Abstract
Coronavirus is a type of RNA-positive single-stranded virus with an envelope, and the spines on its surface derived its official name. Seven human coronaviruses 229E, OC43, SARS, NL63, HKU1, MERS, SARS-CoV-2 can cause both a mild cold and an epidemic of large-scale deaths and injuries. Although their clinical manifestations and many other pathogens that cause human colds are similar, studying the relationship between their evolutionary history and the receptors that infect the host can provide important insights into the natural history of human epidemics in the past and future. In this review, we describe the basic virology of these seven coronaviruses, their partial genome characteristics, and emphasize the function of receptors. We summarize the current understanding of these viruses and discuss the potential host of wild animals of these coronaviruses and the origin of zoonotic diseases.
Keywords: Human coronavirus, Emerging coronavirus, Host, Receptor, SARS-CoV-2, COVID-19
1. Introduction
In the past sixty years, the coronavirus has caused two large-scale public health incidents and several regional influenza incidents. The current epidemic of the new coronavirus has also posed an unprecedented challenge to global health. Many experts in public health hold the unanimous view that the coronavirus lurking in bats may cause large-scale disease outbreaks in the future. This review summarizes the current understanding of coronaviruses such as 229E, OC43, SARS, NL63, HKU1, MERS, and SARS-CoV-2. Based on recently published studies, this review covers the basic research of the origin, host, isolation, and identification of the coronavirus infected human being, including the intermediate host, the epidemiology, and receptor binding. Furthermore, we will discuss the clinical and other features.
1.1. The coronavirus: classification and origin
In 1966 Human respiratory virus 229E was discovered. In 2007, a homologous sequence of 229E was founded in a kind of alpaca in California. It named Alpaca Coronavirus (ACoV). The ACoV was genetically most similar to the common human coronavirus (HCoV) 229E with 92.2% nucleotide identity over the entire genome [1].
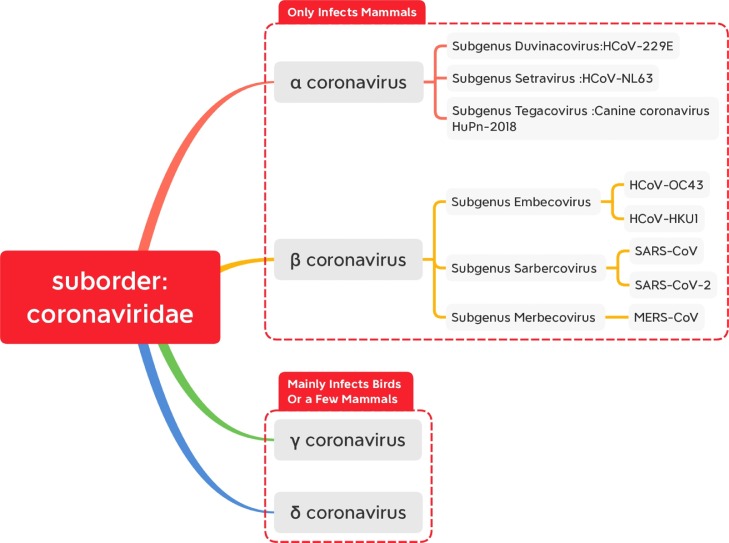
Fig.. 1.
According to the modification of genus of the virus of classification report of 10th ICTV, the coronavirus suborder divides into four-part: α coronavirus,β coronavirus,γ coronavirus,δ coronavirus. The first two only infect mammals, and the latter two mainly infect birds or a few mammals.
Follow-up research about the virus carried by Adrian bats demonstrated that 229E also has a high degree of similarity with the coronavirus in bats [2]. It means alpaca is likely to be the intermediate host of the original strain of the 229E virus from bats to humans [3].
Human coronavirus OC43 is a kind of β-coronavirus [4,5]. Also, an analysis of the molecular clock suggested that the recent common ancestor of OC43 and bovine coronavirus (BCOV) was isolated from 1890. It means that around 1890, BCoV has jumped the species barrier and infected humans. It could be the origin of the birth of OC43 [6].
