Background:
To investigate the correlations between mutations in the telomerase reverse transcriptase (TERT) promoter and isocitrate dehydrogenase (IDH) 1 and 2 mutations or 1p/19q deletion in human gliomas.
Methods:
TERT promoter gene and IDH gene mutations in 110 glioma specimens were evaluated using first generation Sanger sequencing. The 1p/19q status was determined with fluorescence in situ hybridization. The relationship between TERT promoter mutations and IDH gene mutations as well as 1p/19q deletion was analyzed using the χ2 test and Spearman rank correlation test.
Results:
The TERT promoter mutation rate in 110 glioma specimens was 39.09% (43/110), with a rate of 32.56% (14/43) for C228T mutation and 67.44% (29/43) for C250T mutation. The IDH gene mutation rate in all specimens was 31.82% (35/110), with a rate of 52.78% (19/36) in low-grade gliomas and 21.62% (16/74) in high grade gliomas. The 1p/19q deletion rate was 28.18% (31/110) in all specimens. Correlation analysis revealed that TERT promoter mutation was positively correlated with 1p/19q deletion (relative precision (rp) = 0.244, P = .015). In lower-grade glioma with IDH mutation, TERT promoter mutation was positively correlated with 1p/19q deletion (rp = 0.856, P = .000). The prognosis for gliomas with IDH mutation/TERT mutation/1p/19qdeletion was good. Mutation of the TERT promoter was negatively correlated with IDH gene mutation (rp = −0.290, P = .004), except in 10 cases of oligodendroglioma and 1 case of anaplastic oligodendroglioma.
Conclusion:
There may be a complex inter-regulatory relationship between the mutations of the TERT promoter and IDH gene as well as 1p/19q abnormalities in human gliomas.
Keywords: 1p19q, glioma, isocitrate dehydrogenase, telomerase reverse transcriptase
1. Introduction
Glioma is an environment-induced multifactorial disease and has been recognized as a complex disease involving multiple genes.[1,2] Traditional surgery and chemoradiotherapy are not effective and the prognoses might differ after the same treatment for the same grade or pathological subtype of gliomas.[3] Some studies have shown that this difference in prognoses is mainly due to the different molecular regulatory mechanisms within the tumors.[1,2]
The discovery of molecular markers provides various alternative ideal targets for targeted therapy for tumors and has played a vital role in the development of tumor molecular typing.[4] There are 3 molecular markers that warrant special attention because they are ubiquitous in gliomas and are closely related to the occurrence and development of gliomas and their prognosis. Telomerase reverse transcriptase (TERT) is a rate-limiting enzyme for telomerase. Mutated TERT can generate 2 promoter-binding domains with the same structure, multiplying the transcription activity. TERT can reversely catalyze the telomerase production and can also add repeated sequences of telomeres to the end of chromosomes, which play a very important role in the occurrence and development of immortalized cells and malignant tumors.[5,6] The second most common molecular markers are isocitrate dehydrogenase 1 and 2 (IDH1/2), which are closely related to the metabolism of tumor cells and a class of molecular markers with survival advantages.[7] Another common molecular marker is 1p/19q. Loss of heterozygosity in 1p/19q is considered a characteristic molecular marker for oligodendroglioma. Glioma patients with 1p/19q loss of heterozygosity have a significantly higher sensitivity to chemotherapy.[8,9] As more molecular markers are discovered, the relationships between the markers have become more complicated. The discovery of gene mutations related to the incidence, development, and diagnosis of gliomas at the molecular level through a combination of bioinformatic and molecular technologies has played a very important role in understanding the biological process in gliomas, screening biomarkers, improving the molecular typing of gliomas, predicting the clinical prognosis, and selecting targets for gene therapy.[10] In the 4th edition of the central nervous system tumor classification guidelines revised by the World Health Organization (WHO) in 2016, IDH mutation and 1p/19q co-deletion were included in the “comprehensive diagnosis”. The relationship between TERT promoter mutation and the above 2 gene mutations is not clear.
