycosylation sites present in 70 glycoproteins from coronavirus GX_P2V-treated Vero E6 cells. To study these sites holistically, we performed functional enrichment clustering analysis of these differential glycoproteins using Metascape (Fig. 2A), and the results were graphically presented using the Sangerbox mapping toolbox (Fig. 2B and C).
Upon the treatment of GX_P2V, the functional alteration related to differentially expressed proteins were shown in Fig. 2A and B. The circle diagram of protein function enrichment analysis demonstrated that ITGB1 was involved in the most relevant function and the extracellular matrix organization was enriched for the most genes. Besides, in response to coronavirus GX_P2V infection, the top 5 aberrant glycoproteins most involved relevant functions include ITGB1, AM17, MYH9, LRP1 and EPHB2, and the top 5 most aberrant functions involved most genes were those related to extracellular matrix organization, response to wounding, regulation of cell adhesion, lymphocyte activation and VEGFA-VEGFR2 signaling pathway. Other important functions such as protein maturation, viral entry into host cell and a platelet degranulation were also affected.
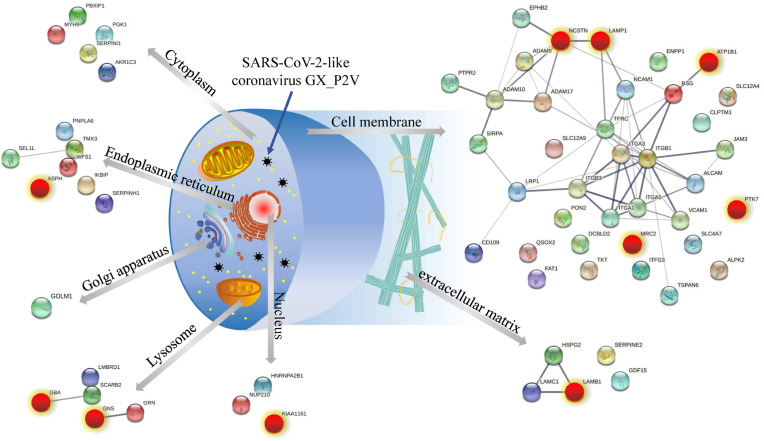
To have an overview about the effects of coronavirus GX_P2V treatment on host cells, we summarized the subcellular localization of differentially expressed glycosylated proteins and PPI networks by UniProt database [24], and STRING software (Fig. 3 ).
Fig. 3.
Schematic diagram of subcellular location and PPI of differentially expressed glycoproteins in GX_P2V treated Vero E6 cells as well as the intervention targets of CEP. A. The subcellular location of aberrant glycoproteins was determined using UniProt. The PPI network maps of these dysregulated glycoproteins were generated using STRING. The red color highlights CEP- regulated proteins. Upon GX_P2V treatment in presence of CEP, 10 out of 66 glycoproteins dysregulated by coronavirus GX_P2V remains in normal glycosylation state, suggesting that these proteins and glycosylation sites might be the targets of CEP. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Aberrant alteration of protein glycosylation can be seen in cytoplasm, endoplasmic reticulum, Golgi apparatus, lysosome, nucleus, cell membrane and extracellular matrix. The PPI networks of differentially expressed glycoproteins in each subcellular distribution were also highlighted (Fig. 3). These results suggested that coronavirus-induced alterations in glycosylation of host cell proteins are widespread in infected cells and have multiple subcellular localizations. The most involved glycoproteins locate in cell membrane, extracellular matrix, lysosomes, endoplasmic reticulum system and cytosol.
3.3. Effects of CEP on GX_P2V-induced aberrant glycosylation of proteins in Vero E6 cells
We have reported that CEP could effectively inhibit coronavirus infestation [3]. To identify the potential targets of CEP in coronavirus-infected cells in response to GX_P2V treatment, we compared the aberrant glycosylation sites and glycoproteins with and without coronavirus treatment in presence or absence of CEP and perform Reactome Pathway Analysis (Fig. 4 ).
