Abstract
Background.
Noninvasive biomarkers distinguishing early immune activation before acute rejection (AR) could more objectively inform immunosuppression management in liver transplant recipients (LTRs). We previously reported a genomic profile distinguishing LTR with AR versus stable graft function. This current study includes key phenotypes with other causes of graft dysfunction and uses a novel random forest approach to augment the specificity of predicting and diagnosing AR.
Methods.
Gene expression results in LTRs with AR versus non-AR (combination of other causes of graft dysfunction and normal function) were analyzed from single and multicenter cohorts. A 70:30 approach (61 ARs; 162 non-ARs) was used for training and testing sets. Microarray data were normalized using a LT-specific vector.
Results.
Random forest modeling on the training set generated a 59-probe classifier distinguishing AR versus non-AR (area under the curve 0.83; accuracy 0.78, sensitivity 0.70, specificity 0.81, positive predictive value 0.54, negative predictive value [NPV] 0.89; F-score 0.61). Using a locked threshold, the classifier performed well on the testing set (accuracy 0.72, sensitivity 0.67, specificity 0.73, positive predictive value 0.48, NPV 0.86; F-score 0.56). Probability scores increased in samples preceding AR versus non-AR, when liver function tests were normal, and decreased following AR treatment (P < 0.001). Ingenuity pathway analysis of the genes revealed a high percentage related to immune responses and liver injury.
Conclusions.
We have developed a blood-based biologically relevant biomarker that can be detected before AR-associated graft injury distinct from LTR never developing AR. Given its high NPV (“rule out AR”), the biomarker has the potential to inform precision-guided immunosuppression minimization in LTRs.
INTRODUCTION
Before the 1980s, outcomes of liver transplant recipients (LTRs) were discouraging primarily because of nonspecific immunosuppression therapy (IST) and high rates of acute rejection (AR) graft injury and related complications. This changed with the institution of calcineurin inhibitor (CNI) therapies that lowered AR rates resulting in significant improvements in graft and patient survival.1-4 Recent data have however continued to show the impact of AR on survival, stimulating a resurgence of interest in rejection types, management, and outcomes.5-7 Thus, this ongoing concern may influence clinicians to maintain a higher level of IST to mitigate this risk. Conversely, IST poses significant risk even at standard doses, compounded by a rising demographic of older recipients with multiple comorbidities. Malignancy, cardiovascular events, infections, and chronic kidney disease, all exacerbated by IST, are the leading causes of death in this patient population.8-10 Importantly, CNIs specifically contribute to the development of chronic kidney disease, observed in a high percentage of LTRs.11
Therefore, LTR outcomes could be enhanced if IST exposure in general, and CNI in particular, could be safely minimized early post-LT when the potential benefit is greatest. In essence, patients should be given “just enough” IST to prevent AR and not “too much” to increase complications (ie, optimized IST). Unfortunately, given narrow therapeutic windows, achieving this tight control of IST has become more art than science in clinical practice. Routine serial monitoring with IST drug levels and liver function tests (LFTs) constitute nonspecific, insensitive tools to assess or predict immune status, specifically the onset of early immune activation preceding AR versus sustained immune quiescence.12-15 Consequently, transplant clinicians practice “trial-and-error” IST adjustments, informed by their own center-specific protocols or reactive to complications from either under- or over-immunosuppression.
Thus, there is a clear need to improve our ability to achieve the delicate balance of safety and efficacy through informed, proactive IST optimization in LTRs. Recent developments in molecular biomarkers in transplantation have brought us closer to the long-standing objective to better assess and predict alloimmune reactivity before the onset of AR graft injury. Several genomic biomarkers have been reported to distinguish various causes of graft dysfunction in LTRs but until recently have not been subjected to validation or analyzed before events.16-29
With this in mind, we have reported a 36-gene biomarker of AR distinct from healthy graft function (Transplant eXcellence [TX]) in LTRs.30 This gene model performed well in serially predicting AR versus TX but did not include patient phenotypes with other causes of liver injury (acute dysfunction no rejection [ADNR]) in the discovery phase. Extending the profile to include all patients not experiencing rejection (non-AR: TX + ADNR) would provide further certainty in confirming immune quiescence versus activation (pre-AR) during IST tapering. To this end, we used a novel modeling approach early in the discovery phase to further enhance the clinical and biological specificity of the gene expression profile for AR. In essence, we have developed a model that is inclusive of the at-risk LT population as a whole, broadening its utility in tailoring IST.
MATERIALS AND METHODS
Cohorts and Subjects
LT patients and samples included in this study were similar to the single-center discovery (Northwestern University [NU] biorepository, 2010–2015) and multicenter validation (Clinical Trials in Organ Transplant [CTOT-14, NCT01672164], 2012–2014) cohorts reported in our previous study.30 Written informed consent was obtained from each patient included in the study and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the human research committee of the institution. The cohorts were merged and randomly split 70:30 into training and testing sets given their differences—single-center, later post-LT (NU) versus multicenter, earlier post-LT (CTOT-14). Inclusion and exclusion criteria were identical and consisted of adult (≥18 y old) recipients of the first LT from either a deceased or living donor. Prior or multiorgan transplants, HIV-infected recipients, and viremic hepatitis B and C virus patients were excluded. Blood samples were collected at the time of “for cause” graft biopsies and, in CTOT-14 only, serially at week 2 and months 1, 2, 3, 6, 9, 12, 15, 18, 21, and 24 following LT. All biopsies were read locally for clinical care and later sent for independent, blinded central review using Banff criteria, as previously reported.30,31 Nonrejection causes, such as steatohepatitis, cholestasis, ischemia, or other, were grouped together as ADNR.
We also collected samples from LTRs with long-standing normal liver tests (Clinical Phenotypes [CPs] below) and labeled them TX (“virtual biopsy”).30 We recognize this as a limitation as the centers participating do not perform surveillance biopsies in patients with normal liver tests that could be paired with blood samples to most accurately define TX. While recent studies have demonstrated subclinical graft injury in LTRs,32,33 few LT centers actually perform such surveillance biopsies as standard practice. Thus, despite the limitation, we had to use a clinical definition that would parallel a healthy “normal” recipient generalizable and relevant to current clinical practice.
