ABSTRACT
Nrf2 activates cytoprotective gene expression, and Nrf2 activity is regulated through at least two protein degradation pathways: the Keap1-mediated and β-TrCP-mediated pathways. To address the relative contributions of these pathways, we generated knock-in mouse lines expressing an Nrf2SA mutant that harbored two substitution mutations of serine residues interacting with β-TrCP. The homozygous (Nrf2SA/SA) mice grew normally, with Nrf2 levels comparable to those of wild-type (WT) mice under unstressed conditions. However, when Keap1 activity was suppressed, high levels of Nrf2 accumulated in Nrf2SA/SA macrophages compared with that in WT macrophages. We crossed Nrf2SA/SA mice with mice in which Keap1 was knocked down to two different levels. We found that the Nrf2SA/SA mutation induced higher Nrf2 activity when the Keap1 level was strongly reduced, and these mice showed severe growth retardation. However, activation and growth retardation were not evident when Keap1 was moderately suppressed. These increases in Nrf2 activity induced by the Nrf2SA mutation caused severe hyperplasia and hyperkeratosis in the esophageal epithelium but did not cause abnormalities in the other tissues/organs examined. These results indicate that the β-TrCP-mediated pathway cooperates with the Keap1-mediated pathway to regulate Nrf2 activity, which is apparent when the Keap1-mediated pathway is profoundly suppressed.
KEYWORDS: β-TrCP, Keap1, Nrf2, degradation
INTRODUCTION
Nrf2 (nuclear factor erythroid 2-related factor 2) is a transcription factor (TF) that activates the expression of a series of cytoprotective genes, including antioxidant enzymes, xenobiotic-detoxifying enzymes, and drug transporters, in response to exogenous and endogenous stresses (1–3). It has been shown that elaborate and rigorous control of Nrf2 activity in response to stress stimuli is critical to protect cells from stress (4–6). On the other hand, perturbation of Nrf2 homeostasis, especially constitutive and high-level activation of Nrf2, has been shown to cause severe adverse effects (7–10). It has been shown that Nrf2 activity is determined mainly at the protein level. While it is constitutively generated, the Nrf2 protein rapidly turns over under unstressed conditions. The Nrf2 protein has been shown to be degraded through at least two types of ubiquitin-proteasome pathways: the Keap1 (Kelch-like ECH-associated protein 1)-mediated pathway and the β-TrCP (β-transducin repeat-containing protein)-mediated pathway (11). Keap1 and β-TrCP are adapter proteins of the E3 ubiquitin ligases Cullin3/Rbx1 and Cullin1/Rbx1, respectively (12–14).
Keap1 regulates Nrf2 degradation in an electrophilic and oxidative stress-dependent manner. Electrophiles and reactive oxygen species (ROS) modify the cysteine residues in Keap1, which inactivates Keap1-dependent Nrf2 degradation (15, 16). Keap1 binds the DLGex and ETGE motifs in the Neh2 (Nrf2-ECH homology 2) domain of Nrf2 via the DC domain and induces proteasomal degradation of the Nrf2 protein in the cytoplasm (17–19). In addition to electrophiles and ROS, the p62 (SQSTM1) protein, which accumulates through impaired selective autophagy, has been shown to suppress the interaction between Keap1 and Nrf2 by binding to the Keap1 DC domain (20, 21).
Studies based on pharmacological and genetic Keap1 suppression, which cause transient and constitutive accumulation of Nrf2, respectively, revealed that a moderate level of Nrf2 activation benefits the animal body in various ways. We previously generated a mouse line harboring a floxed wild-type (WT) Keap1 allele and knocked-in the green fluorescent protein (GFP) gene downstream of the Keap1 gene (Keap1FA allele). We discovered that the Keap1FA is a knockdown allele; Keap1 mRNA is transcribed from this allele at lower levels than from the unfloxed WT allele but at higher levels than from the knockout (Keap1–) allele (22–24). In the latter knockout allele, residues 8 to 204 of Keap1 are replaced with a lacZ gene (7). We have demonstrated that both types of homozygous mice with the knockdown Keap1FA allele (Keap1FA/FA mice) exhibited a decrease in Keap1 expression to approximately 10% to 50% of the Keap1 expression observed in WT mice, depending on the tissue/organ. Both Keap1FA/FA mice and mice hemizygous for the Keap1FA and Keap1 knockout alleles (Keap1FA/– mice), in which Keap1 expression is even lower than that in Keap1FA/FA mice, do not show substantially altered phenotypes and survive into adulthood (22, 23). By crossing Keap1FA/– mice with mouse models of diseases such as diabetes mellitus or sickle cell disease, we showed that the genetic activation of Nrf2 attenuates the symptoms of these diseases (25, 26).
In contrast, systemic Keap1-null mice exhibit severe growth retardation and die in the postnatal period due to malnutrition resulting from hyperkeratosis of the esophagus, but this lethality is completely rescued by systemic Nrf2 knockout (7). In addition, combinatory and conditional knockout of Nrf2 in keratin 5-positive epithelial cells, which rescues systemic Keap1 knockout mice from the juvenile lethality, leads to Keap1 deficiency that causes nephrogenic diabetes insipidus and bone hypoplasia (8, 9). Considering these studies, we have learned that constitutive hyperactivation of Nrf2 caused by genetic Keap1 deficiency impairs homeostasis in many tissues and organs.
The β-TrCP-mediated pathway leads to the degradation of the Nrf2 protein in a signaling pathway-dependent manner. β-TrCP recognizes Nrf2 via the DSGIS (-Asp-Ser-Gly-Ile-Ser-) motif in the Neh6 domain of Nrf2 (27, 28). To recognize Nrf2, β-TrCP requires phosphorylation of two serine residues within the DSGIS motif. These two serine residues are phosphorylated by glycogen synthase kinase-3 (GSK-3) (28–30), and GSK-3 is competent for catalysis under basal conditions. However, phosphorylation at the N-terminus of GSK-3 represses GSK-3 activity, and AKT is the major protein kinase that phosphorylates the N-terminal serine residue in GSK-3 (31–33). The phosphatidylinositol 3-kinase (PI3K) signaling pathway activates AKT, leads to the phosphorylation of GSK-3, and suppresses GSK-3 activity, which results in the repression of β-TrCP-mediated Nrf2 degradation.
Although the impacts of Keap1-dependent Nrf2 degradation on tissue homeostasis have been well characterized, the physiological significance of the β-TrCP-mediated Nrf2 degradation pathway and how the β-TrCP-mediated pathway contributes to the maintenance of tissue homeostasis remain to be clarified. Therefore, in this study, we prepared an Nrf2 mutant in which the two serine residues interacting with β-TrCP are replaced with alanine residues (Nrf2SA) and generated a knock-in mouse line expressing homozygous Nrf2SA alleles.
We addressed the relative contributions of the β-TrCP-mediated and Keap1-mediated pathways to Nrf2 expression levels by studying Nrf2SA mutant mice. As we found neither significant Nrf2 accumulation nor apparent abnormalities in the homozygous mutant (Nrf2SA/SA) mice per se, we crossed the Nrf2SA/SA mice with mice in which Keap1 was knocked down to two different levels. We found that when the expression level of Keap1 was moderately reduced, no additional Nrf2 activation was observed in the Nrf2SA/SA mice. In contrast, when the expression level of Keap1 was severely reduced, the mice showed additional Nrf2 protein accumulation and significant growth retardation compared with the control mice. Importantly, the esophagus of the mice showed hyperkeratosis and hyperplasia. These results demonstrate that the β-TrCP-mediated pathway indeed cooperates with the Keap1-mediated pathway in cellular Nrf2 degradation in vivo, and in the esophageal epithelium, the β-TrCP-mediated pathway appears to serve as an auxiliary system when Keap1-mediated degradation is profoundly suppressed.
RESULTS
Generation of Nrf2SA knock-in mice.
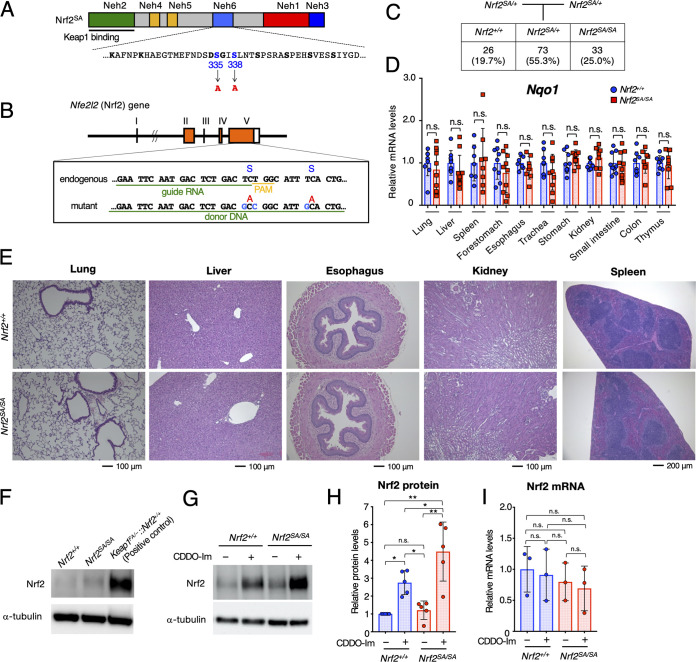
To address the physiological significance of the β-TrCP-mediated degradation of Nrf2, we generated a knock-in mouse line expressing an Nrf2 mutant resistant to β-TrCP-mediated degradation. For this purpose, serine (S) residues at positions 335 and 338 of Nrf2 were both replaced by the CRISPR/Cas9 genome-editing system with alanine (A) residues (hereafter, Nrf2SA) (Fig. 1A). It has been reported that the Nrf2SA mutation abolishes the interaction between β-TrCP and Nrf2 (29). We designed a guide RNA (gRNA) containing 20 nucleotides capable of recognizing the Nrf2 target site and added a protospacer adjacent motif (PAM) after it to recruit Cas9 to the target site (Fig. 1B). We prepared a plasmid expressing both Cas9 and the gRNA and a 67-mer oligonucleotide DNA (oligo-DNA [the donor DNA]) in which TCT at S335 and TCA at S338 were replaced with GCC and GCA, respectively, resulting in substitutions of S residues by A residues. We coinjected both the Cas9-gRNA plasmid and donor DNA into C57BL/6 fertilized eggs and found that two mice (mice 10 and 13) had the Nrf2SA mutation. To obtain mouse lines carrying the Nrf2SA mutation, we crossed these mice with WT mice. After sequencing the pup genomic DNA directly, we confirmed the presence of heterozygous Nrf2SA offspring derived from mice 10 and 13. These results demonstrate the establishment of two Nrf2SA mutant mouse lines. Performing initial characterizations, we found that these two mouse lines exhibited a similar appearance, so we mainly used the mouse 10 line for experiments.
FIG 1.