Fig. 4.
Effects of CEP treatment on GX_P2V-induced aberrant glycosylation of proteins in Vero E6 cells. A. Venn diagram shows the effect of CEP on the aberrant glycosylation of proteins induced by GX_P2V. The number in purple circle indicates the glycosylation sites those are differentially expressed (with Vero_P/Vero Fc ≥ 2) in response to GX_P2V treatment (GX_P2V). The number in light red circle shows the glycosylation sites are in normal state (with Vero_C_P/Vero, 0.667 ≥ Fc ≤ 1.5) in presence of CEP (CEP + GX_P2V). The number in the overlap of the two circle shows that there are 13 site (on 10 proteins) out of 81 sites (on 66 proteins) GX_P2V-induced aberrant glycosylation sites in normal state in presence of CEP. B. Reactome Pathway Analysis of glycosylation sites on 10 glycoproteins affected by CEP. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The Venn diagram showed the effect of CEP on the aberrant glycosylation sites of proteins induced by GX_P2V (Fig. 4A). Coronavirus GX_P2V treatment can cause abnormal glycosylation at 81 sites (on 66 proteins) (Vero_P/Vero ratio, Fc ≥ 2), whereas the intervention of CEP can keep 13 glycosylation sites (on 10 glycoproteins) out of 81 GX_P2V-induced aberrant glycosylation sites at normal state (Vero_C_P/Vero ratio, 0.667 ≤ Fc ≤ 1.5) (Table S1, yellow background). For the subcellular location of these CEP-affected proteins, aspartate beta-hydroxylase (ASPH) is located in endoplasmic reticulum; glucosylceramidase Beta (GBA) and glucosamine (N-acetyl)-6-sulfatase (GNS) are lysosomal proteins; zinc finger C4H2-type containing (KIAA1161, also named MYORG) is in nucleus; nicastrin (NCSTN), lysosomal associated membrane protein 1 (LAMP1), protein tyrosine kinase 7 (PTK7) and mannose receptor C type 2 (MRC2) and ATPase Na+/K+ transporting subunit beta 1 (ATP1B1) are related to cell membrane and laminin subunit beta 1 (LAMB1) are the key proteins related to extracellular matrix (Fig. 3).
We then carried out Reactome Pathway Analysis on the 10 CEP-affected glycoproteins to understand intervention mechanism of CEP (Fig. 4B). The results showed that CEP exerted its antiviral and cell-protective effects by targeting multiple pathways such as Neutrophil degranulation, Ion homeostasis, Degradation of the extracellular matrix, and etc. Among these 10 proteins, LAMB1 is also the pivotal protein in the key interaction module that consists of 8 aberrant glycoproteins in response to coronavirus GX_P2V infestation (Fig. 1D), indicating that protein LAMB1 might be the key targets of CEP against GX_P2V infestation (Fig. 3).
In addition, several proteins were over-regulated by CEP against GX_P2V (Table S1, green background). There are two aberrant glycosylation sites on MRC2 induced by GX_P2V infection. In the presence of CEP, the site N69 on MRC2 protein remained normally glycosylation state, while the other site N1134 was shifted from virus-induced up-regulation to down-regulation. MRC2 may contribute to cellular uptake, remodeling and degradation of extracellular collagen matrices. Over-regulated glycosylation by CEP can also be seen at the site N277 of granulin precursor (GRN), which is a secreted glycoprotein that acts as a key regulator of lysosomal function and as a growth factor involved in inflammation, wound healing and cell proliferation [29]. It is involved in regulating protein trafficking to lysosomes and, also the activity of lysosomal enzymes [29,30]. Alteration of glycosylation GRN may contribute to lysosome-related functions. Therefore, CEP might also perform its anti-viral effect via regulation of glycosylation of MRC2 and GRN.