SARS-CoV causes severe acute respiratory syndrome. This kind of disease was named SARS by World Health Organization in 2003. The research found that the pandemic was caused by a coronavirus which was the first coronavirus to result in severe public health events, and its natural origin could be the civet because the sequence of virus isolated from them is a similarity to the human SARS virus as high as 99.8%. Therefore, human SARS-CoV seems to be an animal virus that crosses the barrier of the intermediate host to infect human beings [7]. Seroepidemiologic data also proved this argument [8], [9], [10].
In January 2003, a 7-month-old kid was sent to the hospital in Amsterdam for fever, conjunctivitis, and coryza [11]. After the first description of HCoV-NL63, a second research group described the same virus named HCoV-NL in Vero-E6 cell culture supernatant [12,13].
Human coronavirus HKU1 (Hong Kong University) was discovered in 2004 in a patient infected by the virus. There is no specific sequence from any other member of the same species [14].
On June 13, 2012, a local private hospital in Jeddah received a man. After 11 days of admission, the infected patient died from renal failure and progressive respiratory [15]. In an outbreak of a hospital in 2013 in Al-Hasa, Saudi Arabia, 23 patients were infected by a novel virus. In 1983, MERS-CoV infection was detected in camel serum samples, which means MERS-CoV could present in camels 30 years ago. [16] The genomic sequence analysis revealed that the MERS-CoV, Pipistrellus bat coronavirus HKU5 and Tylonycteris bat coronavirus HKU4 are phylogenetically related (denoted as betacoronavirus lineage C). The structure of the virus isolated from bats has a high correlation with MERS-CoV. It supports the hypothesis that MERS-CoV originated from bats.
In December 2019, a case of pneumonia was reported. After the report, the local hospital used the surveillance mechanism to identify 4 cases of novel pneumonia. All of them are originated from the Huanan (the southern area of China) seafood market and had direct contact with the trade in the market.
In 2018, researchers analyzed nasal swabs from 301 pneumonia patients treated in a hospital in East Malaysia. It was found that eight patients, except for one child, were infected with the newly discovered coronavirus, which the researchers of the study named CCOV-HuPn-2018. In a study on May 20, 2021, researchers described the genetic characteristics of CCOV-HuPn-2018, indicating that it is a new coronavirus that has been transferred from infected dogs to infected people [17] (Fig.2 ).
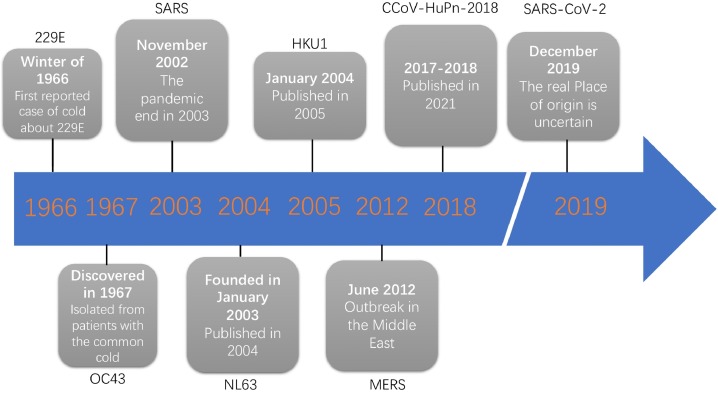
Fig. 2.
It is the timeline of coronavirus that was first discovered in human history. The 229E was first founded in 1966, and it was the first invented coronavirus of which seven ones could infect people. Secondly is OC43, which was isolated from patients with cold. The next one is SARS, the outbreak of SARS was discovered in November 2002 and ended in 2003. In the same year, NL63 was detected using the VIDISCA and PCR method. In 2004, HKU1 was reported by HKU and named. Seven years later, an outbreak was founded in the Middle East, caused by a novel coronavirus named MERS-CoV. In 2018, researchers surprisingly analyzed nasal swabs from 301 pneumonia patients treated in a hospital in East Malaysia. It was found that eight patients, except for one child, were infected with the newly discovered coronavirus, which the researchers of the study named CCOV-HuPn-2018. In December 2019, a case of SARS-CoV-2 was reported in Asia. Nevertheless, there is also evidence in NEJM that revealed that the 2019-nCoV infection could be diagnosed in Germany and transmitted outside to Asia. The authentic original place of SARS-CoV-2 is still unknown.
1.2. Animal host and its spillover
Follow-up research about the virus carried by Adrian bats demonstrated that 229E also has a high degree of similarity with the coronavirus in bats.[2]. Furthermore, it means alpaca is likely to be the intermediate host of the original strain of 229E virus from bats to humans, but the specific role in human infection is unknown [3].