This article aims to evaluate the relationship between TERT promoter mutations, IDH mutations, and 1p/19q deletion, the correlation between glioma histology and grade, and the interactions between the genes.
2. Methods
This research was approved by the Ethics Committee of Guangdong Provincial People’s Hospital (approval number GDREC2018508H(R1)) and written informed consent was obtained from all patients before the experiments. One hundred ten paraffin surgical glioma specimens obtained from patients who did not receive chemotherapy before surgery from April 2017 to April 2018 were randomly selected from the Department of Pathology, Guangdong Provincial People’s Hospital. The study design is cross-sectional.
2.1. Extraction of genomic DNA
Ten pieces of tissue with a thickness of 10 μm were sliced from each specimen and placed onto a clean glass slide. After xylene was dewaxed into water, the tumor tissue was carefully scraped into a 1.5-mL Eppendorf Tube with a sterile blade according to the tumor area revealed by hematoxylin and eosin staining. DNA was extracted using the QIAamp DNA FFPE Tissue Kit (56404, Qiagen, Germany). The extracted DNA was tested to determine the concentration and optical density (OD) value with a Nanodrop 2000 ultra-micro spectrophotometer (ThermoFisher Scientific, USA) and the quality was determined with 0.8% agarose electrophoresis.
2.2. First generation Sanger gene sequencing
The TERT promoter was amplified using polymerase chain reaction (PCR) (ABI Veriti, USA). The primers were: TERT: F: 5’-GTCCTGCCCCTTCACCTT-3’, R: 5’-CAGCGCTGCCTGAAACTC-3’; IDH1: F: 5“-ACCAAATGGCACCATACGA-3”, R: 5’-TTCATACCTTGCTTAATGGGTGT-3’;IDH2: F: 5’-GCTGCAGTGGGACCACTATT-3’, R: 5’-TGTGGCCTTGTACTGCAGAG-3’.
All primers were prepared by Bao Biological Engineering, China. The PCR components and reagents comprised 12.5 uL Go TaqColorless Master Mix (Promega, USA), 0.5 uL of 20 pmol/L primers, and 100 ng DNA template with ionized water in a total volume of 25 uL. Amplification procedure: 7 minutes at 95 °C, 45 seconds at 95 °C; 45 seconds at 550 °C; 90 seconds at 720 °C; 35 cycles. The products were kept for 10 minutes at 72 °C and the DNA was stored at 4 °C.
The loading system for the purification of PCR products contained 5 uL of unpurified PCR product, 0.5 uL SAP, 10.5 uL Exonuclease, and 4 uL deionized water. The reaction proceeded for 1 hour at 37 °C and 20 minutes at 80 °C.
After purification, the PCR products were subjected to a labeling reaction. The loading system consisted of 4 uL of deionized water, 1 uL of the purified PCR product, 3 uL of the primer (2 umol/L), and 2 uL of BigDye (ABI, USA). The reaction was performed for 1 min at 96 °C, followed by 25 cycles of 10 seconds at 96 °C, 5 seconds at 50 °C, and 4 minutes at 60 °C. Each PCR product was sequenced in both directions.
Gene mutations and genetic polymorphisms in the samples were detected through comparisons with the BLAST program and the sequencing map in NCBI. Samples with genetic polymorphisms were excluded from subsequent experiments.
