Abstract
Tuberculosis (TB) and HIV co-infection claims many lives every year. This study assessed immune responses in Mycobacterium tuberculosis–infected lymph node tissues from HIV-negative and HIV-positive patients compared with the peripheral circulation with a focus on myeloid cells and the cell-signaling enzymes, inducible nitric oxide synthase, and arginase (Arg)-1. Methods included immunohistochemistry or confocal microscopy and computerized image analyses, quantitative real-time PCR, multiplex Luminex, and flow cytometry. These findings indicate enhanced chronic inflammation and immune activation in TB/HIV co-infection but also enhanced immunosuppressive responses. Poorly formed necrotic TB granulomas with a high expression of M. tuberculosis antigens were elevated in TB/HIV–co-infected lymph nodes, and inducible nitric oxide synthase and Arg-1 expression was significantly higher in TB/HIV–co-infected compared with HIV-negative TB or control tissues. High Arg-1 expression was found in myeloid cells with a phenotype characteristic of myeloid-derived suppressor cells (MDCS) that were particularly abundant in TB/HIV–co-infected tissues. Accordingly, Lin−/HLA-DRlow/int/CD33+/CD11b+/CD15+ granulocytic myeloid-derived suppressor cells were significantly elevated in blood samples from TB/HIV–co-infected patients. CD15+ myeloid-derived suppressor cells correlated with plasma HIV viral load and M. tuberculosis antigen load in tissue but were inversely associated with peripheral CD4 T-cells counts. Enhanced chronic inflammation driven by M. tuberculosis and HIV co-infection may promote Arg-1–expressing MDSCs at the site of infection thereby advancing TB disease progression.
HIV infection is a major comorbidity of tuberculosis (TB) that is present in 15% of all TB deaths and currently affects approximately 15 million people around the world.1 Cellular immunity is gradually destroyed in Mycobacterium tuberculosis as well as HIV infection, manifesting in severe depletion of CD4+ T cells and dysfunctional activity of macrophages and dendritic cells (DCs). Activation of macrophages and DCs results in elaboration of proinflammatory cytokines, induction of antimicrobial effector pathways, and priming of specific Th1 and cytolytic effector T cells that are required to kill intracellular M. tuberculosis. Th1 cytokines, such as IL-1β, tumor necrosis factor-α, and interferon (IFN)-γ, are instrumental in activation, recruitment, and organization of immune cells at the site of M. tuberculosis infection, which results in TB granuloma formation. TB granulomas are a hallmark of human TB and defined as dynamic cellular clusters of M. tuberculosis–infected macrophages, which are surrounded by a peripheral cuff of T cells and other immune cells.2,3 Although productive granulomas are considered to isolate and limit spread of M. tuberculosis infection,4 their formation may fail in individuals with a compromised immune system, including TB/HIV–co-infected patients.5 This process is likely dependent on the degree of HIV-induced immunosuppression from mild or moderate to more advanced immunosuppression characterized by reduced cell recruitment to the TB granuloma and release of large numbers of M. tuberculosis bacilli.6
Myeloid cells have a central role in TB and HIV infection, including a highly heterogenous population of mononuclear myeloid cells, such as not only monocytes, macrophages, and DCs but also granulocytic myeloid cells and myeloid-derived suppressor cells (MDSCs).7 MDSCs are pathologically activated myeloid cells with immunosuppressive traits that were originally described in the tumor immunology field,7 although these cells are also involved in chronic inflammatory conditions, including TB8 and HIV infection.9 MDSCs comprise a heterogenous population of monocytic or granulocytic subsets that also contain immature progenitors or early-stage MDSCs.10 Such myeloid cell subsets impair protective T-cell responses and may contribute to the inability to eradicate these intracellular pathogens at mucosal sites and instead promote persistent infection.11
It has been proposed that amino acid metabolism in myeloid cells provides important cues that regulate both innate and adaptive immune responses in different diseases.12 Mouse data suggest that classically activated proinflammatory macrophages (M1) produce nitric oxide (NO) and engage in bacterial killing, as opposed to alternatively activated anti-inflammatory macrophages (M2) that instead use arginase (Arg)-1 to induce fibrosis and prohealing responses.13,14 The inducible nitric oxide (iNOS) and Arg-1 regulate L-arginine levels in tissue by competing for cellular L-arginine, which is the substrate for both of these enzymes.15 M1 or M2 polarized macrophages stimulate Th1/cellular or Th2/antibody responses, respectively, which further amplify M1/M2 responses via production of IFN-γ or IL-4.16 Macrophages and DCs could also express the immunosuppressive enzyme indoleamine 2,3-dioxygenase (IDO) that is involved in tryptophan degradation, which may impair both innate and adaptive immune responses and thus assist microbes to escape eradication by the immune system. Tissue deprivation of L-arginine17 or tryptophan18 contributes to not only reduced proliferation and activation of T cells but also induction of regulatory T (Treg) cells. iNOS, Arg-1, and IDO are also factors implicated in MDSC-mediated immunosuppression, with the main target being effector T cells.7,19
TB/HIV co-infection often manifests as cervical lymphadenopathy, a finding that is also common in patients infected by TB or HIV alone.20 Accordingly, human lymph node tissue and blood from HIV-negative or HIV-positive TB-infected individuals were used to explore the effect of HIV co-infection in TB disease, particularly in TB granuloma formation. Specific subsets of myeloid cells and effector molecules as well as immunosuppressive pathways in the local environment of M. tuberculosis–infected lymph nodes were studied.
Material and Methods
Study Participants and Diagnosis
Participants were recruited at the Tikur Anbessa University Hospital in Addis Ababa, Ethiopia, after providing signed informed consent. Inclusion criteria were individuals >18 years of age who were HIV negative or HIV positive and had cervical lymph node enlargement and normal chest radiographic findings. Common clinical signs of TB infection, such as fever, sweating, or weight loss, were not used to stratify patients. Exclusion criteria were a history of previous TB or >1 week of antimicrobial chemotherapy, presence of pulmonary involvement consistent with TB, current or prior antiretroviral therapy, or lack of consent to HIV screening. Individuals with nonspecific reactive enlargement of the cervical lymph nodes with no signs of TB disease were recruited as controls. Ethical approval of the study was obtained from the national ethical review boards in Ethiopia and Sweden.
The study participants (n = 23) were divided into three groups: i) TB-single infection (n = 8), ii) TB/HIV co-infection (n = 8) and iii) HIV-negative controls (n = 3) and HIV-positive controls (n = 4) without TB disease (Table 1). TB diagnosis was based on a combination of histopathological evidence as well as clinical symptoms, including clinical response after completed anti-TB chemotherapy, which was defined as resolution of lymph node enlargement. TB-positive specimens (n = 16) revealed a granulomatous inflammation with epithelioid cell clusters, multinucleated giant cells, and caseating necrosis consistent with TB. The histologic features of TB-negative control lymph node samples (n = 7) typically demonstrated a reactive lymphoid hyperplasia with no granuloma formation.
Table 1.
Clinical and Laboratory Characteristics of Study Participants
| Characteristic∗ | TB/HIV-negative patients (n = 8) | TB/HIV-positive patients (n = 8) | P value† | Controls (n = 7; 4 HIV positive) | P value‡ | P value§ |
|---|---|---|---|---|---|---|
| Age, years | 30 (20–46) | 34 (25–49) | >0.999 | 28 (20–40) | >0.999 | 0.423 |
| Male/female sex | 5/3 | 5/3 | 3/3 (1 NA) | |||
| Histology, n (%)¶ | 8 (100) | 8 (100) | 0 (0) | |||
| TST, mm | 17 (13–25) | 17 (0–25) | 10 (0–20) | |||
| QFT, IU | 11 (0.32–23.4) | 3.3 (0.34–7.9) | 0.152 | ND | ||
| CD4 T-cell counts, cells/mm3 | 677 (448–1277) | 203 (52–488) | 0.001 | 457 (230–886) | 0.121 | 0.021 |
| CD8 T-cell counts, cells/mm3 | 419 (168–670) | 818 (368–1528) | 0.072 | 1048 (389–1961) | 0.007 | 0.367 |
| CD4:CD8 ratio | 1.61 (0.68–2.67) | 0.24 (0.10–0.34) | 0.0003 | 0.53 (0.13–1.13) | 0.002 | 0.114 |
| Viral load, copies/mL | ND | 254,000 (20,000–550,000) | 30,500 (12,000–72,000) | 0.024 | ||
| Hemoglobin, g/dL | 15 (12.4–21.4) | 11 (8.0–13.7) | 0.021 | 14 (10.6–16.7) | 0.977 | 0.046 |
| WBC count, mm3 | 5851 (4100–7100) | 9400 (3800–13,300) | 0.050 | 6450 (4300–8900) | 0.628 | 0.132 |
| ESR, mm/h | 28 (5–50) | 81 (30–130) | 0.011 | 22 (1–56) | 0.609 | 0.008 |
ESR, erythrocyte sedimentation rate; NA, not available; ND, not determined; QFT, QuantiFERON-TB Gold in-Tube; TB, tuberculosis; TST, tuberculin skin test; WBC, white blood cell.
Data are presented as mean (range) unless otherwise stated.
TB versus TB/HIV (Kruskal-Wallis test and Dunn posttest).
TB versus controls (Kruskal-Wallis test and Dunn posttest).
TB/HIV versus controls (Kruskal-Wallis test and Dunn posttest).
Histologically confirmed TB diagnosis of excisional biopsy of cervical lymph nodes showing typical morphological changes consistent with TB.
Clinical Samples and Procedures
An excisional biopsy of a single enlarged cervical lymph node was obtained from each study participant, along with matched peripheral blood. Lymph node biopsy specimens were divided into two parts for future analysis: tissue fixed in formalin (Sigma-Aldrich, St. Louis, MO) and embedded in paraffin blocks and tissue preserved in RNAlater (BioRad, Hercules, CA) at −85°C. Hematoxylin and eosin (H&E) (Sigma-Aldrich)–stained tissue sections were used for histopathological diagnosis of the lymph node samples. Plasma samples were used for HIV screening and determination of plasma HIV viral load, according to the national guidelines in Ethiopia. Blood chemistry analyses included assessment of hemoglobin, white blood cell counts , and erythrocyte sedimentation rate (ESR). Immunologic status was assessed using the tuberculin skin test (Statens Serum Institut, København, Denmark), whereas whole blood samples were used for peripheral CD4/CD8 T-cell counts (FACSCount; BD Biosciences, Franklin Lakes, NJ), and IFN-γ production was measured with the QuantiFERON-TB Gold In-Tube (Cellestis, Victoria, Australia) according to the manufacturer's instructions. A tuberculin skin test reaction measured at 48 to 72 hours after tuberculin (Statens Serum Institute) injection was considered a positive result when the transverse induration was ≥10 mm for TB and ≥5 mm for TB/HIV–co-infected patients, whereas QuantiFERON-TB Gold In-Tube results above a cut-off of 0.35 IU/mL were considered positive.
Antigen Retrieval in Paraffin-Embedded Lymph Node Tissues
Formalin-fixed, paraffin-embedded tissue biopsy specimens were cut into 3-μm-thick sections and mounted on Superfrost Plus microscope slides (Thermo Fisher Scientific/Menzel-Gläser, Waltham, MA). The tissue was deparaffinised using xylene (Thermo Fisher Scientific) for 3 × 5 minutes followed by rehydration with decreasing concentrations of ethanol (Solveco, Stockholm, Sweden) at 100% for 2 × 5 minutes, 95% for 5 minutes, and 70% for 5 minutes. Dewaxed slides were rinsed with Tris-buffered saline (TBS; Sigma-Aldrich), and endogenous peroxidase was blocked with 3% H2O2 (Sigma) for 30 minutes before antigen retrieval was performed by microwave boiling in 0.01 mol/L citrate buffer (pH 6) (Sigma-Aldrich) for 2 × 2.5 minutes. After cooling and washing in TBS, sectioned tissues were used for immunostains as described below.
Immunostaining of Lymph Node Tissues
Deparaffinized and rehydrated tissue sections were blocked for 30 minutes with 5% fetal calf serum (Sigma-Aldrich) in TBS before staining with immunohistochemistry (IHC) or immunofluorescence (IF). IHC was performed using the Vectastain Elite ABC-kit (Vector Laboratories, Burlingame, CA) as previously described,21, 22, 23 and positive stain was developed using a diaminobenzidine substrate (Vector Laboratories) and hematoxylin (Sigma-Aldrich) for nuclear counterstaining. For co-localization of immune molecules, 2- to 4-color stains were performed using indirect IF and DAPI mounting media (Vector Laboratories) for visualization of cell nuclei.
