Abstract
Background
Imatinib reduced 90-day mortality in hospitalised coronavirus disease 2019 (COVID-19) patients in a recent clinical trial, but the biological effects that cause improved clinical outcomes are unknown. We aimed to determine the biological changes elicited by imatinib in patients with COVID-19 and what baseline biological profile moderates the effect of imatinib.
Methods
We undertook a secondary analysis of a randomised, double-blind, placebo-controlled trial of oral imatinib in hospitalised, hypoxaemic COVID-19 patients. Mediating effects of changes in plasma concentration of 25 plasma host response biomarkers on the association between randomisation group and 90-day mortality were studied by combining linear mixed effect modelling and joint modelling. Moderation of baseline biomarker concentrations was evaluated by Cox regression modelling. We identified subphenotypes using Ward's method clustering and evaluated moderation of these subphenotypes using the aforementioned method.
Results
332 out of 385 participants had plasma samples available. Imatinib increased the concentration of surfactant protein D (SP-D), and decreased the concentration of interleukin-6, procalcitonin, angiopoietin (Ang)-2/Ang-1 ratio, E-selectin, tumour necrosis factor (TNF)-α, and TNF receptor I. The effect of imatinib on 90-day mortality was fully mediated by changes in these biomarkers. Cluster analysis revealed three host response subphenotypes. Mortality benefit of imatinib was only present in the subphenotype characterised by alveolar epithelial injury indicated by increased SP-D levels in the context of systemic inflammation and endothelial dysfunction (hazard ratio 0.30, 95% CI 0.10–0.92).
Conclusions
The effect of imatinib on mortality in hospitalised COVID-19 patients is mediated through modulation of innate immune responses and reversal of endothelial dysfunction, and possibly moderated by biological subphenotypes.
Short abstract
The effect of imatinib on mortality in hospitalised COVID-19 patients is mediated through modulation of innate immune responses and reversal of endothelial dysfunction, and possibly moderated by biological subphenotypes https://bit.ly/3QWQyvT
Introduction
Acute hypoxaemic respiratory failure is the most common reason for hospitalisation in patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. The introduction of different treatment strategies, including steroids, interleukin (IL)-6 inhibitors and therapeutic anticoagulation [2–4], has resulted in improved clinical outcomes in hospitalised non-critically ill coronavirus disease 2019 (COVID-19) patients. However, with the best standard of care, mortality in hospitalised patients remains substantial with a rate of ∼6.5% [5].
Observational studies have linked unfavourable outcomes in COVID-19 patients to dynamic changes in the plasma concentrations of biomarkers reflecting modulation of the innate immune response, endothelial barrier protection and epithelial injury [6, 7]. Imatinib, an ABL tyrosine kinase inhibitor, has been shown to improve the endothelial barrier by reversing the loss of cell matrix adhesion and adherens junctions in vitro and in animals in vivo [8, 9], and in addition has immunomodulatory effects [10]. A randomised controlled trial of imatinib in hospitalised, hypoxaemic patients with COVID-19 did not show statistical improvement in the primary end-point (i.e. duration of oxygen therapy), but revealed a large decrease in 28-day mortality [11]. Imbalances in baseline characteristics between treatment arms were suggested to drive part of the protective effect, which could have led to a type I error. In an extended follow-up study, the survival benefit of imatinib at day 90 remained statistically significant in both the unadjusted and adjusted analysis, increasing the likelihood of a true protective effect [12].
Our current understanding of how pharmacological interventions improve outcomes in COVID-19 is limited. There is a general conception that immunomodulation of the innate immune response and endothelial protection are important in the treatment of severe COVID-19 [13]. It is therefore assumed 1) that a change in these biological processes is required for the drug to work and 2) that patients with higher baseline activation of these pathways are more likely to respond [14–16]. However, this assumption has not been formally tested. Mediation analysis can be used to evaluate if a drug only results in improved outcomes when it elicits specific biological effects, thus testing if an intermediate response is required for the drug to work. Moderation analysis can be used to study whether the relationship between two variables is dependent on the value of a third variable, e.g. a baseline biomarker concentration.
In this study, we aimed to describe what biological changes are elicited by imatinib and how these changes relate to clinical outcomes. We hypothesised that the effect of imatinib on 90-day mortality was mediated by reversal of endothelial dysfunction and modulation of innate immune responses. We also postulated that the baseline biological profile of a patient moderated the effect of imatinib on 90-day mortality.
Methods
Study design and patient selection
This is a pre-specified secondary analysis of clinical data and biological material obtained from a randomised, double-blind, placebo-controlled, clinical trial that was performed at 13 hospitals in the Netherlands. Details on study design and patient selection are described elsewhere [11]. In short, patients were eligible for inclusion if they were aged ≥18 years, had been admitted to the hospital with a SARS-CoV-2 infection (confirmed with a PCR test) and required supplemental oxygen to maintain peripheral oxygen saturation >94%.
The trial was approved by the Medical Ethics Committee of Amsterdam UMC (location VUmc, Amsterdam, Netherlands), and was performed in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. All patients provided written informed consent before randomisation.
Study procedures
Patients were randomly assigned (1:1) to either placebo or oral imatinib treatment. After randomisation, patients in the imatinib group received a loading dose of 800 mg imatinib on day 0, followed by 400 mg once daily on days 1–9. Patients in the placebo group received placebo tablets in a similar dosing scheme. Heparin anticoagulated blood was collected immediately before first study drug administration (baseline), and at day 2, day 3 and day 5 thereafter. Plasma was harvested and stored at −80°C within 4 h after blood draw. Plasma was obtained from 20 healthy volunteers to obtain reference normal values.
Data collection
Measurements were done in heparin anticoagulated plasma. 25 biomarkers were measured by Luminex multiplex assay (R&D Systems, Minneapolis, MN, USA), using the Bio-Plex 200 System (Bio-Rad, Hercules, CA, USA) in one batch at the end of the study (supplementary table S1). Ferritin and C-reactive protein (CRP) were not measured, due to differences in dilution. The data quality assessment is described in the supplementary methods.
End-points
For this secondary analysis focusing on the biological effects of imatinib, we used 90-day mortality as the primary end-point and 28-day mortality as the secondary end-point.