An analysis of the molecular clock suggested that the recent common ancestor of OC43 and bovine coronavirus (BCOV) was isolated from 1890. It means that around 1890, BCoV has jumped the species barrier and infected humans. The supporting evidence of this kind of hypothesis is the finding of the absence of 290-nucleotide compared with BCoV because this extra sequence fragment is present in murine hepatitis virus (MHV) and rat sialodacryoadenitis virus (SDAV). Therefore, the assumption that the spread from bovines to humans sounds like much credibility [6].
What is the actual host of SARS? A study unveils this secret. From the study of Wendong Li, his team reported that ‘species of bats are a natural host of coronavirus closely related to those responsible for the SARS outbreak. In 2017, a team found all the genomic components of the SARS virus in two caves (Swallow Cave and Stone Cave) in Xiyang Yi Township, Jinning County, Yunnan Province, China, and pinpointed the source. They also Visited 218 villagers in Xiyang Yi Township, 81.2% raised or owned livestock or pets, and 9.1% witnessed bats flying near houses [14,[18], [19], [20]].
Up to date, some researchers found three different sequences of CoVs(family Hipposideridae) from Kenya that related closely with HCoV-NL63. The report also includes one strain (BtKYNL63–9a, GenBank ACC no. KY073744) with more than 90% similar amino acid sequence identity threshold about species typing in three of seven conserved domains [21]. Significantly, HCoV-229E and HCoV-NL63 could be from sister species. The recombination procedure involved the spike gene through two breakpoints. One is located near the gene's 5′-end and another near 200 nucleotides upstream of the 3′-end [21]. Among SARS- and SARS-like CoV and HCoV-OC43 have shown similar breakpoints [22], [23], [24]. The existence of recombination could implicate incomplete speciation; therefore, the existence of BtKYNL63–9a could be regarded as evidence of the primordial host of HCoV-NL63. Its ancestor could exist in hipposiderid or rhinolophid bats.
The exact source of MERS transmission to human beings remains unknown. Initial investigation revealed that MERS-CoV could originate from bats: some MERS-CoV-related sequences were founded from several bat species [25,26]. However, MERS-CoV has never been isolated directly from bats. Therefore if MERS-CoV was transmitted to people directly or indirectly is unknown. From an early study, all of the dromedary camels in Oman and 14% in the Canary Islands (in Spain) suggested positive anti-MERS-CoV antibodies [27].
Although the bats are significant, some facts revealed that some other animals play the role of intermediate host between humans and bats. First of all, when the pandemic was reported (in the winter of 2019), most bats were hibernating. Secondly, non-aquatic animals are sold at the Huanan seafood market, not including bats. Finally, bats are the natural host between MERS-CoV and SARS-CoV. Masked palm civet is the intermediate host of SARS-CoV, and dromedary camels are the intermediate host for MERS-CoV; humans are their terminal hosts [7,28]. Since the high sequence similarity between SARS-like bats coronavirus (Hipposideros bats in China) and SARS-CoV-2, it seems verified this hypothesis that the natural host of SARS-CoV-2 could be the Hipposideros bat. Another discovery also revealed that the coronavirus genomes of Malayan pangolin have 85.5∼92.4% sequence similarity to SARS-CoV-2. It also represents two sub-lineages of the related viruses of SARS-CoV-2 in the phylogenetic tree. It is said that Malayan pangolins could be the possible intermediate host [29]. Furthermore, the original host could be bats because of the isolation of a highly related coronavirus (RaTG13) from bats [30].
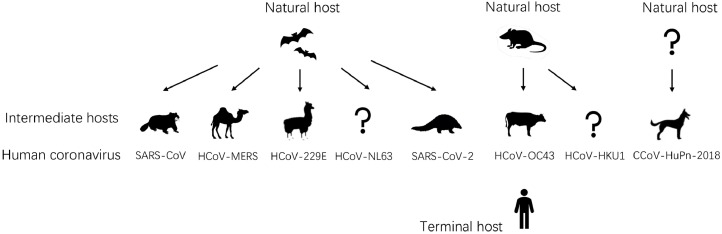
Fig. 3.
