ABSTRACT
The advent of pneumococcal conjugate vaccines led to the near disappearance of most of the included serotypes in high-income settings but also the rise of nonvaccine-type colonization and disease. Alternative strategies, using genetically conserved proteins as antigens, have been evaluated preclinically and clinically for years, so far unsuccessfully. One possible explanation for the failure of these efforts is that the choice of antigens may not have been sufficiently guided by an understanding of the gene expression pattern in the context of infection. Here, we present a targeted transcriptomic analysis of 160 pneumococcal genes encoding bacterial surface-exposed proteins in mouse models, human colonization, and human meningitis. We present the overlap of these different transcriptomic profiles. We identify two bacterial genes that are highly expressed in the context of mouse and human infection: SP_0282, an IID component of a mannose phosphotransferase system (PTS), and SP_1739, encoding RNase Y. We show that these two proteins can confer protection against pneumococcal nasopharyngeal colonization and intraperitoneal challenge in a murine model and generate opsonophagocytic antibodies. This study emphasizes and confirms the importance of studies of pneumococcal gene expression of bacterial surface proteins during human infection and colonization and may pave the way for the selection of a protein-based vaccine candidate.
KEYWORDS: colonization, invasive disease, pneumococcus, transcriptome
INTRODUCTION
The World Health Organization (WHO) estimates that Streptococcus pneumoniae kills over 300,000 children under 5 years old worldwide every year, mostly in developing countries (1). Pneumococcal conjugate vaccines have nearly eradicated the included serotypes in high-income countries that have instituted infant immunization programs (2–7). The success of these vaccines has been somewhat mitigated by the rise in nonvaccine serotypes, which have become more common causes of invasive disease in both infants and the older adult population in high-income countries (8). In response, several vaccine manufacturers are developing newer pneumococcal conjugate vaccines, which include greater numbers of serotypes (9–12). But even this strategy may not suffice because of the diversity of serotypes (13) and the lack of efficacy against serotype 3 (14).
These considerations highlight the need to study the feasibility of new types of capsule-independent vaccines such as protein-based vaccines (15). Ideally, a protein-based vaccine could provide broad, serotype-independent protection against mucosal and invasive pneumococcal disease by virtue of containing antigens that are expressed on most circulating clinical isolates in the context of human infection (16, 17). However, this is not how most pneumococcal antigens that have been included in vaccine studies have been selected. Instead, most protein antigens have been chosen on the basis of the ability to confer protection in mouse models of disease or colonization (17). It is unknown whether these antigens are well expressed during human colonization or human infection. Therefore, in this study, we decided to evaluate the gene expression of a number of likely surface-exposed antigens during human infection or colonization and compare their expression to that observed in mouse models of disease and colonization. To circumvent the problem of limited amounts of bacterial RNA from clinical samples, we used the NanoString platform (18, 19). The NanoString technique counts the specific RNA transcripts with no need for preliminary RNA amplification, avoiding the generation of biases in RNA ratios induced by other methods (20). Here, we were able to identify highly expressed pneumococcal genes during human colonization and human cases of meningitis and compare the relative ranking of expression to that observed in mouse models. We identified two genes, SP_0282, a mannose-specific phosphotransferase system (PTS) component IID protein, and SP_1739, a protein potentially involved in RNA degradation, that are highly expressed in every model evaluated. Antibodies directed against these 2 proteins encoded by these genes were able to induce opsonophagocytic killing; immunization with antigen SP_0282 reduced nasopharyngeal colonization and, combined with SP_1739, conferred protection against invasive disease in mice.
RESULTS
Generation of a NanoString code set for pneumococcal genes.
We analyzed how well primers matched the different strains by evaluating the readouts from control cultures of serotype 4, serotype 1 from Malawi, and serotype 6 grown in broth. Any gene with no significant reads for one (or more) of the three serotypes was discarded from the study. Ultimately, we were able to analyze 4 genes coding for choline-binding proteins, 40 genes coding for proteins with a signal peptide, 92 genes coding for transmembrane proteins, and 23 genes coding for transmembrane proteins with a signal peptide, which were included in the code set.
Mouse model RNA counts.
Colonization data were collected from 2 separate pools of 10 mice after 1 day of colonization and from 3 separate pools of 10 mice after either 3 or 7 days of colonization. After normalization, we generated a list of the 30 most commonly expressed genes across the 3 collection days of colonization as shown in Fig. 1A to C. A Venn diagram shows the 30 most highly expressed genes under every condition (Fig. 1D). For the lung infection model, we recognized the possibility that some of the bacteria recovered from bronchoalveolar lavage (BAL) fluid could be the result of intercurrent bacteremia. With this concern in mind, we performed a lung infection model where the CFU in BAL fluid and peripheral blood samples following infection were monitored over time, to select a time point when the concentration of bacteria in the lung greatly exceeded that in the blood at the same time. BALs were performed at 9 h postinfection due to the relatively high ratio of bacterial CFU found in the lung to those found in the blood, keeping potential contamination from the bloodstream to a minimum (Fig. 2A). The list of the 30 most highly expressed genes in bronchoalveolar lavage fluid samples represents the results combined from 3 pools of 10 mice (Fig. 2B). Blood samples were collected 2 days after lung infection, and the list of the 30 most highly expressed genes represents the pool of data for 10 individual mice (Fig. 2C). A Venn diagram shows that genes SP_1739, SP_1891, SP_2216, SP_0366, SP_1500, SP_0107, SP_0757, SP_0845, SP_1032, SP_0348, SP_1400, SP_2203, SP_0282, SP_0807, and SP_1650 are consistently highly expressed in all 3 mouse models studied (Fig. 2D). Of note, the total RNA concentration in each sample did not predict a successful NanoString run, likely due to the fact that the majority of the RNA in the sample is host derived.
FIG 1.
