ABSTRACT
Hepatocellular carcinoma (HCC), a major primary liver cancer, is one of the most lethal malignancies worldwide. Increasing evidence has demonstrated that chromobox protein homolog 3 (CBX3) functions as an oncogene in different cancers. However, its expression profiles and biological functions in HCC remain unknown. Data on CBX3 expression in HCC acquired from the GEO and TCGA databases were analyzed. The biological functions of CBX3 in HCC were examined by in vitro experiments. Bioinformatics analysis, qRT-PCR and western blotting were performed to explore the mechanism of CBX3 in HCC. CBX3 mRNA was upregulated in HCC tissues, and overexpression of CBX3 mRNA was negatively correlated with malignancies and poor prognosis in HCC patients. CBX3 knockdown decreased growth, migration and invasion of HCC cells in vitro. Moreover, bioinformatics analysis and experimental observation indicated that CBX3 expression was correlated with cell cycle regulatory proteins in HCC cells. Finally, starBase predicted that miR-139 could directly target CBX3 in HCC. Confirmatory experiments verified that miR-139 overexpression attenuated HCC cell proliferation and migration, and these effects could be reversed by overexpressing CBX3. Our results showed that the miR-139/CBX3 axis may be involved in HCC development by regulating cell cycle progression and may be a promising target in the treatment of HCC.
KEYWORDS: HCC, CBX3, miR-139, cell cycle progression
Background
Hepatocellular carcinoma (HCC) is the sixth most common malignancy and the fourth leading cause of cancer-related mortality worldwide [2]. Surgery, such as liver resection, liver transplantation and liver ablation, may provide benefits for individuals with early liver cancer [3]. However, the five-year survival of HCC is usually less than 30% because most people are diagnosed with advanced liver cancer [4]. Therefore, elucidation of the molecular mechanisms and development of effective therapeutic targets of HCC are urgently needed.
Chromobox homolog protein 3 (CBX3), corresponding to HP1γ, is a member of the heterochromatin protein 1 (HP1) family [5]. CBX3 plays a critical role in many biological processes, such as heterochromatin formation and gene silencing, DNA damage repair, RNA splicing, transcriptional activation, stem cell differentiation, and reprogramming of somatic cells [6]. Given its wide distribution and different roles in gene regulation, CBX3 has been reported to be closely involved in the development of pancreatic cancer, prostate cancer and breast cancer. However, the mechanisms and roles of CBX3 in HCC are still unclear and require further clarification.
miRNAs are a group of endogenous noncoding small RNAs that regulate the expression of their target mRNAs by binding to their 3’-untranslated regions (3’-UTRs) [7]. Increasing evidence has confirmed that miRNAs play an important role in the pathogenesis, proliferation and other biological processes of cancers [8]. Hence, we examined whether miRNAs can serve as post-transcriptional regulators of CBX3 in the development of HCC.
Our study suggested that CBX3 was overexpressed and could be a marker of poor prognosis in HCC. We also found that downregulation of CBX3 inhibited the progression of HCC in vitro. Further bioinformatics analysis and validation experiments confirmed that CBX3 promoted the development of HCC by regulating the cell cycle and acted as a direct target of miR-139 in the progression of HCC. In conclusion, the miR-139/CBX3 axis is involved in the progression of HCC by regulating cell cycle progression.
Materials and methods
Collection of public HCC datasets
Differential gene expression profiles in HCC and nontumor liver tissues were acquired from The Cancer Genome Atlas (TCGA) (http://tcga-data.nci.nih.gov/) [9], and the survival data of HCC patients were also collected and analyzed. The LIRIJP (Liver Cancer-RIKEN, JP project) dataset was obtained from the International Cancer Genome Consortium (ICGC), and 18 liver cancer mRNA microarray datasets were acquired from the Gene Expression Omnibus (GEO) databases (http://www.ncbi.nlm.nih.gov/geo). The differential expression between non tumorous tissues and HCC tissues was identified by the BRB-array tool.
Cell culture
HCC cell lines (Hep3B and SMMC-7721) and immortalized human normal liver cell lines (L02 and Changliver) were purchased from the Cell Bank of Institutes for Biological Sciences (Shanghai, China). Cells were cultured at stationary 37°C in Dulbecco’s modified Eagle’s medium (Gibco, NY, USA) with 10% fetal bovine serum in an atmosphere of 5% CO2. All of the above cell lines in culture were passaged for less than 6 months when our experiments began.
Polymerase chain reaction (PCR) assay
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was applied to collect total RNA in accordance with the manufacturer’s instructions. Quantitative reverse transcription-PCR (RT-qPCR) was performed following the manufacturer’s protocol. Actin or U6 expression was used to normalize the relative expression of the target genes. The 2−ΔΔCT method was used to analyze the data. Every experiment was independently performed three times.
Western blotting assay
RIPA protein extraction reagent (KeyGEN, Nanjing, China) was used to extract proteins from whole cells on ice. Each well of 12% a SDS-PAGE gel was loaded in equivalent proteins. After electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After blocking, the membranes were incubated with 1:1000 anti-CBX3, 1:2000 anti-c-Myc, 1:500 anti-E2F, 1:1000 anti-CCK1, 1:1000 anti-cyclin D1, and 1:1000 anti-β-actin antibodies overnight at 4°C. Next, the membranes were incubated with goat anti-mouse secondary antibodies (1:1000; Abcam) at room temperature for 2 hours. Bands were visualized and analyzed by the Odyssey infrared imaging system and Odyssey 3.0 software (Thermo Scientific), respectively.