3.4. Effect of CEP on the N-glycosylation of GX_P2V viral proteins
Coronavirus viral proteins are normally extensively glycosylated, especially coronavirus spike proteins where it encodes around 66-87 N-linked glycosylation sites per trimeric spike. It has been known that the extensive glycosylation of viral protein plays a role in protein folding and shielding immunogenic epitopes, resulting in immune evasion [31,32]. Remarkably, SARS-CoV and SARS-CoV-2 recognize the human angiotensin converting enzyme-2 (ACE2) receptor via their glycosylated spike proteins [33].
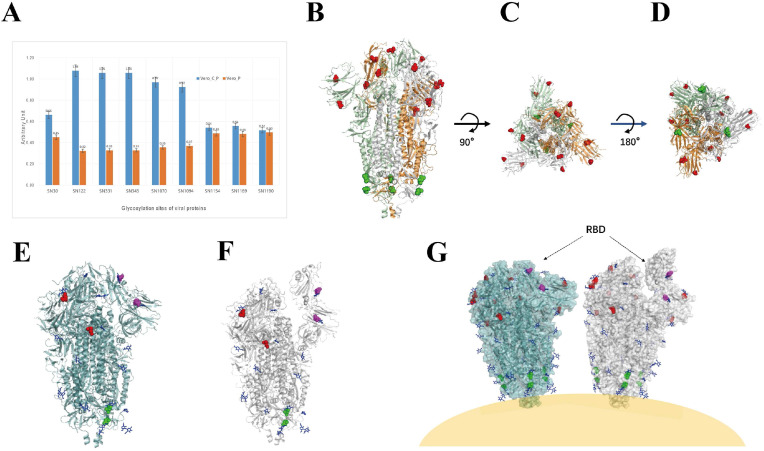
Interestingly, we found that CEP can up-regulate the glycosylation of the N-terminal sites of the viral S protein (including the N30, N122, N331 and N343 sites) and the sites N1070 and N1094 sites in the S2 region (N1070 and N1094, the corresponding positions on SARS-CoV-2 spike protein are N1074 and N1098, respectively), but no effect can be seen in the glycosylation of the C-terminal sites N1154, N1169, and N1190 of S proteins under CEP treatment (Fig. 5 A).
Fig. 5.
Effect of CEP on the N-Glycosylation of GX_P2V viral Proteins. A. Expression levels of N-glycosylation of viral proteins at indicated sites in presence or absence of CEP. B, C and D, indicate the side, top and bottom views of viral Spike protein (in closed state, PDB: 6VXX), respectively. All the N-glycosylation sites on the three chains of S protein affected by CEP are labelled. The N-glycosylation sites close to virus body are labelled in green, and those sites located at far-end of virus body are labelled in red. E and F. Side views of viral Spike protein (the N-glycosylation sites affected by CEP are labelled on B chain (Fig. 5E, PDB: 6VXX, closed state; Fig. 5F, PDB: 6VYB, open state)). Blue, NAG, N-linked glycan to the B chain of viral Spike protein; Magentas, the N-glycosylation sites located on the RBD of viral S protein; Green, the N-glycosylation sites affected by CEP on B chain of S protein close to virus body. Red, the CEP-affected N-glycosylation sites located on N-terminus of B chain. G. Cartoon display of CEP-affected N-glycosylation sites on Spike protein on the coronavirus surface. Note: Since the 3D structure of GX_P2V viral S proteins is not available, considering the high sequence similarity between SARS-CoV-2 and GX_P2V, we adopted the 3D structure of SARS-CoV-2 (6VXX, closed state; 6VYB, closed state) to draw the pictures. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To observe and analyze the location of these affected glycosylation sites on S proteins and their possible roles, we compared the sequences of spike protein of SARS-CoV-2 and GX_P2V viruses. Since the structure of GX_P2V viral S protein is not available by far, and considering the high sequence similarity between SARS-CoV-2 and GX_P2V, we adopted the 3D structure of SARS-CoV-2 (6VXX, closed state; 6VYB, closed state) to label the affected glycosylation sites (Fig. 5B–G). Among the sites affected by CEP on spike protein, N30 and N122 are located at NTD region, N331 and N343 are in CTD/RBD region, and N1070 and N1094 are in S2 region [34]. As can be seen from the cartoon image (Fig. 5G), the glycosylation sites of CEP-affected spike protein are located at both ends of the S protein. Four of the sites (including those located in the NTD and CTD/RBD regions) are at one end away from the virion, while the sites affected by the two sites affected locate in the S2 region at one end close to the virion. This suggests that one of the effects of CEP on viral S proteins might be to interfere with viral assembly and viral recognition or binding to the S protein receptors to exert its antiviral effects.