Statistical analysis
All statistical analyses were performed in R version 4.1.0 (www.r-project.org) using RStudio version 1.4.766.
Mediation analysis
All biomarkers were log10 transformed to better approach a normal distribution. The association between randomisation to imatinib and the longitudinal biomarker values was estimated using linear mixed effects models (with the lme4 package) [17]. The randomisation group, the measurement day and their interaction were included as fixed effects. Random intercepts were given to each subject. The effect of imatinib over time was determined by evaluating the interaction term and the 95% confidence interval of this term was calculated using bootstrapping. In a sensitivity analysis, baseline differences (i.e. age, body mass index (BMI), diabetes and cardiovascular disease) were included as covariates.
Biomarkers that were statistically significant in the earlier described linear mixed effects models were subsequently studied using Baron and Kenny's approach for mediation [18]. First, the abovementioned model was used to describe the effect of treatment on biomarker concentration (nlme package) [19]. Second, a joint model that combines a linear mixed effects model and a Cox proportional hazards model was used to describe the effect of a change in biomarker concentration on mortality (using the survival and JM packages) [20, 21]. Third, a Cox proportional hazards model was used to describe the association between randomisation group and mortality. Next, the isolated effect of imatinib on mortality (i.e. the effect explained if no change in biomarker was observed) was calculated. As a sensitivity analysis, a mediation analysis using natural effects as described in the medflex package was performed [22]. All model assumptions are described in the supplementary methods.
Moderation analysis
To estimate the moderation of the baseline biological profile on the association between randomisation group and outcome, we performed Cox regression modelling (with the survival package) [20]. Randomisation group, baseline biomarker concentration and its interaction term were used as independent variables and 90-day mortality as time-to-event variable. Resulting p-values were corrected for multiple testing using the Benjamini–Hochberg false discovery rate. A significant interaction term indicates that the effect between imatinib and mortality is influenced by the baseline biomarker concentration. In a secondary analysis, the baseline biomarker concentrations were dichotomised by maximally selected rank statistics (survminer package) [23]. This dichotomised variable was included in the aforementioned Cox models.
Lastly, we identified subphenotypes of patients with a similar baseline biological status using Ward's method clustering. For this, baseline (pre-treatment) host response biomarker concentrations were used. IL-10 and IL-17 were excluded from this analysis since these disproportionally affected the clustering due to a high proportion of values below the lower limit of quantification (supplementary table S2). The optimal number of clusters was determined using a majority ruling as described in the NbClust package [24]. This approach has been used previously to identify and validate subphenotypes of acute respiratory distress syndrome (ARDS) [25]. To evaluate the effect of imatinib treatment on mortality within each cluster subgroup, a Cox regression model with randomisation group as only covariate was performed for patients within each cluster. A Cox regression model with randomisation group, age, BMI, diabetes and cardiovascular disease as covariates was performed as a sensitivity analysis.
Results
Patients
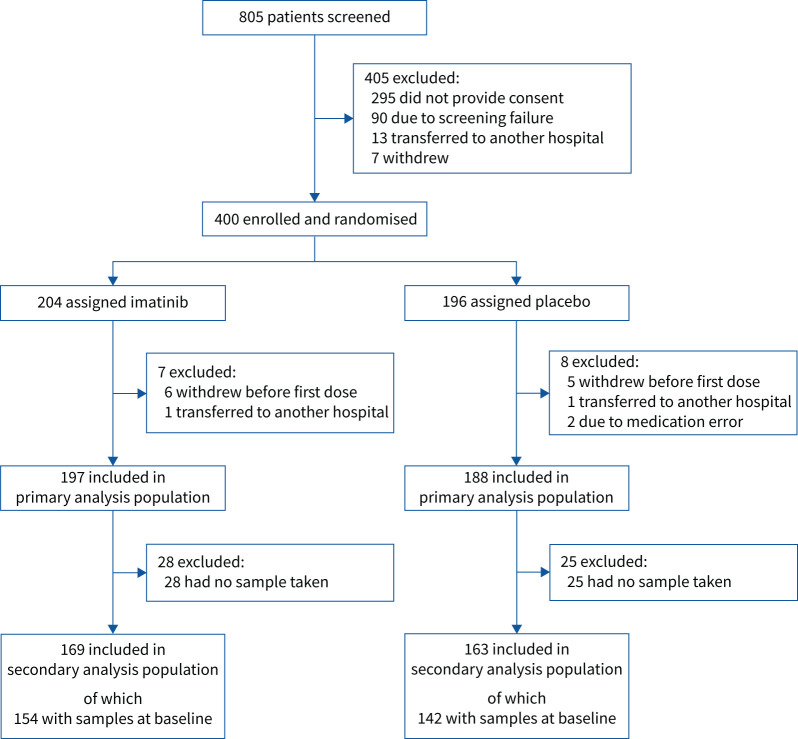
Between March 2020 and January 2021, 385 patients were included in the final analysis population, of which 197 patients were randomised to the imatinib group and 188 patients to the placebo group (figure 1). Baseline biomarker data were available for 154 imatinib patients (78%) and 142 placebo patients (76%). 169 imatinib patients (86%) and 163 placebo patients (87%) had at least one measurement during the study period (supplementary table S3). Patients included in the secondary analysis were comparable in terms of age and sex (table 1). Patients in the placebo group more often had obesity, diabetes mellitus and cardiovascular disease. Baseline routine laboratory values and medical treatments, both chronic medication use and medication initiated at hospital admission, in particular dexamethasone, were comparable between the groups. In line with the analysis of the total population [12], 90-day mortality was significantly lower in the imatinib group (unadjusted HR 0.49, 95% CI 0.26–0.92; adjusted HR 0.47, 95% CI 0.24–0.94). The primary end-point of the clinical trial (i.e. time to discontinuation of ventilation and supplemental oxygen for >48 consecutive hours, while being alive during a 28-day period after randomisation) was also comparable to the analysis of the total population (unadjusted HR 0.99, 95% CI 0.77–1.26). The comparison between patients included in the secondary analysis versus patients excluded in the secondary analysis did not demonstrate meaningful differences between the two groups, indicating that the patients in the secondary analysis were a representative reflection of the full study cohort (supplementary table S4).