Transfection
Three CBX3-targeting siRNAs (si-CBX3-1, si-CBX3-2 and si-CBX3-3) were utilized to knock down CBX3; a nonsilencing siRNA oligonucleotide (si-NC) was used as a negative control (Invitrogen, CA, USA). CBX3 cDNA was subcloned into a pcDNA3.1 vector as a plasmid (obtained from Invitrogen, CA, USA) to overexpress CBX3, and an empty vector was used as a negative control (pcDNA3.1-NC). MiR-139 mimics, miR-139 inhibitor and the respective negative controls (mimic-NC, inhibitor-NC) and miR-NC were synthesized by GenePharma (Shanghai, China). For transient transfection, HCC cell lines were transfected using Lipofectamine 3000 (Invitrogen, Carlsbad, USA) following the manufacturer’s guidelines.
Dual-luciferase reporter assay
Both wild-type and mutant 3’-UTR fragments of the CBX3 mRNA were cloned into pmirGLO vectors (Promega, Madison, WI, USA), yielding Wt-CBX3 3’-UTR and Mut-CBX3 3’-UTR, respectively. Then, Hep3B and SMMC-7721 cells were cotransfected with the Wt-CBX3 or Mut-CBX3 vectors and miR-139 mimics or miR-NC with Lipofectamine 3000 reagent (Invitrogen). 48 hours later, the Dual-Luciferase Reporter Assay System was used to analyze the luciferase activity (Promega, Madison, USA). The results of the experiments are shown as the mean value ± SD of three measurements.
Cell growth and colony formation assays
A Cell Counting Kit-8 (CCK-8) assay was applied to examine cell growth [10]. Transfected and nontransfected cells were cultured in six 96-well plates. Then, the 96-well microplates were placed in the incubator. Ten milliliters of CCK-8 reagent was added to each well in a 96-well plate every 24 hours for 5 days, after which the plate was incubated for 2 hours at 37°C. Subsequently, the absorbance was recorded at 450 nm by using a spectrophotometer (Molecular Devices, CA, USA).
A colony formation assay was performed to evaluate HCC cell growth. A total of 1,000 cells per well were placed in 6-well plates. After 14 days of incubation, the cells were fixed with paraformaldehyde (4%), stained with 2% crystal violet, counted and imaged.
Migration assay
Wound healing assays were utilized to evaluate the migration of HCC cells. Hep3B and SMMC-7721 cells were seeded in six-well plates, and when the cells reached approximately 90% confluence, we made a gap using a 10 μL pipette tip. The gaps were photographed at 0, 24 and 48 hours with a microscope (magnification, ×100).
Invasion assay
Transwell assays were performed to assess the cell invasion. A total of 1 × 104 indicated cells were plated in the Transwell insert (the upper chamber) with serum-free medium, and DMEM with 10% FBS was added to the bottom chamber. 24 hours later, the invasive cells on the bottom chamber were fixed and stained. Finally, the migratory cells were imaged and counted.
Bioinformatics analysis
StarBase predicted that miR-139 binds to the 3ʹUTR of CBX3 mRNA. Gene set enrichment analysis (GSEA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were utilized to elucidate the individual gene sets associated with CBX3 expression in the LIHC TCGA data, and |NES| >1, FDR q value < 0.25 and P-value < 0.05 were considered significant set points for GSEA. Moreover, gene set variation analysis (GSVA) was performed to further identify significant differences in genome-regulated biological processes.
Statistical analysis
We analyzed the data by using SPSS software (version 23.0, SPSS, Inc., Chicago, IL) and GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). The association of CBX3 expression with clinicopathological parameters in HCC was verified using the χ2 test. The continuous variables were analyzed by the Mann-Whitney U test and an unpaired t test. Pearson’s correlation test was applied to analyze the correlation between CBX3 and Ki-67 mRNA expression, CBX3 and PCNA expression, and CBX3 and miR139 mRNA expression. The log-rank tests and Kaplan-Meier methods were applied to evaluate patient survival. P < 0.05 was defined as significant for all analyses.
Results
CBX3 is usually unregulated and acts as an unfavorable prognostic marker in HCC
To identify the CBX3 expression profiles in HCC, we extracted data from the GEO, TCGA, and the LIRIJP databases and analyzed the datasets. Figure 1a shows that the CBX3 mRNA level was commonly upregulated in HCC tissues compared with normal liver tissues. Moreover, analysis of GSE6764 (Figure 1b) revealed that CBX3 mRNA expression was overexpressed in the HCC tissues compared with the nontumor liver tissues (early HCC or advanced HCC vs the average of normal liver, cirrhotic, low-grade dysplastic liver and high-grade dysplastic liver, all P < 0.001). Analysis of GSE84598 (Figure 1c) revealed that CBX3 was elevated in the HCC tissues compared with the normal liver tissues (P < 0.01) and liver border tissues (P < 0.05). As shown in Figure 1d-1e, the CBX3 mRNA level was substantially higher in the HCC tissues (P < 0.01) and portal vein tumor thrombosis (P < 0.01) than in the normal liver tissues. To further study the clinical prognostic significance of CBX3 mRNA overexpression in HCC, we extracted information on pathological parameters and patient survival from TCGA. We found that CBX3 mRNA expression was positively correlated with malignant tumor growth factors, including Ki67 (P < 0.001, figure 1f) and PCNA (P < 0.001, Figure 1g). Furthermore, gene set enrichment analysis (GSEA) revealed that CBX3 overexpression was negatively correlated with increased survival (Figure 1h) and positively correlated with decreased survival (Figure 1i) in the TCGA-LIHC dataset.