4. Conclusions
In this paper, N-glycosylation modification proteomics analysis were carried out to study the mechanisms of SARS-CoV-2 infection and the anti-coronavirus effects of CEP. To investigate the protein-protein interaction (PPI) networks and the interaction modules of these differentially expressed proteins in Vero E6 cells in response to GX_P2V treatment, we conducted STRING and Cytoscape analysis. It was found that the key interaction module obtained from PPI network with MCODE plugin involved in 8 aberrant glycoproteins named 4 up-regulated glycoproteins namely ITGA1, ITGB3, LAMC1 and HSPG2, and 3 down-regulated glycoproteins named ITGB1, LAMB1, and ITGA5. Functional enrichment analysis of the key modules revealed that these modules fall into the functional interaction categories of ECM-receptor interaction, laminin interactions, PID integrin1 pathway, integrin cell surface interactions and non-integrin membrane-ECM interactions. The heavily affected ECM and cell surface interactions might give a reasonable explanation on the gross glass-like changes in the clinical lung image. Furthermore, there are more than 75% of aberrant glycosylation of proteins after GX_P2V infection, which can be seen in cytoplasm, endoplasmic reticulum, Golgi apparatus, lysosome, nucleus, cell membrane and extracellular matrix. The PPI networks of differentially expressed glycoproteins in each subcellular distribution were also highlighted (Fig. 3). These results suggested that coronavirus-induced alterations in glycosylation of host cell proteins are widespread in infected cells and have multiple subcellular localizations. The most involved glycoproteins locate in cell membrane, extracellular matrix, lysosomes, endoplasmic reticulum system and cytosol. These abnormal changes in glycoproteins caused by coronaviruses may play an important role in viral invasion of host cells, resulting in cellular dysfunction.
Coronavirus viral proteins are normally extensively glycosylated [6], especially coronavirus spike protein where it encodes around 66-87 N-linked glycosylation sites per trimeric spike. From the data set for N-glycoproteomics profiling, we found that one of the effects of CEP on viral S protein might be to interfere with viral assembly and viral recognition or binding to the S protein receptors to exert its antiviral effects. It was displayed that coronavirus GX_P2V leaded to aberrant protein glycosylation, whereas CEP partially maintained GX_P2V-induced aberrant glycoproteins at homeostasis. The antiviral mechanism of CEP is also diverse, as it exerts its antiviral function by regulating the glycosylation of about 10 proteins which locate in endoplasmic reticulum, lysosomal, nucleus, cell membrane and ECM, respectively. Further study revealed that protein LAMB1 was pivotal in counteracting coronavirus-induced aberrant protein glycosylation by CEP. Furthermore, CEP dramatically regulate the glycosylation of viral proteins S, which may be the root cause of the virus affecting cell entry. This study provides a landscape of N-glycoproteomic profiling using the established SARS-CoV-2 cellular model, suggesting that it is of great importance in therapeutic target screening for drug discovery. The results indicated that coronavirus can cause alterations in the glycosylation of proteins at multiple levels in infected cells, whereas CEP can partially maintain GX_P2V-induced aberrant N-glycoprotein targets as well as partially regulate the glycosylation of viral proteins. However, at least 57 aberrant glycoproteins caused by coronavirus were not significantly improved by CEP treatment. Therefore, multi-target drug use in combination with CEP is essential for the treatment of coronavirus infection. These aberrant N-glycosylation of GX_P2V infestation might be potential targets for combination therapy.