Homozygous Nrf2SA mice show no apparent abnormality in the maintenance of Nrf2 homeostasis under unstressed conditions. (A) Structure of the Nrf2SA protein. The 335th and 338th serine residues (S) positioned in the Neh6 domain were replaced with alanine (A). (B) Cas9/gRNA-targeting site in the Nrf2 (Nfe2l2) gene. Sequences of gRNA and donor DNA coinjected for targeting mutagenesis are underlined. The PAM sequence is indicated in yellow. The S335 and S338 residues and corresponding A residues are shown. (C) Schematic showing the mating strategy. The numbers (percentages) of pups at weaning that bear the three possible genotypes are depicted. Nrf2SA/SA mice were born following the expected Mendelian percentages. (D) mRNA levels of the Nrf2 target gene Nqo1 in the lung, liver, spleen, forestomach, esophagus, trachea, stomach, kidney, small intestine, colon, and thymus. Hprt was used as the internal control. The mRNA levels in Nrf2+/+ mice were set to 1. The bar graphs show the mean ± standard deviation (SD). n.s., not significant by Mann-Whitney test. (E) HE staining of the lung, liver, esophagus, kidney, and spleen of Nrf2+/+ and Nrf2SA/SA mice; (F) representative immunoblot showing Nrf2 and α-tubulin in Nrf2+/+ and Nrf2SA/SA macrophages under unstressed conditions. Keap1FA/–::Nrf2+/+ macrophages were used as the positive controls. (G) Representative immunoblot showing Nrf2 and α-tubulin in Nrf2+/+ and Nrf2SA/SA macrophages treated with (+) or without (–) 10 nM CDDO-Im for 3 h; (H) protein levels of Nrf2 in Nrf2+/+ and Nrf2SA/SA macrophages treated with or without 10 nM CDDO-Im for 3 h. α-Tubulin was used for the internal control. The Nrf2 level in Nrf2+/+ macrophages without CDDO-Im treatment was set to 1. (I) mRNA levels of Nrf2 in Nrf2+/+ and Nrf2SA/SA macrophages treated with or without 10 nM CDDO-Im for 3 h. Hprt was used as the internal control. The Nrf2 level in Nrf2+/+ macrophages without CDDO-Im treatment was set to 1. The bar graphs show the means ± SD. **, P < 0.01; *, P < 0.05 (one-way analysis of variance [ANOVA] with Tukey’s multiple-comparison test).
Homozygous Nrf2SA mice show no apparent abnormality in the maintenance of Nrf2 homeostasis under unstressed conditions.
To examine whether homozygous mutant Nrf2SA/SA mice were born in Mendelian ratios, we crossed male and female heterozygous mutant Nrf2SA/+ mice and analyzed the genotypes of the pups. We found that 26 (19.7%), 73 (55.3%), and 33 (25.0%) pups were of the Nrf2+/+, Nrf2SA/+, and Nrf2SA/SA genotypes, respectively, indicating that Nrf2SA/SA mice were born in the expected Mendelian ratios (Fig. 1C).
To examine whether Nrf2 is activated in Nrf2SA/SA mouse organs, we analyzed the mRNA levels of Nqo1, an Nrf2 target gene whose mRNA levels closely reflect Nrf2 activation levels. We found that the Nqo1 mRNA levels were comparable between various Nrf2+/+ and Nrf2SA/SA tissues, including lung, liver, spleen, forestomach, esophagus, trachea, stomach, kidney, small intestine, colon, and thymus (Fig. 1D). These results indicate that Nrf2 is not activated in Nrf2SA/SA mouse tissues or organs under steady-state conditions. Consistent with this observation, we found no significant abnormalities in the histology of various Nrf2SA/SA mouse tissues and organs, including the lung, liver, esophagus, kidney, and spleen (Fig. 1E). Considering these results, we conclude that the β-TrCP-mediated Nrf2 degradation pathway is dispensable for the maintenance of Nrf2 homeostasis under unstressed conditions.
The β-TrCP pathway regulates cellular Nrf2 protein levels under Keap1-inactivated conditions.
We next examined whether inactivation of the β-TrCP-mediated degradation pathway induces Nrf2 protein accumulation. For this purpose, we designed an ex vivo system utilizing peritoneal macrophages derived from Nrf2SA/SA mice. We collected thioglycolate-elicited peritoneal macrophages from Nrf2+/+ and Nrf2SA/SA mice and analyzed the Nrf2 protein levels.
We found that the Nrf2 protein levels in Nrf2SA/SA mouse-derived peritoneal macrophages were comparable to those in Nrf2+/+ mouse-derived peritoneal macrophages. Nrf2 protein levels are regulated by at least two ubiquitin-proteasome-dependent pathways: i.e., the Keap1-mediated and β-TrCP-mediated pathways. Therefore, we compared the Nrf2 protein levels in peritoneal macrophages derived from Keap1 knockdown (Keap1FA/–::Nrf2+/+) mice with those in peritoneal macrophages derived from Nrf2SA/SA mice. We found that the level in the Nrf2SA/SA mouse-derived cells was significantly lower than that in Keap1 knockdown mouse-derived cells, indicating that the suppression of the β-TrCP-mediated degradation pathway alone does not lead to significant Nrf2 protein accumulation (Fig. 1F).
Considering these results, we hypothesized that because the Keap1-mediated degradation pathway is intact in Nrf2+/+ and Nrf2SA/SA cells, most of the Nrf2 protein is degraded through the Keap1 pathway. Therefore, we next examined whether the Nrf2 protein accumulates when Keap1-mediated degradation is suppressed. To this end, we treated the cells with CDDO-Im {1-[2-cyano-3,12-di-oxooleana-1,9(11)-dien-28-oyl]imidazole}, which is known to inactivate the Keap1-mediated degradation pathway by modifying a cysteine residue in Keap1. We found that the Nrf2 protein level was increased by CDDO-Im treatment of Nrf2+/+ cells (Fig. 1G and H), showing very good agreement with previous reports (15). We also found that the Nrf2 protein level was increased by CDDO-Im treatment of Nrf2SA/SA cells. Notably, the Nrf2 protein level in Nrf2SA/SA cells was significantly higher than that in Nrf2+/+ cells, and this result was highly reproducible (Fig. 1G and H). The Nrf2 mRNA levels were comparable between Nrf2+/+ and Nrf2SA/SA cells treated with and without CDDO-Im treatment, indicating that the accumulation of Nrf2 protein did not occur due to an increase in Nrf2 transcription (Fig. 1I). These results demonstrate that the contribution of the β-TrCP-mediated degradation pathway to the increase in the Nrf2 protein levels is apparent only when the Keap1-mediated degradation pathway is suppressed in peritoneal macrophages.
Mice without Keap1- and β-TrCP-mediated Nrf2 degradation pathways exhibit severe growth retardation.
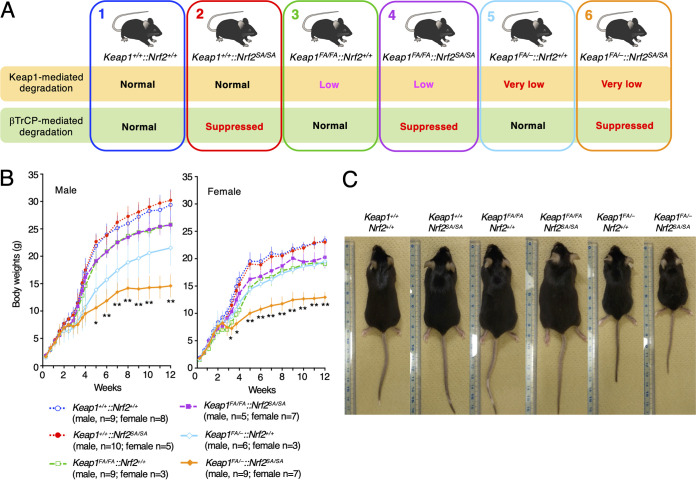
After performing ex vivo experiments utilizing peritoneal macrophages, we examined the contribution of β-TrCP-mediated Nrf2 degradation in vivo in Keap1 expression-suppressed mice. We crossed Nrf2SA/SA mice with two types of Keap1 knockdown mice, namely, Keap1FA/FA and Keap1FA/– mice. The Keap1 expression levels in the Keap1FA/FA mice were decreased to approximately 10% to 50% of those observed in the WT mice, depending on the tissues/organs, and the Keap1 levels in the Keap1FA/– mice were even lower than those in the Keap1FA/FA mice (22). The mating of Nrf2SA/SA mice with Keap1FA/FA or Keap1FA/– mice resulted in six genotypes with various degrees of suppressed Keap1 activity (Fig. 2A). Graded suppression of Keap1-mediated Nrf2 degradation was expected based on these genotypes.
FIG 2.
Mice without Keap1- and β-TrCP-mediated Nrf2 degradation pathways exhibit severe growth retardation. (A) Schematic showing the mutant mice with Keap1 knocked down and the Nrf2SA mutation. We crossed the Nrf2SA/SA mice with two types of Keap1 knockdown mice: Keap1FA/FA and Keap1FA/– mice. (B) Body weight changes in Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA male (left) and female (right) mice. Keap1FA/–::Nrf2SA/SA mice exhibit severe growth retardation. The line graphs show the means ± SD. **, P < 0.01, and *, P < 0.05, compared with Keap1FA/–::Nrf2+/+ mice by one-way ANOVA with Dunnett’s multiple-comparison test. (C) Representative images of Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mice.
We first analyzed body weight changes in male and female mice with the six genotypes from birth to 12 postnatal weeks (Fig. 2B). We found that the body weight gains of male and female Keap1+/+::Nrf2SA/SA mice were comparable to those of control Keap1+/+::Nrf2+/+ mice (Fig. 2B). These results indicate that homozygous Nrf2SA mutant mice grow normally under normal unstressed conditions.
While the body weights of the male and female Keap1FA/FA::Nrf2+/+ mice were lower than those of the respective Keap1+/+::Nrf2+/+ mice, the body weights of the male and female Keap1FA/FA::Nrf2SA/SA mice were comparable to those of the respective Keap1FA/FA::Nrf2+/+ mice. In contrast, whereas body weight curves of female Keap1FA/–::Nrf2+/+ mice were comparable to those of female Keap1FA/FA::Nrf2+/+ mice, the body weight curve of male Keap1FA/–::Nrf2+/+ mice was lower than that of male Keap1FA/FA::Nrf2+/+ mice (Fig. 2B). Importantly, both male and female Keap1FA/–::Nrf2SA/SA mice exhibited severe growth retardation compared with the growth of the respective Keap1FA/–::Nrf2+/+ mice. However, despite the lower weight gain, the Keap1FA/–::Nrf2SA/SA mice did not show apparent abnormalities (Fig. 2C). These results demonstrate that the loss of β-TrCP-mediated Nrf2 degradation activity severely affects the weight gain of mice when the Keap1 pathway is strongly suppressed.