origins of human coronavirus. Severe acute respiratory syndrome coronavirus is a novel coronavirus related to bats with recombination. Before the SARS epidemic outbreak, the bats' virus infected civets and evolved to adapt to infect humans. The MERS-CoV could be originated from bats and spread to camels about 30 years ago and has been prevalent in dromedary camels. HCoV-NL63 and HCoV-229E, these two viruses do not cause severe respiratory damage and usually only bring a mild cold. The ancestor of both of them has been founded in African bats recently, and the camelids could be the intermediate host of HCoV-229E. Discovery revealed that the coronavirus genomes of Malayan pangolin have high sequence similarity to SARS-CoV-2. It means that Malayan pangolins could be thought to be the possible intermediate host. HCOV-OC43 and HCOV-HKU1 could be originated from rodents. Black arrows showed the direction of propagation. In this figure, humans are the ultimate hosts of transmission.
Table 1.
Summary of discussed coronaviruses.
| Virus | Virus species | Natural host | Intermediate host | Receptor | Clinical manifestation | Tropism | Available vaccine | reference |
|---|---|---|---|---|---|---|---|---|
| TGEV | Alpha-CoV | unknown | Pig | APN | / | enteric | LAV, PRCV as natural vaccine | [31] |
| BCoV | Beta-CoV | unknown | Bovine | SIALIC ACIDS | / | Enteric, respiratory | Inactivated virus, live attenuated vaccine | [32] |
| MHV | Beta-CoV | unknown | Mice | CEACAM1 | / | Strain dependent (enteric, hepatic, respiratory, CNS) | / | [33] |
| HCoV-229E | Alpha-CoV | bats | Alpaca | APN | General malaise, headache, nasal discharge, sneezing, sore throat, fever and cough | respiratory | / | [34, 35] |
| HCoV-NL63 | Alpha-CoV | bats | Unknown | ACE2 | cough, fever, sore throat, rhinitis, expectoration, and upper and lower respiratory tract infection, such as bronchitis, bronchiolitis or pneumonia | respiratory | / | [36], [37], [38], [39] |
| HCoV-OC43 | Beta-CoV | mice | Bovine | SIALIC ACIDS | Upper respiratory tract infection, headache, rhinorrhea and sore throat, coughing and fever | respiratory | / | [35, [40], [41], [42]] |
| HCoV-HKU1 | Beta-CoV | mice | Unknown | SIALIC ACIDS | rhinorrhea, fever, coughing, and wheezing, and can result in bronchiolitis and pneumonia if left untreated | respiratory | / | [43], [44], [45] |
| SARS-CoV | Beta-CoV | bats | Civet cats | ACE2 | ARDS, DAD, pulmonary fibrosis, Fever, Myalgia,Headache, Malaise, Chills, Nonproductive cough, Dyspnea, Respiratory distress | respiratory | multiple phase 1 trials | [46], [47], [48], [49], [50] |
| MERS-CoV | Beta-CoV | bats | Camels | DPP4 | ARDS, multi-lobar airspace disease, pulmonary fibrosis | respiratory | three recently concluded phase 1 trials | [47, 51, 52] |
| SARS-CoV-2 | Beta-CoV | bats | pangolins | ACE2 | ARDS, DAD, pulmonary fibrosis, fever, myalgia,headache, malaise, Chills, nonproductive cough, dyspnea, respiratory distress fatigue, chest pain, cognitive disturbances, arthralgia | respiratory | several ongoing clinical trials, RNA-based vaccine, Adenovirus vector (nonreplicating),protein subunit | [53], [54], [55], [56], [57], [58], [59], [60] |
| CCoV-HuPn-2018 | Alpha-CoV | unknown | canine | APN | respiratory illness that does not include the gastrointestinal issues | respiratory | / | [17] |
HCoV, human coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MERS-CoV, Middle East respiratory syndrome coronavirus; CCoV-HuPn-2018, Canine coronavirus HuPn-2018; DAD, diffuse alveolar damage.
1.3. Host and its receptor
1.3.1. APN receptor (HCoV-229E)
Aminopeptidase N (APN) is the primary cell surface receptor for group 1 coronaviruses. A region in the feline APN (fAPN) molecule (amino acids 135–297) is required to function as a receptor for HCV 229E. As in the human APN (hAPN, also known as CD13) protein, the aminoacid sequences required for the HCV 229E receptor function of fAPN are located within the amino-terminal part of the protein. Therefore, recognizing different determinants of the APN protein may reflect both functional and structural differences between the HCV 229E surface glycoprotein and the surface glycoproteins of another group 1 coronavirus [61].