Primary antibodies used for IHC were monoclonal mouse anti-human CD3, CD4, and CD8 (BD Biosciences, San Jose, CA), CD20 and collagen type I (Abcam, Cambridge, UK), CD68 and CD163 (Novocastra, Leica Biosystems, Wetzlar, Germany), monoclonal rabbit anti-human CD11b and CD11c (Abcam) and polyclonal rabbit anti-human iNOS (Thermo Fisher Scientific), Arg-1 and CD163 (GeneTex, Irvine, CA), and IDO-1 (Sigma). A polyclonal rabbit anti–M. tuberculosis bacteria antibody (Invitrogen, Waltham, MA) was used for detection of M. tuberculosis, whereas a polyclonal rabbit anti-HIV-1 p55+p24+p17 antibody (BIOSS Antibodies, Boston, MA) was used for detection of HIV antigens in tissue. Biotinylated secondary antibodies for IHC included goat anti-mouse IgG and swine anti-rabbit F(ab’)2 from Dako Cytomation (Jena, Germany). Tissue sections stained with secondary antibodies only were used as negative controls. Primary antibodies used for IF were monoclonal mouse anti-human CD68, CD163, CD33, CD3, CD56, CD123 (Novocastra), CD15, CD20, MAC387 (Dako), HLA-DR (Abcam), polyclonal rabbit anti-human iNOS (Thermo Fisher Scientific), Arg-1, and CD163 (GeneTex). Primary antibodies were added and incubated for 90 minutes (polyclonal antibodies) or overnight (monoclonal antibodies) in room temperature. Before addition of the secondary antibodies, tissue sections were washed with TBS and blocked for 30 minutes with 1% normal goat serum (Sigma-Aldrich) in TBS. The secondary antibodies were added and incubated for 30 minutes (IF) or 1 hour (IHC) in the dark. Fluorophore-conjugated secondary antibodies used for IF included goat anti-mouse IgG Alexa-488, goat anti-mouse IgG2b Alexa/Fluor-555, and goat anti-mouse IgG1 Alexa/Fluor-647 (Invitrogen), goat pAb to Ms IgM Daylight-488 (Abcam), as well as goat anti-rabbit IgG Alexa-594 (Molecular Probes, Eugene, OR).
Quantitative Image Analysis of Lymph Node Tissues
Acquired computerized image analysis was used to quantify IHC staining by transferring digital images of the stained tissue samples from a DMR-X microscope to a computerized Quantimet 5501W image analyzer (Leica Microsystems). Positive immunostaining was quantified at the single-cell level in 10 to 50 high-power fields with a mean total area of 1.5 × 105 μm/field, using the Qwin 550 v.2 software program (Leica Imaging Systems, Wetzlar, Germany). Approximately 50 microscopy fields were assessed in TB- and TB/HIV-infected lymph nodes and approximately 10 to 30 fields for control lymph nodes, which were mostly smaller compared with the TB-infected lymph nodes. Protein expression was determined as the percentage positive area of the total relevant cell area defined as the nucleated and cytoplasmic area within the tissue biopsy, excluding fibrotic and necrotic tissue areas. Occasionally, positive immunostaining determined inside the TB granulomas was compared with tissue outside the granuloma using a tissue excluder function of the software.
Confocal Analysis of Lymph Node Tissues
For quantification of IF stains, a Nikon A1R confocal microscope coupled to a 32-channel spectral detector was used (Nikon Instruments, Tokyo, Japan). Reference spectra were collected for each fluorophore by imaging samples single stained with the respective probes, using a single excitation source. The reference spectra were stored in a spectral library and used for off-line unmixing of all samples imaged by the spectral unmixing algorithm in the NIS elements AR software version 4.50.00 (Nikon Instruments). For analysis of colocalization, the freeware ImageJ version 2.0.0 (NIH, Bethesda, MD; http://imagej.nih.gov/ij) was used. In short, each unmixed channel was thresholded and segmented. Next, the segmented area was measured and masked before the masked areas were transformed into 8-bit images and colocalization assessed using the colocalization plugin, which highlights colocalized points of two 8-bit images. The area of the colocalized points was measured and, in the case of triple-colocalization, masked and transformed into a new 8-bit image. The new image was subsequently analyzed for colocalization against a third channel, using the same plugin.
Light Microscopy Analysis of H&E-Stained Lymph Node Tissues
H&E stain and light microscopy were used to assess the different types of granulomas in M. tuberculosis–infected lymph node tissues (ie, n = 8 TB-infected and n = 8 TB/HIV–co-infected lymph nodes). Specific granulomas in the lymph nodes were counted and visually graded using developmental stages 1 to 4 (Table 2). Accordingly, a total of 164 granulomas in TB-infected and 117 granulomas in TB/HIV lymph node biopsy specimens were manually counted and graded using a modified version of a previously described histopathological staging of mycobacterial granulomas.24
Table 2.
Histopathological Staging of Granulomas in Lymph Node Tuberculosis
| Granuloma | Appearance | Necrosis |
|---|---|---|
| Stage 1 | Irregular clusters of epithelioid cells interspersed with lymphocytes | Absent |
| Stage 2 | Epithelioid cell aggregates with well-defined borders | Absent or minimal necrotic areas |
| Stage 3 | Epithelioid cell aggregates with well-defined borders | Limited areas of central caseous necrosis |
| Stage 4 | Multicentric epithelioid cell clusters with irregular borders | Extensive areas of caseous necrosis |
Lymph node granulomas were examined and graded using a modified version of a previously described histopathologic staging of mycobacterial granulomas in hematoxylin and eosin–stained tissue sections.24
BCG-Specific IgG Enzyme-Linked Immunosorbent Assay
The release of mycobacteria-specific IgG antibodies in plasma samples or in vitro peripheral blood mononuclear cell (PBMC) cultures obtained from the study participants was quantified using a BCG-specific enzyme-linked immunosorbent assay. Briefly, a BCG vaccine antigen (Japan BCG Laboratories, Tokyo, Japan) was used to coat Maxisorb plates (Nunc, Roskilde, Denmark) overnight at 4°C. The plates were washed with 0.05% Tween-20 (Sigma-Aldrich) and blocked with 10% fetal calf serum in phosphate-buffered saline. Diluted plasma samples or PBMC culture supernatants (from PBMCs cultured without exogenous stimulation for 48 hours) from patients and controls were added (100 μL per well) and incubated for 2 hours at 37°C before washing and adding a rabbit anti-human IgG horseradish peroxidase conjugate (Jackson Immunoresearch Laboratories, West Grove, PA) for 2 hours at room temperature. Dilution buffer was used as a negative control. The enzyme-substrate reaction was developed after 20 minutes using an O-phenylenediamine (Sigma-Aldrich) substrate solution. BCG-specific IgG titers were expressed as 492 OD and multiplied by the dilution factor.
Multiplex Luminex Assay
Inflammatory mediators [IL-1 receptor antagonist (IL-1RA), CXCL9, and CXCL10] in plasma samples were assessed using a human standard cytokine 25-plex panel (LHC0009) from Invitrogen according to the manufacturer's instructions using the Bio-Plex Luminex detection system and the Bio-Plex manager 4.0 curve fitting software (Bio-Rad Laboratories, Hercules, CA).
mRNA Extraction and Quantitative Real-Time PCR
PBMCs or lymph node tissue were saved in RNAlater until mRNA was extracted using the Ambion RiboPure extraction kit (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. RNA was reverse transcribed to cDNA using superscript reverse transcriptase (SuperScript VILO cDNA master mix, Invitrogen). Amplification of target genes in the cDNA samples was performed using the ABI PRISM 7700 Sequence Detection System, including the 7500 software version 2.0.6 (Applied Biosystems, Foster City, CA). Commercial FAM dye–labeled Taq-Man MGB probes and primers for Tim-3, Tbet, and GATA3 were from Sigma-Aldrich, whereas the other primers and probes were from Applied Biosystems. Cycle threshold values for the target genes were normalized to the CT value for the housekeeping gene 18S, and the results were analyzed by using the relative standard method as previously described.22,25 Data were presented as mRNA fold change in TB-infected or TB/HIV–co-infected samples compared with uninfected control samples (n = 5, matched PBMC and lymph nodes tissue samples).
Flow Cytometry
Frozen PBMC samples were thawed and washed with FACS buffer [PBS containing 0.5% (v/v) fetal calf serum and 0.5 mmol/L EDTA] before cells were counted and stained. A total of 1 × 106 cells were stained for 30 minutes at 4°C with fluorochrome-conjugated anti-human antibodies: CD33 (phosphatidylethanolamine), CD11b (phosphatidylethanolamine-Cy7), HLA-DR (phosphatidylethanolamine-Cy5), CD15 (fluorescein isothiocyanate), CD14 (Brilliant Violet 711), and the death cell marker (Zombie UV), whereas CD3, CD19, and CD56 were assessed together in the DUMP channel (Brilliant Violet 510). CD33 and Zombie UV were obtained from Biolegend (San Diego, CA), whereas the other antibodies were from BD Pharmingen (San Diego, CA). Unstained cell samples as well as fluorescence minus one controls were used to determine the background fluorescence and to set the appropriate gates. Cells were washed twice with FACS buffer after staining and fixed with 4% formaldehyde (Sigma-Aldrich) at room temperature for 10 minutes. After fixation, cells were washed twice with FACS buffer, and 50,000 cells were acquired from each sample by using BD LSR Fortessa (BD Biosciences) and analyzed with FlowJo version 9 (BD, Ashland, OR).
Statistical Analysis
Data are presented as medians ± interquartile ranges or medians ± ranges. Nonparametric analyses were used to calculate indicated P values and included a Kruskal-Wallis test and Dunn posttest (when comparing more than two groups) or a Mann-Whitney U-test (when comparing two unmatched groups) or a Wilcoxon signed-rank test (when comparing two matched groups). The Spearman correlation test was used for the correlation analyses. A value of r = 1 indicates a perfect positive correlation, whereas r = −1 indicates a perfect negative correlation. Statistical analyses were performed in GraphPad Prism software version 5 (GraphPad Software, San Diego, CA).
Results
Enhanced Systemic Inflammation and Elevated Proportion of Poorly Formed and Necrotic Granulomas in Lymph Node Tissue from TB/HIV–Co-Infected Patients
Clinical and laboratory features of HIV-negative and HIV-positive patients with lymph node TB and controls are summarized in Table 1. As expected, TB/HIV–co-infected patients had significantly lower CD4+ T-cell counts (P = 0.001) and relatively higher CD8+ T-cell counts in peripheral blood, resulting in a lower CD4:CD8 ratio (P = 0.0003) compared with HIV-negative patients with TB. Furthermore, hemoglobin levels were lower (P = 0.02) and white blood cell counts (P = 0.05) and erythrocyte sedimentation rates (P = 0.01) were higher in HIV-positive compared with HIV-negative patients with TB. Plasma HIV viral load was higher in TB/HIV–co-infected compared with HIV-positive controls (P = 0.02), which may suggest that viral replication is less controlled in TB/HIV co-infection.
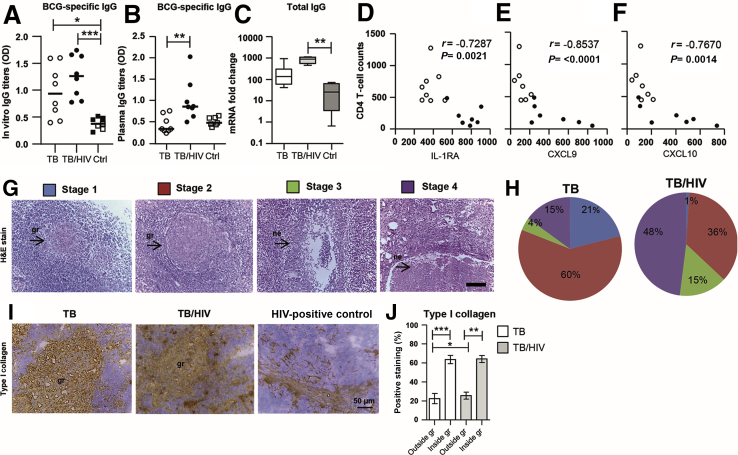
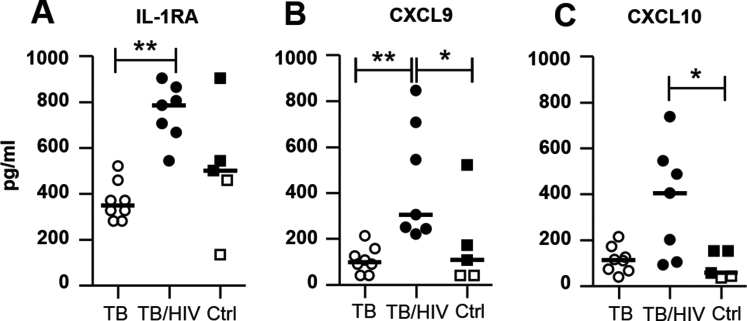
Elevated mycobacteria-specific IgG production from PBMCs cultured in vitro was observed in both TB-infected (P = 0.02) and TB/HIV–co-infected (P = 0.0006) patients compared with the controls (Figure 1A). TB/HIV–co-infected patients also had enhanced mycobacteria-specific IgG responses in plasma (P = 0.0031) as well as elevated total IgG mRNA in lymph node tissue (P = 0.002) compared with HIV-negative patients with TB or controls, respectively. Multiplex profiling of plasma samples revealed an altered systemic cytokine/chemokine milieu in TB/HIV–co-infected patients, characterized by significantly elevated levels of the anti-inflammatory IL-1RA (P = 0.0038), and increased levels of the IFN-γ–stimulated chemokines CXCL9 (P = 0.004) and CXCL10 (P = 0.041) compared with HIV-negative patients with TB as well as the controls (Supplemental Figure S1, A–C). Accordingly, there was a significant inverse correlation between CD4+ T-cell counts and IL-1RA (P = 0.002), CXCL9 (P < 0.0001), and CXCL10 (P = 0.0014) (Figure 1, D–F), supportive of dysregulated immune activation in TB/HIV–co-infected patients.