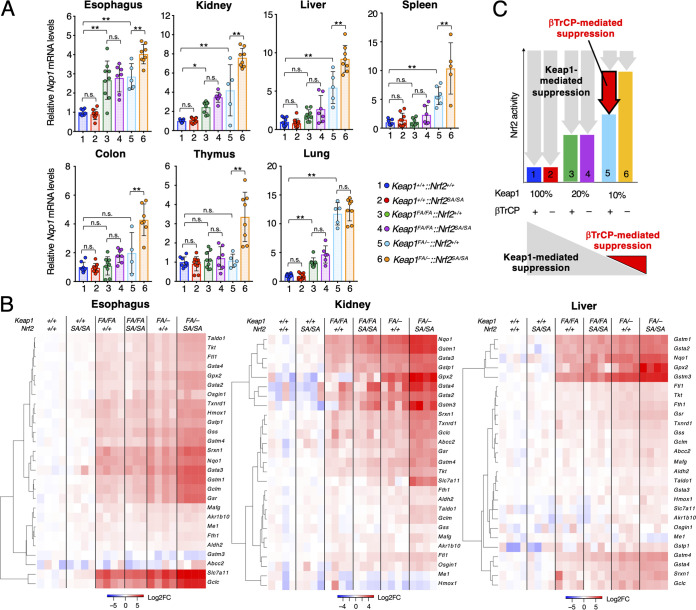
Nrf2 activity is regulated by the β-TrCP pathway under Keap1-suppression conditions.
We next addressed the question of how Nrf2 activity is regulated by the β-TrCP pathway under Keap1-suppressed conditions. For this purpose, we examined the mRNA levels of Nqo1, a tightly regulated Nrf2 target gene, in the esophagus, kidney, liver, spleen, colon, thymus, and lung of mice with the six genotypes at 12 weeks of age (Fig. 3A). The mice with the six genotypes (i.e., Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mice) are numbered from 1 to 6 in the following figures. In all seven organs that we analyzed, the Nqo1 mRNA levels were comparable between the Keap1+/+::Nrf2SA/SA mice (red) and Keap1+/+::Nrf2+/+ mice (blue). We also examined Keap1FA/FA::Nrf2+/+ (green) and Keap1FA/FA::Nrf2SA/SA (purple) mice and found that the Nqo1 mRNA levels in the esophagus, kidney, and lung were increased compared with those in the Keap1+/+::Nrf2+/+ mice, but the differences between the Keap1FA/FA::Nrf2+/+ and Keap1FA/FA::Nrf2SA/SA mice were not significant. The Nqo1 mRNA levels in the other organs of Keap1FA/FA::Nrf2+/+ and Keap1FA/FA::Nrf2SA/SA mice did not change substantially compared with those in the organs of the Keap1+/+::Nrf2+/+ mice. These results indicate that the activity of the β-TrCP pathway is dispensable or not significant for Nrf2 regulation under conditions where Keap1 activity is greater than approximately 20% of that of the WT mice.
FIG 3.
Nrf2 activity is regulated by the β-TrCP pathway under Keap1-suppressed conditions. (A) mRNA levels of the Nrf2 target gene Nqo1 in the esophagus, kidney, liver, spleen, colon, thymus, and lung. Hprt was used as the internal control. The mRNA levels in the Keap1+/+::Nrf2+/+ mice were set to 1. The bar graphs show the means ± SD. **, P < 0.01, and *, P < 0.05, by one-way ANOVA with Tukey’s multiple-comparison test. (B) Heat map of the expression levels of representative Nrf2 target genes in the esophagus, kidney, and liver from Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mice. Colors indicate the log2 fold change (FC) value relative to the mean expression level of each gene in Keap1+/+::Nrf2+/+ samples. (C) Schematic of Keap1-mediated (gray arrows) and β-TrCP-mediated (red arrow) suppression of Nrf2 activity. The decrease in Nqo1 mRNA levels in Keap1FA/–::Nrf2SA/SA (orange bar) mice compared to that in Keap1FA/–::Nrf2+/+ (light blue bar) mice reflects the ability of the β-TrCP pathway to degrade Nrf2 (red arrow, upper panel).
Consistent with the results from our previous study (22), Nqo1 mRNA levels were increased in the esophagus, kidney, liver, spleen, and lung of Keap1FA/–::Nrf2+/+ (light blue) mice compared with those of Keap1+/+::Nrf2+/+ mice (Fig. 3A). Notably, the Nqo1 mRNA levels in the six organs (i.e., esophagus, kidney, liver, spleen, colon, and thymus) of Keap1FA/–::Nrf2SA/SA (orange) mice were significantly higher than those of Keap1FA/–::Nrf2+/+ mice. On the other hand, in the lung, there was no further increase in Nqo1 mRNA levels in Keap1FA/–::Nrf2SA/SA mice compared to Keap1FA/–::Nrf2+/+ mice, although there was a significant increase in Keap1FA/–::Nrf2+/+ mice compared to Keap1+/+::Nrf2+/+ mice.
To precisely assess Nrf2 activity, we comprehensively examined the expression levels of Nrf2 target genes in mice with the six genotypes, assuming that changes en bloc in the Nrf2 target genes would correlate with cellular Nrf2 activity. To this end, we performed RNA sequencing analysis using RNA samples from the esophagus, kidney, and liver of mice with these six genotypes. We selectively analyzed genes that encode enzymes and transcription factors related to detoxication and antioxidative responses that are known to be Nrf2 target genes (34, 35). We found that the expression levels of approximately 2/3, 1/2, and 1/3 of the Nrf2 target genes were upregulated in a graded manner, similar to Nqo1, in the esophagus, kidney, and liver, respectively (Fig. 3B). Similar to Nqo1 mRNA expression, upregulated mRNA levels were comparable between the Keap1+/+::Nrf2SA/SA and Keap1+/+::Nrf2+/+ mice but increased in the Keap1FA/FA::Nrf2+/+ and Keap1FA/FA::Nrf2SA/SA mice compared with the Keap1+/+::Nrf2+/+ mice. The mRNA level was highest in the Keap1FA/–::Nrf2SA/SA mice. These results support our contention that Nrf2 activity is also upregulated in this manner.
As summarized in Fig. 3C, we interpret the decrease in the Nqo1 mRNA levels between Keap1FA/–::Nrf2SA/SA (orange) mice and Keap1FA/–::Nrf2+/+ (light blue) mice as indicative of the effect of the β-TrCP pathway on Nrf2 degradation (red arrow). Thus, these results unequivocally demonstrate that the β-TrCP pathway is truly functional in vivo. These results also demonstrate that the activity is apparent under conditions in which the activity of the Keap1 pathway is suppressed. The β-TrCP pathway must contribute to the regulation of Nrf2 activity under pathological conditions in which Keap1 is profoundly inactivated by electrophilic chemicals and/or oxidative stresses or by genome mutations.
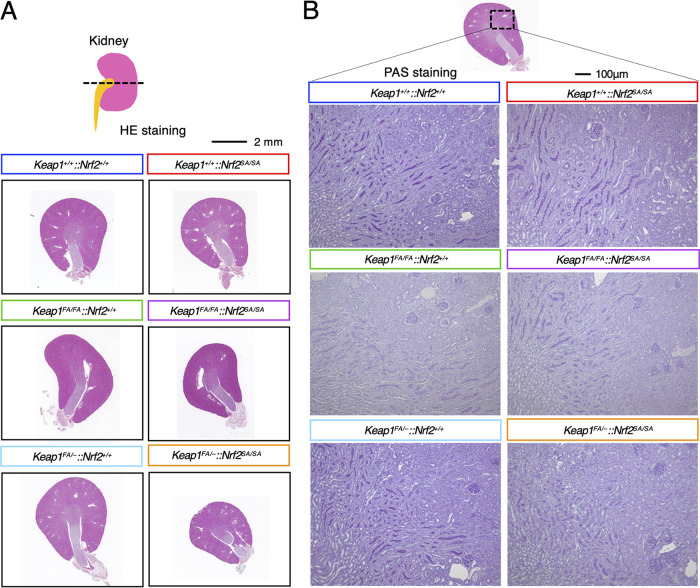
The kidney abnormalities in Keap1-null mice are not found in Keap1FA/–::Nrf2SA/SA mice.
We next analyzed how additional Nrf2 accumulation caused by suppression of the β-TrCP pathway, in addition to that induced by suppression of the Keap1 pathway, affects the mouse phenotype. Mice with the Keap1 gene systemically knocked out die in the juvenile stage due to hyperkeratosis and obstruction of the esophagus caused by Nrf2 activation in the esophageal squamous cell epithelium (7). In NEKO mice (Keap1–/–::Nrf2F/F::Keratin 5-Cre), Nrf2 accumulation in the keratin 5-positive squamous cell layers is canceled by simultaneous keratin 5-positive cell-specific knockout of the Nrf2 gene (8). However, the kidneys of NEKO mice showed bilateral hydronephrosis characterized by an enlarged renal pelvic space at 8 weeks of age or later, a phenotype that resembled diabetes insipidus (8).
Considering these findings, we examined whether Nrf2 hyperactivation in the Keap1FA/–::Nrf2SA/SA mice causes kidney damage similar to that in NEKO mice. To this end, we analyzed the morphology of the kidneys in Keap1FA/–::Nrf2SA/SA mice and mice with related genotypes by performing hematoxylin-eosin (HE) and periodic acid-Schiff (PAS) staining in mouse tissues taken at 12 weeks of age. We found that the Keap1FA/–::Nrf2SA/SA mice, as well as the mice with the other genotypes, did not show an enlarged renal pelvic space due to hydronephrosis (Fig. 4A). As PAS staining of the NEKO mouse kidney showed severe dilation of collecting ducts, indicating elevated pressure in the kidney (8), we performed PAS staining of the kidneys in mice with the six genotypes, but we did not find dilated collecting ducts or any apparent abnormalities in the kidneys of Keap1FA/–::Nrf2SA/SA mice or the mice with other genotype (Fig. 4B). These results indicate that Nrf2 hyperactivation in Keap1FA/–::Nrf2SA/SA mice is insufficient to cause the severe kidney damage observed in Keap1-null mice.
FIG 4.
Kidney abnormalities are not found in Keap1FA/–::Nrf2SA/SA mice. (A) HE staining of kidney cross sections of Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mice; (B) PAS staining of kidney cross sections from Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mice.
Liver abnormalities in Keap1 and Pten double-knockout mice were not observed in Keap1FA/–::Nrf2SA/SA mice.