FIGURE 1.
Flowchart of patient selection.
TABLE 1.
Clinical characteristics of patients included in the cohort that was used for the presented secondary analyses
| Imatinib group (n=169) | Placebo group (n=163) | |
| Demographics | ||
| Age, years | 65 (57–73) | 64 (55–74) |
| Male | 127 (75.1) | 107 (65.6) |
| BMI, kg·m−2 | 27.3 (25.2–31.1) | 29.8 (25.6–33.0) |
| Comorbidities# | ||
| Current or ex-smoker | 63 (38.7) | 67 (43.5) |
| BMI >30 kg·m−2 | 42 (28.2) | 71 (49.0) |
| Diabetes | 36 (21.3) | 52 (31.9) |
| Cardiovascular disease¶ | 32 (18.9) | 45 (27.6) |
| Hypertension | 56 (33.1) | 66 (40.5) |
| COPD or asthma | 30 (17.8) | 32 (19.6) |
| Venous thromboembolism | 3 (1.8) | 2 (1.2) |
| Renal failure | 5 (3.0) | 7 (4.3) |
| Hepatic disease | 1 (0.6) | 1 (0.6) |
| Rheumatic disease | 8 (4.7) | 15 (9.2) |
| Heart failure | 8 (4.7) | 3 (1.8) |
| Medical treatments+ | ||
| Glucose-lowering drugs | 35 (20.7) | 48 (29.4) |
| Antihypertensive treatment | 78 (46.2) | 92 (56.4) |
| ACE or ARB | 41 (24.3) | 63 (38.7) |
| Statins | 51 (30.2) | 57 (35.0) |
| Platelet inhibitors | 35 (20.7) | 37 (22.7) |
| Oral anticoagulants | 15 (8.9) | 18 (11.0) |
| Laboratory values on admission | ||
| Haemoglobin, mmol L−1 | 8.4 (7.8–9.1) | 8.6 (7.9–9.1) |
| Leukocytes, ×109 L−1 | 7.7 (5.6–10.5) | 7.8 (5.9–10.0) |
| Neutrophils, ×109 L−1 | 6.0 (4.2–8.6) | 5.9 (4.4–8.3) |
| Lymphocytes, ×109 L−1 | 0.86 (0.60–1.10) | 0.91 (0.62–1.28) |
| Thrombocytes, ×109 L−1 | 244 (185–321) | 235 (190–311) |
| Urea, mmol·L−1 | 6.3 (4.5–8.5) | 6.7 (5.0–8.9) |
| Creatinine, µmol·L−1 | 76 (65–88) | 78 (66–94) |
| C-reactive protein, mg·L−1 | 104 (48–158) | 92 (46–150) |
| Medication initiated on admission | ||
| Low-molecular-weight heparin | 143 (84.6) | 128 (78.5) |
| Oral anticoagulants | 11 (6.5) | 16 (9.8) |
| Antibiotics | 68 (40.2) | 63 (38.7) |
| Dexamethasone | 125 (74.0) | 117 (71.8) |
| Remdesivir | 32 (18.9) | 34 (20.9) |
| (Hydroxy)chloroquine | 13 (7.7) | 13 (8.0) |
| Disease severity on admission | ||
| qSOFA score | 0 (0–1) | 0 (0–1) |
Data are presented as median (interquartile range) or n (%). BMI: body mass index; ACE: angiotensin-converting enzyme; ARB: angiotensin receptor blocker; qSOFA: Quick Sequential Organ Failure Assessment. #: comorbidities as reported at admission or present in the patient's medical record; ¶: cardiovascular diseases included arrhythmias (predominantly atrial fibrillation), valvular disease, coronary artery disease and conduction disorders; +: medical treatment (or home medication) as reported at admission or present in the patient's medical record. No p-values are shown for baseline data, since data were obtained from a randomised controlled trial.
Effect imatinib on biomarkers
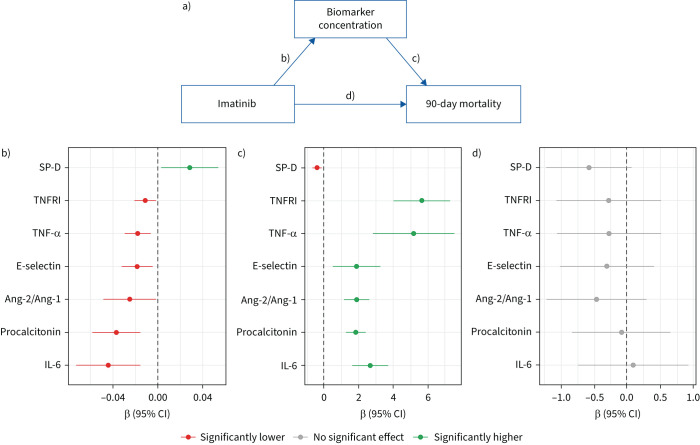
In the linear mixed effect models, imatinib was found to increase the log10-transformed concentration of the epithelial injury marker SP-D by 0.028 (95% CI 0.003–0.054) per day (figure 2b and supplementary figure S1). Imatinib caused a decrease in the log10-transformed concentration of pro-inflammatory markers: IL-6 by 0.044 (95% CI 0.015–0.073) per day, procalcitonin by 0.037 (95% CI 0.016–0.058) per day, tumour necrosis factor (TNF)-α by 0.018 (95% CI 0.006–0.030) per day and TNF receptor I (TNFRI) by 0.011 (95% CI 0.001–0.021) per day. Imatinib also resulted in a decrease of endothelial markers: angiopoietin (Ang)-2/Ang-1 ratio by 0.025 (95% CI 0.001–0.049) per day and E-selectin by 0.018 (95% CI 0.005–0.032) per day. The concentrations of other biomarkers were not affected by imatinib treatment (supplementary figure S2a). In a sensitivity analysis, corrected for age, BMI, cardiovascular disease and diabetes, the estimated effects remained the same (supplementary figure S2b), confirming that the results were not caused by baseline differences in these variables.
