Abstract
An exo-arabinanase, designated Abnx, was purified from a culture filtrate of Penicillium chrysogenum 31B by ammonium sulfate precipitation, anion-exchange chromatography, and hydrophobic chromatography. Abnx had an apparent molecular mass of 47 kDa. The enzyme released only arabinobiose from the nonreducing terminus of α-1,5-l-arabinan and showed no activity towards p-nitrophenyl-α-l-arabinofuranoside and α-1,5-l-arabinofuranobiose. Abnx is the first enzyme with this mode of action.
Arabinose residues are found in arabinans, arabinogalactans, or arabinoxylans in many plant cell walls. In sugar beet arabinan, l-arabinose residues are linked to form α-1,5-l-arabinan to which l-arabinofuranose units are attached mainly at position 3 in the α-configuration as side chains. Soybean arabinogalactans have a linear chain of β-1,4-d-galactan to which α-1,5-l-linked arabinofuranooligosaccharides are bound in side chains. Arabinogalactan in larch wood consists of β-1,3-galactan to which β-1,3-linked arabinooligosaccharides or β-1,6-linked galactooligosaccharides are attached at position 6 as side chains.
Arabinose-containing polymers are degraded by various enzymes, which have been classified into six types depending on their mode of action and substrate specificity by Beldman et al. (3), as follows: (i) α-l-arabinofuranosidase (EC 3.2.1.55), which is not active with polymers (10, 22); (ii) α-l-arabinofuranosidase, which is active with polymers (9, 16); (iii) α-l-arabinofuranohydrolase, which is specific for arabinoxylans (11, 20); (iv) exo-α-l-arabinanase, which is not active with p-nitrophenyl-α-l-arabinofuranoside (8, 13); (v) β-l-arabinopyranosidase (4); and (vi) endo-1,5-α-l-arabinanase (EC 3.2.1.99) (7, 21). Of these enzymes, little is known about exo-α-l-arabinanases. This study dealt with isolation and characterization of an exo-arabinanase, designated Abnx, that is produced by Penicillium chrysogenum 31B, which was isolated from rotten sugar beet. Interestingly, filtrate from a culture of this microorganism contained at least five different arabinan-degrading enzymes. The work described here was the first step in characterizing these enzymes and should be followed by elucidation of the mode of degradation of sugar beet arabinan by this strain.
Three liters of a liquid medium consisting of 0.2% NH4NO3, 0.1% K2HPO4, 0.05% MgSO4 · 7H2O, 0.05% KCl, 0.001% FeSO4, 0.1% peptone, 0.1% glucose, and 2% sugar beet pulp (pH 5.0) was inoculated with precultured P. chrysogenum 31B and incubated at 30°C for 12 days under static conditions in a 5-liter Erlenmayer flask. The culture filtrate was concentrated by ultrafiltration (10-kDa cutoff), dialyzed against 20 mM acetate buffer (pH 5.0), and used for enzyme purification. A typical assay for arabinan-degrading activity was performed by measuring the release of reducing groups in a reaction mixture containing 195 μl of 0.1% debranched arabinan (Megazyme International Ireland Ltd., Wicklow, Ireland) in 20 mM acetate buffer (pH 5.0) and 5 μl of enzyme sample at 37°C. Reducing sugars were measured by the method of Somogyi (18). One unit of enzyme activity was defined as the amount of enzyme that formed reducing groups corresponding to 1 μmol of l-arabinose in 1 min. For the first step, pulverized crystal ammonium sulfate was added to the enzyme solution to 80% saturation at a rate of approximately 2 g/min. The precipitate was recovered by centrifugation at 6,000 × g for 15 min, dissolved in 20 mM acetate buffer (pH 5.0), and dialyzed against the same buffer. The enzyme solution was loaded onto a DEAE-Toyopearl 650M column (4 by 10 cm; Tosoh Corp., Tokyo, Japan) equilibrated with the dialysis buffer. Although 60% of the initial arabinan-degrading activity did not bind to this column, this fraction is not described further here. The bound enzymes were eluted by a 500-ml linear 0 to 0.4 M NaCl gradient in the same buffer. The arabinan-degrading activity eluted from the DEAE-Toyopearl 650M column as two peaks. The peak eluting at a lower NaCl concentration was designated Abnx and purified further. The enzyme solution was then dialyzed against the buffer described above and concentrated under reduced pressure. Pulverized crystal ammonium sulfate was added to the concentrate to 30% saturation, and the enzyme solution was loaded onto a Phenyl Superose HR 5/5 column (Amersham Pharmacia) equilibrated with 20 mM acetate buffer (pH 5.0) containing ammonium sulfate at 30% saturation. The adsorbed proteins were eluted by a linear ammonium sulfate gradient (30 to 0% saturation) at a flow rate of 0.5 ml/min. The arabinanase-containing fractions were pooled, dialyzed against 20 mM acetate buffer (pH 5.0), concentrated, and put on a Mono Q HR 5/5 column (Amersham Pharmacia) equilibrated with the dialysis buffer. The bound proteins were eluted by a linear 0 to 0.15 M NaCl gradient at a flow rate of 1 ml/min.