Clinical Phenotypes
As previous, we used clinical, biochemical, and biopsy criteria to define 4 diagnostic CPs in both cohorts: AR, ADNR, TX, and non-AR (ADNR + TX): (1) AR: “for cause biopsy” consistent with AR. (2) ADNR: “for cause biopsy” consistent with nonrejection cause. (3) TX: normal LFTs at the time of and 3 mo before and after “virtual biopsy” (total bilirubin ≤1.5 mg/dL, direct bilirubin [DB] <0.5 mg/dL, alkaline phosphatase [AP] ≤200 U/L, and alanine transaminase [ALT] ≤60 U/L in males ≤36 U/L in females). (4) Non-AR: ADNR and TX combined. For the serial prediagnosis analysis before each CP (CTOT-14 only), we used pre–non-AR samples as controls for pre-AR samples to enhance the specificity of detecting AR. Importantly, we required at least 2 of the 3 liver tests (DB, AP, and ALT) to be normal at each presample collection, and ALT >100 U/L were excluded even if AP and DB were normal. For postdiagnosis, we focused only on gene expression changes following AR therapy.
Biomarker Development
Discovery and validation phases were performed in accordance with Institute of Medicine guidelines.34 Blood samples in PAXgene tubes (BD BioSciences, San Jose, CA) were processed, as described previously.30,35,36 Raw expression (.CEL) data files from Affymetrix GeneChip HT HG-U133 + PM Array plates were used as input for normalization using a custom frozen robust multiarray analysis37 vector from a cohort of 560 CEL files previously generated from LTRs.30 To maintain the distribution of the phenotypes and pertinent metadata in the cohorts, the NU and CTOT-14 samples were merged and then split into the 70% training and 30% testing groups. This split-sample approach provides accurate estimations of phenotypic gene expression profiles while retaining a sufficiently large validation cohort to have confidence in the performance metrics.38 Thus, we generated the final discovery model from the 70% training set and then applied it to the 30% testing set reserved for validation purposes.
All probes on the GeneChip were filtered to retain the genes with median expression >6 in 50% of the samples and in the top 40th percentile of variance across all samples. Five independent classification algorithms (nearest shrunken centroid,39 partial least squares discrimination analysis,40 support vector machine,41 random forest,42 and elastic net43) were used to calculate a multivariate score for each probe in the filtered data set based on metrics (unique for each algorithm) that reflect the relative contribution of a probe toward classification of out of bag samples for a total of 1000 resamplings.44 As mentioned above, the final discovery model was generated from the training set using random forest, and a performance threshold was selected favoring negative predictive value (NPV) over positive predictive value (PPV), which was then locked in validation on the 30% testing cohort. Analyses were performed using R version 3.5.1 (RStudio, Boston, MA). Ingenuity Core Analysis (Qiagen, Inc., Hilden, Germany) was used to generate enriched pathways and comparison with the literature. A detailed methodology is provided in the Supplementary Methods (SDC, http://links.lww.com/TP/C268).
RESULTS
Patient Cohorts
Forty-six AR, 38 ADNR, and 45 TX patients in NU and 14 AR, 28 ADNR, and 50 TX patients in CTOT-14 were analyzed at the diagnostic time points, similar to our initial study but now including ADNR.30 ADNR biopsies consisted of the following: 23 biliary obstruction/cholestasis, 16 nonspecific minimal inflammation, 14 steatosis, 6 ischemia/reperfusion injury, and 7 other causes. Table 1 displays comparisons of the merged cohorts (60 ARs versus 161 non-ARs). AR subjects were more often female and had LFT differences compared with non-AR, as expected.
TABLE 1.
Patient characteristics
| AR (n = 60) | Non-AR (n = 161) | P | |
|---|---|---|---|
| Age at transplant (y) | 51.69 (30.42, 63.05) | 56.37 (46.00, 62.00) | 0.199 |
| Caucasian race (%) | 43 (71.7) | 132 (82.0) | 0.092 |
| Male sex (%) | 27 (45.0) | 101 (62.7) | 0.026 |
| Primary liver diagnosis (%) | |||
| Hepatitis C (nonviremic) | 5 (8.5) | 4 (3.1) | 0.248 |
| Alcohol | 8 (13.6) | 30 (23.4) | |
| Nonalcoholic fatty liver or cryptogenic | 10 (16.9) | 50 (39.1) | |
| Immune-mediated (PSC, AIH, PBC) | 10 (16.9) | 18 (14.1) | |
| Other | 26 (44.1) | 26 (20.3) | |
| Months from LT | 11.62 (5.44, 29.91) | 10.43 (5.70, 34.37) | 0.977 |
| Immunosuppression | |||
| CNI therapy | 47 (78.3) | 131 (81.4) | 0.752 |
| Mycophenolic acid therapy | 37 (61.7) | 91 (56.5) | 0.592 |
| Laboratory values | |||
| ALT (U/L) | 201.00 (136.00, 369.75) | 27.00 (18.00, 76.00) | <0.001 |
| Alkaline phosphatase (U/L) | 205.00 (142.25, 381.00) | 95.00 (67.00, 164.00) | <0.001 |
| Total bilirubin (mg/dL) | 1.20 (0.50, 4.40) | 0.70 (0.40, 1.10) | 0.001 |
| Creatinine (mg/dL) | 1.21 (0.57) | 1.26 (0.61) | 0.562 |
| Rejection characteristics | |||
| Mild (RAI 3–4) (%) | 26 (43%) | — | — |
| Moderate–severe (RAI 5–9) (%) | 34 (57%) | — | — |
AIH, autoimmune hepatitis; ALT, alanine aminotransferase; AR, acute rejection; CNI, calcineurin inhibitors; LT, liver transplant; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis; RAI, rejection activity index.