Figure 1.
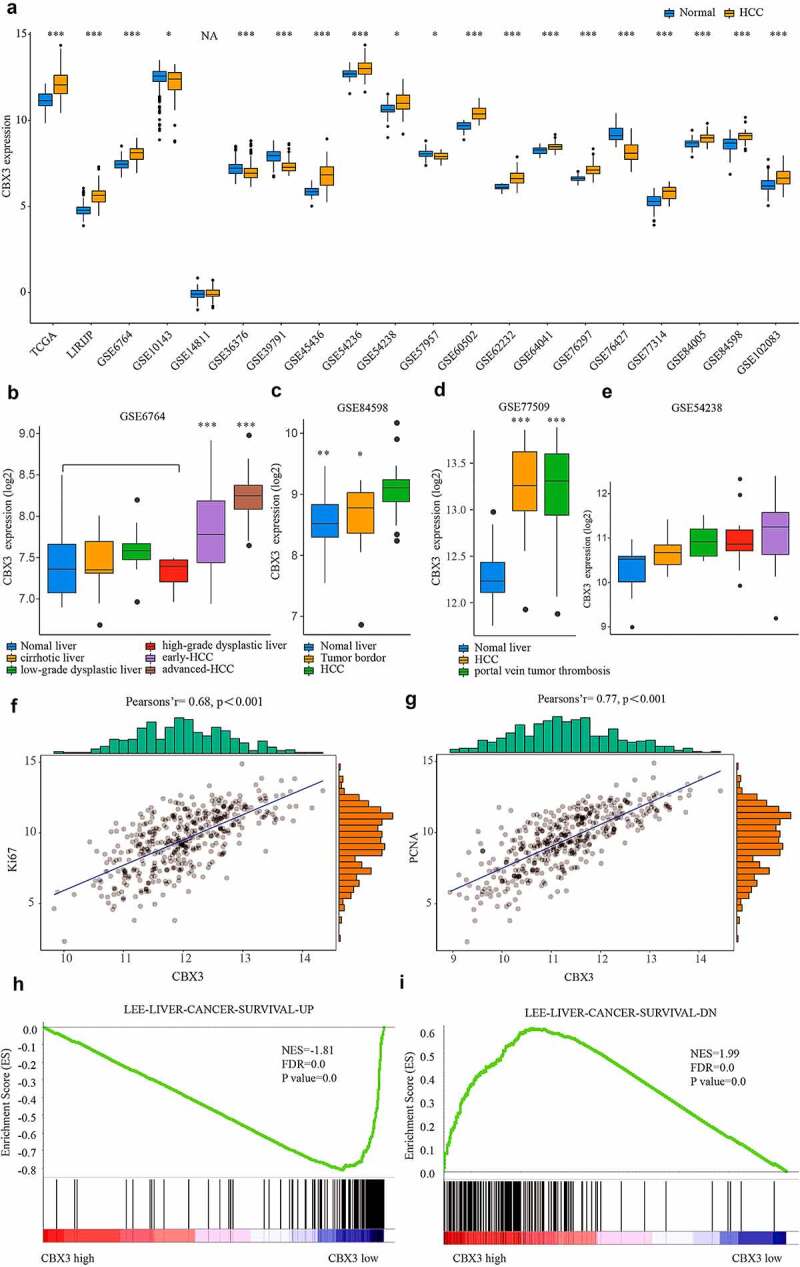
CBX3 mRNA was upregulated in HCC tissues and was positively correlated with malignant pathological features of HCC. (a) Data from the GEO, TCGA, and the LIRIJP databases showed that the CBX3 mRNA levels tended to be high in HCC tissues. (b, c) CBX3 mRNA expression was higher in HCC tissues than in nontumor liver tissues. (d, e) Elevated CBX3 expression was found in HCC and portal vein tumor thrombosis. (f, g) Spearman’s correlation showed that CBX3 mRNA expression was positively correlated with Ki67 (f) and PCNA (g) in HCC patients. (h, i) GSEA showed that CBX3 mRNA expression was positively correlated with increased liver cancer survival (h) and negatively associated with decreased liver cancer survival (i).
Kaplan-Meier analysis demonstrated that high expression of CBX3 predicted short overall survival (P < 0.001, Figure 2a) and short progression-free survival (P < 0.001, Figure 2b) in the HCC patients. After further stratification by TNM stage, high expression of CBX3 was also a marker of poor overall survival (Figure 2c) and progression-free survival (Figure 2d) of HCC patients, regardless of TNM stage (TNM I–II or TNM III–IV). These results indicated that high CBX3 expression was a marker of poor prognosis for HCC patients.
