ABSTRACT
This report describes 28 genome sequences from a new clade within subtype 1 of Betaarterivirus suid 1, formerly known as porcine reproductive and respiratory syndrome virus 1. All share a potential recombinant pattern, with a highly pathogenic Italian strain as the putative major parental sequence and three other possible parents.
ANNOUNCEMENT
Currently, the International Committee on Taxonomy of Viruses (ICTV) designates porcine respiratory and reproductive syndrome virus 1 (PRRSV1) as Betaarterivirus suid 1, within the Variarterivirinae subfamily (1). PRRSV1 is one of the most important pathogens of pigs and causes serious economic losses (2). Within PRRSV1, at least 3 subtypes are recognized. Subtype 1 is the most widely distributed (3), while subtypes 2 and 3 were described in Western Siberia and Eastern Europe, respectively (4). Within each subtype, the genetic diversity is considerable. For PRRSV1, recombinant strains have been repeatedly reported in European countries, occasionally involving different modified live virus (MLV) vaccines (5).
Within the scope of a series of longitudinal studies aiming to evaluate the diversity of PRRSV1 quasispecies and the utility of genome segments as epidemiological markers, serum samples were collected between February and November 2021 from sows and piglets on 11 breeding farms located in Catalonia (northeast Spain), owned by different companies. All farms were suffering severe PRRS outbreaks characterized by high abortion rates and mortality in the sows and increased mortality in the weaners and growers (>20%). Briefly, PRRSV1 was isolated from individual serum samples after a single passage in porcine alveolar macrophages (PAMs). The PAMs were inoculated into Eagle’s minimal essential medium (EMEM), supplemented with penicillin-streptomycin and 7.5% bovine calf serum, and maintained. If a cytopathic effect was observed, supernatant was collected, and total RNA was extracted using TRIzol. Libraries were prepared using the NEBNext RNA library prep kit, without an initial amplification step, and sequenced on the Illumina MiSeq platform (paired-end 251-bp reads). Next-generation sequencing (NGS) reads were trimmed using Trimmomatic v0.36 (6) and mapped against the PRRSV1 prototype strain Lelystad (GenBank accession number NC_043487), using the Burrows-Wheeler Aligner (BWA-MEM algorithm). The SAMtools view (7) was used to remove reads with a mapping quality lower than Q20. LoFreq-Star v2.0 (8) was used to call variants. Bam-readcount v1.0.1 (9) was used to determine the frequency of each base at each position, and this frequency was used to infer a quasispecies population of 100 genomes in FASTA format. Variants with a minimum base-call quality score lower than 20 and coverage less than 5× were discarded. From this quasispecies, a consensus sequence was generated using Consensus Maker (https://www.hiv.lanl.gov/content/sequence/CONSENSUS/consensus.html). All tools were run with default parameters. The whole-genome sequences obtained (Table 1) were compared with other PRRSV1 sequences available in GenBank, as well as in the database of the Veterinary Laboratory for the Diagnosis of Infectious Diseases of the Universitat Autònoma de Barcelona. Phylogenetic analyses were performed using MEGA X. Recombination was detected using RDP5 software and GARD.
TABLE 1.
Sample and sequencing data for the 28 PRRSV1 genomes described
| Sample IDa | GenBank accession no. | SRA accession no. | No. of reads | Length (bp) | %GC | Coverage (×) |
|---|---|---|---|---|---|---|