Discovery and Validation of AR Versus Non-AR Genomic Model (at Diagnosis)
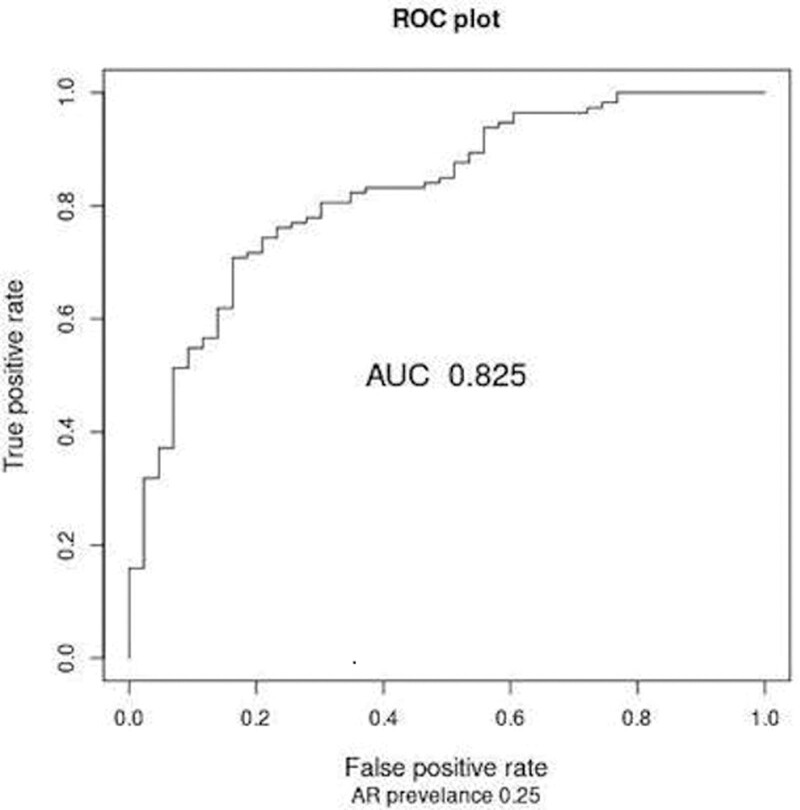
In the initial 70% training set (n = 156), random forest modeling generated a classifier on the training set distinguishing AR versus non-AR (Figure 1—area under the curve 0.825; accuracy 0.78, sensitivity 0.70, specificity 0.81, PPV 0.54, NPV 0.89; F-score 0.61). Using the same locked probability threshold, the final 59-gene probe model performed well on the 30% testing set (accuracy 0.72, sensitivity 0.67, specificity 0.73, PPV 0.48, NPV 0.86; F-score 0.56.) Given that the NU cohort was not a prevalent population being enrolled only at biopsies later after LT, we used the overall rejection prevalence averaged from the literature (25%) to report an adjusted PPV and NPV.5,45-47
FIGURE 1.
The ROCs—AR vs non-AR. The AUC is displayed as well as the performance characteristics (25% AR prevalence adjustment) at the 0.34 threshold. AR, acute rejection; AUC, area under the curve; ROC, receiver operating curve.
Serial Analysis of the AR Versus Non-AR Genomic Model (Pre- and Post-diagnosis)
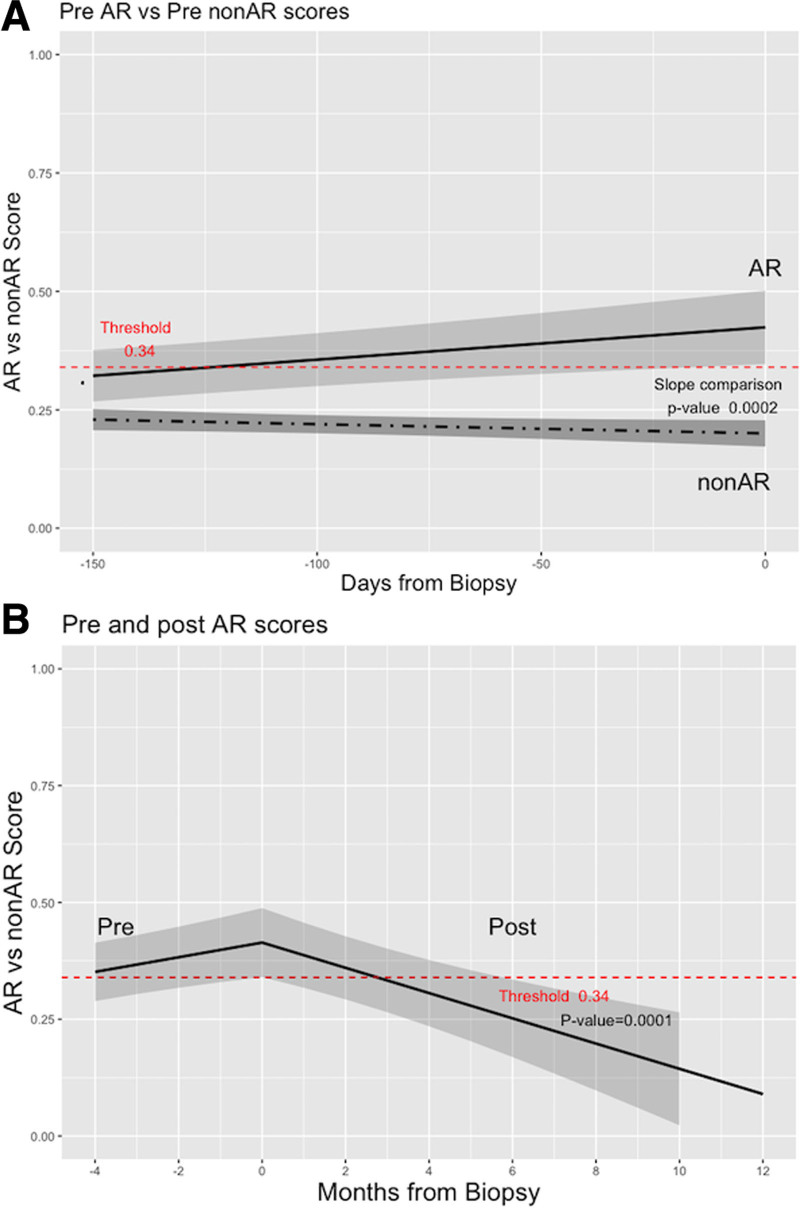
To test the ability of the 59 gene probes to detect AR versus non-AR before these diagnoses, 33 and 179 CTOT-14 serial samples were analyzed before 12 AR and 58 non-AR diagnoses, respectively. To compare changes in gene expression over time prediagnosis, we compared the slopes of each line of scores between the phenotypes. This demonstrated opposing directions of the line slopes before AR (positive—increasing) versus non-AR (negative—decreasing) (Figure 2A). The slopes were statistically different in the mixed model (P = 0.0002). We performed a similar analysis for 53 samples following treatment of 12 AR cases. All responses to treatment were defined by normalization of LFTs within 90 d. The results showed that the slope following treatment of AR was negative (decreasing scores) (P = 0.0001) (Figure 2B) even more abruptly in our AR versus TX model.30
FIGURE 2.