Taken together, the results highlight the differentially expressed glycoproteins targets and related pathways in coronavirus infection and CEP intervention. Generally, coronavirus GX_P2V-induced aberrant N-glycosylation alteration in infected cells are prevalent. CEP can reduce the N-glycosylation of proteins located on cell membrane surface, indicating that CEP may achieve the protective effect against virus invasion by interfering potential N-glycoprotein targets in the affected cells. Besides, CEP can regulate the glycosylation of viral Spike protein, revealing targets or pathways relevant for viral pathogenicity. CEP inhibited SARS-CoV-2 entry through the blocking of viral binding to target cells [5], the enhancement of glycan shield on the viral proteins S might weaken coronavirus' binding to its recognition targets. There is an interested urgency to discovery new drug targets against COVID-19 [[35], [36], [37]]. This paper provides important insights into the mechanism of coronaviruses infection and the identification of drug targets for anti-coronavirus of multi-target drug combination usage.
Author contributions
Conceptualization, W. An and Y. Tong; methodology, W. An; software, J. Li.; validation, F. Tian, and J. Li; formal analysis, F. Tian, and J. Li; investigation, W. An and J. Chen; resources, W. An and J. Chen; data curation, F. Tian, and J. Li; writing—original draft preparation, F. Tian, and J. Li; writing—review and editing, W. An, J. Chen and Y. Tong; visualization, F. Tian, and J. Li; supervision, W. An, J. Chen and Y. Tong; project administration, W. An, J. Chen and Y. Tong; funding acquisition, W. An, J. Chen and Y. Tong. All authors have read and agreed to the published version of the manuscript.
Funding sources
This research was funded by the National Key Research and Development Program of China (2018YFA0903000), the National Natural Science Foundation of China (81672001), China MoST Emergency Project on COVID-19 (2020YFC0840800), Guangdong Basic and Applied Basic Research Foundation (2021A1515110619) and the Fundamental Research Funds for the Central Universities (KG16052901).
Data availability statement
All data generated or analyzed during this study are presented in this paper or in the Supplementary Information. The MS proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021297. Please visit http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD021297 via the Username: reviewer40579@ebi.ac.uk and Password: DM8k8f9F.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.medntd.2022.100156.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184(1):92–105. doi: 10.1016/j.cell.2020.10.030. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S., Liu W., Chen Y., Wang L., An W., An X., et al. Transcriptome analysis of cepharanthine against a SARS-CoV-2-related coronavirus. Briefings Bioinf. 2021;22(2):1378–1386. doi: 10.1093/bib/bbaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan H.H., Wang L.Q., Liu W.L., An X.P., Liu Z.D., He X.Q., et al. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin Med J. 2020;133(9):1051–1056. doi: 10.1097/CM9.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huo D., An W., Xu H., Yan A., Tong Y. Investigation of cepharanthine binding with viral proteins reveal its potential targets against coronavirus. Virol Curr Res. 2021;5:124. [Google Scholar]
- 5.Ohashi H., Watashi K., Saso W., Shionoya K., Iwanami S., Hirokawa T., et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience. 2021;24(4) doi: 10.1016/j.isci.2021.102367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Zhao W., Mao Y., Chen Y., Wang S., Zhong Y., et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol Cell Proteomics : MCP. 2021 doi: 10.1074/mcp.RA120.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng B.G., Freeze H.H. Perspectives on glycosylation and its congenital disorders. Trends Genet : TIG. 2018;34(6):466–476. doi: 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanova N., Shrimal S., Gilmore R. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr Opin Cell Biol. 2016;41:57–65. doi: 10.1016/j.ceb.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther R., Zelikin A.N. Chemical (neo)glycosylation of biological drugs. Adv Drug Deliv Rev. 2021;171:62–76. doi: 10.1016/j.addr.2021.01.021. [DOI] [PubMed] [Google Scholar]