Figure 1.
Characterization of systemic inflammation and tissue pathology in tuberculosis (TB)–infected and TB/HIV–co-infected patients. A–C: BCG-specific and total IgG were assessed using enzyme-linked immunosorbent assay of peripheral blood mononuclear cell (PBMC) culture supernatants (A) and serum samples (B), respectively, as well as real-time PCR of freshly isolated PBMCs (C) from TB-infected patients, TB/HIV–co-infected patients, and controls (Ctrls). D–F: Peripheral CD4 T-cell counts were inversely correlated to inflammatory mediators in plasma, including IL-1 receptor antagonist (IL-1RA) (D), CXCL9 (E), and CXCL10 (F) as assessed using the Spearman correlation test. G and H: Hematoxylin and eosin (H&E) stain of lymph node tissue sections from a TB/HIV–co-infected patient where the arrows indicate solid or necrotic TB granulomas (G) that were manually counted and graded as stage 1 to 4 granulomas in TB-infected and TB/HIV–co-infected tissues (H). I and J: Type I collagen staining in TB-infected, TB/HIV–co-infected, and HIV-positive Ctrl tissue (I) was used to determine the level of fibrosis in TB- and TB/HIV–co-infected lymph nodes (J). G and I: Representative images of H&E as well as type I collagen staining are shown. Data are expressed as medians ± ranges or interquartile ranges. n = 8 TB-infected patients (open circles and white bars); n = 8 TB/HIV–co-infected patients (closed circles and light gray bars); n = 3 HIV-negative Ctrls (open squares); n = 4 HIV-positive Ctrls (closed squares). ∗P < 0.05, ∗∗P < 0.005, and ∗∗∗P < 0.001 (Kruskal-Wallis and Dunn multiple comparisons tests or Mann-Whitney U-test or Wilcoxon signed-rank test). Scale bar = 50 μm. Original magnification, ×25. gr, granuloma; ne, necrosis.
Next, microscopy and computerized image analysis were applied to assess the morphology of a total of 281 TB granulomas (164 TB infected and 117 TB/HIV co-infected) in M. tuberculosis–infected lymph node tissues. Four major types of granulomas were observed and presented as stages 1 to 4, with and without necrosis (Table 2 and Figure 1G). Overall, 81% of the TB granulomas in HIV-negative lymph nodes had well-defined borders and little necrosis (stage 1 to 2), whereas disrupted cellular aggregates with extensive necrosis (stage 3 to 4) were more common in TB/HIV co-infection and observed in 63% of the granulomas (Figure 1H). Quantification of tissue fibrosis using type I collagen showed that both HIV-negative TB-infected and TB/HIV–co-infected lymph nodes contained enhanced fibrosis compared with control tissues (Figure 1I). Fibrosis was particularly high in the granulomatous areas (Figure 1I), and image analyses confirmed that granulomas in both groups demonstrated significantly elevated levels of type I collagen (P = 0.0006 and P = 0.008, respectively) (Figure 1J) compared with the surrounding tissue. Type I collagen expression in tissue outside the granulomas was also significantly higher (P = 0.02) in TB/HIV co-infection compared with TB infection (Figure 1I). Altogether, these results suggested that persistent immune activation and chronic inflammation, including poorly formed and necrotic lymph node granulomas, were more common in TB/HIV–co-infected patients compared with HIV-negative patients with TB.
Elevated Expression of M. tuberculosis Antigen in M. tuberculosis–Infected Tissues
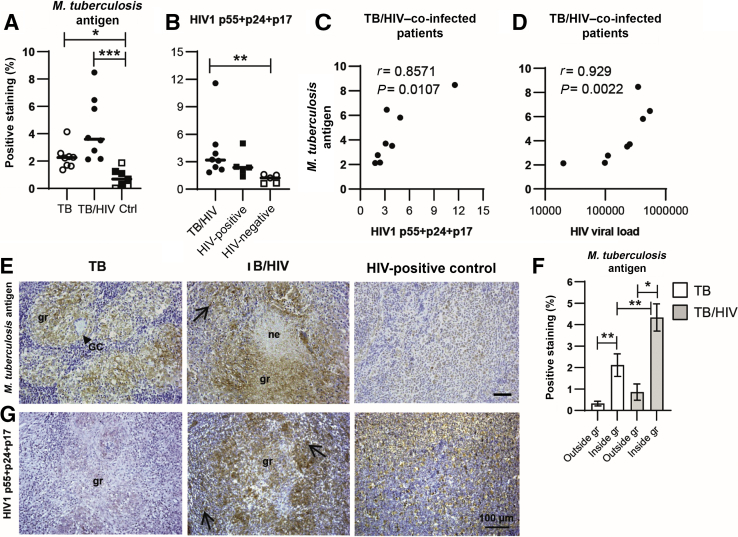
To quantify local bacterial burden as well as viral load in the study participants, M. tuberculosis and HIV-1 antigen expression was assessed in the lymph node tissues. M. tuberculosis antigen was significantly higher in both TB-infected (P = 0.05) and TB/HIV–co-infected (P = 0.0004) tissues compared with the controls, whereas M. tuberculosis antigen load was relatively higher in TB/HIV–co-infected compared with HIV-negative TB-infected tissues (Figure 2A). In situ quantification of the HIV-1 antigens p55+p24+p17 showed a significantly (P = 0.006) higher expression in TB/HIV–co-infected compared with HIV-negative tissues (Figure 2B). In TB/HIV–co-infected patients, M. tuberculosis antigen expression in tissue correlated with HIV-1 antigen load in tissue (r = 0.86, P = 0.0107) as well as HIV viral load in plasma (r = 0.93, P = 0.0022) (Figure 2, C and D). Microscopy and image analysis revealed that M. tuberculosis antigen expression in both TB-infected and TB/HIV–co-infected lymph nodes was significantly higher in the granulomatous areas compared with the surrounding tissue (P = 0.0078 and P = 0.016, respectively) (Figure 2E and F). M. tuberculosis antigen was also significantly higher in TB/HIV–co-infected granulomas compared with HIV-negative TB granulomas (P = 0.0095) (Figure 2F). Tissue expression of HIV-1 antigens was detected in TB/HIV granulomas but also in the lymphoid areas of both TB/HIV–co-infected and HIV-positive study participants (Figure 2G). These results support the notion that HIV can fuel M. tuberculosis replication and antigen deposition in co-infected macrophages at the site of infection.6
Figure 2.
In situ quantification of Mycobacterium tuberculosis and HIV1 antigen load in tuberculosis (TB)–infected and TB/HIV–co-infected lymph node tissues. A and B: Immunohistochemistry and computerized image analyses was used to determine the expression (percentage of positive staining in the total cell area) and distribution of M. tuberculosis antigen (A) and HIV1 p55+p24+p17 (B). Note that HIV antigen stain in TB/HIV co-infection is compared with HIV-positive and HIV-negative controls (Ctrls) (3 TB-infected patients and 2 HIV-negative Ctrls). C and D:M. tuberculosis antigen expression in TB/HIV–co-infected tissue correlated to HIV antigen load in tissue (C) and to HIV viral load in plasma (D) as assessed using the Spearman correlation test. E and F: Representative images of M. tuberculosis–positive staining in TB-infected, TB/HIV–co-infected, and HIV-positive Ctrl tissue (E) and image analyses of M. tuberculosis antigen expression outside and inside the granulomatous areas in TB-infected and TB/HIV–co-infected patients is shown (F). G: Representative images of HIV1 p55+p24+p17–positive staining in TB-infected, TB/HIV–co-infected, and HIV-positive Ctrl tissue. E and G: Arrows in the images indicate positively stained cells, and arrowheads indicate giant cells (GCs). Data are expressed as medians ± ranges or interquartile ranges. n = 8 TB-infected patients (open circles and white bars); n = 8 TB/HIV–co-infected patients (closed circles and light gray bars) (A, B and F); n = 3 HIV-negative Ctrls (open squares); n = 4 HIV-positive Ctrls (closed squares). ∗P < 0.05, ∗∗P < 0.005, and ∗∗∗P < 0.001 (Kruskal-Wallis and Dunn multiple comparisons tests or Mann-Whitney U-test or Wilcoxon signed-rank test). Scale bar = 100 μm. Original magnification, ×16. gr, granuloma; ne, necrosis.
Altered Cellular Composition in TB/HIV–Co-Infected Lymph Nodes
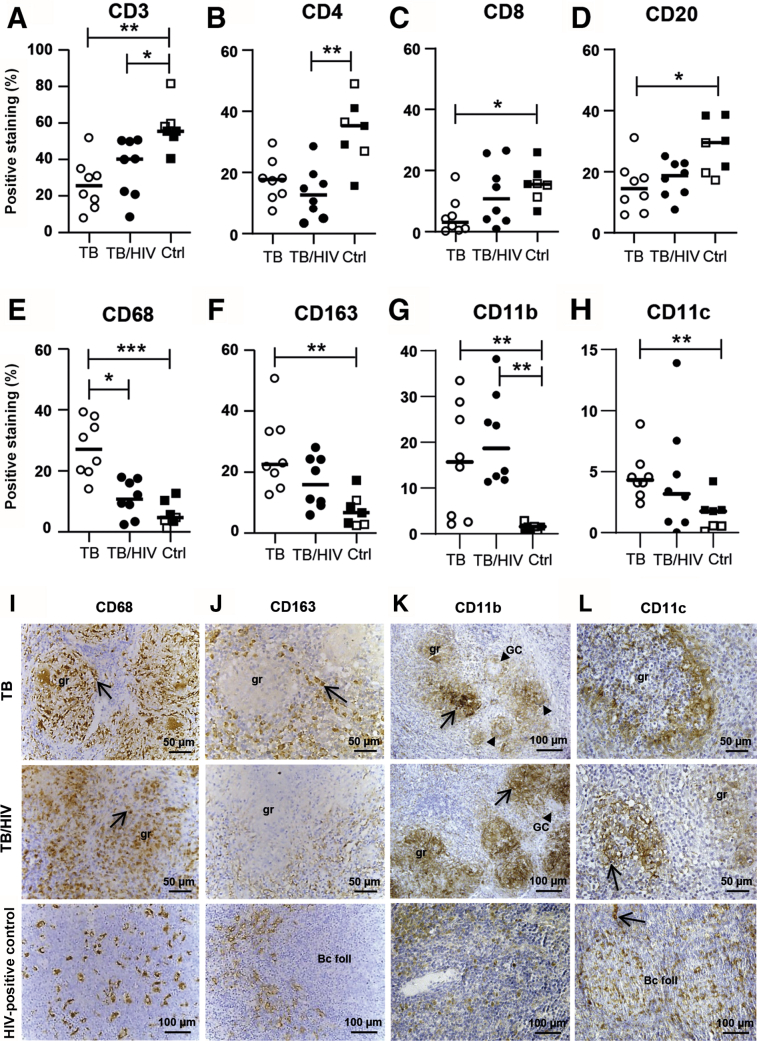
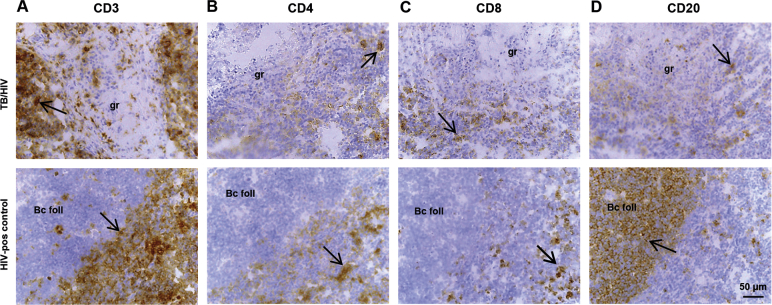
IHC (Supplemental Figure S2, A–D) and image analyses revealed a significant reduction (P = 0.002) of CD3+ T cells in both TB-infected and TB/HIV–co-infected lymph nodes (Figure 3A). Although CD4+ T cells were significantly reduced in TB/HIV co-infection (P = 0.007) (Figure 3B), CD8+ T cells were significantly reduced in HIV-negative TB tissues (P = 0.04) (Figure 3C). TB-infected (P = 0.03) and TB/HIV–co-infected patients had reduced levels of CD20+ B cells in lymph node tissue compared with the controls (Figure 3D), and these reduced levels were particularly pronounced in HIV-negative TB-infected tissues.