Serial changes in AR vs non-AR gene expression scores using line slopes. A, Pre-AR vs pre–non-AR (P = 0.0002). B, Pre-AR vs post-AR (P = 0.0001). AR, acute rejection.
Ingenuity Pathway Analysis and Differential Gene Expression Plots
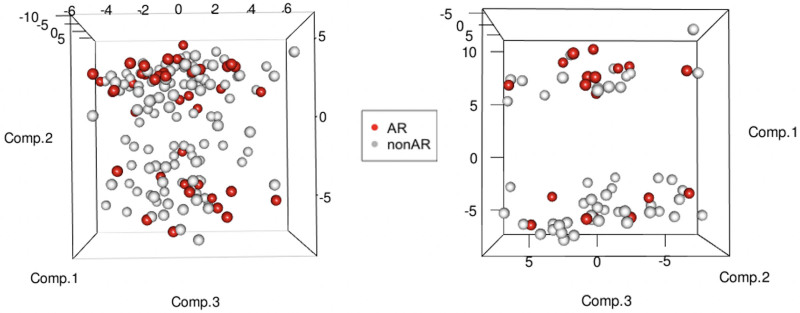
To evaluate the biological relevance, we tested the 59 probes using Ingenuity Pathway Analysis. Table 2 displays the fold changes and identifications of the 59 genes and the direction and magnitude of gene expression. Canonical pathways analysis (Supplemental Table S1, SDC, http://links.lww.com/TP/C268) displays the biological pathways that appear most significantly affected by the genes in the test set. Nearly half of the significant pathways were associated with immune responses and liver-related function including allograft rejection signaling, T-cell receptor, and liver X receptor-retinoid X receptor. Toxicity analysis (Supplemental Table S1, SDC, http://links.lww.com/TP/C268) indicated that the highest percentage of genes were previously reported to be involved in liver toxicity, including liver X receptor-retinoid X receptor activation, liver necrosis/cell death, and hepatic fibrosis. The differential gene expression between AR and non-AR based on the random forest probes is also displayed in a 3-dimensional principal component analysis plot (Figure 3) showing distinct clustering in the training and testing sets, as well as an overall hierarchical clustering plot (Supplemental Figure S1, SDC, http://links.lww.com/TP/C268).
TABLE 2.
AR vs non-AR 59-gene probe model—gene name and expression values
| Model probe | Symbol | Gene name | Log FC (AR/non-AR) | Average log exp |
|---|---|---|---|---|
| 224588_PM_at | XIST | X inactive-specific transcript | 1.606 | 6.363 |
| 227671_PM_at | XIST | X inactive-specific transcript | 1.492 | 5.951 |
| 208792_PM_s_at | CLU | Clusterin | 0.428 | 7.688 |
| 201005_PM_at | CD9 | CD9 molecule | 0.342 | 8.017 |
| 206150_PM_at | CD27 | CD27 molecule | 0.286 | 7.91 |
| 205849_PM_s_at | UQCRB | Ubiquinol-cytochrome c reductase-binding protein | 0.259 | 7.937 |
| 236422_PM_at | Unmapped | 0.251 | 7.543 | |
| 211372_PM_s_at | IL1R2 | Interleukin 1 receptor type 2 | 0.211 | 9.116 |
| 206804_PM_at | CD3G | CD3g molecule | 0.184 | 8.777 |
| 221558_PM_s_at | LEF1 | Lymphoid enhancer-binding factor-1 | 0.182 | 9.098 |
| 216950_PM_s_at | FCGR1A | Fc fragment of IgG receptor Ia | 0.176 | 9.248 |
| 200859_PM_x_at | FLNA | Filamin A | 0.109 | 7.927 |
| 209480_PM_at | HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 0.074 | 6.263 |
| 1552583_PM_s_at | ABCC13 | ATP-binding cassette subfamily C member 13 | 0.06 | 7.283 |
| 222139_PM_at | Unmapped | 0.057 | 7.813 | |
| 205033_PM_s_at | DEFA1 | Defensin alpha 1 | 0.055 | 11.816 |
| 219799_PM_s_at | DHRS9 | Dehydrogenase/reductase 9 | 0.043 | 8.197 |
| 235157_PM_at | Unmapped | 0.04 | 8.846 | |
| 205863_PM_at | S100A12 | S100 calcium-binding protein A12 | 0.03 | 11.579 |
| 205789_PM_at | CD1D | CD1d molecule | 0.011 | 8.07 |
| 203936_PM_s_at | MMP9 | Matrix metallopeptidase 9 | −0.003 | 9.743 |
| 206110_PM_at | H3C10 | H3 clustered histone 10 | −0.025 | 7.681 |
| 224558_PM_s_at | MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 | −0.044 | 10.18 |
| 202912_PM_at | ADM | Adrenomedullin | −0.054 | 8.963 |
| 242943_PM_at | ST8SIA4 | ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 4 | −0.064 | 7.794 |
| 218066_PM_at | SLC12A7 | Solute carrier family 12 member 7 | −0.064 | 8.147 |
| 225123_PM_at | SESN3 | Sestrin 3 | −0.076 | 7.837 |
| 209374_PM_s_at | IGHM | Immunoglobulin heavy constant mu | −0.089 | 7.851 |
| 204909_PM_at | DDX6 | DEAD-box helicase 6 | −0.103 | 7.793 |
| 235693_PM_at | Unmapped | −0.139 | 8.49 | |
| 205171_PM_at | PTPN4 | Protein tyrosine phosphatase nonreceptor type 4 | −0.146 | 8.887 |
| 214470_PM_at | KLRB1 | Killer cell lectin like receptor B1 | −0.165 | 7.907 |
| 206061_PM_s_at | DICER1 | Dicer 1, ribonuclease III | −0.167 | 7.794 |
| 225177_PM_at | RAB11FIP1 | RAB11 family interacting protein 1 | −0.172 | 9.754 |
| 1555446_PM_s_at | TRAPPC10 | Trafficking protein particle complex 10 | −0.172 | 8.125 |
| 220000_PM_at | SIGLEC5 | Sialic acid-binding Ig like lectin 5 | −0.173 | 8.574 |
| 236685_PM_at | Unmapped | −0.182 | 8.43 | |
| 243395_PM_at | Unmapped | −0.188 | 8.674 | |
| 208003_PM_s_at | NFAT5 | Nuclear factor of activated T-cells 5 | −0.191 | 7.928 |
| 1552480_PM_s_at | PTPRC | Protein tyrosine phosphatase receptor type C | −0.199 | 7.79 |
| 206170_PM_at | ADRB2 | Adrenoceptor beta 2 | −0.209 | 7.786 |
| 1555745_PM_a_at | LYZ | Lysozyme | −0.211 | 11.702 |
| 230332_PM_at | ZCCHC7 | Zinc finger CCHC-type containing 7 | −0.216 | 8.29 |
| 244414_PM_at | Unmapped | −0.224 | 8.551 | |
| 243819_PM_at | Unmapped | −0.225 | 8.904 | |
| 236545_PM_at | Unmapped | −0.232 | 8.352 | |
| 1556185_PM_a_at | STEAP4 | STEAP4 metalloreductase | −0.234 | 8.007 |
| 203021_PM_at | SLPI | Secretory leukocyte peptidase inhibitor | −0.237 | 8.437 |
| 243109_PM_at | MCTP2 | Multiple C2 and transmembrane domain-containing 2 | −0.24 | 10.006 |
| 223578_PM_x_at | TALAM1 | TALAM1 transcript, MALAT1 antisense RNA | −0.247 | 7.963 |
| 242827_PM_x_at | Unmapped | −0.264 | 7.803 | |