The purification procedure for Abnx is summarized in Table 1. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the purified enzyme showed that there was a single protein band at a molecular mass of 47 kDa. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by the method of Laemmli (12) with a discontinuous 10% polyacrylamide gel.
TABLE 1.
Purification of Abnx from P. chrysogenum 31B
| Purification step | Protein (mg) | Activity (U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Culture filtrate | 473.0 | 228.0 | 0.48 | 1 | 100 |
| (NH4)2SO4 | 199.0 | 211.0 | 1.06 | 2.2 | 92.5 |
| DEAE-Toyopearl | 31.7 | 18.9 | 0.60 | 1.3 | 8.3 |
| Phenyl Superose | 2.6 | 5.2 | 2.00 | 4.2 | 2.3 |
| Mono Q | 0.6 | 2.5 | 4.17 | 8.7 | 1.1 |
To study the effects of pH and temperature on enzyme activity, the enzyme reaction was performed at various pHs by using 20 mM acetate buffer (pH 3 to 5) and 20 mM phosphate buffer (pH 6 to 7) at 37°C and at various temperatures in 20 mM acetate buffer (pH 5.0). Optimum activity occurred at pH 4.0 and 40°C. Temperature stability was evaluated by measuring the residual activity after 1 h of preincubation of the enzyme (158 μg/ml) at temperatures between 30 and 70°C in 20 mM acetate buffer (pH 5.0). The enzyme was stable at temperatures up to 50°C. pH stability was studied by preincubating the enzyme (75 μg/ml) at 30°C for 16 h at various pHs, using 100 mM HCl-KCl buffer (pH 1 to 2), acetate buffer (pH 3 to 5), phosphate buffer (pH 6 to 8), and Na2CO3-NaHCO3 buffer (pH 9 to 11). More than 80% of the initial enzyme activity remained at pH 3 to 8. The sensitivity of Abnx to metals was examined by adding compounds at a concentration of 1 mM to the reaction mixture for the standard arabinanase assay. HgCl2 caused a loss of 70% of the enzyme activity. No effect on activity was detected with the chloride salts of Ba2+, Ca2+, Cd2+, Co2+, Fe3+, K+, Mg2+, Na+, Ni2+, and Zn2+, AgNO3, and CuSO4. NaN3 (1 mM) had no effect on the activity.
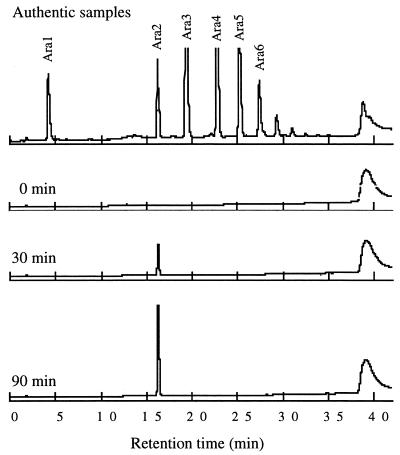
To determine the mode of action of the enzyme with linear arabinan, 0.08 mU of Abnx was incubated with 200 μl of 0.2% reduced debranched arabinan in 20 mM acetate buffer (pH 5.0) at 37°C, and after different times the products were analyzed by high-performance anion-exchange chromatography using a Carbopac PA-1 column (Dionex). Sugars were eluted at a flow rate of 1 ml/min with 0.1 M NaOH for 5 min and then with a 30-ml linear 0 to 0.45 M sodium acetate gradient in 0.1 M NaOH. The effluent was monitored with pulsed amperometric detection. Only arabinobiose was detected during the early stage of the enzyme reaction. Moreover, no arabinofuranosyl arabitol was formed during degradation of reduced debranched arabinan (Fig. 1). These results indicated that Abnx cleaved α-1,5-l-linked arabinofuranose residues at the nonreducing terminus in an exo manner. Reducing ends of debranched arabinan were reduced by treatment with 10 mM NaBH4 in 25 mM NaOH at 30°C for 4 h (19).