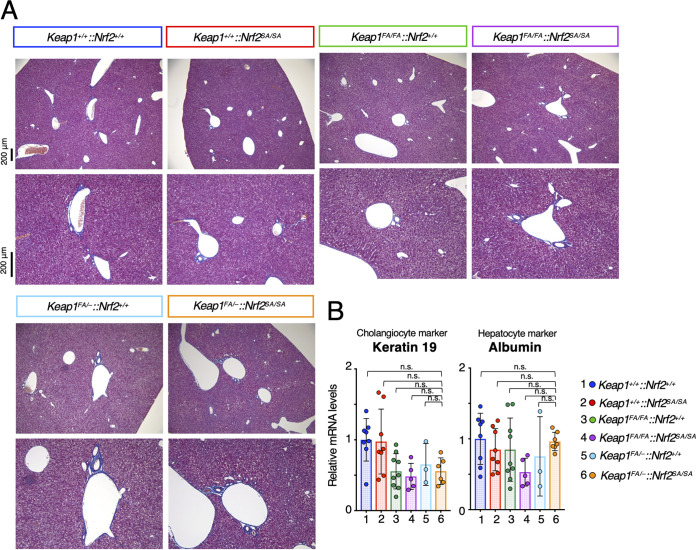
We next compared the liver phenotypes of Keap1FA/–::Nrf2SA/SA mice with that of liver-specific Keap1 and Pten double-knockout mice. The liver-specific Keap1 and Pten double-knockout mice (Keap1FA/FA::Nrf2+/+::PtenF/F::Albumin-Cre) displayed severe liver damage due to abnormal expansion of the ductal structures composed of cholangiocytes (10). Pten deletion causes activation of the AKT signaling pathway, which suppresses β-TrCP-mediated Nrf2 degradation (30). Since both Keap1-mediated and β-TrCP-mediated pathways are suppressed in liver-specific Keap1 and Pten double-knockout mice, we assumed that the Keap1FA/–::Nrf2SA/SA liver might show a phenotype similar to that of Keap1 and Pten double-knockout mice.
To examine this possibility, we performed Masson’s trichrome staining of the livers of Keap1FA/–::Nrf2SA/SA mice and mice with related genotypes to detect collagen fibers and tubular structures. We found that the livers of Keap1FA/–::Nrf2SA/SA mice, as well as those of mice with the other genotypes examined in this study, did not show abnormal increases in the bile duct or any other abnormal phenotype (Fig. 5A). It has been found that the expression of cholangiocyte genes, including Keratin 19, is increased significantly and that of hepatocyte genes, including Albumin, is decreased in liver-specific Keap1 and Pten double-knockout mice, and these changes are abrogated by the simultaneous knockout of Nrf2 (10). In contrast, we found no significant change in the mRNA levels of the cholangiocyte marker Keratin 19 or the hepatocyte marker Albumin in Keap1FA/–::Nrf2SA/SA mice and mice with the other genotypes (Fig. 5B). These results support the notion that the activation levels of Nrf2 in the liver of Keap1FA/–::Nrf2SA/SA mice are much more moderate than those in mouse models with pathologically high Nrf2 activation due to Keap1-null::Pten-null mutation.
FIG 5.
Liver abnormalities were not observed in Keap1FA/–::Nrf2SA/SA mice. (A) Masson’s trichrome staining of Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mouse livers; (B) mRNA levels of keratin 19 and albumin in the liver. Hprt was used as the internal control. The mRNA levels in the Keap1+/+::Nrf2+/+ mice were set to 1. The bar graphs show the means ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple-comparison test.
Hyperkeratosis and hyperplasia of the esophagus in Keap1FA/–::Nrf2SA/SA mice.
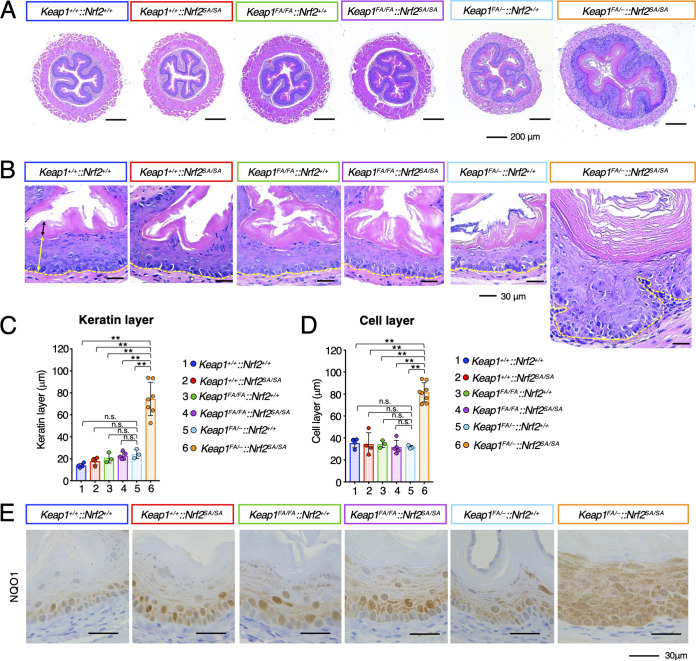
Systemic Keap1-null mice exhibit severe growth retardation and die in the postnatal period due to malnutrition resulting from hyperkeratosis of the esophagus (7). Therefore, we analyzed the esophageal morphology of 12-week-old Keap1FA/–::Nrf2SA/SA mice and mice with related genotypes. We found that the esophagi of the Keap1FA/–::Nrf2SA/SA mice were markedly enlarged, with diameters much greater than those in the mice with the other genotypes (Fig. 6A). Notably, both the cell layer and keratin layer of the Keap1FA/–::Nrf2SA/SA mouse esophagus were greatly increased compared with those of the esophagi of the other genotype mice. The thickness of the outer muscle layers was comparable in mice of all genotypes.
FIG 6.
Hyperkeratosis and hyperplasia of the esophagus in Keap1FA/–::Nrf2SA/SA mice. (A, B) HE staining of mouse esophageal cross sections. The yellow and black double-edged arrows in the left panel show the epithelial cell and keratinous layers, respectively. The yellow dotted lines show the boundary of the epithelial cell layer. Hyperkeratosis and hyperplasia of the esophagus are specifically induced in Keap1FA/–::Nrf2SA/SA mice. (C, D) Thickness of keratinous layers (C) and cell layers (D) of the esophageal epithelium. The bar graphs show the means ± SD. **, P < 0.01 by one-way ANOVA with Tukey’s multiple-comparison test. (E) Immunohistochemistry of NQO1.
We measured the thickness of keratin layers and cell layers of the esophagi of mice of all genotypes (black and yellow double-headed arrows, respectively, in Fig. 6B) and found that the keratin layer in the Keap1FA/–::Nrf2SA/SA mouse esophagus was significantly thicker than that of the mice of other genotypes (Fig. 6B and C). The thickness of the cell layer of the Keap1FA/–::Nrf2SA/SA mouse esophagus was also markedly increased compared with that of esophagi of the mice of other genotypes (Fig. 6B and D). A histological analysis revealed that the basal layer in the esophagus of Keap1FA/–::Nrf2SA/SA mice was distorted, while that in the esophagi of the mice with other genotypes was almost straight (dotted yellow lines in Fig. 6B). Moreover, the borderline between the cell layer and the keratin layer was straight in the Keap1FA/–::Nrf2SA/SA mouse epithelium. These results indicate that the differentiation and proliferation of esophageal epithelial cells are imbalanced in the Keap1FA/–::Nrf2SA/SA mice.
To assess whether Nrf2 is activated in the esophagus of Keap1FA/–::Nrf2SA/SA mice, we decided to measure the expression of the Nqo1 gene, one of the most tightly linked target genes of Nrf2, and performed NQO1 immunohistochemical analyses. In the Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, and Keap1FA/–::Nrf2+/+ mouse esophagi, NQO1 was highly expressed in the lower cell layers (Fig. 6E). In contrast, in the Keap1FA/–::Nrf2SA/SA mice, NQO1 was highly expressed in all the cell layers, indicating that Nrf2 accumulated not only in basal layer cells but also in upper layer cells, which are also known as suprabasal cells in Keap1FA/–::Nrf2SA/SA mice.
Thus, we found that perturbation in Nrf2 homeostasis caused by concomitant inactivation of the Keap1 and β-TrCP pathways induces imbalanced esophageal epithelial cell differentiation and proliferation. This perturbation occurs when Keap1 activity is severely suppressed and with complete loss of β-TrCP pathway activity. These results support our contention that Nrf2 homeostasis is critically important for the maintenance of the squamous cell epithelium in the esophagus and that Nrf2 activity is likely regulated much more strictly in the esophagus than in the liver or kidney. To the best of our knowledge, this is the first demonstration of the contribution of the β-TrCP pathway to the regulation of physiological/pathological functions in vivo, showing that the dysfunction of the β-TrCP pathway leads to the acquisition of abnormal phenotypes.
Abnormality of the Keap1FA/–::Nrf2SA/SA mouse esophageal epithelium.
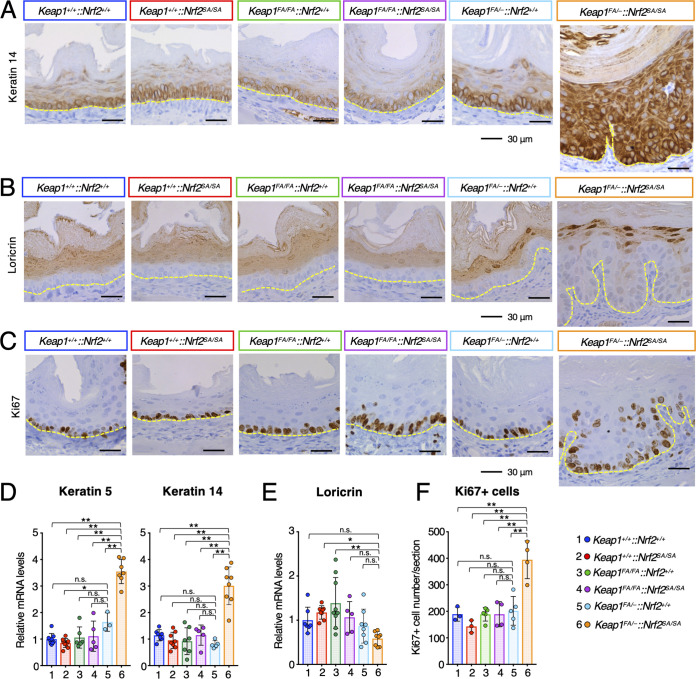
In the squamous cell epithelium of the esophagus, basal cells proliferate and differentiate into suprabasal cells (36, 37). Suprabasal cells further differentiate and move to the top layers, where they form keratin layers. To assess whether differentiation of the esophageal epithelium is impaired in Keap1FA/–::Nrf2SA/SA mice, we examined the expression profiles of differentiation markers of the squamous cell epithelium in the mouse esophagus. We first performed immunohistochemical analyses of a basal cell marker, keratin 14, and found that, although only basal cells and some second-layer cells were keratin 14-positive in the mice with other genotypes, all the epithelial layers were keratin 14 positive in the Keap1FA/–::Nrf2SA/SA mouse esophagi, which were very thick (Fig. 7A).
FIG 7.