2.3. Fluorescence in situ hybridization (FISH) detection
Two consecutive 4-μm sections were cut from each specimen, placed onto a coated antistrip sheet and warmed for 2 hours at 65 ○C. The slides were immersed in xylene for 10 minutes twice. Afterward, the slices were dehydrated in ethanol for 5 minutes and the process was repeated. The sections were left to dry at room temperature. Afterward, the slices were placed in a preheated pretreatment solution (80 ○C) for 10 minutes and soaked in ionized water for 5 minutes at room temperature. Two hundred milligrams of gastric enzyme was added to 50 mL of gastric enzyme buffer to create the working solution. Two slides from each specimen were immersed in the gastric enzyme working solution and incubated for 10 minutes at 37 °C, and another 5 minutes after the addition of deionized water. The slices were immersed in ethanol with different gradient concentrations (70%, 80%, 90%, and 100%) for dehydration. After the specimens were suitably dehydrated, 1p/1q and 19q/19p probes (04N60-020, Vysisi, USA) were dropped onto the sections. All the slices were sealed and placed in a hybridizer (S500-24, ThermoBrite, USA) for denaturation for 5 minutes at 82 °C, and incubated overnight at 37 °C. The sealant was removed and the slides were soaked in postHybridization Wash Buffer Kit (01N31-005, Vysisi, USA) with 2 × saline sodium citrate (SSC) to remove the coverslips. The sections were washed for 2 minutes in 0.3% NP40/ 2 × SSC at 72 ○C, and then air-dried. Following 4,6-diamidino-2-phenylindole (DAPI) counterstaining, the signal was observed under a fluorescence microscope (BX51, Olympus, Japan).
2.4. Interpretation of FISH results
The 1p/19q probe contained 2 sets of DNA probes: 1p36/1q25 and 19q13/19p13 probes. The 1p36/1q25 probe hybridized to human chromosome 1. 1p36 was marked orange and 1q25 was green. The 19q13/19p13 probe hybridization signal was located at human chromosome 19. The 19q13 fluorescence signal was orange and the 19p13 fluorescence signal was green. The radio value was counted and calculated in 100 cells (total orange signals in 100 nuclei/total green signals in 100 nuclei) (Table 1). We conducted telephone follow-up for patients with IDH mutant lower-grade glioma. The last follow-up was in October 2020, and the longest follow-up duration was 38 months. The survival date was from the day of surgery to the day of death or the end of follow-up.
Table 1.
Interpretation of 1p/19q FISH results.
| Group | 1p/1q | 19p/19q |
|---|---|---|
| Cell | 100 | 100 |
| 1p/19q deletion | Ratio value < 0.75 | Ratio value < 0.75 |
2.5. Immunohistochemistry
The detection of IDH protein was performed by immunohistochemistry with antibody against IDH R132H mutation (H09, ZSGB-BIO, China) and hematoxylin-eosin (HE) staining, according to the protocol of manufacturer.
2.6. Statistical analysis
All data were analyzed with SPSS 18.0 software. The χ2 test and Spearman rank correlation test were conducted, and the Kaplan-Meier method was used for survival analysis. P < .05 was considered statistically significant.
3. Results
The specimens were obtained from patients with an average age of 42.22 ± 17.12 years. There were 64 specimens from male patients and 46 from female patients including 40 cases of lower grade glioma, 50 cases of glioblastoma, 8 cases of hair cell astrocytoma, 3 cases of pleomorphic yellow astrocytoma, 5 cases of diffuse midline glioma, and 4 cases of ganglioglioma. According to the WHO classification of central nervous system (CNS) tumors, 36 specimens were grades I-II and 74 specimens were grades III-IV.