FIG. 1.
Analysis of the enzymatic products of reduced debranched arabinan obtained with Abnx. Authentic samples Ara1 to Ara6 represent arabinose to arabinohexaose, respectively.
Substrate specificity data for the enzyme are summarized in Table 2. Arabinosidase activity was tested by incubating 5 μl (10 mU) of the enzyme with 195 μl of 0.2% p-nitrophenyl-α-l-arabinofuranoside or p-nitrophenyl-α-l-arabinopyranoside (Sigma) in 20 mM acetate buffer (pH 5.0) at 37°C overnight. p-Nitrophenol release was monitored spectrophotometrically at 420 nm after 1.8 ml of 0.2 M Na2CO3 was added to the reaction mixture. To determine degradation activity with α-1,5-l-arabinofuranobiose (Megazyme), triose (Megazyme), and various polysaccharides, 4 mU of Abnx was incubated with 200 μl of 0.2% substrate in 20 mM acetate buffer (pH 5.0) for 1 h at 37°C, and then the reaction products were analyzed by high-performance anion-exchange chromatography under the conditions described above. The enzyme released significant amounts of arabinobiose from debranched arabinan and α-1,5-l-arabinofuranotriose. Minor activity was detected with sugar beet arabinan (Megazyme) or soybean arabinogalactan, which was prepared as previously described (14). The enzyme did not cleave the following substrates: larch wood arabinogalactan (Sigma), p-nitrophenyl-α-l-arabinofuranoside, p-nitrophenyl-α-l-arabinopyranoside, and α-1,5-l-arabinofuranobiose. Considering the structure of the substrates, these results indicated that Abnx degraded α-1,5-l-linked arabinofuranose residues more than it degraded trimers.
TABLE 2.
Enzyme activity of Abnx with various substrates
| Substrate | Amt of sugar released (μg/ml)a |
|---|---|
| p-Nitrophenyl-α-l-arabinofuranoside | 0 |
| p-Nitrophenyl-α-l-arabinopyranoside | 0 |
| Sugar beet debranched arabinan | 455 |
| Sugar beet arabinan | 42 |
| Soybean arabinogalactan | 57 |
| Larch wood arabinogalactan | 0 |
| α-1,5-l-Arabinofuranobiose | 0 |
| α-1,5-l-Arabinofuranotriose | 252 |
Concentration of arabinobiose released into the reaction mixture. Details of the methods used for the enzyme assays are described in the text. The values are averages based on three experiments.
Two exo-arabinanases have been found previously, one in Erwinia carotovora IAM 1024 (8) and one in Pseudomonas fluorescens subsp. cellulosa (13). The former produces only arabinotriose from sugar beet arabinan but not from linear arabinan. In contrast, the latter is not active with sugar beet arabinan, but with linear arabinan it produces arabinotriose. Abnx was similar to the exo-arabinanase from P. fluorescens in terms of substrate specificity, except that Abnx produced arabinobiose from linear arabinan. Enzyme Nomenclature includes numbers for the following five enzymes that catalyze the release of dimeric sugars from polysaccharides in an exo manner: β-amylase (EC 3.2.1.2) (2), exo-poly-α-galacturonosidase (EC 3.2.1.82) (6), cellulose 1,4-β-cellobiosidase (EC 3.2.1.91) (5), glucan 1,6-α-isomaltosidase (EC 3.2.1.94) (17), and mannan 1,4-β-mannobiosidase (EC 3.2.1.100) (1). Furthermore, Nakano et al. have isolated exo-1,4-β-d-galactanase (which does not have an EC number) from Bacillus subtilis; this enzyme releases predominantly β-galactobiose from soybean arabinogalactan (15). No arabinanases which produce arabinobiose from arabinan have been found previously. The Abnx described here may be the first enzyme with its mode of action.
Acknowledgments
We are grateful to S. Nishimura for isolating the microorganism from rotten sugar beet.