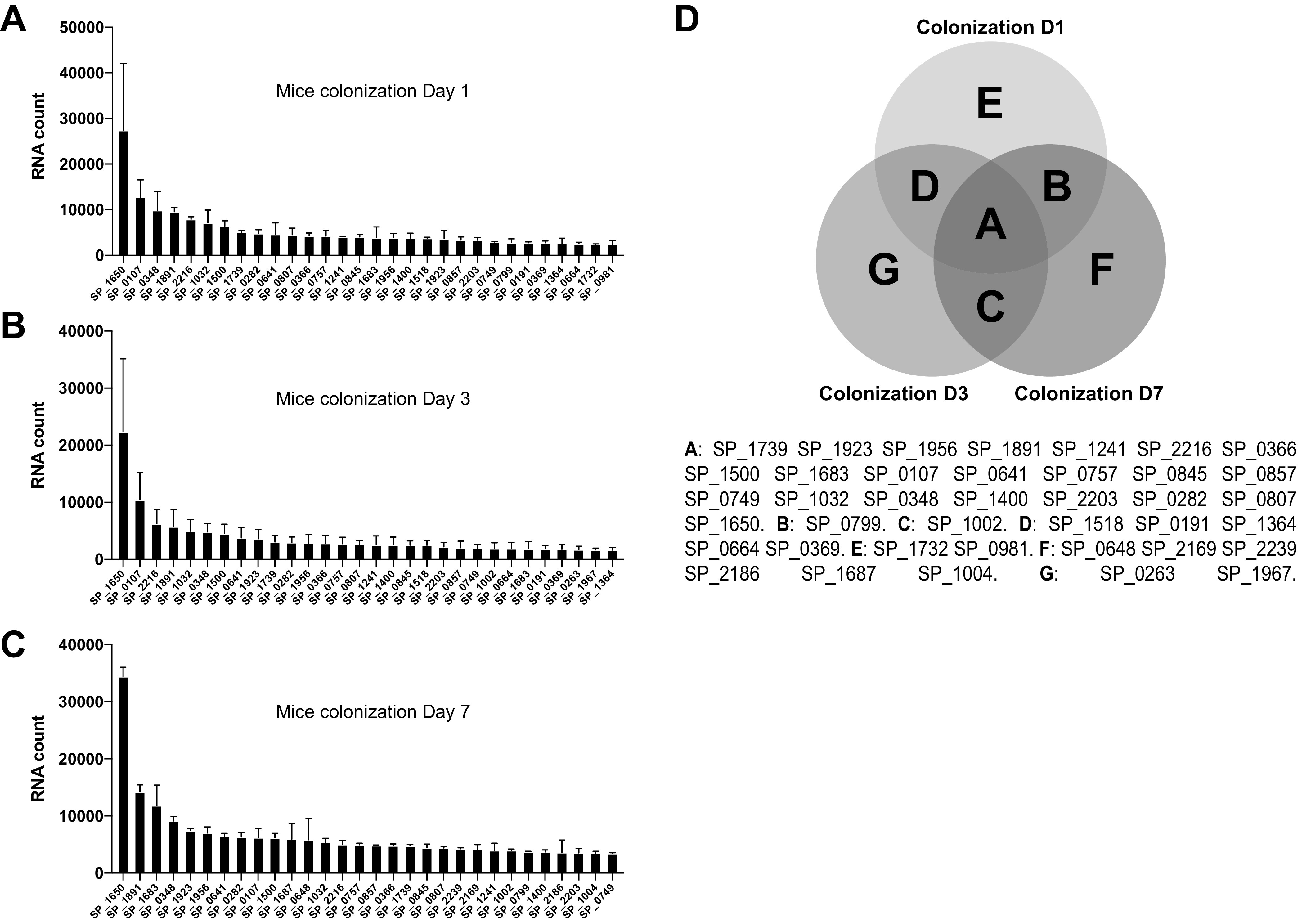
NanoString data representing highly expressed bacterial genes in a mouse colonization model. (A to C) The 30 most highly expressed pneumococcal genes from bacteria recovered on day 1 (A), day 3 (B), and day 7 (C) after initial colonization. (D) Venn diagram representing the most highly transcribed shared pneumococcal genes at all three time points. D1, D3, and D7 represent sample collection on days 1, 3, and 7, respectively.
FIG 2.
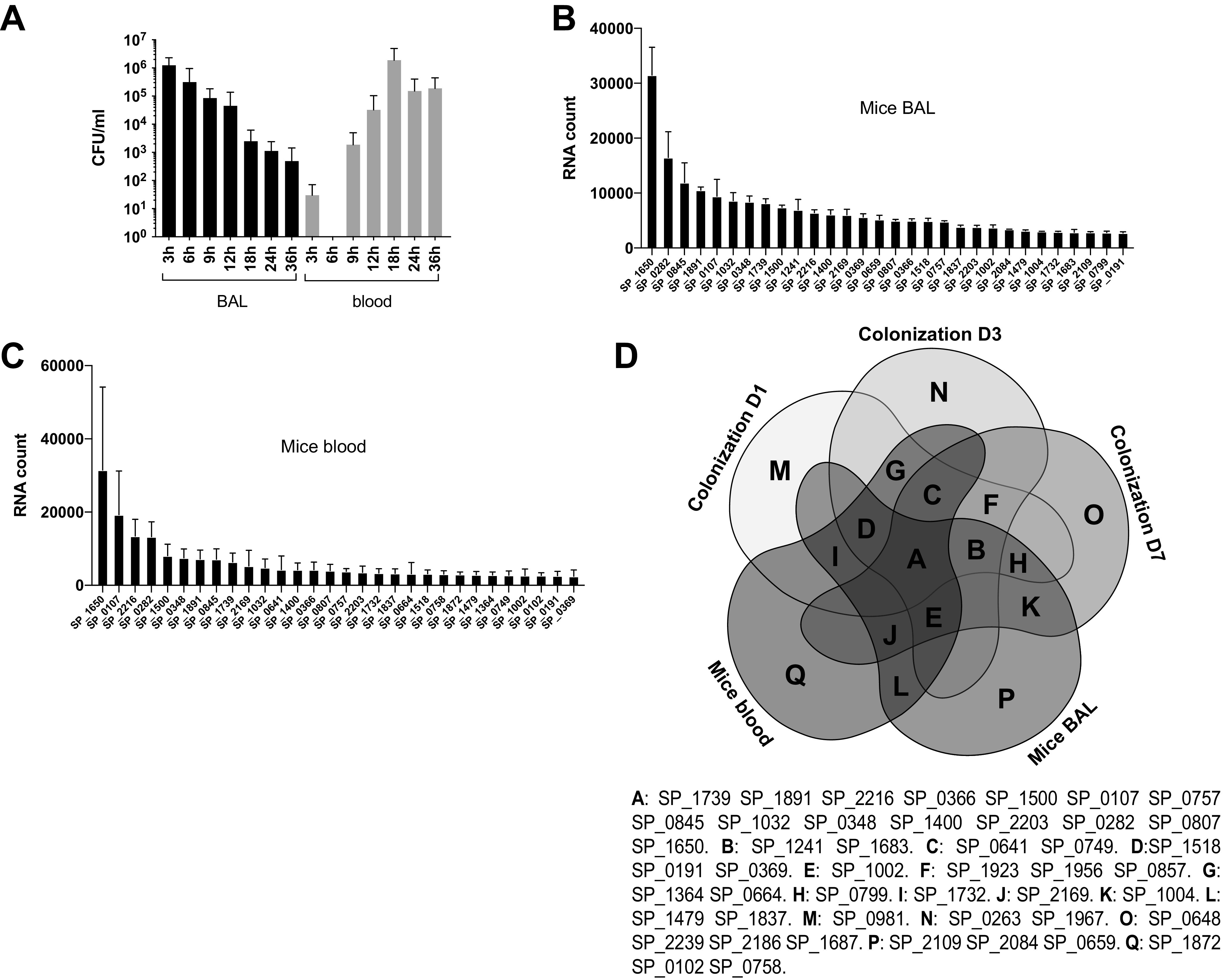
NanoString data representing the most highly expressed bacterial genes from bacteria recovered from the bronchoalveolar lavage (BAL) fluid in a mouse pneumococcal pneumonia or a septicemia model (blood). (A) Infected mice were euthanized at different time points postinoculation. BAL fluid and blood were collected from each mouse. CFU are reported for each condition tested. Each bar represents data from pools of 5 mice. (B) The 30 most highly expressed genes from pneumococci recovered from mouse BAL fluid at 9 h postinfection. (C) The 30 most highly expressed genes from pneumococci recovered from blood collected 2 days after lung infection. (D) Venn diagram representing the most highly transcribed shared pneumococcal genes under all five conditions (three colonization time points, blood, and BAL fluid). D1, D3, and D7 represent sample collection on days 1, 3, and 7, respectively.
RNA counts from clinical samples.