The nature of the interaction between various RBDs and hAPN also provides clues for the in-depth understanding of how HCoV-229E and related bat, camel, and alpaca 229E viruses are transmitted to the corresponding hosts [62].
1.3.2. DPP4 receptor (HCoV-MERS)
The length of 30–31 kb genome could encode many proteins, which could endow the ability to adapt to new environments and cross-species transmission. It means MERS-CoV could originate from the differential exchange of various viral ancestors, including the strains isolated from the camels and its assuming natural host: bats. Since the outbreak of 2012, MERS-related virus (such as HKU4 and HKU5) has been founded in different kinds of bats. The ORF1ab of these viruses are highly similar to MERS-CoV's. Some MERS-related viruses could even use the same receptor (DPP4) of MERS-CoV [63], [64], [65].
1.3.3. ACE2 receptor (The similarities and differences between NL-63, SARS, and SARS-CoV-2)
The human ACE2 receptor is a crucial target of the NL-63, SARS-CoV, and SARS-CoV2 spike (S) protein receptor-binding domain (RBD) that facilitates viral entry into host cells. The S protein drives the infection between coronaviruses of the target cell. The S protein could promote the entry of SARS-CoV-2 into the target cell. Depending on the S1 unit of the surface of S protein can facilitate the combination with the ACE2 receptor. S protein of SARS-CoV-2 shares about 76% amino acid identity with SARS-CoV's. Since SARS-S uses ACE2 as a receptor for cell entry and uses the cellular serine protease TMPRSS2 for S protein initiation, it is not difficult to deduce that SARS-CoV-2-S also has a similar effect. This mechanism makes the degree of fusion of ACE2 and SARS-CoV a decisive factor in virus transmission [66]. During the infection of SARS-CoV, S protein plays a significant role in mediating the production of neutralizing-antibody and T-cell responses, also protective immunity [67]. In addition, the RBD in S1 is responsible for the recombination between virus and host cells [68, 69]. It means ACE2 is the functional receptor of SARS-CoV [69, 70].
Compared with SARS-CoV, ACE2 is also the docking point, but SARS-CoV-2 binds to ACE2 is more robust 2–4 times than SARS-CoV because the changes in RBD stabilize the virus-binding hotspots [69,71,72]. From the sequence analysis, although the SARS-CoV-2 genes shared less than 80% identity of the sequence of nucleotide to SARS-CoV, the amino acid sequences of seven conservation replicase domains in ORF1ab for classification shared 94.4% identification between SARS-CoV-2 and SARS-CoV. That is to say, both of them belong to one same species, SARSr-CoV. From the study, it is said that a short paragraph of the RNA-dependent RNA polymerase (RdRP) of a kind of coronavirus (BatCoV RaTG13) isolated from bats has the identification of sequence to SARS-CoV-2 highly. After the full-length sequencing of the sample, the result revealed that the SARS-CoV-2 is highly similar to RaTG13 in the whole genome sequence. The identity between them is as high as 96.2%. In addition, the known virus in bat SARS-CoV-2 lineage has not used the ACE2 of humans efficiently compared with the SARSr-CoV-2 from pangolin or SARSr-CoV-1 lineage viruses [73]. Compared with SARS-CoV, the amino acid sequence identity between the S protein of them is 76.47% [74]. It is evident that SARS-CoV-2 is a high-speed virus; it could prevent our immune system from identifying and fighting for the infection with its unique ability. When our immune system realize there is virus survives, the immune-response proteins would swarm into the bloodstream with high speed than normal condition and could cause damage. Some infected patients become deteriorate by the overactive immune response to SARS-CoV-2 and the toxic effects of the virus itself [[72], [75]].