| 233690_PM_at | Unmapped | −0.264 | 8.266 | |
| 224681_PM_at | GNA12 | G protein subunit alpha 12 | −0.271 | 8.391 |
| 242197_PM_x_at | CD36 | CD36 molecule | −0.289 | 7.963 |
| 221675_PM_s_at | CHPT1 | Choline phosphotransferase 1 | −0.29 | 10.188 |
| 237330_PM_at | Unmapped | −0.295 | 8.403 | |
| 204467_PM_s_at | SNCA | Synuclein alpha | −0.395 | 10.037 |
| 211781_PM_x_at | Unmapped | −0.44 | 10.706 | |
| 209728_PM_at | HLA-DRB4 | Major histocompatibility complex, class II, DR beta 4 | −0.624 | 5.766 |
AR, acute rejection; ATP, adenosine triphosphate; FC, fold change; IgG, immunoglobulin G.
FIGURE 3.
Three-dimensional principal component analysis score plots for sample clustering, using 59 probes from the random forest classifier between AR and non-AR. The left plot is for training (n = 156) and the right plot is for testing (67) sample sets. AR, acute rejection.
DISCUSSION
In this report, we have used a novel modeling approach to augment the clinical and biological specificity of blood-based genomic biomarkers of AR in LTRs. This resulted in discovery and validation of a 59-gene biomarker that can distinguish LTRs with AR from essentially all other recipients—those with either a stable course or with other causes of graft injury. Similar to our earlier work analyzing well-curated serial samples at time points with no evidence of impending liver graft injury (normal LFTs),30 we have shown that this biomarker signature is detectable long before the AR event (Figure 2A—threshold crossed ~day −120 and error bars fully above at ~day −25) and importantly not seen in LTRs never developing rejection (non-AR).
Other groups have also identified biomarkers of rejection in LTRs.16,17,19,21,22,28,48 Most have had limited sample sizes, lack of validation cohorts, and lack of serial samples preceding graft dysfunction. More recent studies using serial samples have demonstrated increasing levels of microRNAs, donor-specific antibodies, and blood CXCL10 gene expression before rejection, mainly during the course of full IST withdrawal.28,29,49,50 These may have a role in predicting AR in highly select LTRs enrolled in tolerance studies, but they have not been validated in the broader LT population as in our study.
The proposed clinical context of use for this current biomarker is to provide reassurance to the clinician contemplating a preemptive or “for cause” reduction in IST that a patient’s immune status is “quiescent,” reducing the risk of triggering AR. As protocols of immunosuppression minimization are becoming more common in LTRs to mitigate complications from over-immunosuppression, they increase the risk of AR as a consequence and can impact survival.11,49–50 While the most likely cause of graft dysfunction during IST minimization is AR, the current biomarker further reduces the possibility that a positive test indicates causes other than AR and also reduces the likelihood that a negative test indicates AR. Thus, knowing at each IST adjustment consideration whether the immune status is quiescent or activated could help inform safer IST reduction decisions. This is important in terms of the need for the most specificity for detecting immune activation preceding rejection and thus moving toward increasing the number of patients successfully and appropriately minimized. In addition, a signal of immune activation following IST reduction may inform the clinician to consider ceasing reductions until quiescent signals are present.
One of the unique key aspects of this biomarker signature is that it is enriched with immune response genes, particularly those of allograft rejection and signaling pathways, as well as liver injury genes. The most upregulated gene in AR was X inactive specific transcript (XIST), which is a long noncoding RNA involved in X chromosome silencing.51 XIST upregulation in AR may reflect the gender differences in the 2 cohorts and the role of XIST in modulating injury in organ transplantation. XIST silencing has been shown to protect against sepsis-induced acute liver injury via inhibition of the Bromodomain-containing protein 4 expression.52 Bromodomain inhibitors are currently being evaluated in clinical trials for their anti-inflammatory and anticancer properties. The interaction of XIST with microRNAs modulates acute kidney injury.53 Another immune response gene in the panel is alpha-synuclein, which is highly expressed in monocytes and hematopoietic precursor cells, which could reflect impaired hematopoiesis in liver rejection.54 Similarly, protein tyrosine phosphatase receptor type C (CD45), a tyrosine phosphatase required by T, B, and natural killer cells for optimal signal transduction after stimulation, has been shown to be modulated in acute kidney rejection.55 CD9 and CD27 were downregulated in the AR subjects. CD9, a tetraspanin, has been targeted for modulating inflammation, and CD27 has been shown to promote survival and expansion of activated T cells.56 In addition, the rapid resolution of the signature along with LFT normalization with corticosteroid therapy further suggests its specificity for immune-related graft injury and its potential to track disease status as a companion diagnostic.