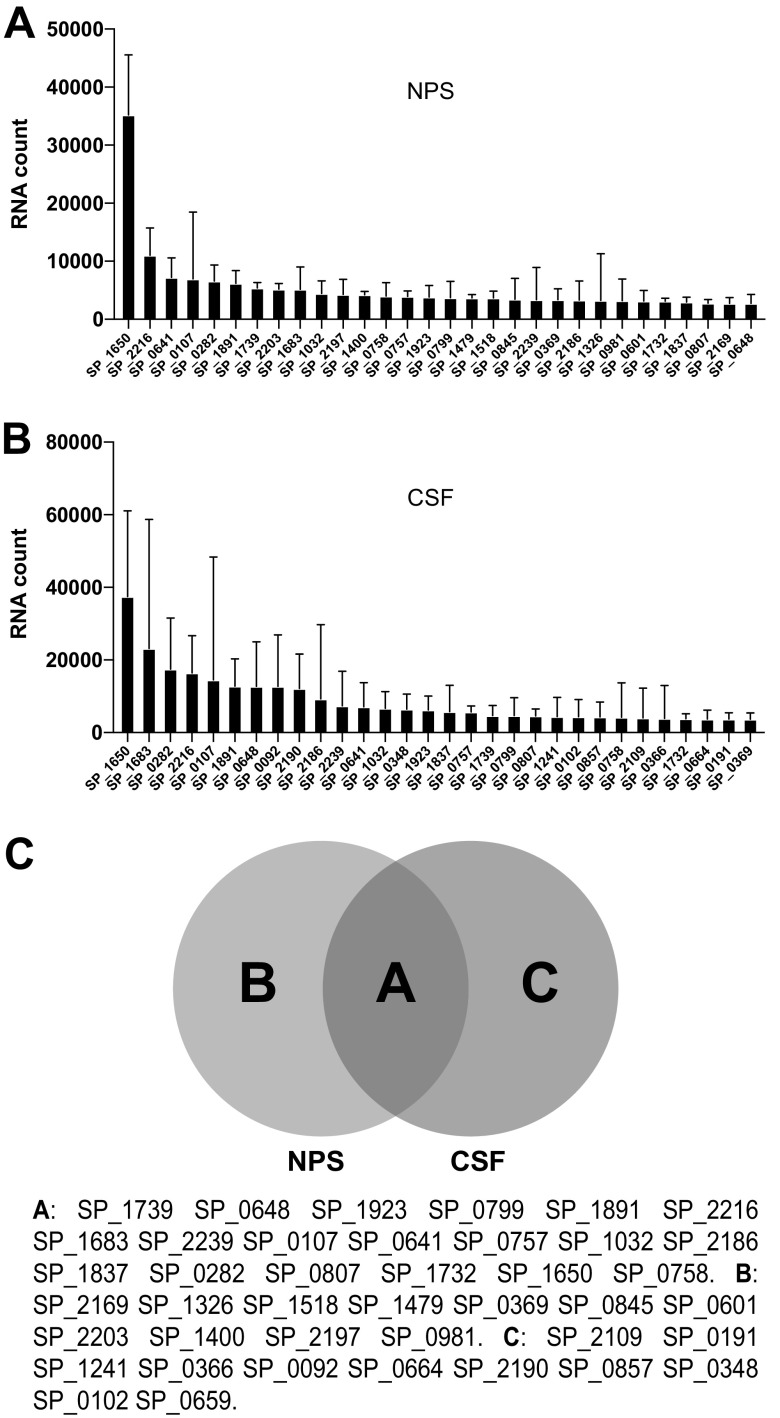
We were able to obtain successful NanoString reads when the total bacterial density from the nasal cultures of children was 105 CFU or higher. The list of the 30 most highly expressed genes in colonized pediatric samples (nasopharyngeal samples [NPSs]) was generated from a pool of 9 swabs, each from children who were naturally colonized with a pneumococcal strain. Some strains recovered were typeable (serotypes 15, 23, and 11), whereas two were nontypeable but reacted with antisera for group E and group I, respectively (Fig. 3A).
FIG 3.
NanoString data representing the most highly expressed bacterial genes during natural pediatric colonization (NPS) or human meningitis (CSF). (A) The 30 most highly expressed genes from pneumococci recovered from children naturally colonized with pneumococcal strains. (B) The 30 most highly expressed genes from pneumococci recovered from human meningitis cases. (C) Venn diagram representing the shared highly transcribed pneumococcal genes under the two conditions (NPS and CSF).
In a subanalysis, we also evaluated nasal curettage samples collected from the inferior turbinate prior to NPS collection. We were able to analyze 5 that we compared to their respective nasal cultures; the only gene that had a >3-fold difference in expression between the two methods of sampling was SP_0945, a lipoprotein. While limited by small numbers, these data suggest that the bacteria collected from NPSs versus more anterior nasal scrapings were likely in similar transcriptomic states.
We also analyzed the results of RNA expression from cerebrospinal fluid (CSF) samples. The list of the 30 most highly expressed genes in cerebrospinal fluid samples represents a pool of data collected from 16 patients, of whom 12 died from pneumococcal meningitis and the remaining patients survived (Fig. 3B). A Venn diagram analysis shows that bacterial genes SP_0107, SP_0282, SP_0641, SP_0648, SP_0757, SP_0758, SP_0799, SP_1032, SP_1650, SP_1683, SP_1732, SP_1739, SP_1837, SP_1891, SP_1923, SP_2136, SP_2186, SP_2216, and SP_2239 are consistently highly expressed in NPS and CSF samples.
Correlations.
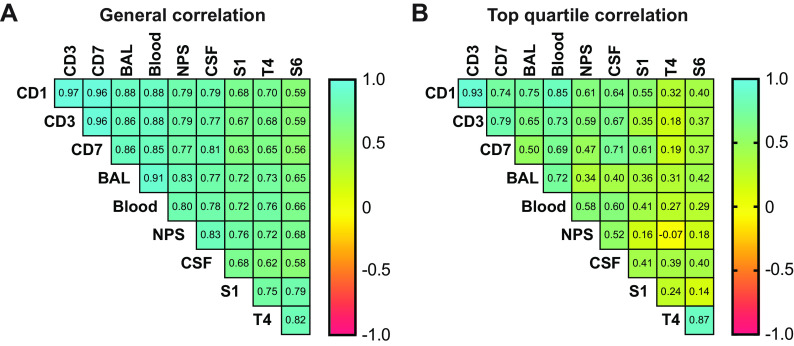
Spearman coefficient correlation (ρ) factors were determined between models and control cultures in Todd-Hewitt broth supplemented with yeast extract (THY) (Fig. 4). When the whole data sets were compared, there was generally an excellent correlation across most transcriptomic profiles, with the average correlation coefficient being 0.76 (with a range of 0.56 to 0.97). However, as we are mostly interested in identifying those genes that are highly expressed, we also performed an analysis where only the top quartile was evaluated, in which case subtle differences emerge across the different models. First, it is noteworthy that there is a poor correlation between samples obtained from in vitro cultures and any of the other samples, highlighting that the mRNA expression evaluated under culture conditions is not very representative of in vivo conditions and confirming the importance of evaluating RNA patterns without an in vitro growth step. In the mouse colonization model, a strong correlation (ρ > 0.7) is seen across days 1, 3, and 7; however, there is a slightly lower correlation over time in the mouse colonization model, potentially suggesting adaptation of the bacterial transcriptome to the mouse nasal mucosal environment. The same appears to be the case when colonization and BAL fluid or blood samples are compared: the correlation coefficient declines with subsequent time points in colonized mice. Samples obtained from the BAL fluid and blood of mice are strongly correlated (correlation coefficient [ρ] = 0.72).
FIG 4.