| PRRSV1/Pig-wt/CAT-Esp/R1/May-2021 | OM893828 | SAMN27285123 | 680,713 | 14,910 | 52.8 | 538 |
| PRRSV1/Pig-wt/CAT-Esp/R2/Feb-2021 | OM893829 | SAMN27285124 | 674,411 | 14,910 | 52.8 | 845 |
| PRRSV1/Pig-wt/CAT-Esp/R3/Feb-2021 | OM893830 | SAMN27285125 | 829,785 | 14,910 | 52.7 | 3,317 |
| PRRSV1/Pig-wt/CAT-Esp/R4/Jun-2021 | OM893831 | SAMN27285126 | 345,101 | 14,910 | 52.6 | 1,147 |
| PRRSV1/Pig-wt/CAT-Esp/R5/Oct-2021 | OM893832 | SAMN27285127 | 1,061,085 | 14,910 | 52.7 | 2,691 |
| PRRSV1/Pig-wt/CAT-Esp/R6/Mar-2021 | OM893833 | SAMN27285128 | 477,811 | 14,910 | 52.8 | 812 |
| PRRSV1/Pig-wt/CAT-Esp/R7/May-2021 | OM893834 | SAMN27285129 | 920,987 | 14,910 | 52.7 | 669 |
| PRRSV1/Pig-wt/CAT-Esp/R8/Jun-2021 | OM893835 | SAMN27285130 | 86,290 | 14,910 | 52.8 | 169 |
| PRRSV1/Pig-wt/CAT-Esp/R9/Jul-2021 | OM893836 | SAMN27285131 | 878,809 | 14,910 | 52.7 | 1,975 |
| PRRSV1/Pig-wt/CAT-Esp/R10/Apr-2021 | OM893837 | SAMN27285132 | 5,653,071 | 14,910 | 52.6 | 2,182 |
| PRRSV1/Pig-wt/CAT-Esp/R11/Apr-2021 | OM893838 | SAMN27285133 | 968,821 | 14,910 | 52.8 | 1,328 |
| PRRSV1/Pig-wt/CAT-Esp/R12/May-2021 | OM893839 | SAMN27285134 | 561,265 | 14,910 | 52.7 | 609 |
| PRRSV1/Pig-wt/CAT-Esp/R13/Jun-2021 | OM893840 | SAMN27285135 | 419,903 | 14,910 | 52.7 | 1,546 |
| PRRSV1/Pig-wt/CAT-Esp/R14/Jul-2021 | OM893841 | SAMN27285136 | 933,866 | 14,910 | 52.7 | 1,547 |
| PRRSV1/Pig-wt/CAT-Esp/R15/Nov-2021 | OM893842 | SAMN27285137 | 1,292,169 | 14,910 | 52.7 | 2,564 |
| PRRSV1/Pig-wt/CAT-Esp/R16/Nov-2021 | OM893843 | SAMN27285138 | 795,071 | 14,910 | 52.7 | 1,855 |
| PRRSV1/Pig-wt/CAT-Esp/R17/Apr-2021 | OM893844 | SAMN27285139 | 1,187,971 | 14,910 | 52.8 | 4,345 |
| PRRSV1/Pig-wt/CAT-Esp/R18/Apr-2021 | OM893845 | SAMN27285140 | 1,260,111 | 14,910 | 52.7 | 4,168 |
| PRRSV1/Pig-wt/CAT-Esp/R19/May-2021 | OM893846 | SAMN27285141 | 440,400 | 14,910 | 52.6 | 353 |
| PRRSV1/Pig-wt/CAT-Esp/R20/May-2021 | OM893847 | SAMN27285142 | 763,771 | 14,910 | 52.6 | 1,477 |
| PRRSV1/Pig-wt/CAT-Esp/R21/Jun-2021 | OM893848 | SAMN27285143 | 301,518 | 14,910 | 52.6 | 263 |
| PRRSV1/Pig-wt/CAT-Esp/R22/Jul-2021 | OM893849 | SAMN27285144 | 790,502 | 14,910 | 52.6 | 1,150 |
| PRRSV1/Pig-wt/CAT-Esp/R23/Mar-2021 | OM893850 | SAMN27285145 | 1,008,440 | 14,910 | 52.7 | 2,128 |
| PRRSV1/Pig-wt/CAT-Esp/R24/Mar-2021 | OM893851 | SAMN27285146 | 886,568 | 14,910 | 52.8 | 2,154 |
| PRRSV1/Pig-wt/CAT-Esp/R25/Apr-2021 | OM893852 | SAMN27285147 | 440,798 | 14,910 | 52.8 | 406 |
| PRRSV1/Pig-wt/CAT-Esp/R26/May-2021 | OM893853 | SAMN27285148 | 830,917 | 14,910 | 52.7 | 761 |
| PRRSV1/Pig-wt/CAT-Esp/R27/Jun-2021 | OM893854 | SAMN27285149 | 352,456 | 14,910 | 52.8 | 993 |
| PRRSV1/Pig-wt/CAT-Esp/R28/Jul-2021 | OM893855 | SAMN27285150 | 716,349 | 14,910 | 52.6 | 839 |
Includes host, origin, strain, and sampling date (mo-yr). Pig-wt, XXX; CAT-Esp, Catalonia, Spain.
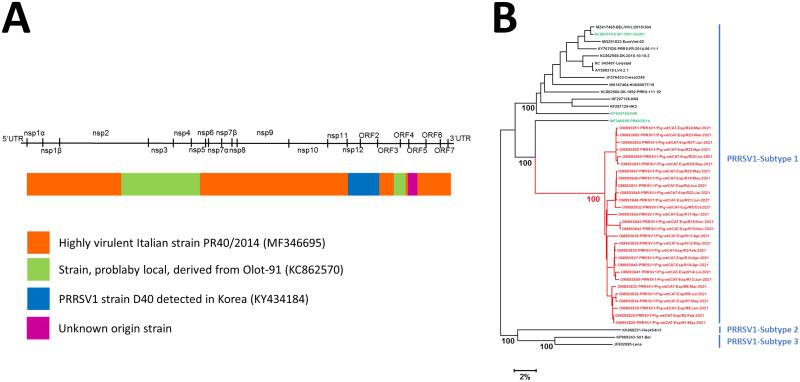
Local similarity analyses conducted using RDP and GARD revealed a common pattern among all isolates (Fig. 1A). A local similarity analysis conducted using BLAST identified a highly pathogenic Italian strain (10) as the putative major parental sequence (PR40/2014; GenBank accession number MF346695; 80% of the genome comprised in 5 segments, with 86 to 89% nucleotide identity) and the following three putative minor parental strains: ESP-1991-Olot91 (KC862570; 11% of the genome comprised in 2 segments, with 86% nucleotide identity), D40 (KY434184; 7% of the genome comprised in a single segment, with 86% nucleotide identity), and an unknown strain (2% of the genome comprised in a single segment, with a nucleotide identity lower than 85%).