Figure 2.
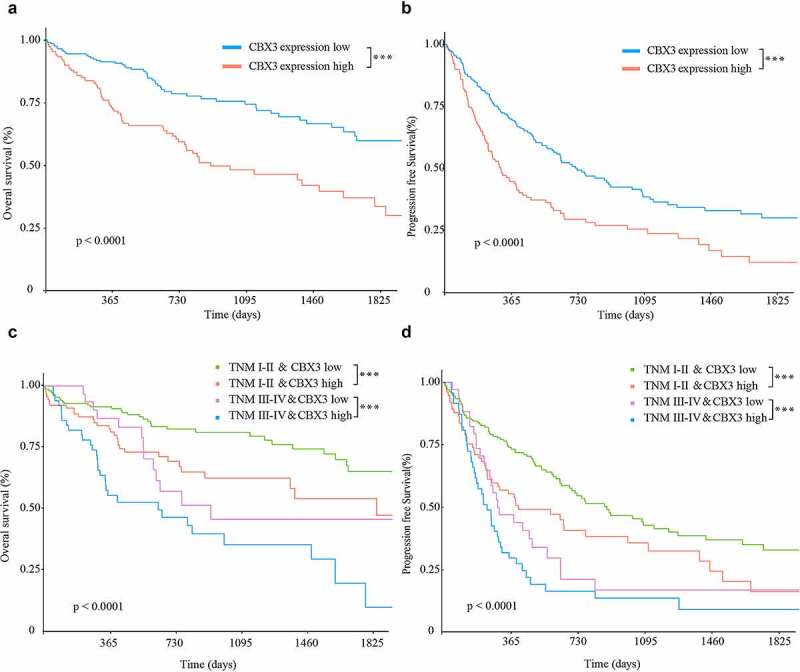
High CBX3 expression was a biomarker of poor prognosis for HCC patients. (a, b) Kaplan‑Meier analysis showed that overexpression of CBX3 was correlated with poor overall survival (OS, A) and progression-free survival (PFS, B) of HCC patients based on analysis of TCGA LIHC database. (c, d) Regardless of the TNM stage (TNM I–II or TNM III–IV), HCC patients with high expression of CBX3 had both a short OS (c) and a short PFS (d).
Biological functions of CBX3 in HCC cells
To illustrate the functional role of CBX3 in the development of HCC cells in vitro, we first conducted qRT-PCR and western blot analysis, and the results showed that the mRNA and protein levels of CBX3 were higher in the HCC cell lines (Herp-3B and SMMC-7721 cells) than in the normal liver cell lines (L02 and Changliver cells) (Figure 3a). Furthermore, we constructed Herp-3B and SMMC-7721 cell lines with CBX3 knockdown by using RNA interference. CBX3 knockdown was confirmed by western blotting (Figure 3b). The CCK-8 assay revealed that the viability of the Herp-3B (Figure 3c) and SMMC-7721 (Figure 3d) cells with CBX3 knockdown was lower than that of the control cells. The wound healing assay showed that CBX3 knockdown repressed the migration of HCC cells (Figure 3e). Decreased CBX3 expression reduced the colony formation and invasive capacity of HCC cells, as determined by colony formation (figure 3f) and Transwell (Figure 3g) assays.
Figure 3.
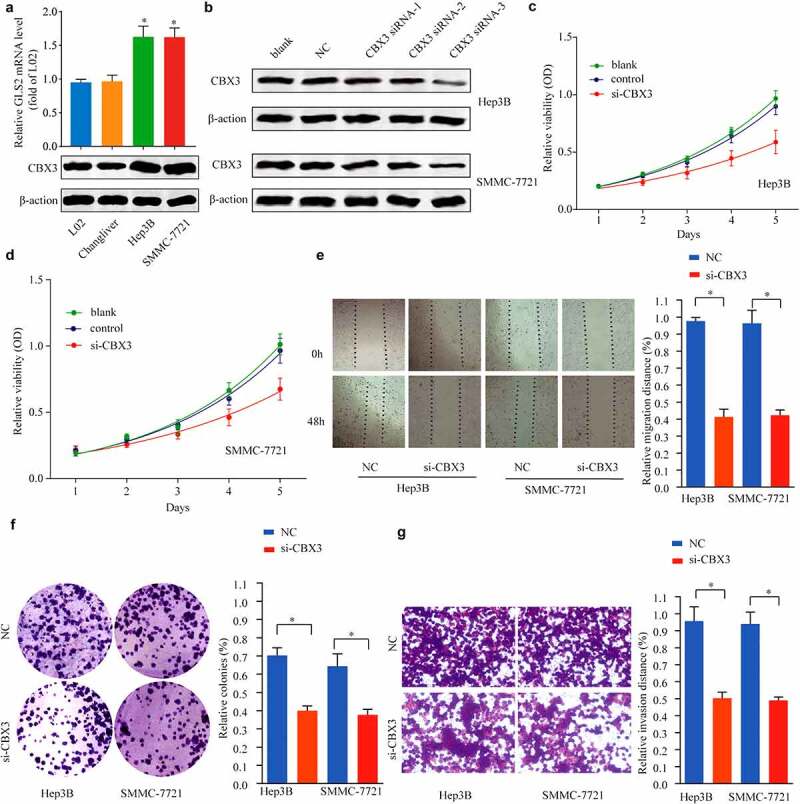
Inhibition of CBX3 suppressed the biological behaviors of HCC cells. (a) CBX3 was overexpressed in HCC cell lines compared with normal liver cell lines, as shown by qRT-PCR and western blots. (b) The knockdown efficiency of CBX3 siRNAs in Hep3B and SMMC-7721 cells was measured by western blots. (c, d) MTT assays showed that the viability of Hep3B (c) and SMMC-7721 cells (d) was decreased by inhibition of CBX3. (e) Wound healing assays indicated that CBX3 knockdown inhibited Hep3B and SMMC-7721 cell migration. (f) Colony numbers of CBX3 knockdown HCC cells were significantly reduced compared to those of the NC. (g) Transwell assays showed that the invasion of HCC cells was attenuated by CBX3 knockdown.