Abnormality of the Keap1FA/–::Nrf2SA/SA mouse esophageal epithelium. (A to C) Immunohistochemistry of keratin 14 (A), loricrin (B), and Ki67 (C). The yellow dotted lines show the boundary of the epithelial cell layer. (D, E) mRNA levels of keratin 5 (D, left panel), keratin 14 (D, right panel), and loricrin (E). Hprt was used as the internal control. The mRNA levels in the Keap1+/+::Nrf2+/+ mice were set to 1. (F) Number of Ki67-positive cells per section. The bar graphs show the means ± SD. **, P < 0.01, and *, P < 0.05, by one-way ANOVA with Tukey’s multiple-comparison test.
We next conducted immunohistochemical analysis of loricrin, a marker of differentiated squamous epithelial cells. The loricrin signal was positive in the upper half of the esophageal epithelium in the WT mice and mice with other genotypes (Fig. 7B). In contrast, the loricrin signal was positive only in the boundary region facing the keratin layers in the esophagus of the Keap1FA/–::Nrf2SA/SA mice (Fig. 7B).
To validate these immunohistochemical results, we carried out quantitative reverse transcription-PCR (qRT-PCR) analysis. The mRNA levels of both keratin 14 and keratin 5, another basal cell marker, were significantly increased only in the Keap1FA/–::Nrf2SA/SA mouse esophagus (Fig. 7D). In contrast, the loricrin mRNA level was comparable or mildly decreased in the Keap1FA/–::Nrf2SA/SA esophagus compared with that in the esophagi in mice with other genotypes (Fig. 7E). These results suggest that the differentiation of esophageal epithelium cells is delayed in Keap1FA/–::Nrf2SA/SA mice.
We next examined whether the proliferation of basal cells is enhanced in the esophagus of Keap1FA/–::Nrf2SA/SA mice. For this purpose, we performed immunohistochemical analysis of Ki67, a marker of proliferating cells, and found Ki67-positive cells in basal layers in the mice with one of the five genotypes, but not in Keap1FA/–::Nrf2SA/SA mice (Fig. 7C). In the esophagus of Keap1FA/–::Nrf2SA/SA mice, Ki67-positive cells were found not only in the basal layers, but also in the upper layers. The number of Ki67-positive cells was significantly increased in the esophagus of Keap1FA/–::Nrf2SA/SA mice but did not change notably in the esophagus of mice with other genotypes (Fig. 7F).
Thus, in the Keap1FA/–::Nrf2SA/SA mouse esophagus, the proliferation of basal cells is markedly enhanced. Basal layers are horizontally enlarged, resulting in turbulence of the esophageal epithelium and a rolling epithelial cell layer boundary. In contrast, the top layers were negligibly perturbed, and the number of loricrin-expressing cells was decreased in the Keap1FA/–::Nrf2SA/SA mouse esophagus. These results support the contention that there is an imbalance between the speed of epithelial cell proliferation and differentiation in the Keap1FA/–::Nrf2SA/SA mouse esophagus.
Promotion of keratin gene expression is associated with epithelial proliferation in Keap1FA/–::Nrf2SA/SA esophageal epithelium.
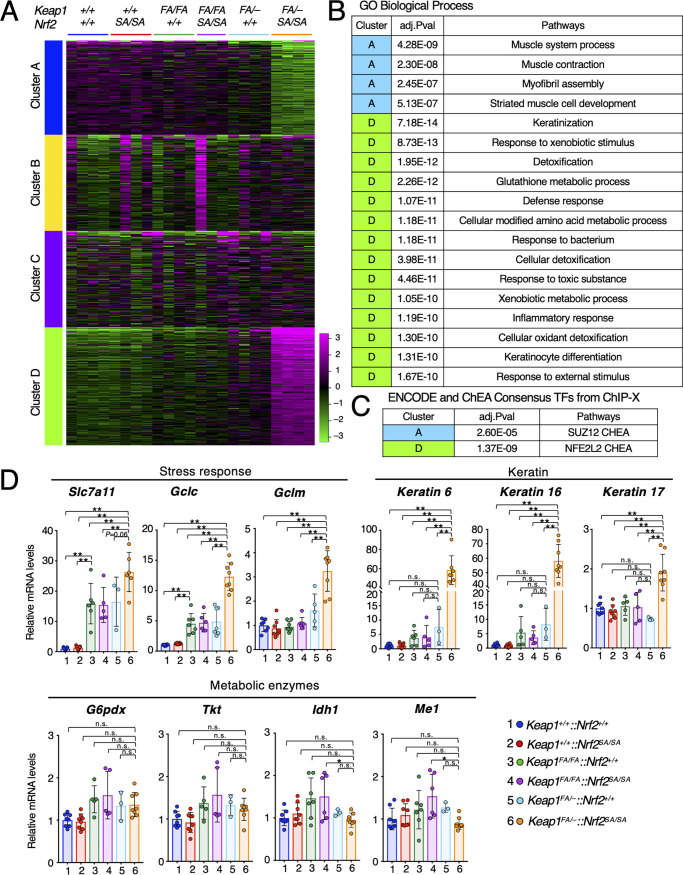
Finally, we examined the mechanisms underlying esophageal hyperplasia in the Keap1FA/–::Nrf2SA/SA mouse esophageal epithelium by analyzing Nrf2 target gene expression. To this end, we reanalyzed the RNA sequencing data of the esophagus described in the previous section. First, we performed k-means clustering. Clusters A and D were clearly identified, and these clusters contained downregulated and upregulated genes, respectively, specifically in Keap1FA/–::Nrf2SA/SA mice (Fig. 8A).
FIG 8.
Promotion of keratin gene expression associated with epithelial cell proliferation in the Keap1FA/–::Nrf2SA/SA esophageal epithelium. (A) k-means clustering of gene expression levels in esophagus samples from Keap1+/+::Nrf2+/+, Keap1+/+::Nrf2SA/SA, Keap1FA/FA::Nrf2+/+, Keap1FA/FA::Nrf2SA/SA, Keap1FA/–::Nrf2+/+, and Keap1FA/–::Nrf2SA/SA mice; (B) enriched pathways in GO Biological Process for clusters A and D. (C) Enriched pathways in ENCODE and ChEA consensus TFs from ChIP-X for clusters A and D. (D) mRNA levels of Nrf2 target genes related to the stress response, metabolic enzymes, and keratin by qRT-PCR. Hprt was used as the internal control. The mRNA levels in the Keap1+/+::Nrf2+/+ mice were set to 1. The bar graphs show the means ± SD. **, P < 0.01 by one-way ANOVA with Tukey’s multiple-comparison test.
We then explored enriched pathways of GO biological processes in clusters A and D. It should be noted that in cluster D, pathways associated with keratinization were enriched, along with canonical Nrf2 target genes in detoxification, oxidative stress response, and glutathione metabolic process pathways (Fig. 8B). While the expression of these genes was markedly upregulated in Keap1FA/–::Nrf2SA/SA mice, pathways related to muscle were enriched in cluster A and downregulated. We surmise that these changes may reflect the observation that muscle cells were relatively decreased in Keap1FA/–::Nrf2SA/SA mice.
To identify transcription factors that regulate the expression of the genes in clusters A and D, we analyzed enriched transcription factor-binding sites by comparing the RNA sequencing data with the chromatin immunoprecipitation database ENCODE and ChEA Consensus TFs from ChIP-X (Fig. 8C). As expected, Nrf2 (Nfe2l2) binding sites were enriched with high probability in the genes in cluster D, supporting the assertion that target genes whose expression is directly activated by Nrf2 are responsible for the esophageal phenotype. Among the genes in cluster A, SUZ12 was enriched with moderate probability, but this relationship remains to be elucidated.
We next examined the mRNA levels of individual direct Nrf2 target genes by qRT-PCR. We first examined the mRNA levels of Nrf2 target genes associated with the stress response and found that the expression of stress response genes Slc7a11, Gclc, and Gclm was significantly increased in Keap1FA/–::Nrf2SA/SA mice (Fig. 8D). Although no histological changes in the esophageal epithelium were observed after Keap1 knockdown alone (Fig. 6 and 7), the mRNA expression of Slc7a11 and Gclc was increased only upon Keap1 knockdown, and the levels were further increased by the additional suppression of β-TrCP-mediated Nrf2 degradation (Fig. 8D). These results indicate that although the expression of oxidative stress response genes is activated, the proliferation of esophageal epithelial cells is not stimulated by Keap1 knockdown alone, demonstrating multiple distinct thresholds for the activation of Nrf2 target genes.
Nrf2 regulates the expression of metabolic enzymes related to the pentose phosphate pathway, which is associated increased cell proliferation (38). We found that the mRNA levels of the metabolic enzyme genes G6pdx, Tkt, Idh1, and Me1 were comparable in mice with all genotypes, suggesting that expansion of esophageal epithelial cells in Keap1FA/–::Nrf2SA/SA mice is not caused by the upregulated expression of these metabolic enzymes (Fig. 8D). The mRNA levels of these enzymes were previously shown to increase in response to Nrf2 activation in the Keap1 knockout mouse esophagus (39) and the Keap1 and Pten double-knockout mouse liver (38). In addition, in the forestomach, upregulated expression of metabolic genes was observed in Keap1FA/–::Nrf2+/+ mice (38). Thus, the thresholds for Nrf2-induced enzyme gene expression are cell type specific and appear to be very high in the esophagus.
It has been reported that Nrf2 is associated with keratin gene regulation, especially the genes coding for keratins 6, 16, and 17 (40). Keratins 6, 16, and 17 are expressed in proliferating epithelial cells (41). We examined the mRNA levels of these keratin genes in the esophagus of mouse esophagi. To our surprise, the mRNA expression levels of both keratins 6 and 16 were induced approximately 60-fold in the esophagus of Keap1FA/–::Nrf2SA/SA mice compared with those in the esophagus of Keap1+/+::Nrf2+/+ mice and 7- to 8-fold higher than those in the esophagus of Keap1FA/–::Nrf2+/+ mice (Fig. 8D). Keratin 17 mRNA levels were increased approximately 2-fold in the esophagus of Keap1FA/–::Nrf2SA/SA mice compared with the level in the esophagus of mice with other genotypes. These results demonstrate that the expression of keratin 6 and keratin 16 genes is under the regulation of Nrf2 and that the thresholds of Nrf2-induced activation of these genes are set in a relatively narrow range and at a very high magnitude compared to those associated with the stress response. These results explain, at least in part, the dramatic increase in keratin expression level in the Keap1FA/–::Nrf2SA/SA mouse esophagus. We surmise that the expression of the genes critical for cell proliferation may be under the same Nrf2 regulatory influences, resulting in the dramatic thickening of the cell layer.
In conclusion, in this study, we demonstrate that Nrf2 activation caused by the suppression of β-TrCP-mediated degradation alone does not induce acquisition of a deleterious phenotype in vivo in Nrf2SA/SA mice. We also found that simultaneous suppression of Keap1-mediated degradation in vivo in Keap1FA/–::Nrf2SA/SA mice does not result in the acquisition of deleterious phenotypes in various organs/tissues, including the kidney and liver. However, the combined suppression of these two Nrf2 degradation pathways specifically promotes the expression of the genes associated with epithelial cell proliferation and keratin production in the esophagus, demonstrating multiple distinct thresholds for gene expression induction in response to an increase in Nrf2 protein in the esophagus.