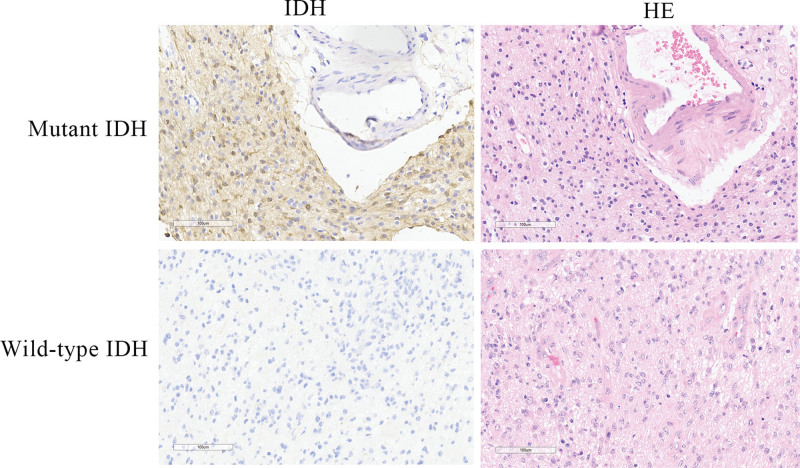
From the 110 specimens, we got the adjacent normal tissues in 23 samples, which were used as control. No IDH, TERT, or 1p/19q mutations were observed in the adjacent normal tissues. For the 110 glioma paraffin specimens, TERT promoter sequencing revealed C228T and C250T mutations. The total number of mutations was 43/110 (39.09%), among which C228T mutation occurred in 14/43 (32.56%) and C250T mutation occurred in 29/43 (67.44%). According to the histological classification, the proportion of C228T and C250T mutations was 22/50 (44%) in glioblastoma samples (IDH wild-type), 8/12 (66.67%) in anaplastic astrocytoma samples (IDH wild-type), 1/6 (16.67%) in anaplastic astrocytoma samples (IDH mutation), 1/9 (11.11%) in diffuse astrocytoma samples (IDH mutation), 10/10 (100%) in oligodendroglioma samples, and 1/1 in the anaplastic oligodendroglioma sample (1 case). There were no mutations (0%) in glioblastoma (IDH mutation, 9 cases), pilocytic (hair-like) cell astrocytoma (8 cases), pleomorphic xanthoastrocytoma (3 cases), and diffuse midline glioma (5 cases) samples, which included ganglioglioma (4 cases) samples (Table 2). By immunohistochemistry, the specimen with IDH mutation showed positive staining for IDH R132H mutation, which locates in the cytoplasm of cells (Fig. 1).
Table 2.
Mutation of TERT promoter and IDH gene, and deletion of 1p/19q in 110 glioma specimens.
| Grading | TERT | 1p/19q | ||||||
|---|---|---|---|---|---|---|---|---|
| C228T | C250T | deletion | TERT/1p/19q co-mutation/deletion |
rp/P | ||||
| WHO grade II-III | Lower-grade Glioma (40) | IDH mutation (26) | Diffuse astrocytoma (9) | 0 | 1 | 2 | 1 | |
| Anaplastic astrocytoma (6) | 0 | 1 | 2 | 1 | ||||
| Anaplastic oligodendroglioma (1) | 0 | 1 | 1 | 1 | ||||
| Oligodendroglioma (10) | 5 | 5 | 10 | 10 | 0.856/0.00 | |||
| IDH wild-type (14) | Diffuse astrocytoma (2) | 0 | 0 | 0 | 0 | |||
| Anaplastic astrocytoma (12) | 2 | 6 | 5 | 4 | 0.344/0.228 | |||
| WHO grade IV | Glioblastom | IDH mutation (9) | 0 | 0 | 1 | 0 | ||
| a (50) | IDH wild-type (41) | 7 | 15 | 9 | 5 | 0.20/0.9 | ||
| WHO grade I- IV |
Other Glioma (20) |
IDH wild-type (20) | Pilocytic astrocytoma (8) | 0 | 0 | 0 | 0 | |
| Diffuse midline glioma (5) | 0 | 0 | 1 | 0 | ||||
| Pleomorphic xanthoastrocytoma (3) | 0 | 0 | 0 | 0 | ||||
| Ganglioglioma (4) | 0 | 0 | 0 | 0 | ||||
| Total number of cases | 14 | 29 | 31 | 22 | ||||
| (32.56%, 14/43) | (67.44%, 29/43) | (28.18%, 31/110) |
(70.97%, 22/31) |
|||||
| rp | -0.290 | 0.244 | ||||||
| P | 0.004 | 0.015 | ||||||
Figure 1.
Immunohistochemistry staining of IDH mutation in specimens with mutant or wild-type IDH. Anaplastic oligodendroglioma specimen with IDH R132H mutation, and glioblastoma specimen with wild-type IDH were chosen for immunohistochemistry by using antibody against IDH R132H mutation (H09, ZSGB-BIO, China).