Evaluation of the correlation between mouse and human data. (A) Table indicating the Pearson correlation coefficients between mouse and human data. (B) Table indicating the Pearson correlation coefficients between the results obtained in mice and humans taking into consideration only the top quartile of the most highly expressed bacterial genes under each condition. CD1, CD3, and CD7, colonization days 1, 3, and 7, respectively; BAL, bronchoalveolar lavage fluid from mice in the pneumonia model; Blood, blood samples from mice in the septicemia model; NPS, natural pneumococcal sample, collected from naturally colonized children; CSF, cerebrospinal fluid sample from patients with pneumococcal meningitis; S1, T4, and S6, RNA samples of strains of serotype 1, TIGR4 (type 4), and 6B grown in Todd-Hewitt broth, respectively.
The correlation between pediatric and mouse colonization is more modest and also appears to decrease with the duration of colonization in the mouse. There is only a moderate correlation (0.52) of the top-quartile expression patterns across clinical NPS or CSF samples.
Evaluation of protein vaccine candidate gene expression in mouse and human samples.
Only 9 genes happened to be consistently highly expressed (top quartile) among the top 30 genes when all models were taken into consideration: SP_0107, SP_0282, SP_0757, SP_0807, SP_1032, SP_1650, SP_1739, SP_1891, and SP_2216 (Fig. 5A). We selected two protein candidates, SP_0282 and SP_1739, for further analysis, as their genes were among the 30 most highly expressed in each of the models studied; additionally, to our knowledge, neither of these antigens had been studied as a protective antigen previously. Both proteins showed strong expression by Western blotting using a collection of pneumococcal strains, including the ones that we used in subsequent animal studies (Fig. 5B). We immunized mice for protection studies and rabbits for serum generation. Following bacterial nasal challenge, those immunized with the positive-control whole-cell vaccine (WCV) (consisting of the killed whole-cell pneumococcal antigen and cholera toxin [CT] [21]) were almost completely protected compared to mice that received CT alone. Mice intranasally immunized with SP_0282 and CT showed a statistically significant, nearly 2-log decrease in bacterial CFU counts compared to those in mice that received CT alone (Fig. 5D), a result that is consistent with the strong interleukin-17 (IL-17) response induced against SP_0282 immunization (Fig. 5C). Immunization with SP_1739 and CT did not provide significant protection against colonization.
FIG 5.
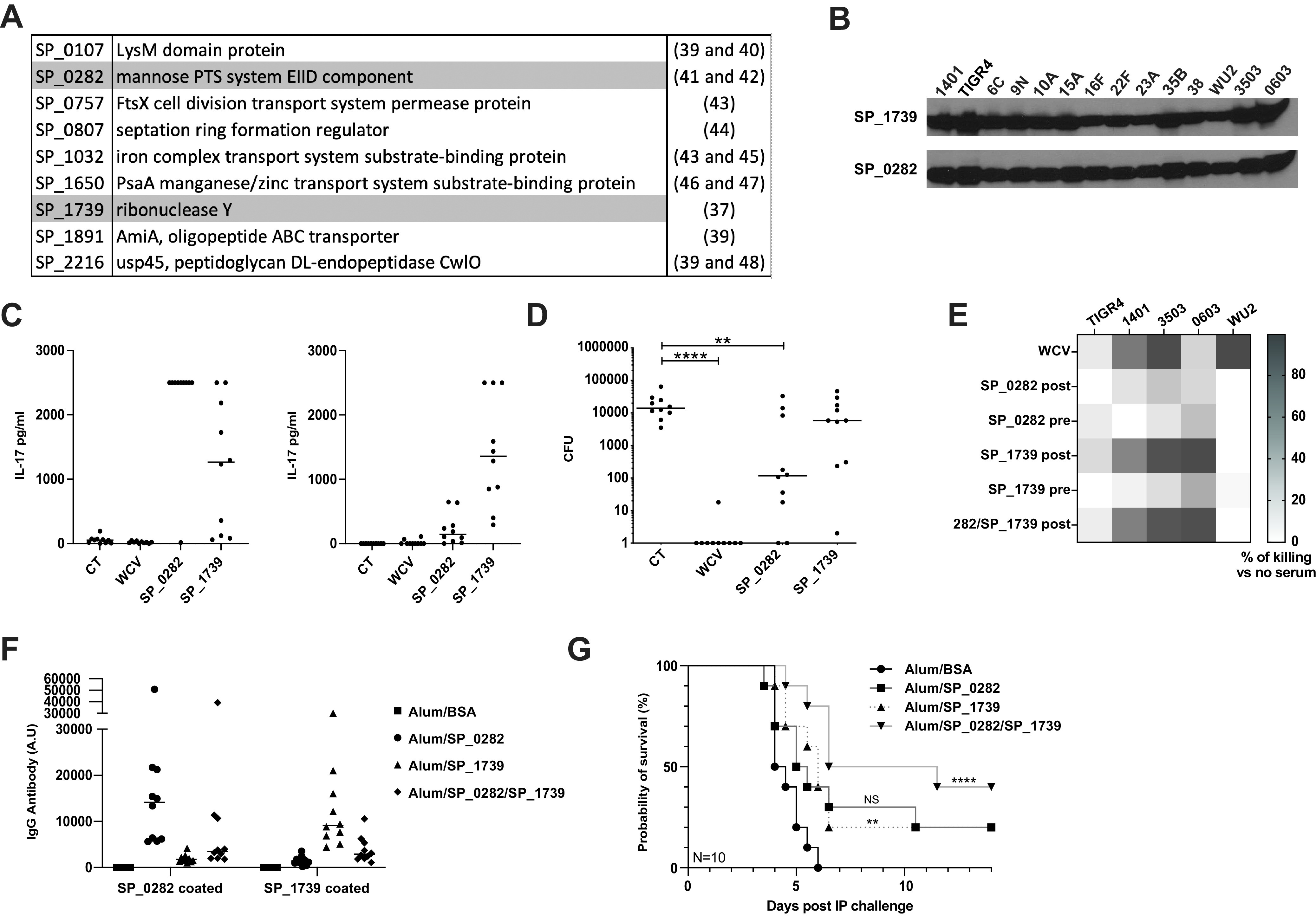
(A) List of the genes most highly expressed (top quartile) in all evaluated models (33). (B) Western blots targeting the SP_1739 and SP_0282 proteins. (C) IL-17 concentrations in the supernatants obtained from blood samples from immunized mice that were stimulated ex vivo with SP_0282 (left) or SP_1739 (right) protein. (D) Mice were immunized intranasally with SP_0282 and SP_1739 (10 μg of each protein and 1 μg of cholera toxin) twice (1 week apart) and challenged 4 weeks after the last immunization with a serotype 6B clinical strain. SP_0282 conferred statistically significant protection against nasal colonization. Plotted are the bacterial CFU recovered following retrograde nasopharyngeal washes. (E) Pneumococcal strains of serotypes 4, 14, 35, 0603, and 3 were used in a modified opsonophagocytosis assay (MOPA) with pre- and postimmunization rabbit serum targeting SP_0282 and SP_1739 antigens. Plotted are percentages of bacteria recovered following MOPAs in immune sera versus nonimmune sera. WCV is used as a positive control. (F) Specific protein antibody ELISA performed on sera collected from subcutaneously immunized mice. The ELISA plate was coated with SP_0282 protein (1 μg/mL) (left) or SP_1739 protein (1 μg/mL) (right). Sera were adsorbed with a nonspecific His-tagged protein purified in the same way as for SP_0282 and SP_1739. A.U, arbitrary units. (G) Mice were challenged with 103 CFU of the WU2AR strain by intraperitoneal (IP) challenge and monitored for 14 days for illness onset or death. Mice that received the mixture of proteins (SP_0282 and SP_1739) were significantly protected compared to the adjuvant-alone group (P = 0.0003 by a Gehan-Breslow-Wilcoxon test). Statistical analysis was performed using an unpaired nonparametric Mann-Whitney test. NS, not significant; *, P < 0.05; ****, P < 0.0001. BSA, bovine serum albumin.