FIGURE 2.
a) Visualisation of mediation analysis. b) Effect of imatinib on the biomarker concentration over time, when compared with placebo. c) Effect of an increased biomarker concentration over time on 90-day mortality. d) Effect of imatinib on 90-day mortality when the effect of the biomarkers is left out. The effect of imatinib on 90-day mortality is completely mediated by changes in surfactant protein D (SP-D), tumour necrosis factor (TNF) receptor I (TNFRI), TNF-α, E-selectin, angiopoietin (Ang)-2/Ang-1 ratio, procalcitonin and interleukin (IL)-6.
Mediation analysis
Mediation analysis was performed by estimating direct and indirect effects of imatinib on mortality (figure 2a). For the aforementioned significant biomarkers, the association between the change in biomarker concentration and 90-day mortality was estimated. Higher concentrations of TNFRI, TNF-α, E-selectin, Ang-2/Ang-1, procalcitonin and IL-6 and lower concentrations of SP-D were associated with a higher mortality (figure 2c). Incorporation of these models in mediation analysis showed that imatinib was not directly related to 90-day mortality when accounting for the indirect effect via IL-6, procalcitonin, Ang-2/Ang-1, E-selectin, TNF-α, TNFRI and SP-D (figure 2d). In a sensitivity analysis using natural effects mediation, complete mediation via IL-6 and TNFRI was confirmed (supplementary figure S3). The aforementioned findings were replicated by a sensitivity analysis modelling 28-day mortality (supplementary figure S4).
Moderation analysis
Baseline characteristics of patients included in the moderation analysis (i.e. patients with baseline biomarker data) were comparable to those of patients in the mediation analysis (supplementary table S5). The baseline biomarker concentrations in this cohort were comparable between the randomisation groups (supplementary table S6). After correction for multiple testing, moderation analysis showed no significant interaction between baseline concentration of any single biomarker and treatment with imatinib on 90-day mortality. In a secondary analysis where patients were categorised by having a high or low baseline biomarker level, no moderation was found either (supplementary figure S5).
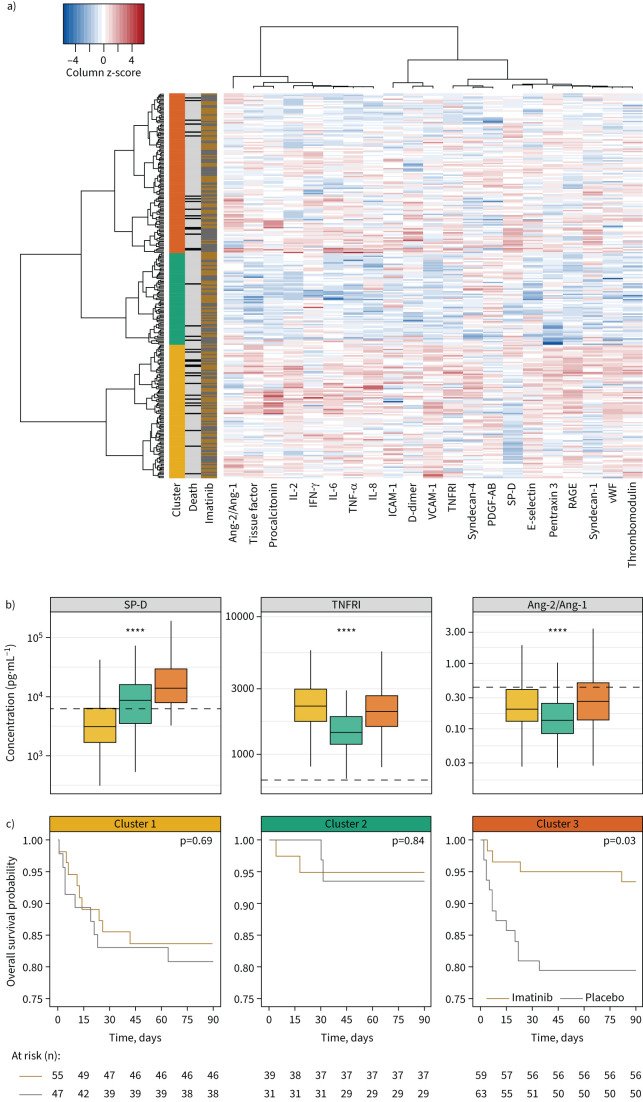
As none of the biomarkers could capture the biological complexity observed in the included patients, hierarchical clustering was used to group patients into biologically similar groups based on baseline plasma biomarker levels. Majority rules showed that three clusters (10 out of 22 classifications) best explained the variation based on 22 plasma biomarkers (figure 3). 90-day mortality was highest in clusters 1 and 3 (17.6% and 13.9%, respectively), and lowest in cluster 2 (5.7%) (supplementary table S7). Plasma concentrations of all biomarkers were generally highest in cluster 1 and lowest in cluster 2, with plasma concentrations of patients in cluster 3 in between (supplementary figure S6). Patients in cluster 3 were distinct in their higher concentration of SP-D in plasma, indicative of alveolar epithelial injury. Longitudinal plasma concentrations of patients within each cluster are visualised in supplementary figures S7–S9. Imatinib resulted in a 90-day mortality reduction only in patients assigned to cluster 3 (HR 0.30, 95% CI 0.10–0.92) (figure 3). There was no mortality difference in patients in cluster 1 or cluster 2. These results remained the same in a sensitivity analysis, correcting for baseline imbalances (supplementary table S8).
FIGURE 3.
a) Heatmap of subphenotypes, based on baseline biological profile. Rows represent patients, columns represent biomarkers. First column: three clusters; yellow is cluster 1, green is cluster 2, orange is cluster 3. Second column: patients that died within 90 days are indicated with black, surviving patients with grey. Third column: patients who received imatinib therapy are indicated with gold, placebo patients with grey. Heatmap: a higher concentration in comparison to the other included patients is indicated with red, while a lower concentration is indicated by blue. b) Baseline plasma concentrations of three biomarkers reflective of cluster analysis, stratified according to subphenotype. Data are presented as box and whisker plots (median, interquartile range and minimum–maximum within 1.5 times the interquartile range). Dotted lines indicate median values obtained in healthy controls. c) Kaplan–Meier curves and risk tables for imatinib and placebo stratified per biological subphenotype identified by cluster analysis shown in (a). A Cox proportional hazards model was used to provide p-values. Ang: angiopoietin; IL: interleukin; IFN: interferon; TNF: tumour necrosis factor; ICAM: intracellular adhesion molecule; VCAM: vascular cell adhesion molecule; TNFRI: TNF receptor I; PDGF: platelet-derived growth factor; SP-D: surfactant protein D; RAGE: receptor for advanced glycation end-products; vWF: von Willebrand factor. ****: p<0.0001, ANOVA.