Three abnormalities in the 1p/19q were detected, namely co-deletion of the 1p/19q, single-deficiency of the 19q, and single-deficiency of the 1p. The proportion of 1p/19q abnormalities was 31/110 (28.18%). Twenty-two cases of 1p/19q deletion were associated with TERT promoter mutations: 10 cases (10/10, 100%) of oligodendroglioma, 5 cases (5/18, 5.56%) of anaplastic astrocytoma, 5 cases (5/50, 10%) of glioblastoma, 1 case (1/11, 9.09%) of diffuse astrocytoma, and 1 case (1/1, 100%) of intervariant oligodendroglioma . Fifteen out of 22 IDH wild-type glioblastoma cases (68.18%) had C250T mutation, while 7 of them (31.82%) had C228T mutation (Table 2).
In the IDH gene, only IDH1 gene mutation (R132H) was detected in this experiment, and the mutation occurred in 35/110 specimens (31.82%). According to the WHO classification of CNS tumors, the number of mutations in the IDH gene in low-grade gliomas (grades I-II) was 19/36 (52.78%) and that in high-grade gliomas (grades III-IV) was 16/74 (21.62%), with a statistically significant difference (P = .001) (Table 3).
Table 3.
IDH gene mutation in 110 cases of glioma (n).
| IDH mutation (R132H) | ||
|---|---|---|
| WHO grade I-II (36) | Oligodendroglioma (10) | 10 |
| Diffuse astrocytoma (11) | 9 | |
| Pilocytic astrocytoma (8) | 0 | |
| Others (7) | 0 | |
| 52.78% (19/36) | ||
| WHO grade III-IV (74) | Glioblastoma (50) | 9 |
| Anaplastic astrocytoma (18) | 6 | |
| Anaplastic oligodendroglioma (1) | 1 | |
| Others (5) | 0 | |
| 21.62% (16/74) | ||
| χ2 | 10.889 | |
| P | 0.001 |
The Spearman rank correlation test showed that TERT promoter mutations were positively correlated with 1p/19q deletion (rp = 0.244, P = .015). In lower-grade gliomas with IDH mutation, TERT promoter mutation was significantly positively correlated with 1p/ 19q deletion (rp = 0.856, P = .000). Survival analysis was performed in the subgroups of TERT mutation and 1p/19q deletion combined with 4 different molecular patterns: IDH mutation/TERT mutation/1p/19q deletion group, IDH mutation/TERT wild-type/1p/19q nondeletion group, IDH mutation/1p/19q deletion group, and IDH mutation/1p/19q nondeletion group (26 cases). The respective median survival times in the 4 groups were 22, 31, 32, and 27 months, which suggested that the prognosis for the IDH mutation/TERT mutation/1p/19q deletion group was better than that for the IDH mutation/TERT wild-type/1p/19q nondeletion group (P = .007, Table 4). Mutation of the TERT promoter was negatively correlated with IDH gene mutation (rp = −0.290, P = .004), except in 10 cases of oligodendroglioma and 1 case of anaplastic oligodendroglioma (Table 2).
Table 4.
Survival analysis of the subgroups of IDH/TERT/1p/19q in lower-grade gliomas (26 cases).
| Group | Case | Median survival time (mo) | P |
|---|---|---|---|
| IDH mutation/TERT mutation/1p/19q deletion | 13 | 32 | 0.007 |
| IDH mutation/TERT wild-type/1p/19q nondeletion | 13 | 22 | |
| IDH mutation/1p/19q deletion | 15 | 31 | 0.249 |
| IDH mutation/1p/19q nondeletion | 11 | 27 |
4. Discussion
Glioma is a tumor that originates from the neuroepithelium and contains different morphological characteristics of glial cells. It is also the most common primary tumor in the CNS, accounting for 45% of intracranial tumors, with characteristics of invasive growth, easy relapse, difficult to cure, and high mortality.[11,12] The WHO 2016 classification of CNS tumors adopted a combination of traditional histomorphology and molecular markers to further clarify the role of related genes in the pathological diagnosis of gliomas.