Multiplex opsonophagocytosis assays (MOPAs) were then performed using rabbit pre- and postimmunization sera for SP_0282 and SP_1739 proteins, using strains of pneumococcal serotype 4, 6B, 14, 35 and 3 (Fig. 5E). The serum from SP_1739-immunized rabbits killed pneumococci of most serotypes, although the effect on the serotype 4 strain (TIGR4) was more modest. No effect was observed against the serotype 3 strain (WU2) for any of the antisera under the conditions tested.
Mice were then subcutaneously immunized with either or both proteins. Specific antibody responses were observed against both single proteins by an enzyme-linked immunosorbent assay (ELISA) (Fig. 5F). Of interest, a reduced antibody response to each protein was noted when a mixture of both proteins was used as the immunogen; despite this, mice immunized with both antigens (SP_0282 and SP_1739) showed statistically significant protection against pneumococcal type 3 intraperitoneal (i.p.) challenge, which was not noted with either antigen alone.
DISCUSSION
Despite decades of research and highly supportive preclinical data, the development of a universal, protein-based pneumococcal vaccine has yet to produce a viable and effective vaccine candidate (15). There are, of course, many possible reasons for this disappointing reality. Certainly, the success of conjugate vaccines and the gradual reduction in the cost of pneumococcal vaccines may have dampened the enthusiasm for the search for alternatives. To complicate matters further, when candidates were tested for evidence of a clinical impact in a pediatric trial, the results were largely disappointing, with no evidence of efficacy (22, 23).
A potential explanation for these failures may be that the selection of the antigen candidates has been dictated mostly by preclinical studies performed in mouse models. There are many reasons why mouse models may not be reflective of human disease; here, we evaluate whether there are differences in protein antigen expression between mouse and human infection, which may explain these results.
There have been several studies of the pneumococcal transcriptome with bacteria grown under different in vitro conditions (24–30). To our knowledge, ours is the first direct study of the pneumococcal transcriptome in human colonization and meningitis. The main technical challenge in these studies is the relatively small amount of RNA and the need to avoid growing the organism in vitro, which will distort the transcriptome, as shown here. The NanoString platform, which we have recently used for the evaluation of the Staphylococcus aureus transcriptome under different conditions (31), allows the quantitation of RNA transcripts from relatively low concentrations of RNA. This platform allowed us to evaluate the transcriptomic profiles of 160 genes, selected for their (presumed) surface location and conservation across the pneumococcal genome. Overall, we were able to obtain 75% successful reads for nasal sampling and 52% with CSF samples from human subjects.
In this study, we compared the RNA expression profiles in mouse and clinical samples. When we focused on the top quartile of expressed genes, we found that mouse in vivo samples were not highly correlated with samples grown in vitro, confirming the need to avoid a step in which isolates obtained from clinical specimens are grown in media prior to RNA harvest. Additionally, we found that in general, samples obtained from mouse colonization, BAL fluid, and blood specimens were strongly correlated. This correlation is higher for day 1 than for day 7 data, suggesting bacterial adaptation to different environments over time. At the same time, due to our need to collect BAL fluid relatively early postinfection (to avoid bacterial contamination coming from the blood), we cannot determine the transcriptomic profile of pneumococci in the lungs over time. It is also notable that the invasive models in mice did not correlate as well with CSF data obtained from humans with pneumococcal meningitis.
Perhaps most importantly, there was little correlation between mouse and human clinical samples. In particular, the genes that were highly expressed in the invasive mouse models did not correlate well with human CSF data, raising questions with respect to the mouse model to screen for potential protein vaccine candidates. For vaccine development, an optimal goal would be to identify a gene that is highly expressed in isolates obtained from relevant human samples and is also relevant in an animal model such that protection by a vaccine consisting of the gene product can be tested. For this reason, we chose to focus our studies on candidates that demonstrated strong expression in both mouse and human infection.
Using the results of our study, we generated a list of candidate genes that are highly expressed across these mouse models and human samples, which will be tested in our mouse models of immunization and opsonophagocytosis assays in the future. In the present study, we focused on the SP_0282 and SP_1739 proteins. SP_0282 is part of a PTS, which was shown to have increased expression after contact with THP-1 cells, and is a virulence factor in pneumococcal meningitis (30, 32). SP_1739 is a protein of hypothetical function that may play a role in biofilm formation (33) and appears to be a target of human T cell responses (34). The SP_0282 protein was able to confer immune protection against pneumococcal intranasal colonization in mice, and the combination of both antigens was able to confer protection against intraperitoneal challenge with a type 3 strain in mice. SP_1739 rabbit antiserum was also able to induce strong opsonophagocytic killing capacities against three different serotypes (serotypes 14, 35, and 0603) and moderate killing against TIGR4. Despite the absence of opsonophagocytosis against WU2, it is interesting that mice immunized with the combination of both proteins had significant protection against WU2AR strain i.p. challenge. The discrepancy in these results could be due to the difference in the states of the transcriptomes and/or specifically capsule production between bacteria grown in media for MOPAs and bacteria during mouse challenge.
In conclusion, we believe that our work represents a potential new strategy to identify pneumococcal antigens with relatively high expression levels under different conditions, which may better predict their efficacy as vaccine antigens. Our results with SP_0282 and SP_1739 suggest that this type of screening may indeed reveal additional antigens worthy of consideration in a broad, serotype-independent, protein-based vaccine.
MATERIALS AND METHODS
Bacterial strains.
Strains TIGR4 (serotype 4); 0603 (serotype 6B); 1401 (serotype 14); 6C, 9N, 10A, 15A, 16F, 22F, 23A, 35B, 38, and WU2 (serotype 3); and 3503 (serotype 35) were used for in vitro and in vivo experiments. For invasive disease experiments in mice, we used strain WU2AR, a mutant strain of WU2 in which a histidine tag sequence is integrated into the pneumolysin gene; this strain is highly virulent in mice but with a delay in the time to illness compared to the wild-type WU2 strain. For challenge studies, the strains were chosen based on their ability to cause invasive disease (TIGR4 and WU2AR) or colonization (0603) in mice. Clinical isolates were obtained from individuals in Blantyre, Malawi, who presented with pneumococcal meningitis and from children from Liverpool, United Kingdom, in whom pneumococcal colonization had been detected. Pneumococci were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) or on tryptic soy agar (TSA) plates with 5% sheep’s blood. Escherichia coli strains were grown in LB medium supplemented with ampicillin at 100 μg/mL as needed. A killed pneumococcal whole-cell vaccine (WCV) was used as a positive control for modified opsonophagocytic killing assays and mouse immunization, as described previously (35).
Animal models.
Four- to six-week-old female C57BL/6J mice from Jackson Laboratories (Bar Harbor, ME) were used in animal experiments.
Mouse immunization. (i) Intranasal immunization.
Four- to six-week-old female C57BL/6J mice were immunized twice, 1 week apart, with 10 μg of protein and 1 μg of cholera toxin (CT) as an adjuvant. At 3 weeks postimmunization, retro-orbital blood draws were performed to obtain whole blood for stimulation and IL-17A ELISAs. At 4 weeks postimmunization, mice were intranasally inoculated with 107 CFU of a pneumococcal strain of serotype 6 (0603). Finally, at 5 weeks postimmunization, mice were euthanized, and nasopharyngeal lavage fluid samples through the trachea were obtained to evaluate the remaining CFU counts. As a positive control, mice were immunized with a killed WCV adjuvanted with cholera toxin as previously described (21).