These findings are important because having a classifier that clinically predicts and has biological relevance to the outcome (eg, AR) provides further validation for its use in clinical practice as a functional biomarker.57 Biological pathway mapping of our previously reported AR versus TX profile revealed less specific hepatic proliferation genes, which is consistent with liver injury (elevated LFTs) being the main difference between these phenotypes.30 In studies of other organ transplant recipients, blood/graft biomarkers of rejection have not reliably demonstrated biological relevance to immune or inflammatory pathways.36,58-62 Even in cancer, the most widely studied disease, there are hundreds of genetic variants that might predict response to radiation, yet the correlation with these markers does not mean causation or represent actionable targets for cancer therapeutics.63 Some biomarkers may be indirectly related to the disease process or dependent on another causative factor. While not representing determinants of disease, they still may be connected to the causal pathway.64 Ultimately, biological relevance can be an important consideration for using functional biomarkers in clinical practice. We submit that our unbiased approach uncovered novel genes and pathways for further validation.
We believe there are reasons our approach identified genes with higher biological relevance. First, using a custom vector,37 we bioinformatically used previously generated LT-specific gene expression profiles to select the most relevant genes to increase confidence in classification accuracy. This approach is ideal for this highly targeted clinical application in comparison with the standard, nonspecific normalization technique that may skew data for homogenous samples such as ours. Second, adding in ADNR phenotypes likely improved the separation of immune (AR) and nonimmune genes. Finally, we chose to combine the populations and divide them 70:30 into training and testing sets because the cohorts had significant clinical differences (NU = single center, late post-LT; CTOT-14 = multicenter, early post-LT) despite being phenotyped the same way clinically and histologically. This issue supported our approach to combine them to enhance the distinction of AR versus non-AR and allow generation of a signature that spanned both the early and late post-LT period. The split-sample approach with bootstrapping is an established methodology that still allows for sufficient discovery and validation of a biomarker in the absence of large sample sizes and gives reasonably valid estimates of the predictive performance of a given model.38,65-67 That said, we recognize that additional external cohorts would be of use in further validation.
Limitations of this study include a relatively modest number of AR and non-AR phenotypic samples, despite emanating from independent single and multicenter studies with prospectively collected samples. These numbers do however reflect the natural prevalence of the phenotypes.5,45-47 As in our previous study,30 in the absence of protocol surveillance biopsies in LTRs, TX was defined clinically by long-term normal LFTs and not by histology. While we recognize that subclinical graft inflammation despite normal LFTs is increasingly being reported in LTRs,32,33 it is unclear if this is normal trafficking (perhaps regulatory T/B cells) controlling alloimmune responses or low-grade immune activation, that is, T cell–, plasma cell–, or antibody–mediated rejection.6 We did not test for donor-specific antibodies, and, given emerging data, they should be included in future immune monitoring studies predicting or diagnosing different rejection types.50,68-70 That said, there were no CPs of AR or ADNR evident for 90 d before and after the TX sample, and the phenotypes all were validated by an NIH Data Coordinating Center (CTOT-14). We do plan to test our profiles in LTRs who are undergoing surveillance biopsies—pediatrics, autoimmune liver disease, and tolerance protocols. Finally, we did not include clinical variables, such as age, gender, cause of liver disease, severity of AR, etc, into our modeling approach as we sought to validate a classifier that could be universally applied to AR rather than specific patient phenotypes. The 70:30 approach combining single and multicenter cohorts randomized the samples to distribute these variables as equally as possible to reduce their potential bias.
In summary, we have developed a blood-based biologically relevant gene expression profile that has focused on detecting an immune quiescent state to advance the success of immunosuppression optimization (mainly minimization) in LT practice. Use of these gene expression biomarkers could enhance patient outcomes by proactively detecting and avoiding adverse events related to both under- and over-immunosuppression.
Supplementary Material
Footnotes
ClinicalTrials.gov identifier: NCT01672164.
J.L., M.A., M.K., S.K., and T.W. participated in conceptualization. M.K., L.Z., S.K., and T.W. participated in methodology. M.K., L.Z., S.K., K.G., and T.W. participated in formal analysis. J.L., M.A., K.G., L.Z., M.K., S.K., and T.W. participated in investigation and writing. J.L. and M.A. participated in funding acquisition.
J.L. and S.K. provided consultancy to Transplant Genomics Incorporated (Eurofins/Viracor). M.A. is a cofounder of Transplant Genomics Incorporated (Eurofins/Viracor). J.L. received research funding from Novartis. The other authors declare no conflicts of interest.
The funding for this study was supported by internal funds from the Northwestern University Comprehensive Transplant Center and a sponsored research grant between Northwestern and Eurofins-Viracor-Transplant Genomics Inc. Although not used for this current submission, the funding for the initial prospective study and sample collections was supported by Clinical Trials in Organ Transplantation-14 U01AI084146 (PI: M.A.; Protocol Chair: J.L.) from the National Institute of Allergy and Infectious Disease.