Herp-3B and SMMC-7721 cells were selected for transfection with CBX3 overexpression plasmids and empty vector, respectively. The efficiency of CBX3 overexpression was confirmed in HCC cells by western blotting (Figure 4a). The CCK-8 assay (Figure 4b) showed that increased expression of CBX3 accelerated HCC cell proliferation. The wound healing assay revealed that overexpression of CBX3 promoted the movement of SMMC-7721 and Herp-3B cells toward the center of the wound (Figure 4c). The colony formation assay showed that the number of colonies formed was increased in the CBX3-overexpressing group (Figure 4d). The Transwell assay demonstrated that the invasive ability of the SMMC-7721 and Herp-3B cells overexpressing CBX3 was enhanced compared with that of the control (Figure 4e). Collectively, these findings demonstrated that CBX3 could promote the biological behaviors of HCC cells.
Figure 4.
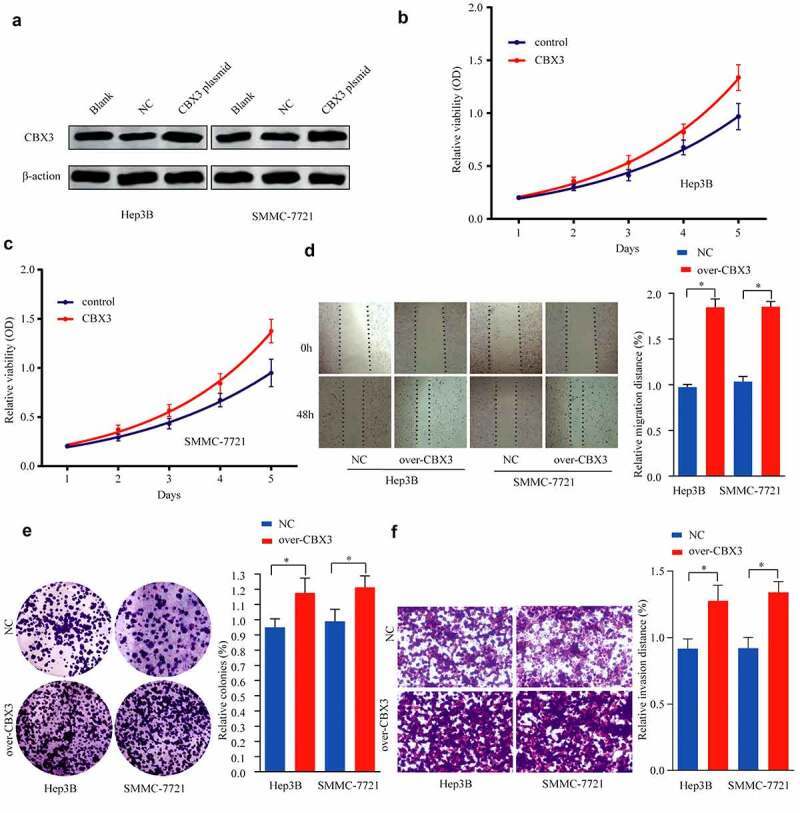
Overexpression of CBX3 played a tumor-promoting role in HCC progression. (a) Hep3B and SMMC-7721 cells transfected with pcDNA3.1-CBX3 or pcDNA3.1-vector were assessed for CBX3 expression by western blots. (b, c) CCK-8 assays were carried out to examine the effects of CBX3 overexpression on the proliferative of Hep3B (b) and SMMC-7721 (c) cells. (d) Wound healing assays were performed to determine the effects of CBX3 overexpression on the migration of HCC cells. (e) Colony formation assays of HCC cells were performed after overexpression of CBX3. (f) The invasion assay was used to examine the effects of CBX3 overexpression on HCC cell invasion. *P < 0.05.
CBX3 may be involved in the regulation of cell cycle progression in HCC
To explore the potential mechanism of CBX3ʹs aggressive activity in HCC, we performed KEGG pathway analysis. Interestingly, this analysis showed that the majority of altered genes were related to cell cycle regulation (Figure 5a). GSEA showed that CBX3 expression was positively correlated with cell cycle progression (Figure 5b). GSVA showed that the differentially expressed genes were related to Myc targets, E2F targets, and the G2/M checkpoint, which are associated with genes in pathways regulating the cell cycle (Figure 5c). Then, we overlapped the altered genes from KEGG analysis and those from GSVA, and E2F, Myc, CDK1 and Cyclin B1 were chosen. To further confirm the major genes correlated with the functional role of CBX3, we performed western blotting to show that the protein levels of E2F, Myc, CDK1 and Cyclin B1 were all decreased in the CBX3 knockdown HCC cells, and the opposite effect was found in the HCC cells overexpressing CBX3 (Figure 5d). Therefore, CBX3 may participate in HCC development by regulating cell cycle progression.