(ii) Subcutaneous immunizations.
Four- to six-week-old female C57BL/6J mice were immunized subcutaneously three times 2 weeks apart with 200 μL containing alum with the protein antigen (10 μg of each protein/mouse). Two weeks after the last immunization, blood was obtained for measurement of serum antibody titers by an ELISA.
Challenge models. (i) Nasopharyngeal colonization.
A total of 105 CFU of live pneumococci was inoculated onto the nares of gently restrained, unanesthetized mice. On day 1, 3, or 7 postinoculation, animals were humanely euthanized, and nasopharyngeal secretions were recovered by retrograde instillation of Tri reagent (MRC) in the transected trachea. Pools of 10 nasal secretion samples were assembled before RNA extraction.
(ii) Intraperitoneal challenge.
A total of 103 CFU of live WU2AR was injected intraperitoneally. Mice were monitored for survival twice daily for 14 days; any ill-appearing animal was humanely euthanized.
Isolation of bacteria from lungs.
Mice were lightly anesthetized with isoflurane and then nasally inoculated with 50 μL of phosphate-buffered saline (PBS) containing 107 CFU of pneumococci. Animals were humanely euthanized at 9 h postinfection. Bronchoalveolar lavages (BALs) were performed using a 1:1 mix of RNA Protect bacteria (Qiagen) and 1× PBS before RNA extraction. Pools of 10 BAL fluid samples were assembled before RNA extraction.
Isolation of bacteria from blood.
Mice that were infected as described above were killed at 2 days postinfection. Heart punctures were performed for blood collection. Blood was immediately mixed with Tri reagent before RNA collection. All animal work was performed in accordance with NIH guidelines and approved by the IACUC at Boston Children’s Hospital.
Isolation of bacteria from colonized children.
Nasopharyngeal swab samples from a previously described pediatric cohort (36) were analyzed for bacterial RNA expression. This study was approved by the National Health Service Research and Ethics Committee (17/NW/0663). Informed consent from a parent was obtained, and the study complied with relevant regulatory standards (Human Tissue Act, 2004). In short, samples were collected from hospitalized children aged 1 to 5 years, directly after the onset of general anesthesia but prior to the start of their planned procedure (e.g., dental extraction or plastic surgery). For the detection of pneumococcal colonization, a nasopharyngeal swab was collected in medium containing skim milk, tryptone, glucose, and glycerin (STGG), and classical culture was performed to assess colonization. A second nasopharyngeal swab was collected in 1 mL RNA Protect (Qiagen) and frozen at −80°C. Nasal curettage samples were also collected from the inferior turbinate using two Rhino-Pro curettes (Arlington Scientific) and collected in 0.5 mL RNA Protect (Qiagen). RNA was extracted using the Split RNA extraction kit (Lexogen), according to the manufacturer’s protocol. RNA quantity and quality were analyzed using the Qubit RNA high sensitivity (HS) assay kit (Thermo Fisher) and a Nanodrop spectrometer (Thermo Fisher). RNA was frozen at −80°C until analysis.
RNA extraction on mouse samples and human CSF samples.
Bacteria were centrifuged and rinsed once in sterile PBS before resuspension in Tri reagent buffer. Bacteria were then lysed 5 times for 30 s using a mini-bead beater from Biospec with acid-washed glass beads of ≤106 μm. RNA was extracted using a Tri reagent-chloroform protocol followed by an ethanol precipitation step. The RNA concentration was quantified using a Thermo Scientific Nanodrop 1000 instrument. Patients presenting to the Queen Elizabeth Central Hospital in Blantyre, Malawi, with bacterial meningitis caused by S. pneumoniae between 2011 and 2013 were included (Current Controlled Trials registration number ISRCTN96218197) (37). RNA was extracted from blood and CSF using the PAXgene blood miRNA (microRNA) kit (Pre-Analytix, Qiagen, USA) according to the manufacturer’s instructions, with an additional mechanical disruption step for the CSF samples to disrupt the pneumococcal cell wall at 6,200 rpm for 45 s in the Precellys evolution tissue homogenizer (Bertin Instruments). The extracted RNA was quantified, and the RNA integrity number (RIN) scores were calculated using the RNA Tapestation 4200 (Agilent, USA) and Nanodrop (Thermo Scientific, USA) systems.
Generation of the pneumococcal code set.
Two hundred four unique molecular fluorescent barcodes linked to reporter tags were used in this study. The JGI website (https://img.jgi.doe.gov/) was used to generate a NanoString code set specifically including surface-exposed proteins. We selected genes encoding secreted proteins, surface-attached proteins, or transmembrane proteins with presumptive extracellular domains (to maximize the chance of accessibility to antibodies). Only one gene per operon was selected to avoid crowding the code set. Code set primers were designed based on the TIGR4 genome sequence targeting the most conserved regions of the genes among pneumococcal strains. In total, 4 genes coding for choline-binding proteins, 40 genes coding for proteins with a signal peptide, 91 genes coding for transmembrane proteins, and 25 genes coding for transmembrane proteins with a signal peptide were included in the code set. Primers for these 160 pneumococcal genes were designed using the TIGR4 reference genome sequence (at https://img.jgi.doe.gov/). For the design of the primers, areas of homology with human and murine genes were avoided using in silico analysis. The Ncounter XT assay protocol provided by NanoString Technologies was performed to analyze each RNA sample. RNA was hybridized for 24 h at 65°C with primer sets and NanoString Elements reagents (30-μL total volume for each hybridization). Hybridized samples were washed and immobilized onto cartridges (NanoString prep station) that were analyzed (Digital Analyzer) for direct counting of the fluorescent molecular barcodes unique to each primer that attached to the hybridized transcript within each flow cell.
Protein purification.
Genes of interest were cloned into the pET21b vector for expression. Bacteria were grown to an optical density at 600 nm of 0.6, and protein expression was induced with 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 16°C. Bacteria were harvested and then suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 1× Halt protease inhibitor cocktail [Thermo Scientific], 0.1% lysozyme, 10 mM MgCl2, silicone, and DNase) before being sonicated six times for 30 s each. Samples were centrifuged for 30 min at 14,000 rpm in the cold, and the supernatants were collected. Five hundred microliters of Ni-nitrilotriacetic acid (NTA) was added to the samples, and the mixture was incubated for 1 h at 4°C. Samples were loaded onto a column, washed twice with buffer (50 mM NaH2PO4, 300 mM NaCl, 30 mM imidazole) at pH 8.0, and then eluted in buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole) at pH 8.0. Proteins were further purified through a Superdex S200 column using an Äkta Pure 25 system. Protein purity was analyzed on a NuPage 4 to 12% Bis-Tris gel (Invitrogen), and protein concentrations were quantified using the bicinchoninic acid (BCA) protein assay (Pierce).
Generation and testing of polyclonal antibodies.
Purified recombinant His6-tagged SP_0282 and SP_1739 were used to immunize New Zealand White rabbits three times at 2-week intervals using alum as an adjuvant (Cocalico Biologicals, Inc.). Blood samples were obtained at least 2 weeks following the third immunization.
IL-17A ELISA.
Retro-orbital blood draws were performed on anesthetized immunized mice using collection tubes containing lithium heparin (BD Microtainer). For IL-17 ELISAs, 25 μL of blood was incubated for 5 days at 37°C with 225 μL of Dulbecco’s modified Eagle’s medium (DMEM)–F-12 medium with 10% fetal bovine serum (FBS), containing 11 μg/mL of the specific pneumococcal antigen. The supernatants were then collected by centrifugation, and an IL-17A ELISA was performed according to the instructions provided by the manufacturer (Thermo Fisher).