From the study, the genome sequence level of SARS-CoV-2 is closer to bat-SL-CoVZXC21 and bat-SL-CoVZC45, but from the receptor-binding domain and external subdomain, it is closer to SARS-CoV. Isolated from the patient species of Wuhan, China, the phylogenetic analysis suggested that SARS-CoV-2 belongs to the Sabecovirus. Although specific antibodies in patients with SARS-CoV can be maintained for two years after infection, the difference is that despite being vaccinated with specific vaccines, SARS-CoV-2 patients still have a greater possibility of re-infection after recovery [76]. Interestingly, Marlin et al. described in the macaque (humanoid) model that infected and convalescent macaques were still at risk of reinfection at six months. This result also means that the levels of neutralizing antibodies in the body tend to weaken over time [77]. Studies have shown that soluble ACE2 (sACE2) can bind to the viral S protein. The sACE2 released from the human airway epithelium will limit the binding of the above virus to the ACE2 receptor on the cell surface, preventing the virus from further replicating in the human body [78,79]. Secondly, studies have shown that CD40L expressed on T cells can be activated and interacted with by expressing CD40. And then regulating the immune function of B cells and T cells, improving the recovery period of patients with secondary infection and effectively increasing the level of neutralizing antibodies. These two mechanisms may serve as a new direction for vaccine development [77,80]. Vaccines at the current stage can only prevent major complications after infection but cannot prevent re-infection and secondary transmission. Many virus spreaders are vaccinated, but they remain asymptomatic and can continue exporting the virus. While building herd immunity, we also need to prevent asymptomatic infections from becoming a source of infection again.
2. Conclusions
To sum up, it is not difficult to conclude that the SARS-CoV-2 could be more infectious than SARS-CoV and MERS-CoV. Is this related to other unknown receptor interactions? Will 2019-nCoV produce adaptive mutations in human-to-human transmission? These issues are still worthy of our in-depth study. Both SARS-CoV-2 and SARS-CoV can activate the body's antiviral immune response and cause uncontrolled inflammatory responses in critical patients. It is characterized by the release of proinflammatory cytokines, leading to lymphopenia, lymphocyte dysfunction, and granulocyte and monocyte abnormalities. The clinical characteristics of the two are different, but the difference lies in the longitudinal factors such as the severity of the onset, the fatality rate, and the infection rate. For their pathological manifestations in the respiratory tract, there seems to be a specific correlation with the utilization rate of ACE2 receptors. In terms of virus grouping, MERS-CoV, which belongs to Beta-CoV, is quite different from the above two viruses in lung manifestations; this is also related to the utilization of the receptor DPP4 of MERS-CoV. The origin of coronaviruses is relatively simple compared to other viruses, but its hosts are primarily distributed in mammals. This result should arouse our attention more than other phenomena because humans are mammals and advanced mammals. The relationship is self-evident. Whether one-day humans will be infected with a new type of virus through excessive contact with mammals, no one will know. We can draw some conclusions from the coronavirus we mentioned above. First of all, from the lessons of MERS-CoV and SARS-CoV-2, promoting early intervention and identification plays a significant role in protecting from deteriorating and preventing aberrant proinflammatory response even if the vaccination strategy is not mainstream. However, form using the strategies of immune evasion could prevent the identification by pattern recognition receptors. Secondly, neutralizing antibody responses exhibit its protection during the infection of coronavirus. It could be considered as the critical point for vaccine strategies. Also, the studies of animals and humans show that vaccines should strongly induce humoral responses and cellular adaptive immune responses because they are the utmost essential mediators of protection. Last but not least, we should also pay more attention to the potential variants of the virus.
3. Future Outlooks
Although the death of SARS-CoV-2 is less than MERE-CoV and SARS, the rapid pandemic outbreak has posed the severest threat to the public health of our world. The pandemic has lasted over two years. when will it end? Nobody knows. The vaccine for the current epidemic has been developed, but the real question is, how should we face the next pandemic of the zoonotic coronavirus? Will the next global pandemic be in 5 years? Ten years? Or did it appear earlier? After we control the pandemic, the next step should focus on virus screening, identification, isolation, and further detecting coronaviruses' presence in wild animals, especially bats. At the same time, researchers should find suitable animal models to conduct in vitro and in vivo studies to assess the risk of future epidemics. Therefore, we may build up appropriate animal models of COVID-19, which could be used to understand the immune operational mechanism. Non-human primates could be best. Due to non-human primates being humans' closest relatives in evolution, they may provide the most relevant data for pathological alteration, the safety and effectiveness of the vaccine, therapy, and preclinical experiment. However, the availability and expense of the non-human animal model should also be taken into account. Finally, the persistent analysis of the full-length genome is necessary because of the high mutability of coronavirus. At present, the novel coronavirus epidemic is undergoing mutations at a high rate; before the arrival of a new round of the epidemic, we must take measures to prevent the occurrence of a new pandemic. It seems to be an excellent decision to choose cd40 or sACE2 as the new direction for vaccines. Under the epidemic prevalent in the world, addressing the problematic issue requires the efforts of every people, government, and nation. Only through cooperation can we overcome common difficulties.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was sponsored by the National Natural Science Foundation of China (81770063), Guangzhou Science and Technology Program Project (202102010343), Health and Medical Collaborative Innovation Project (201704020225), the grant of the State Key Laboratory of Respiratory Disease, Guangdong-Hong Kong-Macao Joint Laboratory of Respiratory Infectious Disease (GHMJLRID-Z-202104) and Guangdong Basic and Applied Basic Research Foundation (2021A1515110550).