Discussion
In this study, we aimed to describe the biological changes elicited by imatinib, and the relationship between these changes and clinical outcomes in hospitalised COVID-19 patients. Our results suggest that the benefit of imatinib is mediated through modulation of innate immune responses and reversal of endothelial dysfunction. None of the individual baseline biomarker concentrations showed evidence for predictive enrichment of patients benefitting from imatinib. Classification of patients into three subphenotypes suggested that a subgroup of patients with profound alveolar injury combined with systemic inflammation and endothelial dysfunction were selectively profiting from imatinib treatment. This information could aid in providing insight into the mechanism of action of COVID-19-related therapies, and the relationship between biomarkers and a clinical intervention in general.
This is the first human study to assess the effect of imatinib on host response and, as far as we know, the first study that links data from a randomised controlled trial to detailed biological profiles. In vitro and in vivo studies showed that imatinib reinforces the endothelial barrier and mitigates alveolar inflammatory responses through NF-κB-mediated chemotaxis, resulting in lower IL-6 concentrations [10, 26, 27]. Anti-inflammatory effects and endothelial barrier protection were therefore a priori likely to mediate imatinib effectiveness in COVID-19. Although previous studies demonstrated that COVID-19 is not specifically associated with a strong cytokine release syndrome [28], therapy strategies targeting the release of cytokines (e.g. steroids [2], IL-6 inhibitors [3], IL-1 receptor antagonists [29], Janus kinase 1/2 inhibitors [30] and granulocyte–macrophage colony-stimulating factor inhibitors [31]) have also been shown to be effective in reducing COVID-19-related mortality. The mortality-mediating mechanisms of these therapeutics remain uncertain, since only clinical outcomes and no biological data were collected in those studies.
The mediation analysis presented here strongly suggested that a reduction in IL-6 concentration completely mediated the mortality reduction of imatinib. In other words, mortality was only reduced when the plasma concentration of IL-6 decreased after imatinib treatment. Yet, baseline plasma IL-6 concentration did not moderate the effect of imatinib on outcomes, so we dispute the hypothesis that patients with a more pro-inflammatory starting position have more benefit. So why would patients with severe COVID-19 only benefit when systemic anti-inflammatory effects are seen, given that there is little evidence for innate immune responses compatible with cytokine release syndrome? A possible explanation is that imatinib primarily restores the endothelial barrier function, compatible with the observed changes in Ang-2/Ang-1 seen in this study, which might have resulted in cytokine leakage from the alveolar compartment to the systemic compartment [32]. It has indeed been suggested that COVID-19 is characterised by an alveolar cytokine storm instead of a systemic cytokine storm [33, 34].
Given that biological complexity is insufficiently captured by single biomarkers, we used an established clustering method to identify three biological subphenotypes in hospitalised COVID-19 patients [25]. Separation into two biological subphenotypes has been described in ARDS [35] and in COVID-19 [36, 37]. Most of these studies relied on clinical data [38] or a combination of clinical data and biomarkers [35], while we used biological data alone to identify subphenotypes. Furthermore, we focused on patients admitted to the ward, while previous studies were restricted to a critically ill population admitted to the intensive care unit. The inclusion of a comprehensive set of biomarkers provided separation by plasma concentration of SP-D within the subset of patients with an inflammatory profile and endothelial dysfunction (subphenotype 1 versus 3). SP-D is a biomarker of alveolar injury and is increased in patients developing ARDS [39], and an increase in plasma concentration is indicative of alveolar permeability [40]. Imatinib only decreased mortality in the subgroup with this biological profile, suggesting that a certain amount of alveolar permeability in the context of systemic inflammation and endothelial dysfunction needs to be present in order for imatinib to have a protective effect. When validated independently, patients with more alveolar injury in the setting of an inflammatory state could therefore preferentially be selected for imatinib treatment. It remains to be explained how a further increase in SP-D mediated the protective effect imatinib on mortality, as this is counterintuitive in light of the moderation analysis.
Our study has important strengths and some limitations. The use of randomised group allocation eliminates most forms of bias and therefore provides the best possible estimate of a causal treatment effect. Although mortality was a secondary end-point and the protective effect of imatinib attenuated after correction for baseline differences, the long-term analysis at day 90 demonstrated a persistent survival benefit of imatinib, even after adjusting for baseline imbalances. Furthermore, in our study, pre-treatment biomarker concentrations did not show any differences between the groups, confirming a comparable baseline biological profile and limiting the explanation that baseline differences were responsible for the observed mediating effects. Because the data were collected systematically with the performed analyses in mind, we obtained biomarker data of a large share of the study population. All patients in the biomarker cohort were alive at the time of the second measurement, excluding immortal time bias as an explanation for our findings. Patients without biomarker data had similar baseline characteristics, but had fewer days of oxygen therapy. We assume that no bias occurred in the selection of patients for whom biomarker data were available. We selected 25 biomarkers representative of host response pathways implicated in COVID-19 and the mechanism of action of imatinib; nonetheless, we could have missed an important mediator. The absence of commonly measured biomarkers (e.g. ferritin or CRP) in our biomarker panel limits the ability to compare our dataset with other studies. Second, only the systemic host response was evaluated and the alveolar environment was not sampled nor studied because obtaining alveolar samples in non-intubated patients is unfeasible. Last but not least, the study is likely underpowered to detect heterogeneity of treatment effects via moderation analysis and a larger sample might have yielded different results [41]. When examining the moderating effects of the three subphenotypes by an interaction term instead of a stratified analysis, the hazard ratios were comparable but confidence intervals were wide, resulting in p-values >0.05, as expected. Therefore, future prospective testing is required to validate our results.