Figure 3.
Immune cell composition and morphology in tuberculosis (TB)-infected and TB/HIV–co-infected lymph node tissues. A–H: Immunohistochemistry and computerized image analyses was used to determine the expression (percentage of positive staining in the total cell area) and distribution of lymphocytes (A–D) and myeloid cells (E–H): CD3 (A), CD4 (B), and CD8 T cells (C), CD20 B cells (D), as well as CD68 (E), CD163 (F), CD11b (G), and CD11c (H) myeloid subsets. I–L: Representative images of CD68-positive (I), CD163-positive (J), CD11b-positive (K), and CD11c-positive (L) staining in TB-infected, TB/HIV–co-infected, and HIV-positive control (Ctrl) tissue. Arrows in the images indicate positively stained cells, and arrowheads indicate giant cells (GCs). Data are expressed as medians ± ranges; n = 8 TB-infected patients (open circles); n = 8 TB/HIV–co-infected patients (closed circles); n = 3 HIV-negative Ctrls (open squares); n = 4 HIV-positive Ctrls (closed squares). ∗P < 0.05, ∗∗P < 0.005, and ∗∗∗P < 0.001 (Kruskal-Wallis and Dunn multiple comparisons tests). Original magnification: ×16 (I, top and middle rows; J, top and middle rows; L, top and middle rows); ×25. Scale bars, 50 μm (I, top and middle rows; J, top and middle rows; L, top and middle rows); 100 μm (I, bottom row; J, bottom row; K; L, bottom row). Bc foll, B-cell follicle; gr, granuloma.
In situ expression and distribution of different myeloid markers showed a significant increase of CD68+ (P = 0.0007) and CD163+ (P = 0.005) macrophages as well as CD11b+ (P = 0.0097) and CD11c+ (P = 0.031) cells in HIV-negative TB-infected lymph nodes tissues compared with controls (Figure 3, E–H). CD68+ macrophages were also higher in TB-infected compared with TB/HIV–co-infected tissues (P = 0.001) (Figure 3E). The only myeloid marker that was up-regulated in TB/HIV–co-infected tissues compared with the control was CD11b (P = 0.0022), whereas CD68, CD163, and CD11c were all relatively lower compared with HIV-negative TB tissues (Figure 3, E–H). CD68+ granulomas with well-defined borders were less frequent in TB/HIV–co-infected lymph nodes (Figure 3I) and CD68+ cells were confined inside the M. tuberculosis granulomas, whereas CD163+ cells were located at the periphery, surrounding CD68+ macrophage clusters (Figure 3, I and J). Most CD11b+ and CD11c+ cells were located in the granulomatous lesions of TB-infected and TB/HIV–co-infected tissues, but some were also distributed in the lymphoid areas of the lymph nodes (Figure 3, K and L). The size and morphology of CD11b+ cells were usually smaller and more rounded compared with CD11c+ cells, which were larger and more DC-like in the patient as well as control tissues (Figure 3, K and L). Overall, altered cellular dynamics in TB-infected and TB/HIV–co-infected lymph nodes involved extensive tissue remodeling, including reduced expression of both CD4+ and CD8+ T cells and CD20+ B cells but elevated myeloid cell subsets. A lower expression of CD68 and the M2-associated CD163 receptor but elevated levels of the cell surface integrin CD11b in TB/HIV–co-infected tissues may suggest the presence of an aberrant myeloid cell population in these patients.
mRNA Expression Profiling Suggests Elevated Inflammation and Increased Immune Inhibition at the Site of Infection in TB/HIV–Co-Infected Patients
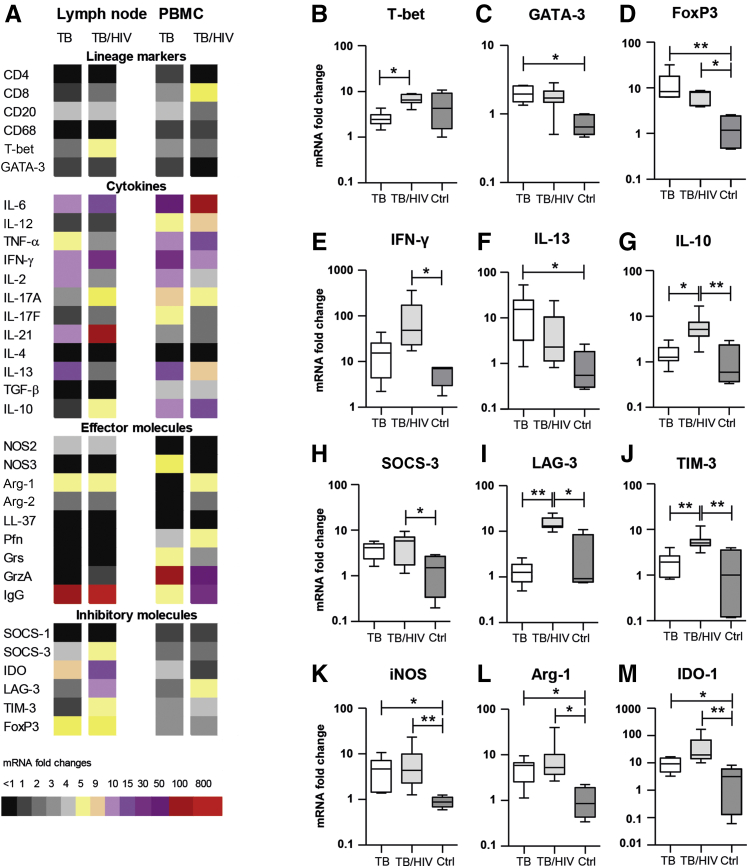
Figure 4A illustrates mRNA expression profiling of lymph node tissue and peripheral blood cells from TB-infected compared with TB/HIV–co-infected patients. Overall, the data demonstrate that the transcriptional landscape at the site of TB infection may be relatively different compared with peripheral blood. Compared with HIV-negative TB-infected patients, PBMCs from TB/HIV–co-infected patients displayed a relative increase of CD8, IL-6, total IgG, Arg-1, and lymphocyte activated gene (LAG)-3 mRNA. In the lymph nodes, there was an increased mRNA expression of IL-6, IFN-γ, IL-21, Arg-1, total IgG, IDO, and forkhead box P3 (FoxP3) in both TB-infected and TB/HIV–co-infected tissues compared with the uninfected controls.
Figure 4.
RNA profiling of lymph node tissues and peripheral blood mononuclear cells (PBMCs) from tuberculosis (TB)–infected and TB/HIV–co-infected patients. A: Heatmap demonstrating mRNA expression (fold change) of a number of immune mediators as determined by quantitative real-time PCR. B–M: mRNA expression of selected immune molecules in lymph node tissues. Data are expressed as medians ± ranges. n = 8 TB-infected patients (white bars); n = 8 TB/HIV–co-infected patients (light gray bars); n = 5 matched PBMC and lymph nodes tissue samples (dark gray bars). ∗P < 0.05, ∗∗P < 0.005 (Kruskal-Wallis and Dunn multiple comparisons tests). Arg, arginase; Ctrl, control; eNOS, endothelial nitric oxide synthase; FoxP3, forkhead box P3; GATA-3, GATA-binding protein 3; Grs, granulysin; GrzA, granzyme A; IDO-1, indoleamine 2,3-dioxygenase-1; IFN-γ, interferon-γ; iNOS, inducible nitric oxide synthase; LAG-3, lymphocyte activated gene-3; LL-37, human cationic antibacterial protein of 18 kDa; NOS, nitric oxide synthase; Pfn, perforin; SOCS, suppressor of cytokine signaling; TIM-3, T-cell immunoglobulin mucin-3; TNF-α, tumor necrosis factor-α.
mRNA expression of selected molecules in TB-infected lymph nodes tissues compared with controls is demonstrated in Figure 4, B–M. TB/HIV–co-infected tissues expressed significantly more T-bet (P = 0.029), IFN-γ (P = 0.05), and IL-10 (P = 0.022) mRNA compared with TB (Figure 4, B, E and G), whereas the HIV-negative TB group displayed higher levels of Th2 markers, such as GATA-binding protein 3 (P = 0.016) and IL-13 (P = 0.014) mRNA (Figure 4, C and F). This finding is consistent with an up-regulation of the inflammatory chemokines CXCL9 and CXCL10 in TB/HIV co-infection (Supplemental Figure S1, B and C) and an enhanced tissue expression of the M2-marker CD163 in HIV-negative TB-infected patients (Figure 3, F and J). Moreover, FoxP3 expression was up-regulated in both TB-infected (P = 0.009) and TB/HIV–co-infected (P = 0.044) patients compared with the controls (Figure 4D). Likewise, the pleiotropic inhibitors suppressor of cytokine signaling (SOCS)-3, LAG-3, and T-cell immunoglobulin mucin (TIM)-3 were all up-regulated in TB/HIV co-infection compared with the controls (P = 0.05, P = 0.02, and P = 0.009, respectively), and LAG-3 and TIM-3 were also significantly higher in TB/HIV co-infection compared with TB infection (P = 0.005 and P = 0.009) (Figure 4, H–J).
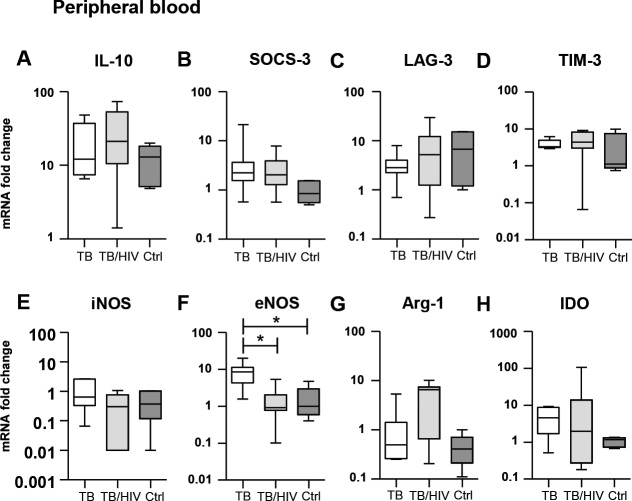
Several T-cell–specific effector molecules as well as the innate antimicrobial peptide human cationic antibacterial protein of 18 kDa (LL-37) were expressed at low levels at the site of infection in TB and TB/HIV lymph node tissues (Figure 4A). Instead, the cell-signaling enzymes iNOS, Arg-1, and IDO were significantly up-regulated in both TB (P = 0.019, P = 0.036, and P = 0.019, respectively) and TB/HIV (P = 0.0072, P = 0.022, and P = 0.0012, respectively) compared with the control group (Figure 4, K–M). Importantly, differences in mRNA expression of most immune molecules shown in Figure 4 were only evident at the site of infection and not in corresponding samples from peripheral blood (Supplemental Figure S3).