TERT promoter mutations are molecular markers of gliomas discovered in recent years. In 2013, Naosuke Nonoguchi et al found that mutations of the TERT promoter were common in 358 glioblastomas, among which 73% were C228T mutations and 27% were C250T mutations. The mutation rate of the TERT promoter in primary glioblastoma was 58% in the study, while the mutation rate in secondary glioblastoma was only 28%.[13] Mutations of C228T and C250T in the TERT promoter region can increase the activity of TERT. The incidence of this mutation in primary glioblastoma varied from 60% to 83%,[14] which was significantly different from that of IDH1 mutation.[15] In this study involving 110 glioma paraffin specimens, the mutation rates of the TERT promoters C228T and C250T were 39.09% (43/110), among which the C228T mutation rate was 32.56% (14/43) and the C250T mutation rate was 67.44% (29/43). Twenty-two glioblastoma samples (IDH wild-type) had TERT mutations. Mutations were mainly detected in C250T (68.18%, 15/22) and a small number was detected in C228T (31.82%, 7/22). No TERT mutations were detected in the glioblastoma samples (IDH mutation). These results were different from previous reports, probably due to the relatively small sample size and the different population composition in this study. The differences in ethnic groups and regions are related to the different locations of mutation sites in the TERT promoter. Further studies should be performed for more advanced exploration. IDH is an important oxidoreductase in the body and one of the key enzymes in the tricarboxylic acid cycle. Tumors with TERT promoter mutation exert high level of TERT mRNA and telomerase activity, contributing to maintaining telomere length and cell proliferation.[16] However, until now, their effect on cell migration and invasion has not been reported and needs more investigation in the future.
IDH mutations have at least 3 subtypes named IDH1 (R132H), IDH2 (R172H), and IDH3. IDH3 mutations have not been found in gliomas.[17,18] Nearly all IDH gene mutations are mutations of IDH1 (90%) or heterozygous mutations of IDH2 (3%)[19,20] IDH gene mutation is a phenomenon that occurs in the early development of gliomas and is currently considered the most important molecular biological marker for low-grade gliomas and secondary glioblastomas.[21] It has been shown that IDH mutations can enhance cell migration and invasion.[22] In this study, it was found that IDH mutation mainly occurred in WHO grade II glioma, and the mutation rate of the IDH1 gene in low-grade glioma (grade II) was significantly higher than that in high-grade glioma (grade III and IV) (P = .001).
The results of this study also indicated that there were both TERT promoter and IDH1 gene mutations in 13 low-grade gliomas (WHO grades II and III) including oligodendroglioma. The Spearman rank correlation test showed that with the exception of the oligodendroglioma and 1 case of variant oligodendrocyte tumor, TERT promoter mutation was negatively associated with IDH mutation (rp = −0.290, P = .004). This may be due to functional redundancy. Besides the oligodendrocyte tumor (10 cases) and variant oligodendrocyte tumor (1 case), TERT and IDH co-mutation was not detected in 50 cases of glioblastoma from 99 cases of glioma. There was repulsion between TERT promoter and IDH mutations, consistent with previous reports of a mutex between TERT promoter and IDH mutations. The other 49 cases of glioma included 2 cases of low-grade glioma (1 case of diffuse astrocytoma and 1 case of variant astrocytoma) with TERT promoter and IDH gene mutations, which suggested the existence of repulsion between TERT promoter mutation and IDH mutation in various types of gliomas.