Figure 5.
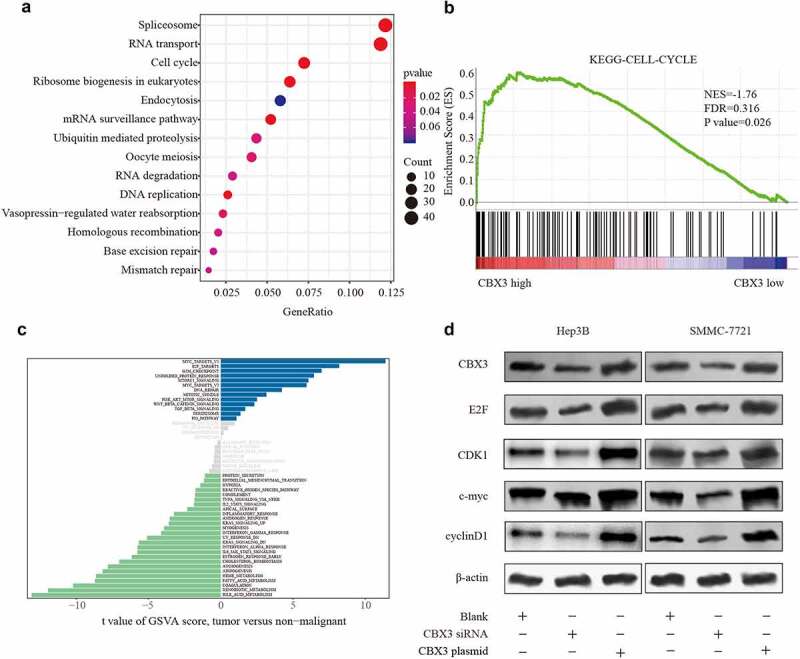
CBX3 regulated HCC progression through the cell cycle. (A, B, C) Cross-sectional analysis of KEGG (a), GSEA (b) and GSVA (c) showed that CBX3 expression was correlated with cell cycle regulation. (d) Western blot analysis was used to detect the levels of E2F, c-Myc, CDK1 and Cyclin B1 in the indicated cells, with β-actin as a loading control.
CBX3 is a direct target of miR-139 in HCC
To identify the upstream mechanism responsible for CBX3-mediated HCC progression, we used starBase and predicted that miR-139 was incompletely complementary to CBX3 (Figure 6a). To determine whether miR-139 could directly bind to the 3ʹUTR of CBX3, we performed double luciferase reporter assays to show that luciferase activity in the SMMC-7721 and Herp-3B cells carrying the wild-type (WT)-CBX3 3ʹUTR was inhibited by miR-139 overexpression compared with that of the cells carrying a mutant (Mut)-CBX3 3’-UTR (Figure 6b). Furthermore, using western blots and qRT-PCR, we found that the CBX3 protein (Figure 6c) and mRNA levels (Figure 6d) were both substantially decreased in the SMMC-7721 and Herp-3B cells transfected with miR-139 mimics compared with the negative control cells. In contrast, inhibiting miR-139 expression in these cells increased the expression of CBX3. Overall, the data suggested that CBX3 is a direct target of miR-139 in HCC. Subsequently, analysis of TCGA dataset indicated that miR-139 expression was negatively correlated with CBX3 (Figure 6e). To demonstrate the relationship between miR-139 expression and prognosis in HCC patients, we used the log-rank test to show that miR-139 expression was positively correlated with favorable overall survival (figure 6f) and progression-free survival (Figure 6g) in HCC patients, regardless of CBX3 expression.
Figure 6.