The data and metadata used for development will be uploaded to an independently managed database dbGAP or GEO and will be made available to research community after publication.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Charlton M, Seaberg E. Impact of immunosuppression and acute rejection on recurrence of hepatitis C: results of the National Institute of Diabetes and Digestive and Kidney Diseases Liver Transplantation Database. Liver Transpl Surg. 1999;5:S107–S114. [DOI] [PubMed] [Google Scholar]
- 2.Wiesner RH, Demetris AJ, Belle SH, et al. Acute hepatic allograft rejection: incidence, risk factors, and impact on outcome. Hepatology. 1998;28:638–645. [DOI] [PubMed] [Google Scholar]
- 3.US Multicenter FK506 Liver Study Group. A comparison of tacrolimus (FK 506) and cyclosporine for immunosuppression in liver transplantation. N Engl J Med. 1994;331:1110–1115. [DOI] [PubMed] [Google Scholar]
- 4.Starzl TE, Klintmalm GB, Porter KA, et al. Liver transplantation with use of cyclosporin a and prednisone. N Engl J Med. 1981;305:266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levitsky J, Goldberg D, Smith AR, et al. Acute rejection increases risk of graft failure and death in recent liver transplant recipients. Clin Gastroenterol Hepatol. 2017;15:584–593.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demetris AJ, Bellamy C, Hübscher SG, et al. 2016 comprehensive update of the Banff Working Group on Liver Allograft Pathology: introduction of antibody-mediated rejection. Am J Transplant. 2016;16:2816–2835. [DOI] [PubMed] [Google Scholar]
- 7.Charlton M, Levitsky J, Aqel B, et al. International liver transplantation society consensus statement on immunosuppression in liver transplant recipients. Transplantation. 2018;102:727–743. [DOI] [PubMed] [Google Scholar]
- 8.VanWagner LB, Serper M, Kang R, et al. Factors associated with major adverse cardiovascular events after liver transplantation among a national sample. Am J Transplant. 2016;16:2684–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubín A, Sánchez-Montes C, Aguilera V, et al. Long-term outcome of ‘long-term liver transplant survivors’. Transpl Int. 2013;26:740–750. [DOI] [PubMed] [Google Scholar]
- 10.Watt KDS, Pedersen RA, Kremers WK, et al. Evolution of causes and risk factors for mortality post-liver transplant: results of the NIDDK long-term follow-up study. Am J Transplant. 2010;10:1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levitsky J, O’Leary JG, Asrani S, et al. Protecting the kidney in liver transplant recipients: practice-based recommendations from the American Society of Transplantation Liver and Intestine Community of Practice. Am J Transplant. 2016;16:2532–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kourkoumpetis T, Levitsky J. Immunosuppressive drug levels in liver transplant recipients: impact in decision making. Semin Liver Dis. 2019;39:414–421. [DOI] [PubMed] [Google Scholar]
- 13.Levitsky J. Next level of immunosuppression: drug/immune monitoring. Liver Transpl. 2011;17(suppl 3):S60–S65. [DOI] [PubMed] [Google Scholar]
- 14.Kowalski RJ, Post DR, Mannon RB, et al. Assessing relative risks of infection and rejection: a meta-analysis using an immune function assay. Transplantation. 2006;82:663–668. [DOI] [PubMed] [Google Scholar]
- 15.Xue F, Zhang J, Han L, et al. Immune cell functional assay in monitoring of adult liver transplantation recipients with infection. Transplantation. 2010;89:620–626. [DOI] [PubMed] [Google Scholar]
- 16.Fan H, Li L-X, Han D-D, et al. Increase of peripheral Th17 lymphocytes during acute cellular rejection in liver transplant recipients. Hepatobiliary Pancreat Dis Int. 2012;11:606–611. [DOI] [PubMed] [Google Scholar]
- 17.Farid WRR, Pan Q, van der Meer AJP, et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18:290–297. [DOI] [PubMed] [Google Scholar]
- 18.Gómez-Mateo J, Marin L, López-Alvarez MR, et al. TGF-beta1 gene polymorphism in liver graft recipients. Transpl Immunol. 2006;17:55–57. [DOI] [PubMed] [Google Scholar]
- 19.Joshi D, Salehi S, Brereton H, et al. Distinct microRNA profiles are associated with the severity of hepatitis C virus recurrence and acute cellular rejection after liver transplantation. Liver Transpl. 2013;19:383–394. [DOI] [PubMed] [Google Scholar]
- 20.Kamei H, Masuda S, Nakamura T, et al. Association of transporter associated with antigen processing (TAP) gene polymorphisms in donors with acute cellular rejection in living donor liver transplantation. J Gastrointestin Liver Dis. 2013;22:167–171. [PubMed] [Google Scholar]
- 21.Karimi MH, Daneshmandi S, Pourfathollah AA, et al. Association of IL-6 promoter and IFN-γ gene polymorphisms with acute rejection of liver transplantation. Mol Biol Rep. 2011;38:4437–4443. [DOI] [PubMed] [Google Scholar]
- 22.Massoud O, Heimbach J, Viker K, et al. Noninvasive diagnosis of acute cellular rejection in liver transplant recipients: a proteomic signature validated by enzyme-linked immunosorbent assay. Liver Transpl. 2011;17:723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moya-Quiles MR, Alvarez R, Miras M, et al. ; Spanish Group of Transplant Immunology and Immunotolerance (G03/104). Impact of recipient HLA-C in liver transplant: a protective effect of HLA-Cw*07 on acute rejection. Hum Immunol. 2007;68:51–58. [DOI] [PubMed] [Google Scholar]
- 24.Sindhi R, Higgs BW, Weeks DE, et al. Genetic variants in major histocompatibility complex-linked genes associate with pediatric liver transplant rejection. Gastroenterology. 2008;135:830–839.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asaoka T, Kato T, Marubashi S, et al. Differential transcriptome patterns for acute cellular rejection in recipients with recurrent hepatitis C after liver transplantation. Liver Transpl. 2009;15:1738–1749. [DOI] [PubMed] [Google Scholar]
- 26.Gehrau R, Maluf D, Archer K, et al. Molecular pathways differentiate hepatitis C virus (HCV) recurrence from acute cellular rejection in HCV liver recipients. Mol Med. 2011;17:824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreekumar R, Rasmussen DL, Wiesner RH, et al. Differential allograft gene expression in acute cellular rejection and recurrence of hepatitis C after liver transplantation. Liver Transpl. 2002;8:814–821. [DOI] [PubMed] [Google Scholar]
- 28.Shaked A, Chang B-L, Barnes MR, et al. An ectopically expressed serum miRNA signature is prognostic, diagnostic, and biologically related to liver allograft rejection. Hepatology. 2017;65:269–280. [DOI] [PubMed] [Google Scholar]
- 29.Bonaccorsi-Riani E, Pennycuick A, Londoño M-C, et al. Molecular characterization of acute cellular rejection occurring during intentional immunosuppression withdrawal in liver transplantation. Am J Transplant. 2016;16:484–496. [DOI] [PubMed] [Google Scholar]
- 30.Levitsky J, Asrani SK, Schiano T, et al. ; Clinical Trials in Organ Transplantation - 14 Consortium. Discovery and validation of a novel blood-based molecular biomarker of rejection following liver transplantation. Am J Transplant. 2020;20:2173–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banff schema for grading liver allograft rejection: an international consensus document. Hepatology. 1997;25:658–663. [DOI] [PubMed] [Google Scholar]
- 32.Londoño M-C, Souza LN, Lozano J-J, et al. Molecular profiling of subclinical inflammatory lesions in long-term surviving adult liver transplant recipients. J Hepatol. 2018;69:626–634. [DOI] [PubMed] [Google Scholar]
- 33.Feng S, Bucuvalas JC, Demetris AJ, et al. Evidence of chronic allograft injury in liver biopsies from long-term pediatric recipients of liver transplants. Gastroenterology. 2018;155:1838–1851.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michell CM, Nass SJ, Ommen GS; Institute of Medicine. Evolution of Translational Omics: Lessons Learned and the Path Forward. The National Academies Press; 2012. [PubMed] [Google Scholar]
- 35.Friedewald JJ, Kurian SM, Heilman RL, et al. ; Clinical Trials in Organ Transplantation 08 (CTOT-08). Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant. Am J Transplant. 2019;19:98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurian SM, Williams AN, Gelbart T, et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant. 2014;14:1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics. 2010;11:242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurian SM, Whisenant T, Mas V, et al. Biomarker guidelines for high-dimensional genomic studies in transplantation: adding method to the madness. Transplantation. 2017;101:457–463. [DOI] [PubMed] [Google Scholar]
- 39.Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thévenot EA, Roux A, Xu Y, et al. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res. 2015;14:3322–3335. [DOI] [PubMed] [Google Scholar]
- 41.Meyer D, Dimitriadou E, Hornik K, et al. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. Available at https://CRAN.R-project.org/package=e1071. Accessed May 23, 2021.