FIG 1.
(A) Local similarity analyses among the 28 PRRSV1 genomes described, including the segments associated with every best BLAST hit of genomic subregions, indicated via the color code (identified using RDP and GARD); (B) maximum likelihood phylogenetic tree built up using the whole-genome sequence and the general time-reversible model. The confidence of the main internal branches, based on 100 bootstrap replicates, is indicated. Three PRRSV1 subtypes are marked in blue, the potential recombinant clade described in this report is highlighted in red, and the three putative parental strains identified are indicated in green. ORF, open reading frame; UTR, untranslated region.
A maximum likelihood (ML) phylogenetic tree placed all the genomes in a monophyletic branch within PRRSV1 subtype 1 (Fig. 1B).
The strains isolated in these outbreaks, characterized by high virulence, potentially originated from the recombination of multiple subtype 1 strains. Its apparent greater pathogenicity should be evaluated in future studies. Furthermore, the regular detection of recombinant PRRSV strains (5) illustrates the plasticity of this virus, together with the need to use whole-genome sequences as an analytical tool.
Data availability.
The 28 whole-genome sequences described in this report have been deposited at GenBank under the accession numbers OM893828 to OM893855; the corresponding fastq files have been deposited in the Sequence Read Archive under the BioSample accession numbers SAMN27285123 to SAMN27285150.
ACKNOWLEDGMENT
M. Cortey was funded by MINECO (Ramon y Cajal grant number RYC-2015-17154).
Contributor Information
Martí Cortey, Email: marti.cortey@uab.cat.
Simon Roux, DOE Joint Genome Institute.
REFERENCES
- 1.Brinton MA, Gulyaeva AA, Balasuriya UBR, Dunowska M, Faaberg KS, Goldberg T, Leung FCC, Nauwynck HJ, Snijder EJ, Stadejek T, Gorbalenya AE. 2021. ICTV virus taxonomy profile: Arteriviridae 2021. J Gen Virol 102:e001632. doi: 10.1099/jgv.0.001632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Wang C, Yeske PE, Mowrer CL, Haley CA. 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod 21:72–84. [Google Scholar]
- 3.Stadejek T, Stankevicius A, Murtaugh MP, Oleksiewicz MB. 2013. Molecular evolution of PRRSV in Europe: current state of play. Vet Microbiol 165:21–28. doi: 10.1016/j.vetmic.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 4.Yuzhakov AG, Raev SA, Shchetinin AM, Gushchin VA, Alekseev KP, Stafford VV, Komina AK, Zaberezhny AD, Gulyukin AM, Aliper TI. 2020. Genetic and pathogenic characterization of a Russian subtype 2 PRRSV-1 isolate. Vet Microbiol 247:108784. doi: 10.1016/j.vetmic.2020.108784. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L, Ge X, Yang H. 2021. Porcine reproductive and respiratory syndrome modified live virus vaccine: a “leaky” vaccine with debatable efficacy and safety. Vaccines (Basel) 9:362. doi: 10.3390/vaccines9040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H. 2011. Statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilm A, Kim Aw PP, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khanna A, Larson DE, Srivatsan SN, Mosior M, Abbott TE, Kiwala S, Ley TJ, Duncavage EJ, Walter MJ, Walker JR, Griffith OL, Griffith M, Miller CA. 2022. Bam-readcount—rapid generation of basepair-resolution sequence metrics. J Open Source Softw 7:3722. doi: 10.21105/joss.03722. [DOI] [Google Scholar]
- 10.Canelli E, Catella A, Borghetti P, Ferrari L, Ogno G, De Angelis E, Corradi A, Passeri B, Bertani V, Sandri G, Bonilauri P, Leung FCC, de Stefano Guazzetti C, Martelli P. 2017. Phenotypic characterization of a highly pathogenic Italian porcine reproductive and respiratory syndrome virus (PRRSV) type 1 subtype 1 isolate in experimentally infected pigs. Vet Microbiol 210:124–133. doi: 10.1016/j.vetmic.2017.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 28 whole-genome sequences described in this report have been deposited at GenBank under the accession numbers OM893828 to OM893855; the corresponding fastq files have been deposited in the Sequence Read Archive under the BioSample accession numbers SAMN27285123 to SAMN27285150.