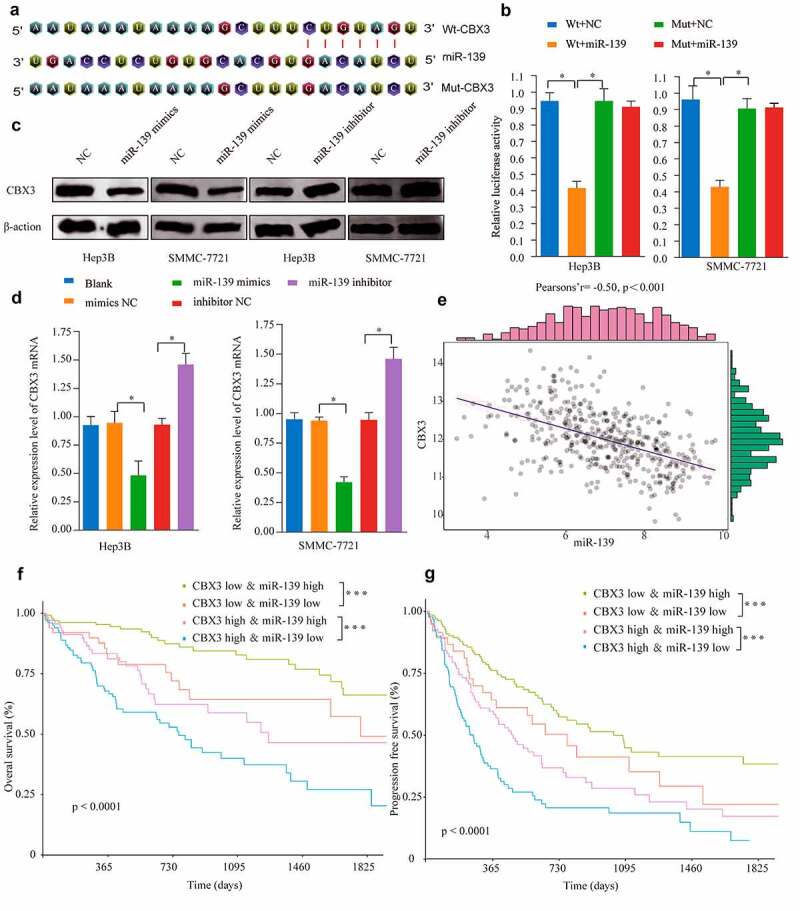
MiR-139 directly targeted CBX3 in HCC. (a) MiR-139 was predicted to directly bind to the 3ʹUTR of CBX3 and the corresponding mutant sites of CBX3. (b) The luciferase activity of wild-type (Wt) CBX3-3ʹUTR and mutant (Mut) CBX3-3ʹUTR was assessed in Hep3B and SMMC-7721 cells. (c, d) After transfection of Hep3B and SMMC-7721 cells with miR-139 mimics or miR-139 inhibitor, western blotting (c) and RT-qPCR (d) analysis were used to examine the protein and mRNA levels of CBX3. (e) Spearman’s analysis revealed that miR-139 expression was inversely associated with CBX3 mRNA expression from TCGA database. (f, g) With stratification by both TNM stage and miR-139 expression levels, Kaplan-Meier analysis showed that miR-139 expression was positively correlated with the overall survival (OS, F) and progression-free survival (PFS, G) of HCC patients. *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-139 regulated HCC progression by directly targeting CBX3
Previous studies have shown that miR-139 can inhibit the growth of HCC cells [11–13]. To determine whether miR-139 is a functional direct regulator of CBX3 in HCC cells, we transfected miR-139 mimics or miR-NC into SMMC-7721 and Herp-3B cells, and the combined effects of miR-139 and CBX3 overexpression were then investigated. The CBX3 protein expression (Figure 7a) in the miR-139 mimic group was dramatically decreased relative to that in the miR-NC group and restored in the combined miR-139 and CBX3 overexpression group. Thereafter, further experiments demonstrated that overexpression of miR-139 alone attenuated the expression levels of cell cycle progression-related proteins (Figure 7g), decreased cell viability and the colony-forming ability and inhibited migration and invasion (Figure 7b-f) (all P < 0.05) in HCC cells; these effects were all significantly antagonized by simultaneously overexpressing miR-139 and CBX3 (Figure 7a-g). Taken together, our data confirmed that miR-139 suppressed the progression of HCC partly by regulating CBX3.
Figure 7.
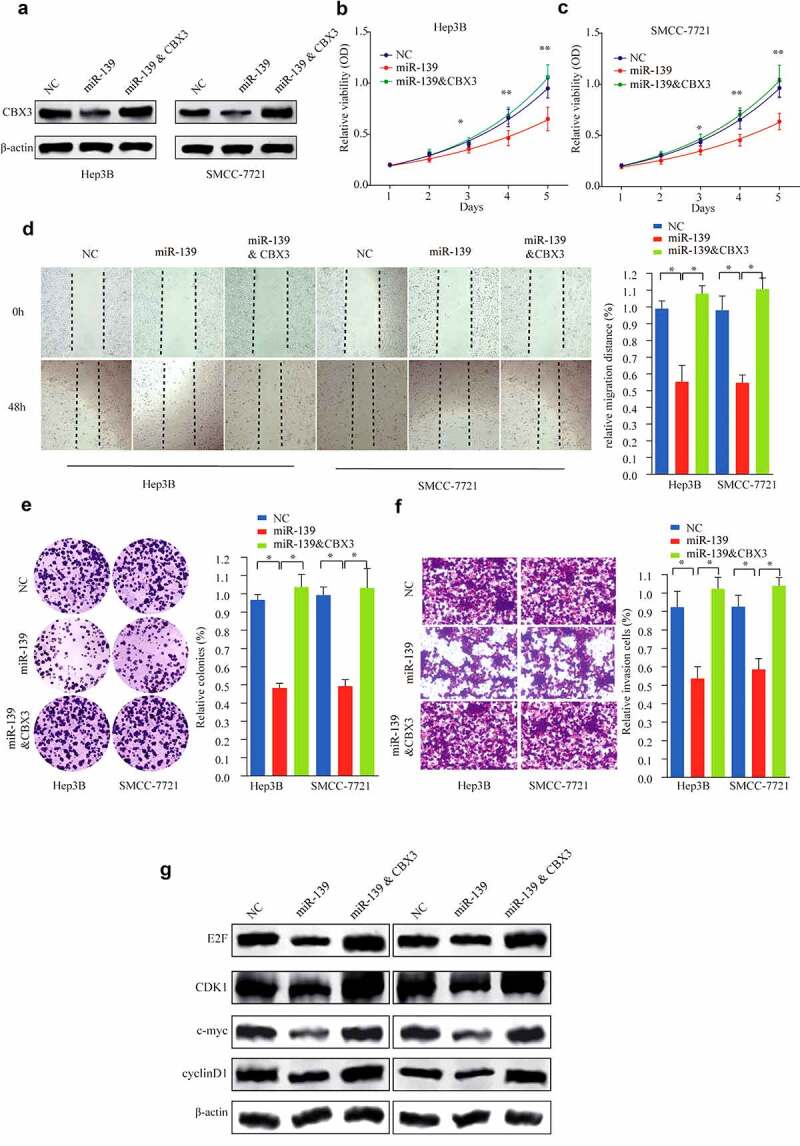
Restoration of CBX3 ameliorated the inhibitory effect of miR-139 on HCC cell lines. (a) Hep3B and SMMC-7721 cells were transfected with NC, miR-139 mimics or miR-139 mimics/CBX3, as shown by western blot analysis. (b, c) Cell viability of Hep3B (b) and SMMC-7721 (c) cells was determined by CCK-8 assays. (d) Wound healing assays were performed to determine the migration of the introduced Hep3B and SMMC-7721 cells. (e) Colony formation assays were performed. (f) The invasive capacities of the cells were assayed as described above. (g) Western blots showed the expression of E2F, c-Myc, CDK1 and Cyclin B1 in the indicated Hep3B and SMMC-7721 cells. *P < 0.05, **P < 0.01.
Discussion
Epigenetic regulation can affect gene transcriptional activity by histone modification without changing DNA sequences [14]. Aberrant epigenetic alteration is a hallmark of cancers [15], and HP1 is an important regulator of histone modification involved in DNA damage repair [16], heterochromatin formation, transcriptional regulation, and chromatin remodeling. CBX3 encodes a protein called HP1γ, which is a member of the HP1 family. CBX3/HP1γ has been reported to play a tumor-promoting role