- 42.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 43.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 44.Van Loon E, Gazut S, Yazdani S, et al. Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: a multicentre, prospective study. EBioMedicine. 2019;46:463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramji A, Yoshida EM, Bain VG, et al. Late acute rejection after liver transplantation: the Western Canada experience. Liver Transpl. 2002;8:945–951. [DOI] [PubMed] [Google Scholar]
- 46.Uemura T, Ikegami T, Sanchez EQ, et al. Late acute rejection after liver transplantation impacts patient survival. Clin Transplant. 2008;22:316–323. [DOI] [PubMed] [Google Scholar]
- 47.Thurairajah PH, Carbone M, Bridgestock H, et al. Late acute liver allograft rejection; a study of its natural history and graft survival in the current era. Transplantation. 2013;95:955–959. [DOI] [PubMed] [Google Scholar]
- 48.Toby TK, Abecassis M, Kim K, et al. Proteoforms in peripheral blood mononuclear cells as novel rejection biomarkers in liver transplant recipients. Am J Transplant. 2017;17:2458–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaked A, DesMarais MR, Kopetskie H, et al. Outcomes of immunosuppression minimization and withdrawal early after liver transplantation. Am J Transplant. 2019;19:1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jucaud V, Shaked A, DesMarais M, et al. Prevalence and impact of de novo donor-specific antibodies during a multicenter immunosuppression withdrawal trial in adult liver transplant recipients. Hepatology. 2019;69:1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130:223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen C, Li J. LncRNA XIST silencing protects against sepsis-induced acute liver injury via inhibition of BRD4 expression. Inflammation. 2021;44:194–205. [DOI] [PubMed] [Google Scholar]
- 53.Cheng Q, Wang L. LncRNA XIST serves as a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in kidney transplant acute kidney injury via sponging hsa-miR-212-3p and hsa-miR-122-5p. Cell Cycle. 2020;19:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grozdanov V, Danzer KM. Intracellular alpha-synuclein and immune cell function. Front Cell Dev Biol. 2020;8:562692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shabtai M, Waltzer WC, Ayalon A, et al. Down regulation of CD45 expression on CD4 T cells during acute renal allograft rejection: evidence of a decline in T suppressor/inducer activity. Int Urol Nephrol. 2002;34:555–558. [DOI] [PubMed] [Google Scholar]
- 56.Hendriks J, Xiao Y, Borst J. CD27 promotes survival of activated T cells and complements CD28 in generation and establishment of the effector T cell pool. J Exp Med. 2003;198:1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kearns GL, Artman M. Functional biomarkers: an approach to bridge pharmacokinetics and pharmacodynamics in pediatric clinical trials. Curr Pharm Des. 2015;21:5636–5642. [DOI] [PubMed] [Google Scholar]
- 58.Modena BD, Kurian SM, Gaber LW, et al. Gene expression in biopsies of acute rejection and interstitial fibrosis/tubular atrophy reveals highly shared mechanisms that correlate with worse long-term outcomes. Am J Transplant. 2016;16:1982–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mengel M, Sis B, Kim D, et al. The molecular phenotype of heart transplant biopsies: relationship to histopathological and clinical variables. Am J Transplant. 2010;10:2105–2115. [DOI] [PubMed] [Google Scholar]
- 60.Pham MX, Teuteberg JJ, Kfoury AG, et al. ; IMAGE Study Group. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–1900. [DOI] [PubMed] [Google Scholar]
- 61.Soma O, Hatakeyama S, Yoneyama T, et al. Serum N-glycan profiling can predict biopsy-proven graft rejection after living kidney transplantation. Clin Exp Nephrol. 2020;24:174–184. [DOI] [PubMed] [Google Scholar]
- 62.Bohne F, Martínez-Llordella M, Lozano J-J, et al. Intra-graft expression of genes involved in iron homeostasis predicts the development of operational tolerance in human liver transplantation. J Clin Invest. 2012;122:368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bien-Willner GA. Biomarkers and cancer: correlation is not causation. Available at https://www.clinicalomics.com/topics/oncology/biomarkers-and-cancer-correlation-is-not-causation. Accessed January 6, 2015.
- 64.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steyerberg EW, Harrell FE, Jr. Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol. 2016;69:245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steyerberg EW, Bleeker SE, Moll HA, et al. Internal and external validation of predictive models: a simulation study of bias and precision in small samples. J Clin Epidemiol. 2003;56:441–447. [DOI] [PubMed] [Google Scholar]
- 67.Harrell FE, Jr, Lee KL, Mark DB. Tutorial in biostatistics. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. [DOI] [PubMed] [Google Scholar]
- 68.O’Leary JG, Demetris AJ, Friedman LS, et al. The role of donor-specific HLA alloantibodies in liver transplantation. Am J Transplant. 2014;14:779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Leary JG, Kaneku H, Jennings LW, et al. Preformed class II donor-specific antibodies are associated with an increased risk of early rejection after liver transplantation. Liver Transpl. 2013;19:973–980. [DOI] [PubMed] [Google Scholar]
- 70.Levitsky J, Kaneku H, Jie C, et al. Donor-specific HLA antibodies in living versus deceased donor liver transplant recipients. Am J Transplant. 2016;16:2437–2444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.