The findings of this study extend our biological understanding of how mortality can be reduced in patients with severe COVID-19. Changes in innate immune responses and endothelial barrier protection appear to mediate the reduction in mortality observed in the imatinib group. We speculate that this may translate to other immunomodulatory treatments as well. Furthermore, we illustrate that we should not assume that patients who have a high concentration of a single biomarker that is considered to be reflective of activation of the pathway that is targeted by the drug results in predictive enrichment within the context of severe COVID-19 pneumonia. Rather, identification of subphenotypes by comprehensive analysis of multiple pathways provided three clusters that responded differently to the tested intervention. This is in line with studies in ARDS [35, 42, 43] and shows that biological profiles should be used for predictive enrichment rather than single biomarker values. The subphenotype that responded favourably to imatinib treatment had a similar severity of illness compared with a subphenotype that did not have survival benefit from imatinib. This is in contrast to previous studies and might suggest that this subphenotype could be used for predictive enrichment rather than prognostic enrichment.
To conclude, we show here that imatinib works as an effective therapy against severe COVID-19 only when circulating biomarkers confirm decreased systemic innate immune response and improved endothelial barrier function after treatment. Changes in these biomarker concentrations may be used as a surrogate end-point when validated as mediators for therapy-related survival in other randomised controlled trials. Three biological subphenotypes were identified and only patients classified as having alveolar injury by increased levels of SP-D in the context of systemic inflammatory response and endothelial dysfunction benefitted from imatinib treatment.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00780-2022.Supplement (4.5MB, pdf)
Shareable PDF
Acknowledgements
The authors would like to thank all the patients and their families who participated in this study. We also thank Barbara Smids-Dierdorp and Tamara Dekker (Amsterdam UMC, location University of Amsterdam, Department of Experimental Immunology (EXIM), Amsterdam, The Netherlands) for their expert technical assistance in the performance of the Luminex multiplex assays.
Footnotes
This article has an editorial commentary: https://doi.org/10.1183/13993003.01808-2022
The CounterCOVID Study Group: Jurjan Aman (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Sara Azhang (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; and Department of Pulmonology, Haaglanden Medisch Centrum, The Hague, The Netherlands), Imke H. Bartelink (Department of Pharmacy, Amsterdam UMC, VUmc, Amsterdam, The Netherlands), Ahmed A. Bayoumy (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; and Department of Pulmonology, Chest Unit, Suez Canal University, Suez, Egypt), Pierre M. Bet (Department of Pharmacy, Amsterdam UMC, VUmc, Amsterdam, The Netherlands), Wim Boersma (Department of Pulmonology, Noordwest Ziekenhuisgroep, Alkmaar, The Netherlands), Harm J. Bogaard (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Peter I. Bonta (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Karin A.T. Boomars (Department of Pulmonology, Erasmus Medisch Centrum, Rotterdam, The Netherlands), Lieuwe D.J. Bos (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands; and Department of Intensive Care, Amsterdam UMC, AMC, Amsterdam, The Netherlands), Liza Botros (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Job J.M.H. van Bragt (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Gert-Jan Braunstahl (Department of Pulmonology, Sint Franciscus Ziekenhuis, Rotterdam, The Netherlands), Lucas R. Celant (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Erik Duijvelaar (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Katrien A.B. Eger (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), J.J. Miranda Geelhoed (Department of Pulmonology Leiden University Medical Center, Leiden, The Netherlands), Yurika L. Evan Glabbeek (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Hans P. Grotjohan (Department of Pulmonology, Isala Ziekenhuizen, Zwolle, The Netherlands), Laura A. Hagens (Department of Intensive Care, Amsterdam UMC, AMC, Amsterdam, The Netherlands), Chris M. Happe (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Boaz D. Hazes (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Leo M.A. Heunks (Department of Intensive Care, Amsterdam UMC, VUmc, Amsterdam, The Netherlands), Michel van den Heuvel (Department of Pulmonology, Radboud UMC, Nijmegen, The Netherlands), Wouter Hoefsloot (Department of Pulmonology, Radboud UMC, Nijmegen, The Netherlands), Rianne J.A. Hoek (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Romke Hoekstra (Department of Pulmonology, Antonius Ziekenhuis, Sneek, The Netherlands), Herman M.A. Hofstee (Department of Internal Medicine, Haaglanden Medisch Centrum, The Hague, The Netherlands), Nicole P. Juffermans (Department of Intensive Care, Amsterdam UMC, AMC, Amsterdam, The Netherlands; and Department of Intensive Care, Onze Lieve Vrouwe Gasthuis, Amsterdam, The Netherlands), E. Marleen Kemper (Hospital Pharmacy, Amsterdam UMC, AMC, Amsterdam, The Netherlands), Azar Kianzad (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Renate Kos (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Peter W.A. Kunst (Department of Pulmonology, Onze Lieve Vrouwe Gasthuis, Amsterdam, The Netherlands), Ariana Lammers (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Ivo van der Lee (Department of Pulmonology, Spaarne Gasthuis, Haarlem, The Netherlands), E. Laurien van der Lee (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Anke-Hilse Maitland-van der Zee (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Frances S. de Man (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Pearl F.M. Mau Asam (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Adinda Mieras (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Mirte Muller (Department of Pulmonology, Catharina Ziekenhuis, Eindhoven, The Netherlands), Elisabeth C.W. Neefjes (Department of Pulmonology, Catharina Ziekenhuis, Eindhoven, The Netherlands), Anton Vonk Noordegraaf (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Esther J. Nossent (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Laurien M.A. Oswald (Department of Pulmonology, Sint Franciscus Ziekenhuis, Rotterdam, The Netherlands), Maria J. Overbeek (Department of Pulmonology, Haaglanden Medisch Centrum, The Hague, The Netherlands), Carolina C. Pamplona (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Nienke Paternotte (Department of Pulmonology, Noordwest Ziekenhuisgroep, Alkmaar, The Netherlands), Niels Pronk (Department of Pulmonology, Gelre Ziekenhuis, Apeldoorn, The Netherlands), Michiel A. de Raaf (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Bas F.M. van Raaij (Department of Pulmonology Leiden University Medical Center, Leiden, The Netherlands), Merlijn Reijrink (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Job R. Schippers (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Marcus J. Schultz (Department of Intensive Care, Amsterdam UMC, AMC, Amsterdam, The Netherlands), Ary Serpa Neto (Department of Critical Care Medicine and Institute of Education and Research, Hospital Israelita Albert Einstein, São Paulo, Brazil), Elise M.A. Slob (Department of Respiratory Medicine, Amsterdam UMC, AMC, University of Amsterdam, Amsterdam, The Netherlands), Patrick J. Smeele (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Frank W.J.M. Smeenk (Department of Pulmonology, Catharina Ziekenhuis, Eindhoven, The Netherlands), Marry R. Smit (Department of Intensive Care, Amsterdam UMC, AMC, Amsterdam, The Netherlands), A. Josien Smits (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Janneke E. Stalenhoef (Department of Internal Medicine, Onze Lieve Vrouwe Gasthuis, Amsterdam, The Netherlands), Pieter R. Tuinman (Department of Intensive Care, Amsterdam UMC, VUmc, Amsterdam, The Netherlands), Arthur L.E.M. Vanhove (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Jeroen N. Wessels (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Jessie C.C. van Wezenbeek (Department of Pulmonary Medicine, Amsterdam Cardiovascular Sciences, Amsterdam UMC, VUmc, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands).