Protein-specific ELISA for measurement of antibodies in serum.
Plates were coated overnight with the protein of interest (1 μg/mL), and heat-inactivated sera were serially diluted. An irrelevant His-tagged protein, purified according to the same protocol as the one used for SP_1739 and SP_0282, was used to adsorb mouse sera at 100 μg/mL, to decrease nonspecific signals.
Multiplex opsonophagocytosis assay.
We used a slightly modified multiplex opsonophagocytosis assay (MOPA) as described previously by Bogaert et al. (38). Briefly, specific sera were heat inactivated at 56°C for 30 min and diluted 4 times in Hanks' balanced salt solution [HBSS] and 10% fetal bovine serum [FBS]. Two thousand CFU were added per well to diluted sera, and the mixture as incubated for 30 min at room temperature under agitation. Next, 40 μL of differentiated HL-60 corresponding to 4 × 105 cells and 10 μL of baby rabbit complement (Pel-freeze) were added, and the mixture was incubated for 1 h at 37°C under agitation. After incubation, CFU were determined by serial dilutions on TSA plates.
Statistical analysis.
Data were normalized using n Solver software analysis 3.0, against data gathered for 3 housekeeping genes (SP_0668, SP_0806, and SP_1243), either by using the geometric mean or by dividing the result for each sample by the respective median for each sample. The top quartiles for each model were compared using both normalization methods, and the ρ factors obtained using the Spearman rank correlation method are listed in Fig. 4. ρ factor values were very high and showed no appreciable difference between both procedures for data normalization (Fig. 4). Ultimately, normalization against the median for each sample was chosen. Bacterial densities from nasopharyngeal wash specimens or pulmonary cultures were compared using a nonparametric Mann-Whitney U test, using Prism (version 4.0a; GraphPad Software, Inc.). Correlations were calculated using a nonparametric Spearman two-tailed test, using Prism (version 4.0a; GraphPad Software, Inc.).
All clinical studies were approved by the relevant Institutional Review Boards. Animal studies were approved by the Animal Care and Use Committee of Boston Children’s Hospital.
ACKNOWLEDGMENTS
R.M. gratefully acknowledges support from the Translational Research Program and discretionary funding from Boston Children’s Hospital. E.W. is supported by a postdoctoral clinical research fellowship from the Francis Crick Institute and the University College London Hospitals Department of Health’s National Institute for Health Research (NIHR) Biomedical Research Centre. The acute meningitis clinical trial was funded by the Wellcome Trust. This work was also supported by the Liverpool School of Tropical Medicine Director Catalyst Fund, which was funded by Wellcome Trust Institutional Strategic Support Fund 3 (204806/Z/16/Z), and Liverpool School of Tropical Medicine internal funding (awarded to S.P.J.). It was in part carried out at the NIHR Alder Hey Clinical Research Facility. This work was also supported by, and we acknowledge, the support of the National Institute for Health Research Clinical Research Network as well as the Human Infection Challenge Network for Vaccine Development (HIC-Vac) funded by the GCRF Networks in Vaccines Research and Development, which was cofunded by the MRC and the Biotechnology and Biological Sciences Research Council (BBSRC), in a grant awarded to Thushan de Silva, Ryan Thwaites, and S.P.J. R.H. is an NIHR Senior Investigator. The views expressed in this article are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care, United Kingdom.
Contributor Information
Richard Malley, Email: richard.malley@childrens.harvard.edu.
Liise-anne Pirofski, Albert Einstein College of Medicine.
REFERENCES
- 1.WHO. 2019. Pneumococcal conjugate vaccines in infants and children under 5 years of age: WHO position paper. Wkly Epidemiol Rec 8:177–183. [Google Scholar]
- 2.Marimon JM, Ardanuy C. 2021. Epidemiology of pneumococcal diseases in Spain after the introduction of pneumococcal conjugate vaccines. Enferm Infecc Microbiol Clin (Engl Ed) 39:142–150. 10.1016/j.eimc.2020.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi N, Naito S, Ohkusu M, Abe K, Shizuno K, Takahashi Y, Omata Y, Nakazawa T, Takeshita K, Hishiki H, Hoshino T, Sato Y, Ishiwada N. 2020. Epidemiology of hospitalised paediatric community-acquired pneumonia and bacterial pneumonia following the introduction of 13-valent pneumococcal conjugate vaccine in the national immunisation programme in Japan. Epidemiol Infect 148:e91. 10.1017/S0950268820000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koelman DLH, van Kassel MN, Bijlsma MW, Brouwer MC, van de Beek D, van der Ende A. 2021. Changing epidemiology of bacterial meningitis since introduction of conjugate vaccines: 3 decades of national meningitis surveillance in The Netherlands. Clin Infect Dis 73:e1099–e1107. 10.1093/cid/ciaa1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnstone SL, Moore DP, Klugman KP, Madhi SA, Groome MJ. 2020. Epidemiology of invasive bacterial infections in pneumococcal conjugate vaccine-vaccinated and -unvaccinated children under 5 years of age in Soweto, South Africa: a cohort study from a high-HIV burden setting. Paediatr Int Child Health 40:50–57. 10.1080/20469047.2019.1623572. [DOI] [PubMed] [Google Scholar]
- 6.Hernstadt H, Cheung A, Hurem D, Vasilunas N, Phuong LK, Quinn P, Agrawal R, Daley AJ, Cole T, Gwee A. 2020. Changing epidemiology and predisposing factors for invasive pneumococcal disease at two Australian tertiary hospitals. Pediatr Infect Dis J 39:1–6. 10.1097/INF.0000000000002489. [DOI] [PubMed] [Google Scholar]
- 7.Heo JY, Seo YB, Jeong HW, Choi MJ, Min KH, Choi WS, Lee J, Noh JY, Cheong HJ, Kim WJ, Song JY. 2020. Epidemiology of community-acquired pneumonia in the era of extended serotype-covering multivalent pneumococcal conjugate vaccines. Vaccine 38:7747–7755. 10.1016/j.vaccine.2020.10.046. [DOI] [PubMed] [Google Scholar]
- 8.Weinberger DM, Malley R, Lipsitch M. 2011. Serotype replacement in disease after pneumococcal vaccination. Lancet 378:1962–1973. 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurley D, Griffin C, Young M, Scott DA, Pride MW, Scully IL, Ginis J, Severs J, Jansen KU, Gruber WC, Watson W. 2021. Safety, tolerability, and immunogenicity of a 20-valent pneumococcal conjugate vaccine (PCV20) in adults 60 to 64 years of age. Clin Infect Dis 73:e1489–e1497. 10.1093/cid/ciaa1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson A, Lamberth E, Severs J, Scully I, Tarabar S, Ginis J, Jansen KU, Gruber WC, Scott DA, Watson W. 2019. Phase 1 trial of a 20-valent pneumococcal conjugate vaccine in healthy adults. Vaccine 37:6201–6207. 10.1016/j.vaccine.2019.08.048. [DOI] [PubMed] [Google Scholar]
- 11.Stacey HL, Rosen J, Peterson JT, Williams-Diaz A, Gakhar V, Sterling TM, Acosta CJ, Nolan KM, Li J, Pedley A, Benner P, Abeygunawardana C, Kosinski M, Smith WJ, Pujar H, Musey LK. 2019. Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine (PCV-15) compared to PCV-13 in healthy older adults. Hum Vaccin Immunother 15:530–539. 10.1080/21645515.2018.1532249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg D, Hoover PA, Vesikari T, Peltier C, Hurley DC, McFetridge RD, Dallas M, Hartzel J, Marchese RD, Coller BG, Stek JE, Abeygunawardana C, Winters MA, MacNair JE, Pujar NS, Musey L. 2018. Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine (PCV15) in healthy infants. Vaccine 36:6883–6891. 10.1016/j.vaccine.2018.02.113. [DOI] [PubMed] [Google Scholar]