Differential Distribution of iNOS, Arg-1, and IDO Expression in M. tuberculosis–Infected Granulomatous Tissue
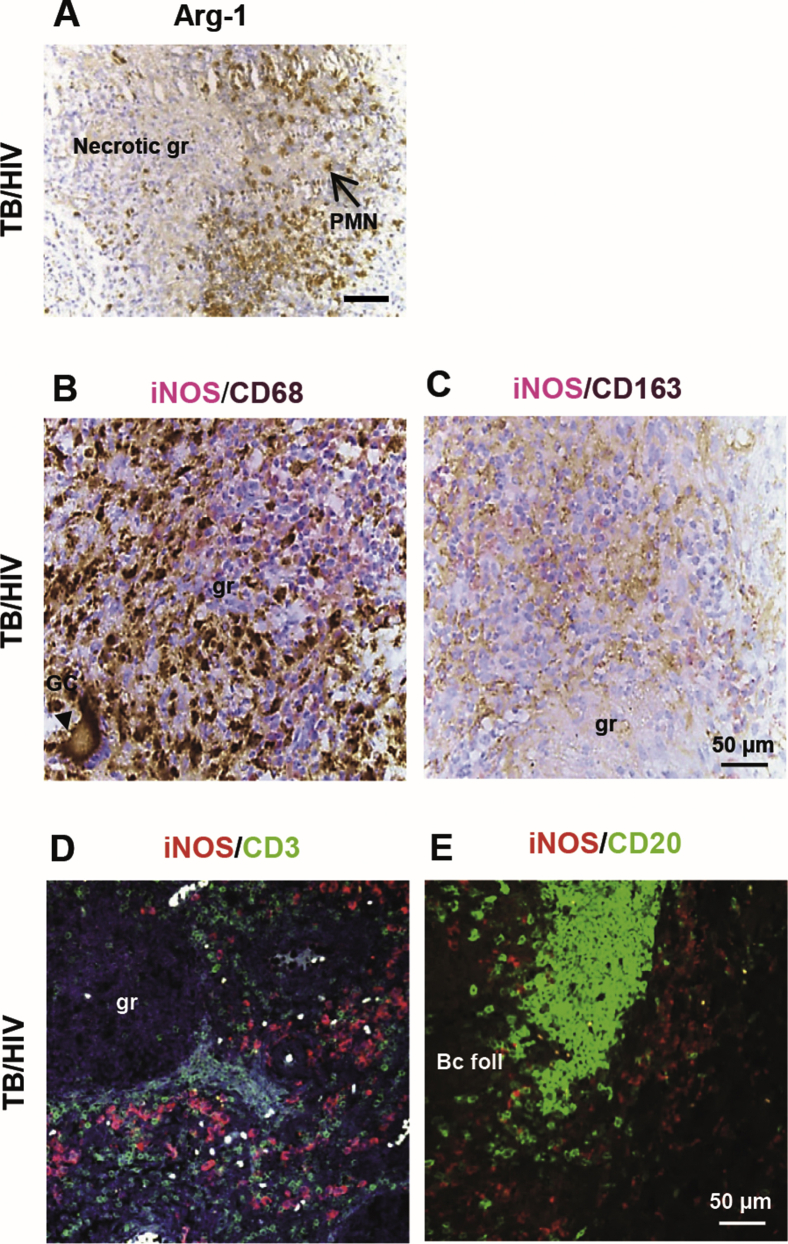
To further explore the phenotype and function of macrophages in the granulomatous tissue, the cell-signaling enzymes iNOS, Arg-1, and IDO were assessed. There was a significant increase of both iNOS (P = 0.003) and Arg-1 (P = 0.004) expression in the TB/HIV–co-infected group, although iNOS was also relatively higher in the TB-infected group compared with the controls (Figure 5, A and B). Notably, both iNOS (median of 4.64% HIV positive versus 0.29% HIV negative) and Arg-1 (median of 0.47% HIV positive versus 0.05% HIV negative) were up-regulated in the HIV-infected controls (Figure 5, A and B). Expression of the inhibitory molecule IDO was strongly enhanced (P = 0.0008) in both patient groups (Figure 5C). Interestingly, iNOS- and Arg-1–producing cells were mostly localized at the periphery of the TB granulomas in cells with a mononuclear morphology (Figure 5, D and E), whereas IDO-1 expression was detected primarily inside the granulomas (Figure 5F). Figure 5, G and H illustrates that iNOS and Arg-1 expression in and around confluent granulomas was mostly confined to the T-cell–rich areas surrounding the cores of the lesions. iNOS-expressing cells were also very few in the B-cell follicles (Figure 5G). Arg-1 expression was generally less stringent than iNOS, and numerous Arg-1–producing cells were found in the peripheral rim of the granulomatous lesions and in the lymphoid areas (Figure 5H). Multinucleated giant cells in the center of the granuloma expressed iNOS at a low intensity compared with the iNOS-positive cells surrounding the granuloma (Figure 5, D and G). Likewise, Arg-1 was expressed at lower intensity in giant cells and polymorphonuclear cells in and around the necrotic areas in the granulomas (Figure 5H and Supplemental Figure S4A).
Figure 5.
In situ expression of cell-signaling enzymes in tuberculosis (TB) infected–and TB/HIV–co-infected lymph node tissues. A–C: Immunohistochemistry and computerized image analyses were used to determine the expression (percentage of positive staining in the total cell area) and distribution of inducible nitric oxide synthase (iNOS) (A), arginase (Arg)-1 (B), and indoleamine 2,3-dioxygenase (IDO) (C). D–F: Representative images of iNOS (D), Arg-1 (E), and IDO (F) staining in TB/HIV–co-infected lymph node tissue. G and H: iNOS (G) and Arg-1 (H) at a lower magnification visualizing staining patterns in confluent granulomas. Arrows indicate positively stained cells, and arrowheads indicate giant cells (GCs). Data are expressed as medians ± interquartile ranges. n = 8 TB-infected patients (open circles); n = 8 TB/HIV–co-infected patients (closed circles); n = 3 HIV-negative controls (Ctrls) (open squares); n = 4 HIV-positive Ctrls (closed squares). ∗P < 0.05, ∗∗P < 0.005 (Kruskal-Wallis and Dunn multiple comparisons tests). Scale bars: 50 μm (D–F); 100 μm (G and H). Original magnification: ×16 (D–F); ×25 (G and H). Bc foll, B-cell follicle; gr, granuloma.
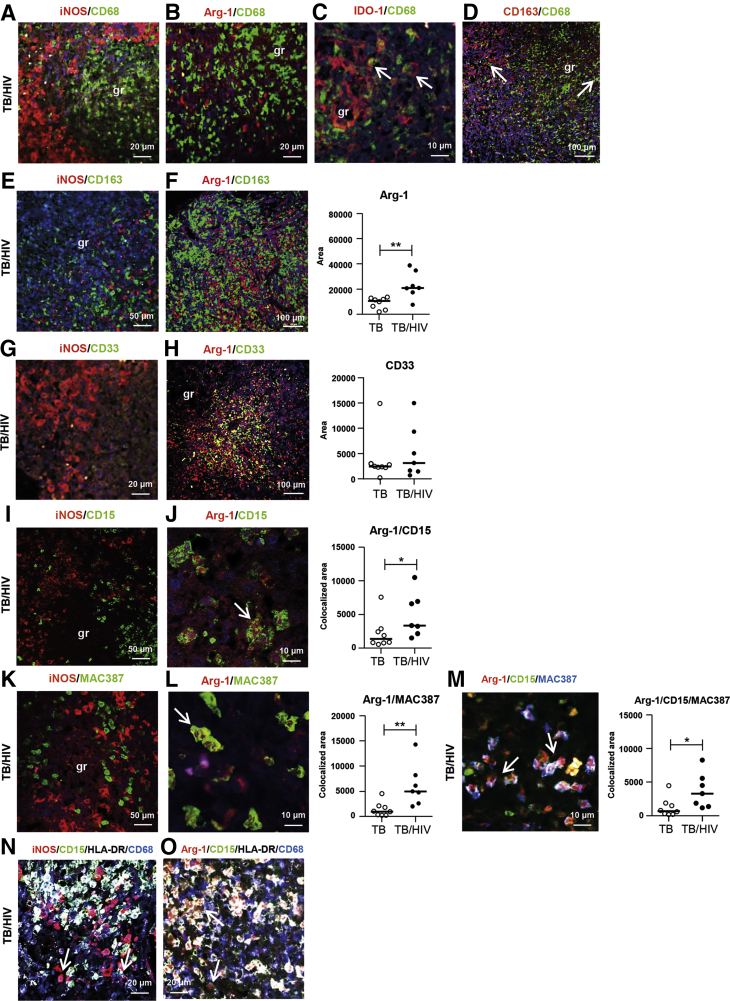
Elevated Expression of Lin−/CD33+CD15+/MAC387+/HLA-DRint/Arg-1+ Cells Characteristic of MDSCs in TB/HIV–Co-Infected Tissue
Multicolour confocal microscopy demonstrated that neither iNOS nor Arg-1 co-localized with CD68-positive or CD163-positive macrophages (Figure 6, A, B, E and F). In contrast, IDO-1 expression co-localized largely with CD68+ macrophages, although IDO was also expressed in some CD68-negative cells (Figure 6C). Consistent with the nonoverlapping expression of CD68 and CD163 detected with IHC (Figure 3, I and J), IF confirmed that CD68 and CD163 co-expression was low in both TB-infected (data not shown) and TB/HIV–co-infected tissues (Figure 6D). Overall, co-expression of iNOS could not be detected in CD68+ or CD163+ cells, CD3+ T cells, or CD20+ B cells (Supplemental Figure S4, B–E) or in subsets, including CD56+ natural killers cells or CD123+ plasmacytoid DCs (data not shown), which were very few in the TB tissue. On testing of myeloid markers, iNOS did not co-localize with CD33+, CD15+, or MAC387+ (Figure 6, G, I and K) but was expressed in CD68-negative cells with low or intermediate HLA-DR expression (Figure 6N). Similar to iNOS, IDO was not expressed in CD15+ or MAC387+ cells (data not shown).
Figure 6.
Confocal imaging and analyses of myeloid cell markers in tuberculosis (TB)–infected and TB/HIV–co-infected lymph node tissues. A–O: Immunofluorescence and confocal microscopy analyses were used to determine the expression (colocalized area) and distribution of the following markers: inducible nitric oxide synthase (iNOS) (red)/CD68 (green) (A), arginase (Arg)-1 (red)/CD68 (green) (B), indoleamine 2,3-dioxygenase (IDO) (red)/CD68 (green) (C), CD163/CD68 (green) (red) (D), iNOS (red)/CD163 (green) (E), Arg-1 (red)/CD163 (green) (F), iNOS (red)/(CD33) (green) (G), Arg-1 (red)/(CD33) (green) (H), iNOS (red)/(CD15) (green) (I), Arg-1 (red)/(CD15) (green) (J), iNOS (red)/(MAC387) (green) (K), Arg-1 (red)/(MAC387) (green) (L), Arg-1 (red)/(CD15) (green)/MAC387 (blue) (M), iNOS (red)/CD15 (green)/HLA-DR (white)/CD68 (blue) (N), and Arg-1 (red)/CD15 (green)/HLA-DR (white)/CD68 (blue) (O). Representative images of iNOS or Arg-1 double, triple, or quadruple stains are shown in TB/HIV–co-infected lymph node tissue. F–M: Positively stained area of Arg-1 (F) or CD33 (H) and colocalized area of Arg-1 and CD15 (J) or MAC387 (L) or Arg-1 and CD15 and MAC387 (M) was quantified using digital image analyses. Arrows indicate double-positive or triple-positive cells. Data are expressed as medians ± ranges. n = 8 TB-infected patients (open circles); n = 7 TB/HIV–co-infected patients (closed circles). ∗P < 0.05, ∗∗P < 0.005 (Mann-Whitney U-test). Original magnification: ×63 (A, B, G, N, and O); ×100 (C, J, L, and M); ×16 (D, F, and H); ×25 (E, I, and K). Scale bars: 20 μm (A, B, G, N, and O); 10 μm (C, J, L, and M); 100 μm (D, F, and H); 50 μm (E, I, and K). gr, granuloma.
The confocal analyses further revealed that Arg-1–expressing cells were negative for lineage-specific markers CD3, CD20, CD56, and CD123 (data not shown). However, Arg-1 co-localized considerably with CD33+, CD15+, and MAC387+ cells with low or intermediate expression of HLA-DR (Figure 6, H, J, L, M, and O), suggestive of an MDSC cell subset. Quantification of co-expression revealed a significantly elevated expression of Arg-1+ (P = 0.006), Arg-1+MAC387+ (P = 0.002), Arg-1+CD15+ double-positive (P = 0.05), and Arg-1+CD15+MAC387+ triple-positive (P = 0.03) cells in TB/HIV co-infection compared with TB infection (Figure 6, F, J, L and M). TB/HIV–co-infected tissues demonstrated a high co-localization of Arg-1 with CD33+ cells located primarily around the core of the TB granulomas, although there was no difference in CD33 expression in TB-infected compared with TB/HIV–co-infected tissues (Figure 6H). Altogether, these results demonstrate the presence of elevated levels of cells characteristic of MDSCs in the microenvironment of lymph nodes granulomas from TB/HIV–co-infected patients.