1p/19q deletion is the combined deletion of the short arm of human chromosome 1 (1p) and the long arm of chromosome 19 (19q) and is abundant in patients with all grades of gliomas, especially those with oligodendroglioma (WHO class II).[23] Although the current diagnosis of low-grade gliomas mainly depends on the histopathology results, these molecular markers with special functions have played an increasingly important role in the diagnosis and treatment of gliomas. 1p/19q deletion is prevalent in glioma patients at all grades, especially in oligodendroglioma patients (WHO grade II). Besides the IDH gene mutation, if 1p/19q combined deletion is detected, the tumor will develop towards oligodendroglioma. In this study, 10 cases of oligodendroglioma and 1 case of inter-oligodendroglioma exhibited TERT promoter mutation in addition to IDH gene mutation combined with 1p/19q deletion. Combined deletion of 1p/19q was also positively correlated with IDH mutation. Only 3 cases of IDH wild-type glioblastoma with combined deletion of 1p/19q (3/14) were detected and one of them exhibited TERT promoter mutation. In IDH wild-type gliomas, TERT promoter mutation coupled with 1p/19q deletion was mainly detected in intervariant astrocytomas. The Spearman rank correlation test showed the TERT promoter mutation was positively correlated with 1p/19q deletion (rp = 0.244, P = .015). Especially in lower-grade gliomas with IDH mutation, TERT promoter mutation was significantly positively correlated with 1p/19q deletion. Data from the follow-up period revealed that patients in the IDH mutation/TERT mutation/1p/19q deletion group had the best prognosis. TERT promoter mutation with 1p/19q deletion occurred more frequently in low-grade gliomas (17/22) and less frequently in glioblastomas (5/22, WHO IV). This finding may be because 1p/19q deletion occurred mainly in lower grade and anaplastic tumors.
A 20-year retrospective study showed that mutations in the TERT promoter have an important prognostic value for gliomas. However, the prognostic value varied in different types of gliomas. When TERT promoter mutation occurs simultaneously with IDH and 1p/19q mutations, the prognosis is better.[24] TERT mutations were widely found in various subtypes of brain glioma and IDH mutations were mostly associated with good prognosis. TERT promoter mutations coupled with IDH gene mutations suggest good prognosis and are limited to the glioma subtypes. The results of this study showed that in most histopathological subtypes of gliomas, TERT promoter mutation was negatively correlated with IDH gene mutation. TERT promoter mutation was positively correlated with 1p/19q deletion. For patients with IDH mutation and 1p/19q deletion in glioma, determination of the TERT promoter mutation status is recommended, as the findings can provide an indication of the prognosis and have clinical application significance.
These results suggest that TERT promoter mutation, IDH gene mutation, and 1p/19q deletion may interact with each other and that there is a complex mutual regulatory effect. Combined analysis of multiple target genes can provide more information for the prognosis of gliomas.
There were some limitations in this study. The number of specimens for some subtype gliomas (such as oligodendroglioma and anaplastic oligodendroglioma) was relatively small. The potential mechanisms underlying the gene mutation and glioma incidence were not explored in the study. In a future study, we will explore the potential mechanism using the established system.
5. Conclusion
TERT promoter mutation, IDH gene mutation, and 1p/19q deletion may interact with each other. There is a complex inter-regulatory relationship between the TERT promoter mutations and IDH gene mutations as well as 1p/19q abnormalities in human gliomas. Combined analysis of multiple target genes can provide more information for the prognosis of gliomas.
Author contributions
Conceived and designed the research: JX and ZL;
Collected the data and conducted the research: JX, FPX, ZHL, and KPZ;
Analyzed and interpreted the data: JX, FPX, and QC;
The initial paper written: JX;
Revised the paper: FPX and ZL;
Primary responsibility for the final content: JX;
All authors read and approved the final manuscript.
Abbreviations:
- CNS =
- central nervous system
- FISH =
- Fluorescence in situ hybridization
- IDH =
- isocitrate dehydrogenase
- TERT =
- telomerase reverse transcriptase
How to cite this article: Xu J, Xu F-P, Liu Z-H, Cui Q, Zhang K-P, Li, Z. The correlation analysis of TERT promoter mutations with IDH1/2 mutations and 1p/19q detected in human gliomas. Medicine 2022;101:29(e29668).