- 13.Vyse A, Campling J, Czudek C, Ellsbury G, Slack M. 2020. The proportion of contemporary invasive pneumococcal disease and pneumococcal pneumonia in UK adults reflected by serotypes included in the 13-valent pneumococcal conjugate vaccine and next generation higher valency pneumococcal conjugate vaccines in development. Vaccine 38:8068–8070. 10.1016/j.vaccine.2020.10.090. [DOI] [PubMed] [Google Scholar]
- 14.Lapidot R, Shea KM, Yildirim I, Cabral HJ, Pelton SI, Massachusetts Department of Public Health . 2020. Characteristics of serotype 3 invasive pneumococcal disease before and after universal childhood immunization with PCV13 in Massachusetts. Pathogens 9:396. 10.3390/pathogens9050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichichero ME. 2017. Pneumococcal whole-cell and protein-based vaccines: changing the paradigm. Expert Rev Vaccines 16:1181–1190. 10.1080/14760584.2017.1393335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malley R, Anderson PW. 2012. Serotype-independent pneumococcal experimental vaccines that induce cellular as well as humoral immunity. Proc Natl Acad Sci USA 109:3623–3627. 10.1073/pnas.1121383109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffitt KL, Malley R. 2011. Next generation pneumococcal vaccines. Curr Opin Immunol 23:407–413. 10.1016/j.coi.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, Gullane P, Irish J, Jurisica I, Kamel-Reid S. 2011. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol 11:46. 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325. 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 20.Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. 2016. The impact of amplification on differential expression analyses by RNA-seq. Sci Rep 6:25533. 10.1038/srep25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D, Anderson P. 2001. Intranasal immunization with killed unencapsulated whole cells prevents colonization and invasive disease by capsulated pneumococci. Infect Immun 69:4870–4873. 10.1128/IAI.69.8.4870-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammitt LL, Campbell JC, Borys D, Weatherholtz RC, Reid R, Goklish N, Moulton LH, Traskine M, Song Y, Swinnen K, Santosham M, O’Brien KL. 2019. Efficacy, safety and immunogenicity of a pneumococcal protein-based vaccine co-administered with 13-valent pneumococcal conjugate vaccine against acute otitis media in young children: a phase IIb randomized study. Vaccine 37:7482–7492. 10.1016/j.vaccine.2019.09.076. [DOI] [PubMed] [Google Scholar]
- 23.Odutola A, Ota MOC, Antonio M, Ogundare EO, Saidu Y, Foster-Nyarko E, Owiafe PK, Ceesay F, Worwui A, Idoko OT, Owolabi O, Bojang A, Jarju S, Drammeh I, Kampmann B, Greenwood BM, Alderson M, Traskine M, Devos N, Schoonbroodt S, Swinnen K, Verlant V, Dobbelaere K, Borys D. 2017. Efficacy of a novel, protein-based pneumococcal vaccine against nasopharyngeal carriage of Streptococcus pneumoniae in infants: a phase 2, randomized, controlled, observer-blind study. Vaccine 35:2531–2542. 10.1016/j.vaccine.2017.03.071. [DOI] [PubMed] [Google Scholar]
- 24.Aprianto R, Slager J, Holsappel S, Veening JW. 2016. Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol 17:198. 10.1186/s13059-016-1054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aprianto R, Slager J, Holsappel S, Veening J-W. 2018. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res 46:9990–10006. 10.1093/nar/gky750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhter F, Womack E, Vidal JE, Le Breton Y, McIver KS, Pawar S, Eichenbaum Z. 2020. Hemoglobin stimulates vigorous growth of Streptococcus pneumoniae and shapes the pathogen’s global transcriptome. Sci Rep 10:15202. 10.1038/s41598-020-71910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afzal M, Shafeeq S. 2017. Impact of aspirin on the transcriptome of Streptococcus pneumoniae D39. Genom Data 12:38–40. 10.1016/j.gdata.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamya MF, Ayoola MB, Shack LA, Swiatlo E, Nanduri B. 2021. The effect of impaired polyamine transport on pneumococcal transcriptome. Pathogens 10:1322. 10.3390/pathogens10101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Mello A, Riegler AN, Martinez E, Beno SM, Ricketts TD, Foxman EF, Orihuela CJ, Tettelin H. 2020. An in vivo atlas of host-pathogen transcriptomes during Streptococcus pneumoniae colonization and disease. Proc Natl Acad Sci USA 117:33507–33518. 10.1073/pnas.2010428117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song XM, Connor W, Hokamp K, Babiuk LA, Potter AA. 2009. Transcriptome studies on Streptococcus pneumoniae, illustration of early response genes to THP-1 human macrophages. Genomics 93:72–82. 10.1016/j.ygeno.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Moffitt K, Cheung E, Yeung T, Stamoulis C, Malley R. 2021. Analysis of Staphylococcus aureus transcriptome in pediatric soft tissue abscesses and comparison to murine infections. Infect Immun 89:e00715-20. 10.1128/IAI.00715-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molzen TE, Burghout P, Bootsma HJ, Brandt CT, van der Gaast-de Jongh CE, Eleveld MJ, Verbeek MM, Frimodt-Moller N, Ostergaard C, Hermans PW. 2011. Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. Infect Immun 79:288–297. 10.1128/IAI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muñoz-Elías EJ, Marcano J, Camilli A. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun 76:5049–5061. 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Gierahn T, Thompson CM, Trzciński K, Ford CB, Croucher N, Gouveia P, Flechtner JB, Malley R, Lipsitch M. 2012. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog 8:e1002989. 10.1371/journal.ppat.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y-J, Leite L, Gonçalves VM, Dias WDO, Liberman C, Fratelli F, Alderson M, Tate A, Maisonneuve J-F, Robertson G, Graca R, Sayeed S, Thompson CM, Anderson P, Malley R. 2010. GMP-grade pneumococcal whole-cell vaccine injected subcutaneously protects mice from nasopharyngeal colonization and fatal aspiration-sepsis. Vaccine 28:7468–7475. 10.1016/j.vaccine.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolaou E, Blizard A, Pojar S, Mitsi E, German EL, Reine J, Hill H, McNamara PS, Collins AM, Ferreira DM, Jochems SP. 2019. Minimally invasive nasal sampling in children offers accurate pneumococcal colonization detection. Pediatr Infect Dis J 38:1147–1149. 10.1097/INF.0000000000002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wall EC, Brownridge P, Laing G, Terra VS, Mlozowa V, Denis B, Nyirenda M, Allain T, Ramos-Sevillano E, Carrol E, Collins A, Gordon SB, Lalloo DG, Wren B, Beynon R, Heyderman RS, Brown JS. 2020. CSF levels of elongation factor Tu is associated with increased mortality in Malawian adults with Streptococcus pneumoniae meningitis. Front Cell Infect Microbiol 10:603623. 10.3389/fcimb.2020.603623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogaert D, Sluijter M, De Groot R, Hermans PW. 2004. Multiplex opsonophagocytosis assay (MOPA): a useful tool for the monitoring of the 7-valent pneumococcal conjugate vaccine. Vaccine 22:4014–4020. 10.1016/j.vaccine.2004.03.049. [DOI] [PubMed] [Google Scholar]