Elevated Levels of Granulocytic MDSCs in the Peripheral Circulation of TB/HIV–Co-Infected Patients
To explore whether MDSCs were present in the peripheral circulation of TB-infected patients, TB/HIV–co-infected patients, and controls, flow cytometry staining was performed using corresponding PBMC samples from the study participants. In humans, MDSCs have been characterized as monocytic CD11b+/CD14+ or granulocytic CD11b+/CD15+ MDSCs and are broadly defined as lymphocyte lineage-negative cells that express the common myeloid marker CD33 together with low or no expression of mature myeloid cell markers, such as HLA-DR, CD40, or CD80.10,19,26 Accordingly, a gating strategy was used for identification of monocytic (CD14+) or granulocytic (CD15+) MDSC subsets in peripheral blood based on lineage-negative (CD3, CD19, CD56) and HLA-DR–negative or low cells expressing the myeloid markers CD33+ and CD11b+ (Supplemental Figure S5).27 Flow cytometry showed that MDSCs identified among Lin−/HLA-DRlow/CD33+/CD11b+ cells were mostly CD15+/CD14−, which is consistent with the phenotype of granulocytic MDSCs (Figure 7A). Indeed, CD15+ granulocytic MDSCs were significantly higher in TB/HIV–co-infected patients compared with TB-infected patients (P = 0.02) and controls (P = 0.0012) (Figure 7B). CD15+ MDSCs were also elevated in HIV-negative TB-infected patients, but this difference was not significant compared with the controls (Figure 7, A and B).
Figure 7.
Flow cytometric analyses of cells characteristic of myeloid-derived suppressor cells (MDSCs) present in peripheral blood from tuberculosis (TB)–infected and TB/HIV–co-infected patients. A: Representative dot plots showing MDSCs present in TB-infected or TB/HIV–co-infected patients as well as HIV-negative and HIV-positive controls (Ctrls) based on gating of peripheral blood mononuclear cells (PBMCs) for Lin-negative, HLA-DRlow, CD33+, and CD11b+ cells that were further divided into CD15+ and CD14+ cells. Frequency (percentage of CD15+ cells) of MDSCs in PBMC samples. B–E: CD15+ MDSCs in TB/HIV–co-infected and HIV-positive controls were positively correlated with plasma HIV viral load (C), whereas CD15+ MDSCs in TB-infected and TB/HIV–co-infected patients and Ctrls were positively correlated with Mycobacterium tuberculosis antigen load in lymph node tissue (D) but inversely correlated to peripheral CD4 T-cell counts (E) as determined using the Spearman correlation test. n = 8 TB-infected patients (open circles); n = 7 TB/HIV–co-infected patients (closed circles); n = 3 HIV-negative Ctrls (open squares); n = 4 HIV-positive Ctrls (closed squares). ∗∗P < 0.01, ∗∗∗P < 0.001 (Kruskal-Wallis and Dunn multiple comparisons tests). Ctrl, control.
Correlation analyses further showed a positive correlation between the percentage of CD15+ MDSCs in peripheral blood and HIV viral load in plasma (HIV-positive participants; r = 0.77, P = 0.0074) or M. tuberculosis antigen load in tissue [HIV-positive participants; r = 0.82, P = 0.0033 (data not shown); all study participants; r = 0.71, P = 0.0002] but an inverse correlation between the percentage of CD15+ MDSCs and CD4+ T-cell counts in the peripheral circulation [HIV-positive participants; r = −0.70, P = 0.0204 (data not shown); all study participants; r = −0.55, P = 0.0082] (Figure 7, C–E). These results suggested that MDSCs are prevalent in TB/HIV–co-infected patients, both at the site of M. tuberculosis infection and in the systemic circulation, and that MDCSs may be associated with progression of TB disease in HIV-positive individuals.
Discussion
This study demonstrates enhanced chronic inflammation with an accompanying immunosuppressive microenvironment in lymph node granulomas from TB/HIV–co-infected compared with HIV-negative TB-infected patients. TB/HIV co-infection was characterized by poorly formed, necrotic granulomas that displayed a particularly high M. tuberculosis antigen expression. Persistent immune activation in TB/HIV co-infection involved not only enhanced production of proinflammatory (IL-6, IFN-γ, CXCL9, CXCL10, and iNOS) as well as anti-inflammatory (IL-1RA, IL-10, Arg-1, and IDO) mediators but also elevated numbers of myeloid cells with an aberrant phenotype and morphology characteristic of MDSCs in TB-infected lymph nodes. Elevated IgG responses in TB, and particularly in TB/HIV–co-infected patients, are indicative of chronic immune activation and active TB disease progression. On the other hand, reduced T-cell numbers and low mRNA expression of antimicrobial and cytolytic effector molecules, such as perforin and granulysin, were evident in both HIV-negative TB-infected and TB/HIV–co-infected tissues, whereas immunoregulatory FoxP3 and IDO expression was enhanced. Furthermore, Arg-1 was higher in MDSC-like cells in TB/HIV–co-infected tissues, and granulocytic MDSCs were significantly increased in peripheral blood from TB/HIV–co-infected compared with HIV-negative TB-infected patients. MDSC counts correlated positively with HIV viral load in plasma as well as M. tuberculosis antigen load in tissue but were inversely correlated with peripheral CD4 T-cell counts, which may indicate a role for MDSCs in TB/HIV disease progression.
Pathological features of chronic inflammatory diseases and solid cancers typically include enhanced levels of proinflammatory mediators, such as cytokines, chemokines, and reactive oxygen and nitrogen species. Emerging evidence supports the concept that chronic inflammation promotes local immunosuppression via induction of proinflammatory mediators and the accumulation and activation of immunosuppressive cells.28 In line with that view, differentiated macrophages also produce anti-inflammatory cytokines, most notably IL1-RA and IL-10, which are up-regulated by proinflammatory mediators and engage in autocrine signaling.29 IL-10 reduces HLA-DR expression and antigen presentation in macrophages and inhibits the production of proinflammatory cytokines.30 Likewise, MDSCs expand during various pathological conditions associated with acute or chronic inflammation.19 This paradox may represent both immune exhaustion and active immune inhibition. In TB, defective activation of myeloid cells8,13 may contribute to impaired TB immunity by suppression of Th1 effector cells and activation of different pathogenic and Treg cell subsets with a large phenotypic and functional heterogeneity, including expression of immune checkpoint molecules, such as IDO, cytotoxic T-lymphocyte-associated protein (CTLA)-4, programmed death ligand (PD-L)1, and LAG-3.14 Although excessive inflammation in TB is harmful and an anti-inflammatory response is required to avoid tissue destruction, local polarization of immunosuppressive subsets with poor antibacterial properties may also delay or prevent the clearance of infection, leading to progressive TB disease. We previously found that high iNOS expression in TB granulomas coincides with impaired Th1 and CD8+ CTL responses but enhance FoxP3+ Treg cells in patients with active TB.21 The antimicrobial peptides LL-37 and granulysin were reduced in TB lesions from human lung22,23 and lymph nodes,21 whereas Th2 (IL-4, IL-13, and CCL4) and Treg (FoxP3, CTLA-4, glucocorticoid-induced tumor necrosis factor receptor–related protein, and transforming growth factor-β) responses were enhanced together with excess expression of suppressors of cytokine signaling (SOCS-1 and -3).21,25 In this study, the localization of Arg-1+ MDSC-like cells outside the core of the granuloma in close contact with CD3+ T cells may partly explain the mixed immune response profile in TB/HIV–co-infected lymph nodes, including induction of both inflammatory and immunosuppressive or anti-inflammatory mediators as determined using mRNA expression analyses.
TB granulomas share many similarities with the microenvironment in solid tumors, including regions of hypoxia and necrosis, extensive fibrosis, and local immunosuppression. Immunosuppressive cell subsets, such as MDSCs, Treg cells, and T cells, expressing immune checkpoint inhibitors are known to play important roles in tumor immune evasion. MDSCs located within tumor tissue are typically highly immunosuppressive.19,31 In particular, both circulating and tumor-infiltrating MDSCs typically express high levels of Arg-1,32,33 and high arginase activity at the tumor site correlates with decreased cytokine production and low levels of the T-cell receptor (TCR) CD3 ζ-chain.34 In patients with non–small cell lung cancer, granulocytic MDCSs in peripheral blood were found to be inversely correlated with CD8+ T-cell frequencies.35 In those with renal or lung carcinoma, either depletion of MDSCs or inhibition of Arg-1 was found to reestablish T-cell proliferation and TCR CD3 ζ-chain expression.34,36 Recent studies also demonstrated that MDSCs mainly exert immunosuppressive functions through the induction of Arg-1 and L-arginine depletion.37, 38, 39 Circulating MDSCs in patients with gastric cancer expressed Arg-1 that was associated with decreased CD8+ T-cell proliferation as well as with reduced IFN-γ production and granzyme B secretion from those cells.40 Similarly, elevated levels of MDCSs in patients with chronic hepatitis C infection are associated with a down-regulation of the TCR ζ-chain on CD8+ T cells.41 MDSCs in this inflammatory environment produce Arg-1 and increased Arg-1+ cells in the liver were closely associated with tissue pathology in patients with chronic hepatitis.41 Recently, it was also reported that monocytic MDSCs expanded in blood from patients with coronavirus disease 2019 and were strongly associated with disease severity, including suppressed T-cell proliferation and IFN-γ production as well as down-regulated expression of the CD3 ζ-chain, partly via an Arg-1–dependent mechanism.42
Despite extensive evidence of MDSCs in different types of cancer, relatively few studies have investigated these cells in either HIV or TB infection, and assessments of MDSCs at the site of infection are scarce. In HIV infection, the original findings suggest that peripheral granulocytic MDSCs were increased in highly active antiretroviral therapy–naive HIV-positive patients but rapidly declined after the start of therapy.9 Similar to the results of this study, MDSC numbers in blood showed a positive correlation with HIV viral loads and a negative correlation with CD4+ T-cell counts.9 Enhanced levels of immunosuppressive monocytic MDSC have also been found in HIV-infected patients despite 2 years of effective antiretroviral therapy.43 Uncontrolled HIV replication has been observed to result in persistent immune activation, including elevated IL-6 levels, that mediates an expansion of MDSC numbers in HIV-infected patients.44,45 Furthermore, HIV-induced MDSCs may increase the expression of inhibitory mediators, including IL-10 and PD-L1 protein,46 restrict proliferation of effector T cells, and instead promote the differentiation of FoxP3+ Treg cells in T-cell co-cultures.9,44,46 Accordingly, granulocytic MDSCs expressing PD-L1 protein could inhibit both proliferation and IFN-γ production of CD8+ T cells from HIV-infected patients.47 Another report observed an inverse correlation between granulocytic MDSCs and expression of the TCR CD3 ζ-chain in HIV-infected patients that was restored on in vitro depletion of MDSCs.48
In human TB, MDCSs that suppress T-cell functions were initially described in the peripheral blood and in pleural fluid from patients with active TB.8 MDSC levels in patients with active TB were comparable to those in patients with cancer and contributed to reduced Th1 cytokine expression, whereas proinflammatory cytokines, such as IL-1β, IL-6, IL-8, and monocyte chemoattractant protein-1, were increased.8 High frequencies of granulocytic MDSCs have been observed in both peripheral blood and bronchoalveolar lavage specimens from patients with active pulmonary TB that suppressed T-cell proliferation, whereas MDSC were rapidly reduced on effective anti-TB treatment.49 Increased MDSC levels in peripheral blood from TB-infected children have also been linked to heightened IDO and Arg-1 levels in matched plasma samples.50 Furthermore, in vitro generated MDSCs promote M. tuberculosis replication by altering granuloma structure and stability and shift the balance of soluble mediators at the site of infection toward suppressive and regulatory cytokines, including a high release of IL-10 and IL-6,51 which is consistent with the mRNA profiling data in the current study. MDSCs from HIV-infected individuals have also demonstrated defective innate immunity to M. tuberculosis and exhibited a higher intracellular replication of M. tuberculosis compared with HLA-DRhi myeloid subsets.52 This finding is consistent with an enhanced M. tuberculosis antigen expression in TB/HIV–co-infected lymph nodes as well as elevated M. tuberculosis antigen load in TB/HIV granulomas compared with HIV-negative TB, which may imply that HIV contributes to an immune cell dysfunction locally in the tissue that will make macrophage subsets more permissive to intracellular growth of M. tuberculosis. In turn, an enhanced bacterial replication may foster the expansion of MDSCs in TB/HIV–co-infected patients.
In TB-HIV–co-infected lymph node granulomas, IDO was mostly expressed in CD68+ macrophages, which is consistent with previous findings.53 In contrast, iNOS was abundantly expressed around the CD68+ core of the granulomas, although we were not able to fully characterize the phenotype of iNOS-producing cells. Immunosuppressive MDSCs from patients with pulmonary TB correlated with depleted serum levels of L-arginine but increased levels of NO, suggesting that MDSCs may exert their suppression by enhanced iNOS activity.49 In line with this finding, BCG vaccination of mice resulted in infiltration of NO-producing MDSCs into the skin that were unable to kill BCG bacteria but instead impaired T-cell priming in the draining lymph nodes.54 iNOS-expressing cells with immunosuppressive functions resembling monocytic MDSCs have also been identified in HIV and simian immunodeficiency virus infection.55 Because NO production by myeloid cells can contribute to suppressive T-cell responses, it is possible that iNOS-expressing cells observed in TB/HIV–co-infected lymph node tissue also represent a subset of suppressive myeloid cells.