Data sharing: De-identified participant data with a data dictionary can be shared after approval of a proposal with a signed data access agreement and always in collaboration with the study group.
Author contributions: The authors designed the study together and were involved in collecting the data with the help of the study collaborators. J. de Brabander and L.D.J. Bos had access to the raw data, did the analyses and drafted the manuscript. The other authors revised the initial draft. All authors approved the final version of the manuscript.
Conflict of interest: L.D.J. Bos reports grants from the Dutch Lung Foundation, the Dutch Lung Foundation and Health Holland (Public–Private Partnership grant), the Dutch Lung Foundation (Dirkje Postma Award), the IMI COVID-19 initiative, and Amsterdam UMC fellowship; and consulting fees from Scailyte and Sobi; outside the submitted work. J. Aman is inventor on a patent (WO2012150857A1; 2011) covering protection against endothelial barrier dysfunction through inhibition of the tyrosine kinase Abl-related gene (ARG); and reports serving as a non-compensated scientific advisor for Exvastat. All other authors declare no conflicting interests.
Support statement: This project was funded by an unrestricted grand from the Amsterdam Medical Center Foundation and a bottom-up grant from Nederlandse Organisatie voor Wetenschappelijk Onderzoek/ZonMW (10430 01 201 0007). In addition, this project received funding from the Innovative Medicines Initiative 2 Joint Undertaking (grant agreement 101005142). The funders of this study had no role in study design, data collection, data analysis, data interpretation or writing of the report. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 2020; 324: 782–793. doi: 10.1001/jama.2020.12839 [DOI] [PubMed] [Google Scholar]
- 2.The RECOVERY Collaborative Group . Dexamethasone in hospitalized patients with Covid-19. N Engl J Med 2021; 384: 693–704. doi: 10.1056/NEJMoa2021436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The REMAP-CAP Investigators . Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N Engl J Med 2021; 384: 1491–1502. doi: 10.1056/NEJMoa2100433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The ATTACC, ACTIV-4a and REMAP-CAP Investigators . Therapeutic anticoagulation with heparin in noncritically ill patients with Covid-19. N Engl J Med 2021; 385: 790–802. doi: 10.1056/NEJMoa2105911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen NT, Chinn J, Nahmias J, et al. Outcomes and mortality among adults hospitalized with COVID-19 at US medical centers. JAMA Netw Open 2021; 4: e210417. doi: 10.1001/jamanetworkopen.2021.0417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruin S, Bos LD, van Roon MA, et al. Clinical features and prognostic factors in Covid-19: a prospective cohort study. EBioMedicine 2021; 67: 103378. doi: 10.1016/j.ebiom.2021.103378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leisman DE, Mehta A, Thompson BT, et al. Alveolar, endothelial, and organ injury marker dynamics in severe COVID-19. Am J Respir Crit Care Med 2022; 205: 507–519. doi: 10.1164/rccm.202106-1514OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aman J, van Bezu J, Damanafshan A, et al. Effective treatment of edema and endothelial barrier dysfunction with imatinib. Circulation 2012; 126: 2728–2738. doi: 10.1161/CIRCULATIONAHA.112.134304 [DOI] [PubMed] [Google Scholar]
- 9.Chislock EM, Pendergast AM. Abl family kinases regulate endothelial barrier function in vitro and in mice. PLoS One 2013; 8: e85231. doi: 10.1371/journal.pone.0085231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzo AN, Sammani S, Esquinca AE, et al. Imatinib attenuates inflammation and vascular leak in a clinically relevant two-hit model of acute lung injury. Am J Physiol Lung Cell Mol Physiol 2015; 309: L1294–L1304. doi: 10.1152/ajplung.00031.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aman J, Duijvelaar E, Botros L, et al. Imatinib in patients with severe COVID-19: a randomised, double-blind, placebo-controlled, clinical trial. Lancet Respir Med 2021; 9: 957–968. doi: 10.1016/S2213-2600(21)00237-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duijvelaar E, Schippers JR, Smeele PJ, et al. Long-term clinical outcomes of COVID-19 patients treated with imatinib. Lancet Respir Med 2022; 10: e34–e35. doi: 10.1016/S2213-2600(22)00052-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Veerdonk FL, Giamarellos-Bourboulis E, Pickkers P, et al. A guide to immunotherapy for COVID-19. Nat Med 2022; 28: 39–50. doi: 10.1038/s41591-021-01643-9 [DOI] [PubMed] [Google Scholar]
- 14.Ingraham NE, Lotfi-Emran S, Thielen BK, et al. Immunomodulation in COVID-19. Lancet Respir Med 2020; 8: 544–546. doi: 10.1016/S2213-2600(20)30226-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelmenson DA, Cron RQ. Who, what, and when-effective therapy for severe COVID-19. Lancet Rheumatol 2022; 4: e2–e3. doi: 10.1016/S2665-9913(21)00353-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Placais L, Richier Q, Noel N, et al. Immune interventions in COVID-19: a matter of time? Mucosal Immunol 2022; 15: 198–210. doi: 10.1038/s41385-021-00464-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. J Stat Softw 2015; 67: 1–48. doi: 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 18.Zheng C, Liu L. Quantifying direct and indirect effect for longitudinal mediator and survival outcome using joint modeling approach. Biometrics 2022; 78: 1233–1243. doi: 10.1111/biom.13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinheiro J. nlme: linear and nonlinear mixed effects models. R package version 3.1–157. 2022. https://cran.r-project.org/web/packages/nlme Date last accessed: 4 April 2022.