Availability of data and materials: The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Funding: This work was supported by the Medical Scientific Research Foundation of Guangdong Province, China (No. A2019559), the Natural Science Foundation of Guangdong Province, China (No. 2017A030313779), and the Medical Scientific Research Foundation of Guangdong Province, China (No. A2017433). The authors declare that the funding body was not involved in study design, data collection, analysis, interpretation and writing of the study.
Conflict of interest: The authors declare that they have no conflict of interest.
Contributor Information
Jie Xu, Email: xfpraider@163.com.
Fang-Ping Xu, Email: xfpraider@163.com.
Zhi-Hua Liu, Email: Liuzhihua410@163.com.
Qian Cui, Email: tracy_2016@126.com.
Ke-Ping Zhang, Email: roomsyar@126.com.
References
- [1].Weller M, Pfister SM, Wick W, et al. Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol. 2013;14:e370–9. [DOI] [PubMed] [Google Scholar]
- [2].Mörén L, Bergenheim AT, Ghasimi S, et al. Metabolomic screening of tumor tissue and serum in glioma patients reveals diagnostic and prognostic information. Metabolites. 2015;5:502–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436. [DOI] [PubMed] [Google Scholar]
- [4].Dang L, Yen K, Attar E. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. [DOI] [PubMed] [Google Scholar]
- [5].Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huang F, Bielski C, Rinne M, et al. TERT promoter mutations and monoallelic activation of TERT in cancer. Oncogenesis. 2015;4:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Van Den Bent MJ, Hartmann C, Preusser M, et al. Interlaboratory comparison of IDH mutation detection. J Neurooncol. 2013;112:173–8. [DOI] [PubMed] [Google Scholar]
- [8].Mosrati MA, Malmström A, Lysiak M, et al. TERT promoter mutations and polymorphisms as prognostic factors in primary glioblastoma. Oncotarget. 2015;6:16663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].van den Bent MJ, Erdem-Eraslan L, Idbaih A, et al. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic Oligodendrogliomas and Oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19:5513–22. [DOI] [PubMed] [Google Scholar]
- [10].Grant R, Kolb L, Moliterno J. Molecular and genetic pathways in gliomas: the future of personalized therapeutics. CNS oncology. 2014;3:123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. [DOI] [PubMed] [Google Scholar]
- [12].Bromberg JE, Van Den Bent MJ. Oligodendrogliomas: molecular biology and treatment. The oncologist. 2009;14:155–63. [DOI] [PubMed] [Google Scholar]
- [13].Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arita H, Narita Y, Fukushima S, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–76. [DOI] [PubMed] [Google Scholar]
- [15].Nonoguchi N, Ohta T, Oh J-E, et al. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126:931–7. [DOI] [PubMed] [Google Scholar]
- [16].Barthel FP, Wei W, Tang M, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49:349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bolduc JM, Dyer DH, Scott WG, et al. Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase. Science. 1995;268:1312–8. [DOI] [PubMed] [Google Scholar]
- [18].Koh J, Cho H, Kim H, et al. IDH2 mutation in gliomas including novel mutation. Neuropathology. 2015;35:236–44. [DOI] [PubMed] [Google Scholar]
- [19].Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–74. [DOI] [PubMed] [Google Scholar]
- [20].Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhu H, Zhang Y, Chen J, et al. IDH1 R132H mutation enhances cell migration by activating AKT-mTOR signaling pathway, but sensitizes cells to 5-FU treatment as NADPH and GSH are reduced. PLoS One. 2017;12:e0169038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Riemenschneider MJ, Reifenberger G. Molecular neuropathology of low-grade gliomas and its clinical impact. In: J. Schramm, ed. Advances and Technical Standards in Neurosurgery. Vol. 35: Low-Grade Gliomas. New York: SpringerWien; 2010:35–64. [DOI] [PubMed] [Google Scholar]
- [24].Iwadate Y, Matsutani T, Hara A, et al. Eighty percent survival rate at 15 years for 1p/19q co-deleted oligodendroglioma treated with upfront chemotherapy irrespective of tumor grade. J Neurooncol. 2019;141:205–11. [DOI] [PubMed] [Google Scholar]