DISCUSSION
While the β-TrCP pathway has emerged as a pathway of Nrf2 protein degradation, the extent and specificity with which this pathway contributes to Nrf2 homeostasis remain elusive. To clarify the contributions of the pathway to Nrf2 homeostasis, in this study, we examined the specific roles played by the β-TrCP pathway, as well as the cooperative roles that the pathway plays with the Keap1 pathway, through a mouse genetics approach combining a newly generated Nrf2SA substitution knock-in mouse line with graded Keap1 knockdown that we had previously generated. Considering the results of our experiments, we concluded that whereas the Keap1 pathway acts as the main system of Nrf2 degradation, the β-TrCP pathway serves as an auxiliary and safeguard system that functions when Keap1 activity is severely suppressed. Thus, this study demonstrates that the two intracellular Nrf2 protein degradation pathways work cooperatively and act to maintain cellular Nrf2 activity under safe and rigorous controls.
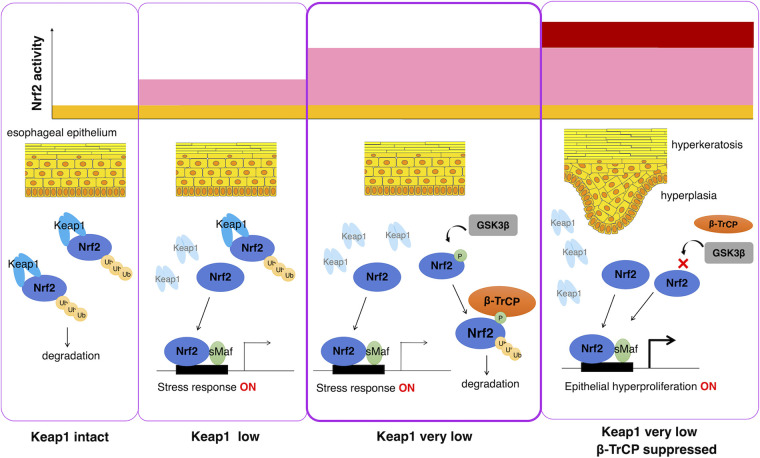
The findings of this study are summarized in Fig. 9. Under normal conditions, in which the Keap1 level remains intact, the majority of Nrf2 protein is degraded through the Keap1 pathway so that Nrf2 activity is maintained at a low level (far left panel). In this situation, loss of β-TrCP pathway activity does not influence cellular Nrf2 activity. When Keap1 activity is moderately suppressed—for instance, by exposure to electrophiles—cellular Nrf2 activity is induced (middle left panel). Even when Keap1 activity is severely suppressed, only the Nrf2-regulated induction of stress response genes was observed, and Nrf2 activation corresponding to the pathological level (middle right panel) was not observed. We surmise that this outcome is a result of the β-TrCP pathway suppression of the Nrf2 protein level, and this level of Keap1 suppression or the corresponding Nrf2 activation level seems to accompany a cellular response to oxidative or electrophilic stress. Therefore, when the β-TrCP pathway is suppressed by Nrf2SA mutation, severe Keap1 pathway suppression causes pathological esophageal hyperkeratosis and hyperplasia (far right panel). These results indicate that the β-TrCP pathway prevents the hyperkeratosis and hyperplasia induced by the severe loss of Keap1. In this regard, the complete loss of Keap1 elicits much more profound Nrf2 activation and acquisition of deleterious phenotypes than were observed in mice with β-TrCP and Keap1 double mutations.
FIG 9.
Contributions of Keap1-mediated and β-TrCP-mediated Nrf2 degradation pathways to Nrf2 activity and esophageal epithelial cell response. Under normal conditions, in which the Keap1 level remains intact, the Keap1-mediated pathway mainly and actively degrades the Nrf2 protein, resulting in low Nrf2 activity (left panel). In this situation, loss of the β-TrCP pathway does not markedly influence cellular Nrf2 activity. When Keap1 is suppressed to low levels, Nrf2 accumulates and activates target gene expression, resulting in a stress response (middle left panel). When Keap1 activity is strongly suppressed to very low levels relative to the control, the β-TrCP-mediated pathway suppresses the Nrf2 protein level. In this situation, Nrf2 activation corresponding to the induction of stress response genes was mainly observed, but Nrf2 activation corresponding to pathological hyperplasia and hyperkeratosis was not observed (middle right panel). When both the Keap1-mediated and β-TrCP-mediated Nrf2 degradation pathways are suppressed, high levels of Nrf2 protein accumulate, resulting in epithelial hyperproliferation and hyperkeratosis (right panel). The β-TrCP-mediated pathway notably prevents the hyperkeratosis and hyperplasia caused by the profound suppression of Keap1 expression.
We surmise that the subcellular localization of the Keap1 and β-TrCP pathways may influence their functional relationship. As Keap1 is localized predominantly in the perinuclear cytoplasm (42), Nrf2 is readily degraded by the Keap1 pathway under unstressed conditions. On the other hand, while both β-TrCP isoforms (β-TrCP1 and β-TrCP2) are localized in the cytoplasm and nucleus (43), components of the ubiquitin-ligating machinery of β-TrCP, such as Cdc34 and Cullin1, are localized mainly in the nucleus. Therefore, β-TrCP-mediated Nrf2 degradation seems to take place, at least to a certain extent, in the nucleus (44). Because the Nrf2 protein is synthesized in the cytoplasm, it seems reasonable to assume that almost all Nrf2 proteins are captured and degraded by the Keap1 pathway, and therefore, the contribution of the β-TrCP pathway may not be necessary in these situations. However, when Keap1-mediated degradation is greatly suppressed (Fig. 9), many Nrf2 proteins are translocated to the nucleus, and the Nrf2 protein is then degraded through the β-TrCP pathway.
We think that another important reason for the differences in the contributions of the Keap1 and β-TrCP pathways is the mechanisms that turn off or turn on these pathways. We have clarified that the Keap1 pathway is constitutively activated by default and that oxidative or electrophilic stresses can turn off Nrf2 degradation through the Keap1 pathway (7). On the other hand, it has been suggested that the β-TrCP pathway can be turned on during cell signaling pathway regulation (45, 46). For instance, GSK3 can phosphorylate the serine 335 and 338 residues of Nrf2, which were replaced by alanine residues in the Nrf2SA mutant, but it has also been shown that phosphorylation of serine 342 and 347 residues is a prerequisite for this reaction (47–49). The kinase critical for phosphorylating the serine 342 and 347 residues is still unknown. In this study, we revealed that additional accumulation of the Nrf2 protein and increased expression of Nrf2 target genes are caused by Nrf2SA substitution mutation under Keap1 knockdown conditions. We speculate that the cascade toward phosphorylation of Nrf2 may be turned on when Keap1 pathway activation is repressed.
The most prominent phenotype in the Keap1FA/–::Nrf2SA/SA mice was observed in the esophageal squamous epithelium, and we did not find deleterious phenotypes in the other tissues, including the kidney or liver. Consistent with this observation, systemic Keap1-null mice show lethal abnormalities in the esophagus (7), and hyperkeratosis and hyperplasia in the esophageal squamous epithelium of systemic Keap1 knockout mice are much more severe than those observed in the esophagus of Keap1FA/–::Nrf2SA/SA mice (7). These observations suggest that the esophageal squamous epithelium is more sensitive to Nrf2 accumulation than other tissues or organs. One plausible reason for this high sensitivity is that the esophagus shows one of the highest expression levels of Nrf2 mRNA (https://www.proteinatlas.org/) (50). Therefore, the Nrf2 protein may accumulate to high levels when the degradation systems cease, making the esophagus sensitive to Nrf2 accumulation.
The esophageal epithelium is directly and continuously exposed to toxic electrophiles and/or oxidative stresses in food, drink, air pollutants, and cigarette smoke. If the stress level is maintained in a mild range, a moderate level of Nrf2 accumulation caused by the regulated suppression of the Keap1-mediated pathway can activate the stress response genes and protect epithelial cells from stress-induced damage. We surmise that, in this situation, the contribution of the β-TrCP pathway to Nrf2 activation is limited or not prevalent. However, when the stress level is high and elicits profound Keap1 suppression, high levels of Nrf2 accumulate. We surmise that some cells in the esophageal epithelium die under these situations, and surviving cells undergo a wound-healing response, which activates AKT signaling (51). Therefore, one possible role that the β-TrCP-mediated pathway plays is that following the activation of AKT signaling, and the β-TrCP-mediated pathway may further activate Nrf2 in collaboration with the suppression of Keap1, leading to regeneration of the esophageal epithelium.
In contrast, somatic mutations in NFE2L2 (encoding NRF2), KEAP1, and CUL3 genes associated with the accumulation of NRF2 protein have been identified in many types of cancers, such as lung adenocarcinoma and squamous cell carcinoma (52). The prognosis of patients with NRF2-accumulating (or NRF2-addicted) cancers has been reported to be poor because Nrf2 accumulation confers resistance to chemo- and radiotherapies as well as conferring proliferative advantages to cancer cells (38). Notably, somatic mutations that have been associated with Nrf2 protein accumulation co-occur with somatic mutations related to the activation of the PI3K/AKT signaling pathway, and this co-occurrence is notable in squamous cell carcinomas of the esophagus, lung, and head-and-neck (53). Therefore, we speculate that in these cancers, inactivation/suppression of both the Keap1-mediated and β-TrCP-mediated pathways contributes to the promotion of cancer malignancy.
In conclusion, our results highlight the cooperative roles of the β-TrCP-mediated and Keap1-mediated degradation pathways in the regulation of Nrf2 activity, especially in the epithelial stress response and resulting pathology.
MATERIALS AND METHODS
Mice.
Keap1FA knockdown (22, 23) and Keap1 knockout (7) mice were described previously. All animal experiments were approved by the Animal Care Committee at Tohoku University.
Generation of Nrf2SA knock-in mice.
A plasmid expressing single-guide RNA (sgRNA) and Cas9 was constructed as described previously (54, 55). The plasmid vector pX330, expressing Cas9 and the sgRNA, was digested with BbsI, and a pair of DNA oligonucleotides recognizing the Nrf2 targeting site (5′-GAA TTC AAT GAC TCT GAC TCT GG-3′) was ligated to a linearized vector. Donor DNA oligonucleotides encoding the replacement of serine with alanine were designed with the following sequence: 5′-AGG CAC AAT GGA ATT CAA TGA CTC TGA CGC CGG CAT TGC ACT GAA CAC GAG TCC CAG CCG AGC GTC C-3′. The plasmid and donor DNA were coinjected into fertilized C57BL/6 mouse eggs.
Screening of Nrf2SA knock-in founder mice.