Recent evidence suggests that pharmacologic exhaustion of MDSCs with the small-molecule inhibitor tasquinimod resulted in reduced FoxP3+ Treg cells but enhanced CD8+ effector T cells in the lung of M. tuberculosis–infected mice, and tasquinimod-mediated depletion of MDSCs also enhanced M. tuberculosis clearance in the lungs.56 In addition, tasquinimod impaired the formation of organized TB granulomas that contain cores of neutrophils with MDSC-like properties.57 In the microenvironment of solid tumors, tasquinimod treatment was also observed to decrease Arg-1 and CD206 mRNA expression in macrophages, indicating a shift from M2-like myeloid cells to the M1 phenotype.58 In a mouse lung tumor model, inhibition of Arg-1 in myeloid cells diminished growth of established tumors by increasing T-cell infiltration into the tumor tissue, which was accompanied by a significant increase in the CD8/Foxp3 ratio and IFN-γ production in T cells isolated from the tumors.59 Similarly, another small-molecule inhibitor of Arg-1 reversed myeloid cell–mediated immunosuppression and restored T-cell proliferation in different solid tumors, supporting the value of Arg-1 as an immunomodulatory target.60 These reports suggest that inactivation or depletion of MDSC subsets or a shift toward proinflammatory myeloid cells might have potential in effective host-directed therapy that could be a powerful adjunct to conventional antibiotic therapy in chronic TB and TB-HIV co-infection.
Acknowledgments
We thank Elisabeth Henriksson for excellent technical assistance on the paraffin-embedded staining protocol and Anette Hofmann for her help with confocal microscopy, Alemayehu Kifle at the Armauer Hansen Research Institute for transportation and storage of the patient samples, and Sr. Alemtsehay, surgical nurse at the Department of Surgery, Tikur Anbessa Specialized Hospital, for recruitment and follow-up of the patients. Finally, we acknowledge the patients who participated in this study.
Footnotes
Supported by Swedish Heart and Lung Foundation grants 2019-0299 and 2019-0302 (S.B.), Swedish Research Council grants 2019-01744 and 2019-04720 (S.B.), and the Foundation to Prevent Antibiotic Resistance. The funders had no role in study design, experimental works, data analysis, decision to publish, or preparation of the manuscript.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2021.12.013.
Author Contributions
S.A., B.L., A.B., G.Ad., M.S., and S.B. designed and initiated the study, including preparation of study protocols and ethical approval; S.B. provided financial support for the study; S.A. was involved in all aspects of the clinical and laboratory work and coordinated the study with support from the co-authors as follows: B.L. recruited and managed all the patients; G.As. analyzed chest radiographic findings and contributed to patient diagnosis; M.Z. and M.C. participated in patient sample preparation and in vitro cell culture work; J.R.M. contributed to flow cytometry analyses and enzyme-linked immunosorbent assay; J.S. and S.R. participated in the confocal microscopy and flow cytometry analyses; A.R. performed quantitative PCR analyses; A.M. assisted with immunohistochemistry; S.B. and S.A. wrote, edited, and revised the manuscript with input from the other authors; all authors approved the final version of manuscript.
Supplemental Data
Supplemental Figure S1.
Assessment of inflammatory mediators in plasma, including IL-1 receptor antagonist (IL-1RA) (A), CXCL9 (B), and CXCL10 (C) using multiplex Luminex. Data are expressed as medians ± ranges. n = 8 tuberculosis (TB)–infected patients (open circles); n = 8 TB/HIV–co-infected patients (closed circles); n = 3 HIV-negative controls (open squares); n = 4 HIV-positive controls (closed squares). ∗P < 0.05, ∗∗P < 0.005 (Kruskal-Wallis and Dunn multiple comparisons tests or Mann-Whitney U-test).
Supplemental Figure S2.
Immunohistochemistry was used to determine the expression and distribution of CD3 (A), CD4 (B), D8 T cells (C), and CD20 B cells (D) in tuberculosis (TB)/HIV–co-infected and HIV-positive control tissue. Representative images of CD3, CD4, CD8, and CD20 immunostainings in TB/HIV–co-infected and HIV-positive control tissues are shown. Arrows indicate positively stained cells. Scale bar = 50 μm. Original magnification, ×25. Bc foll, B-cell follicle; gr, granuloma.
Supplemental Figure S3.
RNA profiling of peripheral blood mononuclear cells (PBMCs) from tuberculosis (TB)-infected and TB/HIV–co-infected patients using quantitative real-time PCR. A–H: mRNA expression of selected immune molecules in lymph node tissues. Data are expressed as medians ± ranges. n = 8 TB-infected patients (white bars); n = 8 TB/HIV–co-infected patients (light gray bars); n = 3 HIV-negative controls 4 HIV-positive controls (dark gray bars). ∗P < 0.05 (Kruskal-Wallis and Dunn multiple comparisons tests). Arg-1, arginase-1; Ctrl, controls; eNOS, endothelial nitric oxide synthase; IDO, indoleamine 2,3-dioxygenase; iNOS, inducible nitric oxide synthase; LAG-3, lymphocyte activated gene-3; SOCS-3, suppressor of cytokine signaling-3; TIM-3, T-cell immunoglobulin mucin-3.
Supplemental Figure S4.
Multicolor microscopy was used to investigate co-expression of inducible nitric oxide synthase (iNOS) in different immune cells present in tuberculosis (TB)/HIV–co-infected lymph node tissue. A:In situ expression of arginase (Arg)-1 visualized in the granulomatous tissue was particularly high in cells with a granulocytic morphology (arrow). B–E: iNOS (pink or red) expression was investigated using immunohistochemistry double staining with CD68 (brown) (B) or CD163 (brown) (C) or using immunofluorescence and confocal microscopy staining together withCD3 (green) (D) or CD20 (green) (E). For immunohistochemistry, iNOS was detected using a horseradish peroxidase–conjugated secondary antibody and double stained with diaminobenzidine. Representative images are shown in TB/HIV–co-infected lymph node tissue. Scale bars = 50 μm. Original magnification, ×25. Bc foll, B cell follicle; gr, granuloma; PMN, polymorphonuclear cell.
Supplemental Figure S5.
Gating strategy used to assess myeloid-derived suppressor cell–like cells present in peripheral blood from tuberculosis (TB)–infected and TB/HIV–co-infected patients. Representative dot plots from a TB/HIV–co-infected patient are shown. Live lymphocytes were gated for lineage-negative cells, and the consecutive gates for CD11b-positive cells, HLA-DR–negative or low cells, and CD33-positive cells are also indicated. The gates were set using unstained cell samples as well as fluorescence minus one controls. SSC, side scatter.
References
- 1.World Health Organization . World Health Organization; Geneva: 2020. WHO Global Tuberculosis Report 2020. [Google Scholar]
- 2.Brighenti S., Andersson J. Local immune responses in human tuberculosis: learning from the site of infection. J Infect Dis. 2012;205(Suppl 2):S316–S324. doi: 10.1093/infdis/jis043. [DOI] [PubMed] [Google Scholar]
- 3.Flynn J.L., Chan J., Lin P.L. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. 2011;4:271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders B.M., Cooper A.M. Restraining mycobacteria: role of granulomas in mycobacterial infections. Immunol Cell Biol. 2000;78:334–341. doi: 10.1046/j.1440-1711.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 5.de Noronha A.L., Bafica A., Nogueira L., Barral A., Barral-Netto M. Lung granulomas from Mycobacterium tuberculosis/HIV-1 co-infected patients display decreased in situ TNF production. Pathol Res Pract. 2008;204:155–161. doi: 10.1016/j.prp.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Lawn S.D., Butera S.T., Shinnick T.M. Tuberculosis unleashed: the impact of human immunodeficiency virus infection on the host granulomatous response to Mycobacterium tuberculosis. Microbes Infect. 2002;4:635–646. doi: 10.1016/s1286-4579(02)01582-4. [DOI] [PubMed] [Google Scholar]
- 7.Gabrilovich D.I. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5:3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.du Plessis N., Loebenberg L., Kriel M., von Groote-Bidlingmaier F., Ribechini E., Loxton A.G., van Helden P.D., Lutz M.B., Walzl G. Increased frequency of myeloid-derived suppressor cells during active tuberculosis and after recent mycobacterium tuberculosis infection suppresses T-cell function. Am J Respir Crit Care Med. 2013;188:724–732. doi: 10.1164/rccm.201302-0249OC. [DOI] [PubMed] [Google Scholar]
- 9.Vollbrecht T., Stirner R., Tufman A., Roider J., Huber R.M., Bogner J.R., Lechner A., Bourquin C., Draenert R. Chronic progressive HIV-1 infection is associated with elevated levels of myeloid-derived suppressor cells. AIDS. 2012;26:F31–F37. doi: 10.1097/QAD.0b013e328354b43f. [DOI] [PubMed] [Google Scholar]
- 10.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., Rodriguez P.C., Sica A., Umansky V., Vonderheide R.H., Gabrilovich D.I. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magcwebeba T., Dorhoi A., du Plessis N. The emerging role of myeloid-derived suppressor cells in tuberculosis. Front Immunol. 2019;10:917. doi: 10.3389/fimmu.2019.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B., Wang X., Ren X. Amino acid metabolism related to immune tolerance by MDSCs. Int Rev Immunol. 2012;31:177–183. doi: 10.3109/08830185.2012.679989. [DOI] [PubMed] [Google Scholar]
- 13.Thomas A.C., Mattila J.T. “Of mice and men”: arginine metabolism in macrophages. Front Immunol. 2014;5:479. doi: 10.3389/fimmu.2014.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brighenti S., Joosten S.A. Friends and foes of tuberculosis: modulation of protective immunity. J Intern Med. 2018 doi: 10.1111/joim.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rath M., Muller I., Kropf P., Closs E.I., Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol. 2014;5:532. doi: 10.3389/fimmu.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills C.D., Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. J Innate Immun. 2014;6:716–726. doi: 10.1159/000364945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munder M., Schneider H., Luckner C., Giese T., Langhans C.D., Fuentes J.M., Kropf P., Mueller I., Kolb A., Modolell M., Ho A.D. Suppression of T-cell functions by human granulocyte arginase. Blood. 2006;108:1627–1634. doi: 10.1182/blood-2006-11-010389. [DOI] [PubMed] [Google Scholar]