References
- 1.Crossley BM, Mock RE, Callison SA, Hietala SK. Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses. 2012;4:3689–3700. doi: 10.3390/v4123689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, et al. Evidence for an ancestral association of human coronavirus 229E with Bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proceedings of the Nat. Acad. Sci. United States of America. 1967;57:933. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 8.Breiman RF, Evans MR, Preiser W, Maguire J, Schnur A, Bekedam H, et al. Role of China in the quest to define and control severe acute respiratory syndrome. Emerg. Infect. Dis. 2003;9:1037. doi: 10.3201/eid0909.030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 11.Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J. Infect. Dis. 2005;191:492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, et al. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U S A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 16.Müller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlasova AN, Diaz A, Damtie D, Xiu L, Toh T-H, Lee JS-Y, et al. Novel canine coronavirus isolated from a hospitalized pneumonia patient, East Malaysia. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 19.Wang L-f, Cowled C. John Wiley & Sons; 2015. Bats and viruses: a New Frontier of Emerging Infectious Diseases. [Google Scholar]
- 20.Wang N, Li SY, Yang XL, Huang HM, Zhang YJ, Guo H, et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, et al. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J. Virol. 2017;91 doi: 10.1128/JVI.01953-16. e01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hon C-C, Lam T-Y, Shi Z-L, Drummond AJ, Yip C-W, Zeng F, et al. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau SK, Li KS, Huang Y, Shek C-T, Tse H, Wang M, et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau SK, Lee P, Tsang AK, Yip CC, Tse H, Lee RA, et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ithete NL, Stoffberg S, Corman VM, Cottontail VM, Richards LR, Schoeman MC, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19:1697. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reusken CBEM, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. The Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5 doi: 10.1128/mBio.00884-14. e00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saif LJ, Wang Q, Vlasova AN, Jung K, Xiao S. Coronaviruses. Dis. Swine. 2019:488–523. [Google Scholar]
- 32.Saif LJ. Bovine respiratory coronavirus. Veterinary Clinics: Food Anim. Practice. 2010;26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mora AL, Woods CR, Garcia A, Xu J, Rojas M, Speck SH, et al. Lung infection with γ-herpesvirus induces progressive pulmonary fibrosis in Th2-biased mice. Am. J. Physiol.-Lung Cellular and Mol. Physiol. 2005;289 doi: 10.1152/ajplung.00007.2005. L711-L21. [DOI] [PubMed] [Google Scholar]
- 34.Pene F, Merlat A, Vabret A, Rozenberg F, Buzyn A, Dreyfus F, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papa A, Papadimitriou E, de Souza Luna LK, Al Masri M, Souliou E, Eboriadou M, et al. Coronaviruses in children, Greece. Emerg. Infect. Dis. 2007;13:947. doi: 10.3201/eid1306.061353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moës E, Vijgen L, Keyaerts E, Zlateva K, Li S, Maes P, et al. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 2005;5:1–10. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastien N, Anderson K, Hart L, Caeseele PV, Brandt K, Milley D, et al. Human coronavirus NL63 infection in Canada. The J. Infect. Dis. 2005;191:503–506. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Der Hoek L, Sure K, Ihorst G, Stang A, Pyrc K, Jebbink MF, et al. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:e240. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu SS, Hung Chan K, Wing Chu K, Kwan SW, Guan Y, Man Poon LL, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monto AS. Medical reviews. Coronaviruses. The Yale J. Biol. Med. 1974;47:234. [PMC free article] [PubMed] [Google Scholar]