Genomic DNA of the founder mice was extracted from a piece of the tail. The targeting region in the genomic DNA was amplified by PCR. The PCR products were inserted into a pGEM-T Easy vector (Promega), and the resulting plasmids were transformed into Escherichia coli. Eleven clones from each mouse were selected, and plasmid DNA was purified using a FastGene plasmid minikit (Nippon Genetics). The targeting region of the plasmids was sequenced to identify the founder mice harboring the Nrf2SA mutant allele.
Genotyping of Nrf2SA knock-in mice.
Genomic DNA was extracted from a piece of the mouse tail. The DNA samples were genotyped by using a TaqMan MGB single nucleotide polymorphism (SNP) detection kit (Thermo Fisher Scientific). The targeted region of the Nrf2 gene was amplified using the following primers: forward, 5′-CGC TGA AGG CAC AAT GGA AT-3′; and reverse, 5′-TGG GTC TCC GTA AAT GGA AGA-3′. The WT alleles were detected with a 6-carboxyfluorescein (FAM)-labeled probe (5′-ACT CTG GCA TTT CA-3′), and mutant alleles were detected with a VIC (2′-chloro-7′-phenyl-1,4-dichloro-6-carboxyfluorescein)-labeled probe (5′-CGC CGG CAT TGC A-3′).
Isolation of thioglycolate-elicited peritoneal macrophages.
Mice were intraperitoneally injected with 2 mL of 4% thioglycolate solution 3 days before peritoneal macrophages were collected (56). Peritoneal lavage fluid was centrifuged and suspended in RPMI 1640 medium containing 10% fetal bovine serum (FBS) and penicillin-streptomycin at 37°C and 5% CO2. Adherent macrophages were used for analyses.
Immunoblotting.
Whole-macrophage extracts were prepared in sample buffer (20% glycerol, 4% SDS, 0.1 M Tris-HCl [pH 6.8], and 12% mercaptoethanol). The protein samples were subjected to 7.5% SDS-PAGE and electrotransferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad) using a Transblot Turbo transfer system (Bio-Rad). The membranes were incubated with 5% skim milk in TBST buffer (137 mM NaCl, 2.68 mM KCl, 25 mM Tris-HCl [pH 7.4], and 0.1% Tween 20) at room temperature for 1 h. After blocking, the membranes were incubated in 5% skim milk–TBST containing an anti-Nrf2 rabbit monoclonal antibody at a 1:500 dilution (Cell Signaling Technology; no. 12721) at 4°C overnight. The membranes were washed three times for 5 min each with TBST buffer and incubated at room temperature for 2 h with an anti-rabbit IgG horseradish peroxidase (HRP)-linked antibody (1:3,000 dilution) prepared in Signal Enhancer HIKARI for Western blotting and enzyme-linked immunosorbent assay (ELISA) solution B (Nacalai Tesque). Membranes were washed three times for 5 min each time with TBST buffer. Protein signals were visualized using chemiluminescence (Western Lightning Plus-ECL; PerkinElmer) and a ChemiDoc Touch MP imaging system (Bio-Rad). The blots were incubated in stripping solution at room temperature for 10 min and washed three times for 5 min each time with TBST buffer. The membranes were incubated at room temperature for 1 h with an anti-α-tubulin antibody at a 1:1,000 dilution (Sigma; no. T9026) prepared in Signal Enhancer Hikari for Western blotting and ELISA solution A. The membranes were washed three times for 5 min with TBST buffer and incubated at room temperature for 1 h with an anti-mouse IgG HRP-linked antibody (1:3,000 dilution) prepared in signal enhancer Hikari for Western blotting and ELISA solution B. The membranes were washed three times for 5 min each with TBST buffer. Protein signals were visualized using chemiluminescence (Chemi-Lumi One L; Nacalai Tesque) and a ChemiDoc Touch MP imaging system (Bio-Rad). Quantification of the immunoblots was performed using Image Lab software version 6.1 (Bio-Rad).
RNA extraction and qRT-PCR.
Total RNA was extracted with Sepazol RNA I Super G reagent (Nacalai Tesque) and reverse-transcribed with a ReverTra Ace qPCR RT kit (Toyobo) according to the manufacturer’s instructions. The resulting cDNA was used as a template for quantitative reverse transcription-PCR (qRT-PCR) using a TaqMan probe or SYBR green with a QuantStudio 6 Real-time PCR analyzer (Thermo Fisher Scientific). The primers and probes are described in Table 1.
TABLE 1.
Primers and probes used for qRT-PCR
| Gene | Primer or probea | Sequence (5′→3′) |
|---|---|---|
| Hprt | F | CTGGTGAAAAGGACCTCTCG |
| R | TGAAGTACTCATTATAGTCAAGGG | |
| P | ATCCAACAAAGTCTGGCCTGTATCCAAC | |
| Nqo1 | F | AGCTGGAAGCTGCAGACCTG |
| R | CCTTTCAGAATGGCTGGCA | |
| P | ATTTCAGTTCCCATTGCAGTGGTTTGGG | |
| Nrf2 | F | CAAGACTTGGGCCACTTAAAAGAC |
| R | AGTAAGGCTTTCCATCCTCATCAC | |
| P | AGGCGGCTCAGCACCTTGTATCTTGA | |
| Keratin 19 | F | CGGTGGAAGTTTTAGTGGGA |
| R | AGTAGGAGGCGAGACGATCA | |
| Albumin | F | GACGTGTGTTGCCGATGAGT |
| R | GTTTTCACGGAGGTTTGGAATG | |
| Keratin 5 | F | CAAGAAGCAGTGTGCCAA |
| R | CCGCGTTGCTCAGC | |
| P | CTCCAGAACGCCATTGCTGA | |
| Keratin 14 | F | CAGCCCCTACTTCAAGACCA |
| R | GGCTCTCAATCTGCATCTCC | |
| Loricrin | F | GCGCCTACCTGGCC |
| R | GCTCTGTTGTCTCCGTTG | |
| P | TGCAAGTAAGGTCACCGGGTT | |
| Slc7a11 | F | TGGGTGGAACTGCTCGTAAT |
| R | AGGATGTAGCGTCCAAATGC | |
| Gclc | F | ATCTGCAAAGGCGGCAAC |
| R | ACTCCTCTGCAGCTGGCTC | |
| P | ACGGGTGCAGCAAGGCCCA | |
| Gclm | F | TGGAGCAGCTGTATCAGTGG |
| R | AAATCTGGTGGCATCACACA | |
| G6pdx | F | GTCCAGAATCTCATGGTGCTGA |
| R | GCAATGTTGTCTCGATTCCAGA | |
| Tkt | F | CGAAACCCTCACAATGATCG |
| R | TTCCTCAGGTTCAGCAGCTC | |
| Idh1 | F | AAAATATCCCCCGGCTAGTG |
| R | TCTCTACTTTTCCAGGCCCA | |
| Me1 | F | GGAGCTCCAGGTCCTTAGAA |
| R | TGAGCACGCTGTAGAAGAGC | |
| Keratin 6 | F | TTGGACCAGTCAACATCTCTGTG |
| R | ACCCCCGGCACTGCC | |
| P | CAGTCCACCGTGTCCAGCGGCTA | |
| Keratin 16 | F | AGCTGATGACTTCAGGACCAAGT |
| R | TGATGTCACCCTCCACGGA | |
| P | CGAGAATGAGCTGTTCTTGCGGCA | |
| Keratin 17 | F | GATGACTTCCGTACCAAGTTTGAGA |
| R | CCATTGATGTCGGCCTCC | |
| P | AGCAGGCTCTGCGCATGAGCG |
F, forward primer; R, reverse primer; P, probe.
RNA sequencing analysis.
Total RNA was extracted with Sepazol RNA I Super G reagent (Nacalai Tesque). RNA quantity and purity were assessed using NanoPhotometer NP80 (Implen). RNA integrity (RIN) was assessed using TapeStation (Agilent). rRNA was removed using an MGIEasy rRNA depletion kit (MGI). Double-stranded DNA (dsDNA) libraries were created from the rRNA-depleted eluate using an MGIEasy RNA directional library prep kit (MGI). The libraries were sequenced on a DNBSEQ-G400RS (MGI) system with a DNBSEQ-G400RS high-throughput sequencing kit V1.0 (MGI) to obtain 150 paired-end reads. The sequencing data were processed on the Galaxy server (https://usegalaxy.org) (57). Transcript per million (TPM) normalized read counts were generated using Salmon quant script (Galaxy version 1.5.1+galaxy0) (58) with mouse reference transcript sequences (version 28) downloaded from GENCODE database (https://www.gencodegenes.org) (59). The heat maps were created using heatmap.2 function in the gplots package (60). The k-means clustering and pathway analysis were conducted on the iDEP platform (http://bioinformatics.sdstate.edu/idep94/).
Histological analyses.
The esophagus, liver, and kidney were fixed with 10% formalin (Mildform 10N; Wako) and embedded in paraffin. The sections were stained with HE and PAS.
Immunohistochemistry.
The esophagus was fixed with 10% formalin (Mildform 10N; Wako) and embedded in paraffin. The sections (4 μm) were rehydrated, autoclaved in 10 mM sodium citrate buffer (pH 6.0) for antigen retrieval, treated with 3% H2O2, blocked in Protein Block Serum-Free Ready-to-Use (Dako; X0909), and sequentially incubated with specific primary antibodies and with the respective secondary antibodies. The following primary antibodies were used: anti-keratin 14, rabbit polyclonal antibody at a 1:1,000 dilution (BioLegend; Poly19053); antiloricrin, rabbit polyclonal antibody at a 1:200 dilution (BioLegend; Poly19051); anti-Ki67, rat monoclonal antibody at a 1:1,000 dilution (BioLegend; 16A8); and anti-NQO1, goat polyclonal antibody at a 1:2,000 dilution (Abcam; ab2346).
Data availability.
Raw data from RNA sequencing analysis are available in the DNA Data Bank of Japan (DDBJ) under accession no. PRJDB13492.
ACKNOWLEDGMENTS
We thank Eriko Naganuma, Hiromi Suda, Keiko Tateno, Haruka Kobayashi, Makiko Hayashi, and the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support.
This work was supported in part by AMED-P-CREATE (JP21cm0106101 to M.Y.) from the Japan Agency for Medical Research and Development (AMED) and KAKENHI 19H05649 (to M.Y.) and 20K08747 (to M.S.) from the Japan Society for the Promotion of Science (JSPS).
Contributor Information
Mikiko Suzuki, Email: suzukimikiko@med.tohoku.ac.jp.
Masayuki Yamamoto, Email: masiyamamoto@med.tohoku.ac.jp.