- 10.Smith B.A.H., Bertozzi C.R. The clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans. Nat Rev Drug Discov. 2021:1–27. doi: 10.1038/s41573-020-00093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23(10):899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang D., Klein J.A., Nalehua M.R., Hackett W.E., Zaia J. Data-independent acquisition mass spectrometry for site-specific glycoproteomics characterization of SARS-CoV-2 spike protein. Anal Bioanal Chem. 2021;413(29):7305–7318. doi: 10.1007/s00216-021-03643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28(4):586–601. doi: 10.1016/j.chom.2020.08.004. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583(7815):282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 16.Meier F., Brunner A.D., Koch S., Koch H., Lubeck M., Krause M., et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol Cell Proteomics : MCP. 2018;17(12):2534–2545. doi: 10.1074/mcp.TIR118.000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesur A., Schmit P.O., Bernardin F., Letellier E., Brehmer S., Decker J., et al. Highly multiplexed targeted proteomics acquisition on a TIMS-QTOF. Anal Chem. 2021;93(3):1383–1392. doi: 10.1021/acs.analchem.0c03180. [DOI] [PubMed] [Google Scholar]
- 18.Loginov D.S., Fiala J., Chmelik J., Brechlin P., Kruppa G., Novak P. Benefits of ion mobility separation and parallel accumulation-serial fragmentation technology on timsTOF Pro for the needs of fast photochemical oxidation of protein analysis. ACS Omega. 2021;6(15):10352–10361. doi: 10.1021/acsomega.1c00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1) doi: 10.1093/nar/gky1131. D607-d13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doncheva N.T., Morris J.H., Gorodkin J., Jensen L.J. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18(2):623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haw R., Hermjakob H., D'Eustachio P., Stein L. Reactome pathway analysis to enrich biological discovery in proteomics data sets. Proteomics. 2011;11(18):3598–3613. doi: 10.1002/pmic.201100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1) doi: 10.1093/nar/gky1049. D506-d15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiro R.G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4):43r–56r. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 26.Rudd P.M., Elliott T., Cresswell P., Wilson I.A., Dwek R.A. Glycosylation and the immune system. Science (New York, NY) 2001;291(5512):2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 27.Raghunathan R., Sethi M.K., Klein J.A., Zaia J. Proteomics, glycomics, and glycoproteomics of matrisome molecules. Mol Cell Proteomics : MCP. 2019;18(11):2138–2148. doi: 10.1074/mcp.R119.001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patarroyo M., Tryggvason K., Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12(3):197–207. doi: 10.1016/S1044-579X(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Sun L., Bracko O., Choi J.W., Jia Y., Nana A.L., et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat Commun. 2017;8 doi: 10.1038/ncomms15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beel S., Moisse M., Damme M., De Muynck L., Robberecht W., Van Den Bosch L., et al. Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet. 2017;26(15):2850–2863. doi: 10.1093/hmg/ddx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science (New York, NY) 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G., et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guruprasad L. Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition. Proteins. 2020;88(11):1387–1393. doi: 10.1002/prot.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang P., Xie J., Sang S., Zhang L., Liu M., Yang L., et al. Multilayered N-glycoproteome profiling reveals highly heterogeneous and dysregulated protein N-glycosylation related to Alzheimer's disease. Anal Chem. 2020;92(1):867–874. doi: 10.1021/acs.analchem.9b03555. [DOI] [PubMed] [Google Scholar]
- 36.Papageorgiou A.C., Mohsin I. The SARS-CoV-2 spike glycoprotein as a drug and vaccine target: structural insights into its complexes with ACE2 and antibodies. Cells. 2020;9(11) doi: 10.3390/cells9112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park B.K., Kim J., Park S., Kim D., Kim M., Baek K., et al. MERS-CoV and SARS-CoV-2 replication can be inhibited by targeting the interaction between the viral spike protein and the nucleocapsid protein. Theranostics. 2021;11(8):3853–3867. doi: 10.7150/thno.55647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are presented in this paper or in the Supplementary Information. The MS proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021297. Please visit http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD021297 via the Username: reviewer40579@ebi.ac.uk and Password: DM8k8f9F.