- 41.Cabeça TK, Granato C, Bellei N. Epidemiological and clinical features of human coronavirus infections among different subsets of patients. Influenza Other Respir Viruses. 2013;7:1040–1047. doi: 10.1111/irv.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dijkman R, Jebbink MF, Koekkoek SM, Deijs M, Jónsdóttir HR, Molenkamp R, et al. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 2013;87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo PC, Lau SK, Tsoi H-w, Huang Y, Poon RW, Chu C-m, et al. Clinical and molecular epidemiological features of coronavirus HKU1–associated community-acquired pneumonia. J. Infect. Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau SK, Woo PC, Yip CC, Tse H, Tsoi H-w, Cheng VC, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peiris J, Lai S, Poon L, Guan Y, Yam L, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. The Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cleri DJ, Ricketti AJ, Vernaleo JR. Severe acute respiratory syndrome (SARS) Infect. Dis. Clin. 2010;24:175–202. doi: 10.1016/j.idc.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peiris JSM, Chu C-M, Cheng VC-C, Chan K, Hung I, Poon LL, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. The Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J, Zhang J-S, Su N, Xu J-G, Wang N, Chen J-T, et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antiviral therapy. 2007;12:1107. [PubMed] [Google Scholar]
- 51.Al-Tawfiq JA, Hinedi K, Ghandour J, Khairalla H, Musleh S, Ujayli A, et al. Middle East respiratory syndrome coronavirus: a case-control study of hospitalized patients. Clin. Infect. Dis. 2014;59:160–165. doi: 10.1093/cid/ciu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arabi YM, Arifi AA, Balkhy HH, Najm H, Aldawood AS, Ghabashi A, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann. Internal Med. 2014;160:389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 53.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JH, Marks F, Clemens JD. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021;27:205–211. doi: 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 58.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 59.Carfì A, Bernabei R, Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenforde MW, Kim SS, Lindsell CJ, Billig Rose E, Shapiro NI, Files DC, et al. Symptom duration and risk factors for delayed return to usual health among outpatients with COVID-19 in a multistate health care systems network - United States, March-June 2020. MMWR Morb Mortal Wkly Rep. 2020;69:993–998. doi: 10.15585/mmwr.mm6930e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolb AF, Hegyi A, Siddell SG. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J. General Virol. 1997;78:2795–2802. doi: 10.1099/0022-1317-78-11-2795. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, Tomlinson AC, Wong AH, Zhou D, Desforges M, Talbot PJ, et al. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife. 2019;8:e51230. doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo C-M, Wang N, Yang X-L, Liu H-Z, Zhang W, Li B, et al. Discovery of novel bat coronaviruses in South China that use the same receptor as Middle East respiratory syndrome coronavirus. J. Virol. 2018;92:e00116–e00118. doi: 10.1128/JVI.00116-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proceedings of the Nat. Acad. Sci. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Q, Qi J, Yuan Y, Xuan Y, Han P, Wan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host & Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:181. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuhn J, Li W, Choe H, Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cellular and Mol. Life Sci. CMLS. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 72.Scudellari M. How the coronavirus infects cells-and why Delta is so dangerous. Nature. 2021;595:640–644. doi: 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- 73.Guo H, Hu B, Si H-R, Zhu Y, Zhang W, Li B, et al. Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor. Emerg. Microbes & Infect. 2021;10:1507–1514. doi: 10.1080/22221751.2021.1956373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu L-P, Wang N-C, Chang Y-H, Tian X-Y, Na D-Y, Zhang L-Y, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marlin R, Godot V, Cardinaud S, Galhaut M, Coleon S, Zurawski S, et al. Targeting SARS-CoV-2 receptor-binding domain to cells expressing CD40 improves protection to infection in convalescent macaques. Nat. Commun. 2021;12:5215. doi: 10.1038/s41467-021-25382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H, Baker A. Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Critical Care (London, England) 2017;21:305. doi: 10.1186/s13054-017-1882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci.(London, England: 1979) 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 80.Pušnik J, Richter E, Schulte B, Dolscheid-Pommerich R, Bode C, Putensen C, et al. Memory B cells targeting SARS-CoV-2 spike protein and their dependence on CD4 T cell help. Cell Reports. 2021;35 doi: 10.1016/j.celrep.2021.109320. [DOI] [PMC free article] [PubMed] [Google Scholar]