REFERENCES
- 1.Yamamoto M, Kensler TW, Motohashi H. 2018. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol Rev 98:1169–1203. 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird L, Yamamoto M. 2020. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol 40:e00099-20. 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki M, Otsuki A, Keleku-Lukwete N, Yamamoto M. 2016. Overview of redox regulation by Keap1-Nrf2 system in toxicology and cancer. Curr Opin Toxicol 1:29–36. 10.1016/j.cotox.2016.10.001. [DOI] [Google Scholar]
- 4.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 5.Ohkoshi A, Suzuki T, Ono M, Kobayashi T. 2013. Roles of Keap1-Nrf2 system in upper aerodigestive tract carcinogenesis. Cancer Prev Res (Phila) 6:149–159. 10.1158/1940-6207.CAPR-12-0401-T. [DOI] [PubMed] [Google Scholar]
- 6.Horiuchi M, Taguchi K, Hirose W, Tsuchida K, Suzuki M, Taniyama Y, Kamei T, Yamamoto M. 2021. Cellular Nrf2 levels determine cell fate during chemical carcinogenesis in esophageal epithelium. Mol Cell Biol 41:e00536-20. 10.1128/MCB.00536-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 35:238–245. 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T, Seki S, Hiramoto K, Naganuma E, Kobayashi EH, Yamaoka A, Baird L, Takahashi N, Sato H, Yamamoto M. 2017. Hyperactivation of Nrf2 in early tubular development induces nephrogenic diabetes insipidus. Nat Commun 8:14577. 10.1038/ncomms14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida E, Suzuki T, Morita M, Taguchi K, Tsuchida K, Motohashi H, Doita M, Yamamoto M. 2018. Hyperactivation of Nrf2 leads to hypoplasia of bone in vivo. Genes Cells 23:386–392. 10.1111/gtc.12579. [DOI] [PubMed] [Google Scholar]
- 10.Taguchi K, Hirano I, Itoh T, Tanaka M, Miyajima A, Suzuki A, Motohashi H, Yamamoto M. 2014. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol Cell Biol 34:900–913. 10.1128/MCB.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139. 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24:8477–8486. 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang DD, Lo S-C, Cross JV, Templeton DJ, Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24:10941–10953. 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito R, Suzuki T, Hiramoto K, Asami S, Naganuma E, Suda H, Iso T, Yamamoto H, Morita M, Baird L, Furusawa Y, Negishi T, Ichinose M, Yamamoto M. 2016. Characterizations of three major cysteine sensors of Keap1 in stress response. Mol Cell Biol 36:271–284. 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T, Muramatsu A, Saito R, Iso T, Shibata T, Kuwata K, Kawaguchi S-I, Iwawaki T, Adachi S, Suda H, Morita M, Uchida K, Baird L, Yamamoto M. 2019. Molecular mechanism of cellular oxidative stress sensing by Keap1. Cell Rep 28:746–758.e4. 10.1016/j.celrep.2019.06.047. [DOI] [PubMed] [Google Scholar]
- 17.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. 2006. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol 26:2887–2900. 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukutomi T, Takagi K, Mizushima T, Ohuchi N, Yamamoto M. 2014. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol Cell Biol 34:832–846. 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horie Y, Suzuki T, Inoue J, Iso T, Wells G, Moore TW, Mizushima T, Dinkova-Kostova AT, Kasai T, Kamei T, Koshiba S, Yamamoto M. 2021. Molecular basis for the disruption of Keap1-Nrf2 interaction via hinge & latch mechanism. Commun Biol 4:576. 10.1038/s42003-021-02100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou Y-S, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S-I, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213–223. 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, Waguri S, Sou Y-S, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee M-S, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. 2013. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51:618–631. 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Taguchi K, Maher JM, Suzuki T, Kawatani Y, Motohashi H, Yamamoto M. 2010. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol Cell Biol 30:3016–3026. 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. 2006. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun 339:79–88. 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi M, Kuga A, Suzuki M, Panda H, Kitamura H, Motohashi H, Yamamoto M. 2020. Microenvironmental activation of Nrf2 restricts the progression of Nrf2-activated malignant tumors. Cancer Res 80:3331–3344. 10.1158/0008-5472.CAN-19-2888. [DOI] [PubMed] [Google Scholar]
- 25.Keleku-Lukwete N, Suzuki M, Otsuki A, Tsuchida K, Katayama S, Hayashi M, Naganuma E, Moriguchi T, Tanabe O, Engel JD, Imaizumi M, Yamamoto M. 2015. Amelioration of inflammation and tissue damage in sickle cell model mice by Nrf2 activation. Proc Natl Acad Sci USA 112:12169–12174. 10.1073/pnas.1509158112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uruno A, Furusawa Y, Yagishita Y, Fukutomi T, Muramatsu H, Negishi T, Sugawara A, Kensler TW, Yamamoto M. 2013. The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol 33:2996–3010. 10.1128/MCB.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. 2004. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 279:31556–31567. 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 28.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. 2011. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31:1121–1133. 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rada P, Rojo AI, Evrard-Todeschi N, Innamorato NG, Cotte A, Jaworski T, Tobón-Velasco JC, Devijver H, García-Mayoral MF, Van Leuven F, Hayes JD, Bertho G, Cuadrado A. 2012. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/β-TrCP axis. Mol Cell Biol 32:3486–3499. 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojo AI, Rada P, Mendiola M, Ortega-Molina A, Wojdyla K, Rogowska-Wrzesinska A, Hardisson D, Serrano M, Cuadrado A. 2014. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid Redox Signal 21:2498–2514. 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stambolic V, Woodgett JR. 1994. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J 303:701–704. 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw M, Cohen P. 1999. Role of protein kinase B and the MAP kinase cascade in mediating the EGF-dependent inhibition of glycogen synthase kinase 3 in Swiss 3T3 cells. FEBS Lett 461:120–124. 10.1016/s0014-5793(99)01434-9. [DOI] [PubMed] [Google Scholar]
- 33.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785–789. 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 34.Kim JW, Botvinnik OB, Abudayyeh O, Birger C, Rosenbluh J, Shrestha Y, Abazeed ME, Hammerman PS, DiCara D, Konieczkowski DJ, Johannessen CM, Liberzon A, Alizad-Rahvar AR, Alexe G, Aguirre A, Ghandi M, Greulich H, Vazquez F, Weir BA, Van Allen EM, Tsherniak A, Shao DD, Zack TI, Noble M, Getz G, Beroukhim R, Garraway LA, Ardakani M, Romualdi C, Sales G, Barbie DA, Boehm JS, Hahn WC, Mesirov JP, Tamayo P. 2016. Characterizing genomic alterations in cancer by complementary functional associations. Nat Biotechnol 34:539–546. 10.1038/nbt.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T, Uruno A, Yumoto A, Taguchi K, Suzuki M, Harada N, Ryoke R, Naganuma E, Osanai N, Goto A, Suda H, Browne R, Otsuki A, Katsuoka F, Zorzi M, Yamazaki T, Saigusa D, Koshiba S, Nakamura T, Fukumoto S, Ikehata H, Nishikawa K, Suzuki N, Hirano I, Shimizu R, Oishi T, Motohashi H, Tsubouchi H, Okada R, Kudo T, Shimomura M, Kensler TW, Mizuno H, Shirakawa M, Takahashi S, Shiba D, Yamamoto M. 2020. Nrf2 contributes to the weight gain of mice during space travel. Commun Biol 3:496. 10.1038/s42003-020-01227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcolea MP, Greulich P, Wabik A, Frede J, Simons BD, Jones PH. 2014. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol 16:612–619. 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doupé DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, Jones PH. 2012. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337:1091–1093. 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22:66–79. 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Fu J, Xiong Z, Huang C, Li J, Yang W, Han Y, Paiboonrungruan C, Major MB, Chen K-N, Kang X, Chen X. 2019. Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J Biol Chem 294:327–340. 10.1074/jbc.RA118.005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Fan X, Cui T, Dang E, Wang G. 2017. Nrf2 promotes keratinocyte proliferation in psoriasis through up-regulation of keratin 6, keratin 16, and keratin 17. J Invest Dermatol 137:2168–2176. 10.1016/j.jid.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Yin M, Zhang L-J. 2019. Keratin 6, 16 and 17—critical barrier alarmin molecules in skin wounds and psoriasis. Cells 8:807. 10.3390/cells8080807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watai Y, Kobayashi A, Nagase H, Mizukami M, McEvoy J, Singer JD, Itoh K, Yamamoto M. 2007. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells 12:1163–1178. 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- 43.Putters J, Slotman JA, Gerlach JP, Strous GJ. 2011. Specificity, location and function of βTrCP isoforms and their splice variants. Cell Signal 23:641–647. 10.1016/j.cellsig.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. 2013. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32:3765–3781. 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayes JD, Ebisine K, Sharma RS, Chowdhry S, Dinkova-Kostova AT, Sutherland C. 2016. Regulation of the CNC-bZIP transcription factor Nrf2 by Keap1 and the axis between GSK-3 and β-TrCP. Curr Opin Toxicol 1:92–103. 10.1016/j.cotox.2016.10.003. [DOI] [Google Scholar]
- 46.Cuadrado A. 2015. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/β-TrCP. Free Radic Biol Med 88:147–157. 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 47.Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. 2001. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105:721–732. 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 48.ter Haar E, Coll JT, Austen DA, Hsiao H-M, Swenson L, Jain J. 2001. Structure of GSK3β reveals a primed phosphorylation mechanism. Nat Struct Biol 8:593–596. 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- 49.Frame S, Cohen P, Biondi RM. 2001. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell 7:1321–1327. 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 50.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 51.Squarize CH, Castilho RM, Bugge TH, Gutkind JS. 2010. Accelerated wound healing by mTOR activation in genetically defined mouse models. PLoS One 5:e10643. 10.1371/journal.pone.0010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502:333–339. 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, Bailey MH, Liang W-W, Foltz SM, Shmulevich I, Ding L, Heins Z, Ochoa A, Gross B, Gao J, Zhang H, Kundra R, Kandoth C, Bahceci I, Dervishi L, Dogrusoz U, Zhou W, Shen H, Laird PW, Way GP, Greene CS, Liang H, Xiao Y, Wang C, Iavarone A, Berger AH, Bivona TG, Lazar AJ, Hammer GD, Giordano T, Kwong LN, McArthur G, Huang C, Tward AD, Frederick MJ, McCormick F, Meyerson M, Van Allen EM, Cancer Genome Atlas Research Network, et al. 2018. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173:321–337.e10. 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M. 2016. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic Biol Med 91:45–57. 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3:3355. 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki T, Kelly VP, Motohashi H, Nakajima O, Takahashi S, Nishimura S, Yamamoto M. 2008. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem 283:2021–2030. 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- 57.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland JE, Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I, Berry A, Bignell A, Boix C, Carbonell Sala S, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Howe KL, Hunt T, Izuogu OG, Johnson R, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Riera FC, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Wolf MY, Xu J, Yang YT, Yates A, Zerbino D, Zhang Y, Choudhary JS, Gerstein M, et al. 2021. GENCODE 2021. Nucleic Acids Res 49:D916–D923. 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. 2020. gplots: various R programming tools for plotting data. R package version 3.1.1. https://CRAN.R-project.org/package=gplots.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data from RNA sequencing analysis are available in the DNA Data Bank of Japan (DDBJ) under accession no. PRJDB13492.