- 20.Therneau TM. A package for survival analysis in R. R package version 3.3–1. 2022. https://cran.r-project.org/web/packages/survival Date last accessed: 4 April 2022.
- 21.Rizopoulos D. JM: an R package for the joint modelling of longitudinal and time-to-event data. J Stat Softw 2010; 35: 1–81. doi: 10.18637/jss.v035.i0921603108 [DOI] [Google Scholar]
- 22.Steen J, Loeys T, Moerkerke B, et al. medflex: an R package for flexible mediation analysis using natural effect models. J Stat Softw 2017; 76: 1–46. doi: 10.18637/jss.v076.i11 [DOI] [Google Scholar]
- 23.Kassambara A, Kosinski M, Biecek P, et al. survminer: drawing survival curves using ‘ggplot2’. 2022. https://cran.r-project.org/web/packages/survminer/index.html Date last accessed: 4 April 2022.
- 24.Charrad M, Ghazzali N, Boiteau V, et al. NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw 2014; 61: 1–36. doi: 10.18637/jss.v061.i06 [DOI] [Google Scholar]
- 25.Bos LD, Schouten LR, van Vught LA, et al. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax 2017; 72: 876–883. doi: 10.1136/thoraxjnl-2016-209719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnthaler T, Jandl K, Sill H, et al. Imatinib stimulates prostaglandin E2 and attenuates cytokine release via EP4 receptor activation. J Allergy Clin Immunol 2019; 143: 794–797. doi: 10.1016/j.jaci.2018.09.030 [DOI] [PubMed] [Google Scholar]
- 27.Letsiou E, Rizzo AN, Sammani S, et al. Differential and opposing effects of imatinib on LPS- and ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol 2015; 308: L259–L269. doi: 10.1152/ajplung.00323.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leisman DE, Ronner L, Pinotti R, et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med 2020; 8: 1233–1244. doi: 10.1016/S2213-2600(20)30404-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyriazopoulou E, Poulakou G, Milionis H, et al. Early treatment of COVID-19 with anakinra guided by soluble urokinase plasminogen receptor plasma levels: a double-blind, randomized controlled phase 3 trial. Nat Med 2021; 27: 1752–1760. doi: 10.1038/s41591-021-01499-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marconi VC, Ramanan AV, de Bono S, et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir Med 2021; 9: 1407–1418. doi: 10.1016/S2213-2600(21)00331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Temesgen Z, Burger CD, Baker J, et al. Lenzilumab in hospitalised patients with COVID-19 pneumonia (LIVE-AIR): a phase 3, randomised, placebo-controlled trial. Lancet Respir Med 2022; 10: 237–246. doi: 10.1016/S2213-2600(21)00494-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meduri GU, Kohler G, Headley S, et al. Inflammatory cytokines in the BAL of patients with ARDS. Persistent elevation over time predicts poor outcome. Chest 1995; 108: 1303–1314. doi: 10.1378/chest.108.5.1303 [DOI] [PubMed] [Google Scholar]
- 33.Saris A, Reijnders TDY, Nossent EJ, et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax 2021; 76: 1010–1019. doi: 10.1136/thoraxjnl-2020-216256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nossent EJ, Schuurman AR, Reijnders TDY, et al. Pulmonary procoagulant and innate immune responses in critically ill COVID-19 patients. Front Immunol 2021; 12: 664209. doi: 10.3389/fimmu.2021.664209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014; 2: 611–620. doi: 10.1016/S2213-2600(14)70097-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha P, Furfaro D, Cummings MJ, et al. Latent class analysis reveals COVID-19-related acute respiratory distress syndrome subgroups with differential responses to corticosteroids. Am J Respir Crit Care Med 2021; 204: 1274–1285. doi: 10.1164/rccm.202105-1302OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranjeva S, Pinciroli R, Hodell E, et al. Identifying clinical and biochemical phenotypes in acute respiratory distress syndrome secondary to coronavirus disease-2019. EClinicalMedicine 2021; 34: 100829. doi: 10.1016/j.eclinm.2021.100829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bos LDJ, Sjoding M, Sinha P, et al. Longitudinal respiratory subphenotypes in patients with COVID-19-related acute respiratory distress syndrome: results from three observational cohorts. Lancet Respir Med 2021; 9: 1377–1386. doi: 10.1016/S2213-2600(21)00365-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Determann RM, Royakkers AA, Haitsma JJ, et al. Plasma levels of surfactant protein D and KL-6 for evaluation of lung injury in critically ill mechanically ventilated patients. BMC Pulm Med 2010; 10: 6. doi: 10.1186/1471-2466-10-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisner MD, Parsons P, Matthay MA, et al. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax 2003; 58: 983–988. doi: 10.1136/thorax.58.11.983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent DM, Steyerberg E, van Klaveren D. Personalized evidence based medicine: predictive approaches to heterogeneous treatment effects. BMJ 2018; 363: k4245. doi: 10.1136/bmj.k4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Famous KR, Delucchi K, Ware LB, et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med 2017; 195: 331–338. doi: 10.1164/rccm.201603-0645OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calfee CS, Delucchi KL, Sinha P, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med 2018; 6: 691–698. doi: 10.1016/S2213-2600(18)30177-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00780-2022.Supplement (4.5MB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00780-2022.Shareable (485.3KB, pdf)