REFERENCES
- 1.Araki T, Kitamikado M. Purification and characterization of a novel exo-β-mannanase from Aeromonas sp. F-25. J Biochem (Tokyo) 1982;91:1181–1186. doi: 10.1093/oxfordjournals.jbchem.a133801. [DOI] [PubMed] [Google Scholar]
- 2.Balls A K, Walden M K, Thompson R R. A crystalline β-amylase from sweet potatoes. J Biol Chem. 1948;173:9–19. [PubMed] [Google Scholar]
- 3.Beldman G, Schols H A, Pitson S M, Searle-van Leeuwen M J F, Voragen A G J. Arabinans and arabinan degrading enzymes. Adv Macromol Carbohydr Res. 1997;1:1–64. [Google Scholar]
- 4.Dey P M. Further characterization of β-l-arabinosidase from Cajanus indicus. Biochim Biophys Acta. 1983;746:8–13. doi: 10.1016/0005-2744(73)90167-8. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson K-E. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 3. Purification and physico-chemical characterization of an exo-1,4-β-glucanase. Eur J Biochem. 1975;51:213–218. doi: 10.1111/j.1432-1033.1975.tb03921.x. [DOI] [PubMed] [Google Scholar]
- 6.Hasegawa H, Nagel C W. Isolation of an oligogalacturonate hydrolase from a Bacillus species. Arch Biochem Biophys. 1968;124:513–520. doi: 10.1016/0003-9861(68)90360-3. [DOI] [PubMed] [Google Scholar]
- 7.Kaji A, Saheki T. Endo-arabinase from Bacillus subtilis F-11. Biochim Biophys Acta. 1975;410:354–360. doi: 10.1016/0005-2744(75)90237-5. [DOI] [PubMed] [Google Scholar]
- 8.Kaji A, Shimokawa K. New exo-type arabinase from Erwinia carotovora IAM 1024. Agric Biol Chem. 1984;48:67–72. [Google Scholar]
- 9.Kaji A, Tagawa K. Purification, crystalization and amino acid composition of α-l-arabinofuranosidase from Aspergillus niger. Biochim Biophys Acta. 1970;207:456–464. doi: 10.1016/s0005-2795(70)80008-3. [DOI] [PubMed] [Google Scholar]
- 10.Komae K, Kaji A, Sato M. An α-l-arabinofuranosidase from Streptomyces purpurascens IFO 3389. Agric Biol Chem. 1982;46:1899–1905. [Google Scholar]
- 11.Kormelink F J M, Searle-van Leeuwen M J F, Wood T M, Voragen A G J. Purification and characterization of an (1,4)-β-d-arabinoxylan arabinofuranohydrolase from Aspergillus awamori. Appl Microbiol Biotechnol. 1991;35:753–758. [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.McKie V A, Black G W, Millward-Sadler S J, Hazlewood G P, Laurie J I, Gilbert H J. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo- mode of action. Biochem J. 1997;323:547–555. doi: 10.1042/bj3230547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita M. Polysaccharides of soybean seeds, part I. Polysaccharide constituents of “hot-water-extract” fraction of soybean seeds and an arabinogalactan as its major component. Agric Biol Chem. 1965;29:564–573. [Google Scholar]
- 15.Nakano H, Takenishi S, Kitahata S, Kinugasa H, Watanabe Y. Purification and characterization of an exo-1,4-β-galactanase from a strain of Bacillus subtilis. Eur J Biochem. 1990;193:61–67. doi: 10.1111/j.1432-1033.1990.tb19304.x. [DOI] [PubMed] [Google Scholar]
- 16.Rombouts F M, Voragen A G J, Searle-van Leeuwen M F, Geraerds C C J M, Schols H A, Pilnik W. The arabinanases of Aspergillus niger—purification and characterisation of two α-l-arabinofuranosidases and an endo-1,5-α-l-arabinanase. Carbohydr Polym. 1988;9:25–47. [Google Scholar]
- 17.Sawai T, Toriyama K, Yano K. A bacterial dextranase releasing only isomaltose from dextrans. J Biochem (Tokyo) 1974;75:105–112. doi: 10.1093/oxfordjournals.jbchem.a130363. [DOI] [PubMed] [Google Scholar]
- 18.Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 19.Takasaki S, Kobata A. Microdetermination of individual neutral and amino sugars and N-acetylneuraminic acid in complex saccharides. J Biochem (Tokyo) 1974;76:783–789. [PubMed] [Google Scholar]
- 20.Van Laere K M J, Beldman G, Voragen A G J. A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl Microbiol Biotechnol. 1997;47:231–235. doi: 10.1007/s002530050918. [DOI] [PubMed] [Google Scholar]
- 21.Voragen A G J, Rombouts F M, Searle-van Leeuwen M F, Schols H A, Pilnik W. The degradation of arabinans by endo-arabinanase and arabinofuranosidases purified from Aspergillus niger. Food Hydrocoll. 1987;1:423–437. [Google Scholar]
- 22.Weinstein L, Albersheim P. Structure of plant cell walls. IX. Purification and partial characterization of a wall-degrading endo-arabanase and an arabinosidase from Bacillus subtilis. Plant Physiol. 1979;63:425–432. doi: 10.1104/pp.63.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]