- 18.Fallarino F., Grohmann U., You S., McGrath B.C., Cavener D.R., Vacca C., Orabona C., Bianchi R., Belladonna M.L., Volpi C., Santamaria P., Fioretti M.C., Puccetti P. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 19.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deveci H.S., Kule M., Kule Z.A., Habesoglu T.E. Diagnostic challenges in cervical tuberculous lymphadenitis: a review. North Clin Istanb. 2016;3:150–155. doi: 10.14744/nci.2016.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman S., Gudetta B., Fink J., Granath A., Ashenafi S., Aseffa A., Derbew M., Svensson M., Andersson J., Brighenti S.G. Compartmentalization of immune responses in human tuberculosis: few CD8+ effector T cells but elevated levels of FoxP3+ regulatory t cells in the granulomatous lesions. Am J Pathol. 2009;174:2211–2224. doi: 10.2353/ajpath.2009.080941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman S., Rehn A., Rahman J., Andersson J., Svensson M., Brighenti S. Pulmonary tuberculosis patients with a vitamin D deficiency demonstrate low local expression of the antimicrobial peptide LL-37 but enhanced FoxP3+ regulatory T cells and IgG-secreting cells. Clin Immunol. 2015;156:85–97. doi: 10.1016/j.clim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Andersson J., Samarina A., Fink J., Rahman S., Grundstrom S. Impaired expression of perforin and granulysin in CD8+ T cells at the site of infection in human chronic pulmonary tuberculosis. Infect Immun. 2007;75:5210–5222. doi: 10.1128/IAI.00624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wangoo A., Johnson L., Gough J., Ackbar R., Inglut S., Hicks D., Spencer Y., Hewinson G., Vordermeier M. Advanced granulomatous lesions in Mycobacterium bovis-infected cattle are associated with increased expression of type I procollagen, gammadelta (WC1+) T cells and CD 68+ cells. J Comp Pathol. 2005;133:223–234. doi: 10.1016/j.jcpa.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Ashenafi S., Aderaye G., Bekele A., Zewdie M., Aseffa G., Hoang A.T., Carow B., Habtamu M., Wijkander M., Rottenberg M., Aseffa A., Andersson J., Svensson M., Brighenti S. Progression of clinical tuberculosis is associated with a Th2 immune response signature in combination with elevated levels of SOCS3. Clin Immunol. 2014;151:84–99. doi: 10.1016/j.clim.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Cassetta L., Baekkevold E.S., Brandau S., Bujko A., Cassatella M.A., Dorhoi A., Krieg C., Lin A., Lore K., Marini O., Pollard J.W., Roussel M., Scapini P., Umansky V., Adema G.J. Deciphering myeloid-derived suppressor cells: isolation and markers in humans, mice and non-human primates. Cancer Immunol Immunother. 2019;68:687–697. doi: 10.1007/s00262-019-02302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayik D., Lauko A.J., Roversi G.A., Serbinowski E., Acevedo-Moreno L.A., Lanigan C., Orujov M., Lo A., Alban T.J., Kim A., Silver D.J., Nagy L.E., Brown J.M., Allende D.S., Aucejo F.N., Lathia J.D. Hepatobiliary malignancies have distinct peripheral myeloid-derived suppressor cell signatures and tumor myeloid cell profiles. Sci Rep. 2020;10:18848. doi: 10.1038/s41598-020-75881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D., DuBois R.N. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis. 2015;36:1085–1093. doi: 10.1093/carcin/bgv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinne R.W., Brauer R., Stuhlmuller B., Palombo-Kinne E., Burmester G.R. Macrophages in rheumatoid arthritis. Arthritis Res. 2000;2:189–202. doi: 10.1186/ar86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isomaki P., Luukkainen R., Saario R., Toivanen P., Punnonen J. Interleukin-10 functions as an antiinflammatory cytokine in rheumatoid synovium. Arthritis Rheum. 1996;39:386–395. doi: 10.1002/art.1780390306. [DOI] [PubMed] [Google Scholar]
- 31.Movahedi K., Guilliams M., Van den Bossche J., Van den Bergh R., Gysemans C., Beschin A., De Baetselier P., Van Ginderachter J.A. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez P.C., Ochoa A.C., Al-Khami A.A. Arginine metabolism in myeloid cells shapes innate and adaptive immunity. Front Immunol. 2017;8:93. doi: 10.3389/fimmu.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren W., Zhang X., Li W., Feng Q., Feng H., Tong Y., Rong H., Wang W., Zhang D., Zhang Z., Tu S. Circulating and tumor-infiltrating arginase 1-expressing cells in gastric adenocarcinoma patients were mainly immature and monocytic Myeloid-derived suppressor cells. Sci Rep. 2020;10:8056. doi: 10.1038/s41598-020-64841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zea A.H., Rodriguez P.C., Atkins M.B., Hernandez C., Signoretti S., Zabaleta J., McDermott D., Quiceno D., Youmans A., O'Neill A., Mier J., Ochoa A.C. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 35.Liu C.Y., Wang Y.M., Wang C.L., Feng P.H., Ko H.W., Liu Y.H., Wu Y.C., Chu Y., Chung F.T., Kuo C.H., Lee K.Y., Lin S.M., Lin H.C., Wang C.H., Yu C.T., Kuo H.P. Population alterations of L-arginase- and inducible nitric oxide synthase-expressed CD11b+/CD14(-)/CD15+/CD33+ myeloid-derived suppressor cells and CD8+ T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:35–45. doi: 10.1007/s00432-009-0634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez P.C., Quiceno D.G., Zabaleta J., Ortiz B., Zea A.H., Piazuelo M.B., Delgado A., Correa P., Brayer J., Sotomayor E.M., Antonia S., Ochoa J.B., Ochoa A.C. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 37.Kong Y.Y., Fuchsberger M., Xiang S.D., Apostolopoulos V., Plebanski M. Myeloid derived suppressor cells and their role in diseases. Curr Med Chem. 2013;20:1437–1444. doi: 10.2174/0929867311320110006. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez P.C., Ochoa A.C. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives. Immunol Rev. 2008;222:180–191. doi: 10.1111/j.1600-065X.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raber P., Ochoa A.C., Rodriguez P.C. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest. 2012;41:614–634. doi: 10.3109/08820139.2012.680634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao F.Y., Zhao Y.L., Lv Y.P., Teng Y.S., Kong H., Liu Y.G., Wu X.L., Hao C.J., Chen W., Duan M.B., Han B., Ma Q., Wang T.T., Peng L.S., Zhang J.Y., Cheng P., Su C.Y., Fu X.L., Zou Q.M., Guo G., Guo X.L., Zhuang Y. CD45(+)CD33(low)CD11b(dim) myeloid-derived suppressor cells suppress CD8(+) T cell activity via the IL-6/IL-8-arginase I axis in human gastric cancer. Cell Death Dis. 2018;9:763. doi: 10.1038/s41419-018-0803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Q.L., Yang B., Sun H.Q., Feng G.H., Jin L., Zou Z.S., Zhang Z., Zhang J.Y., Wang F.S. Myeloid-derived suppressor cells are associated with viral persistence and downregulation of TCR zeta chain expression on CD8(+) T cells in chronic hepatitis C patients. Mol Cells. 2014;37:66–73. doi: 10.14348/molcells.2014.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falck-Jones S., Vangeti S., Yu M., Falck-Jones R., Cagigi A., Badolati I., Osterberg B., Lautenbach M.J., Ahlberg E., Lin A., Lepzien R., Szurgot I., Lenart K., Hellgren F., Maecker H., Salde J., Albert J., Johansson N., Bell M., Lore K., Farnert A., Smed-Sorensen A. Functional monocytic myeloid-derived suppressor cells increase in blood but not airways and predict COVID-19 severity. J Clin Invest. 2021;131:e144734. doi: 10.1172/JCI144734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin A., Cai W., Pan T., Wu K., Yang Q., Wang N., Liu Y., Yan D., Hu F., Guo P., Chen X., Chen L., Zhang H., Tang X., Zhou J. Expansion of monocytic myeloid-derived suppressor cells dampens T cell function in HIV-1-seropositive individuals. J Virol. 2013;87:1477–1490. doi: 10.1128/JVI.01759-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garg A., Spector S.A. HIV type 1 gp120-induced expansion of myeloid derived suppressor cells is dependent on interleukin 6 and suppresses immunity. J Infect Dis. 2014;209:441–451. doi: 10.1093/infdis/jit469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garg A., Trout R., Spector S.A. Human immunodeficiency virus type-1 myeloid derived suppressor cells inhibit cytomegalovirus inflammation through interleukin-27 and B7-H4. Sci Rep. 2017;7:44485. doi: 10.1038/srep44485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L., Zhao J., Ren J.P., Wu X.Y., Morrison Z.D., Elgazzar M.A., Ning S.B., Moorman J.P., Yao Z.Q. Expansion of myeloid-derived suppressor cells promotes differentiation of regulatory T cells in HIV-1+ individuals. AIDS. 2016;30:1521–1531. doi: 10.1097/QAD.0000000000001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z.N., Yi N., Zhang T.W., Zhang L.L., Wu X., Liu M., Fu Y.J., He S.J., Jiang Y.J., Ding H.B., Chu Z.X., Shang H. Myeloid-derived suppressor cells associated with disease progression in primary HIV infection: PD-L1 blockade attenuates inhibition. J Acquir Immune Defic Syndr. 2017;76:200–208. doi: 10.1097/QAI.0000000000001471. [DOI] [PubMed] [Google Scholar]
- 48.Tumino N., Turchi F., Meschi S., Lalle E., Bordoni V., Casetti R., Agrati C., Cimini E., Montesano C., Colizzi V., Martini F., Sacchi A. In HIV-positive patients, myeloid-derived suppressor cells induce T-cell anergy by suppressing CD3zeta expression through ELF-1 inhibition. AIDS. 2015;29:2397–2407. doi: 10.1097/QAD.0000000000000871. [DOI] [PubMed] [Google Scholar]
- 49.El Daker S., Sacchi A., Tempestilli M., Carducci C., Goletti D., Vanini V., Colizzi V., Lauria F.N., Martini F., Martino A. Granulocytic myeloid derived suppressor cells expansion during active pulmonary tuberculosis is associated with high nitric oxide plasma level. PLoS One. 2015;10:e0123772. doi: 10.1371/journal.pone.0123772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du Plessis N., Jacobs R., Gutschmidt A., Fang Z., van Helden P.D., Lutz M.B., Hesseling A.C., Walzl G. Phenotypically resembling myeloid derived suppressor cells are increased in children with HIV and exposed/infected with Mycobacterium tuberculosis. Eur J Immunol. 2017;47:107–118. doi: 10.1002/eji.201646658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agrawal N., Streata I., Pei G., Weiner J., Kotze L., Bandermann S., Lozza L., Walzl G., du Plessis N., Ioana M., Kaufmann S.H.E., Dorhoi A. Human monocytic suppressive cells promote replication of Mycobacterium tuberculosis and alter stability of in vitro generated granulomas. Front Immunol. 2018;9:2417. doi: 10.3389/fimmu.2018.02417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garg A., Spector S.A. Myeloid derived suppressor cells (MDSC) from HIV-infected individuals are defective in innate immunity to Mycobacterium tuberculosis thus increasing the risk of tuberculosis. J Immunol. 2019;202(1 Supplement):62.13. [Google Scholar]
- 53.Gautam U.S., Foreman T.W., Bucsan A.N., Veatch A.V., Alvarez X., Adekambi T., Golden N.A., Gentry K.M., Doyle-Meyers L.A., Russell-Lodrigue K.E., Didier P.J., Blanchard J.L., Kousoulas K.G., Lackner A.A., Kalman D., Rengarajan J., Khader S.A., Kaushal D., Mehra S. In vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2018;115:E62–E71. doi: 10.1073/pnas.1711373114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martino A., Badell E., Abadie V., Balloy V., Chignard M., Mistou M.Y., Combadiere B., Combadiere C., Winter N. Mycobacterium bovis bacillus Calmette-Guerin vaccination mobilizes innate myeloid-derived suppressor cells restraining in vivo T cell priming via IL-1R-dependent nitric oxide production. J Immunol. 2010;184:2038–2047. doi: 10.4049/jimmunol.0903348. [DOI] [PubMed] [Google Scholar]
- 55.Gama L., Shirk E.N., Russell J.N., Carvalho K.I., Li M., Queen S.E., Kalil J., Zink M.C., Clements J.E., Kallas E.G. Expansion of a subset of CD14highCD16negCCR2low/neg monocytes functionally similar to myeloid-derived suppressor cells during SIV and HIV infection. J Leukoc Biol. 2012;91:803–816. doi: 10.1189/jlb.1111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta S., Krug S., Pokkali S., Leanderson T., Isaacs J.T., Srikrishna G., Bishai W.R. Pharmacologic exhaustion of suppressor cells with tasquinimod enhances bacterial clearance during tuberculosis. Am J Respir Crit Care Med. 2019;199:386–389. doi: 10.1164/rccm.201805-0820LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshioka Y., Mizutani T., Mizuta S., Miyamoto A., Murata S., Ano T., Ichise H., Morita D., Yamada H., Hoshino Y., Tsuruyama T., Sugita M. Neutrophils and the S100A9 protein critically regulate granuloma formation. Blood Adv. 2016;1:184–192. doi: 10.1182/bloodadvances.2016000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsson A., Nakhle J., Sundstedt A., Plas P., Bauchet A.L., Pierron V., Bruetschy L., Deronic A., Torngren M., Liberg D., Schmidlin F., Leanderson T. Tasquinimod triggers an early change in the polarization of tumor associated macrophages in the tumor microenvironment. J Immunother Cancer. 2015;3:53. doi: 10.1186/s40425-015-0098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miret J.J., Kirschmeier P., Koyama S., Zhu M., Li Y.Y., Naito Y., Wu M., Malladi V.S., Huang W., Walker W., Palakurthi S., Dranoff G., Hammerman P.S., Pecot C.V., Wong K.K., Akbay E.A. Suppression of myeloid cell arginase activity leads to therapeutic response in a NSCLC mouse model by activating anti-tumor immunity. J Immunother Cancer. 2019;7:32. doi: 10.1186/s40425-019-0504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steggerda S.M., Bennett M.K., Chen J., Emberley E., Huang T., Janes J.R., Li W., MacKinnon A.L., Makkouk A., Marguier G., Murray P.J., Neou S., Pan A., Parlati F., Rodriguez M.L.M., Van de Velde L.A., Wang T., Works M., Zhang J., Zhang W., Gross M.I. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J Immunother Cancer. 2017;5:101. doi: 10.1186/s40425-017-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]