and may be a therapeutic target in many cancers. Previous studies have shown that CBX3 is highly expressed in prostate cancer [17], non–small cell lung cancer (NSCLC) [18], osteosarcoma [19], tongue squamous cell carcinoma (TSCC) [20], colorectal cancer [21] and pancreatic cancer [22]. In addition, Ma Chao et al. demonstrated that CBX3 overexpression is a marker for poor prognosis in osteosarcoma patients [19]. Huayong Zhang et al. found that CBX3/HP1γ upregulation is correlated with a dismal prognosis for tongue squamous cell carcinoma patients. Further cell experiments showed that CBX3 repression inhibited, while CBX3 overexpression promoted TSCC cell growth and colony formation [23]. Recently, Xiaoping Zhong et al. reported that CBX3/HP1γ plays an important role in HCC tumorigenicity and might be a biomarker of poor prognosis for HCC patients [24].
In our study, we first determined the CBX3 mRNA levels in HCC from public databases. Analysis of public datasets showed that CBX3 was significantly upregulated in the HCC tissues relative to the normal liver tissues. Subsequently, these results were further supported by qRT-PCR and western blot analyses of HCC cell lines and IHC staining of HCC tissues. Analysis of a TMA revealed that high CBX3 expression was closely correlated with some malignant clinicopathological characteristics. The log-rank test revealed that high CBX3 expression usually predicted a poor prognosis in HCC patients. In vitro experiments showed that CBX3 knockdown could repress the growth, migration and invasive ability of SMMC-7721 and Herp-3B HCC cells; however, CBX3 overexpression promoted the progression of HCC cells. Our results showed that CBX3 was involved in progression and may be an indicator of poor prognosis in HCC.
Abnormal cell cycle regulation is important in HCC progression [25,26]. Previous research revealed that CBX3 was involved in the progression of multiple cancers by regulating the cell cycle [20,27]. Knockdown of CBX3 expression inhibited cell proliferation via an increase in G0/G1 phase arrest and apoptosis [19]. Zhang HuaYong et al. demonstrated that CBX3 accelerated TSCC cell growth through G1/S phase modulation by suppressing the expression of p21 [23]. Liu Ming et al. demonstrated that HP1γ promoted the proliferation of colorectal cancer cells in vitro and in vivo by directly targeting p21 [21]. We explored the mechanism by which CBX3 regulates HCC progression. Analysis of KEGG pathways and GSEA revealed that altered CBX3 expression was correlated with cell cycle progression. GSEA showed that CBX3 was positively associated with Myc targets, E2F targets and the G2/M checkpoint. Western blotting proved that the HP1 protein was positively correlated with key transcriptional regulators of the cell cycle, such as E2F, Myc, CDK1 and Cyclin B1. Our data showed that CBX3 was involved in HCC development by regulating cell cycle progression.
In recent years, miRNAs have been shown to play tumor-promoting or tumor-suppressing roles in various tumors [28,29]. CBX3 has been reported to be regulated by miR-30a in colorectal cancer. In addition, HP1γ functions as an oncogene that directly represses miR-451a expression in prostate cancer progression [17]. However, upregulation of HP1γ may be controlled by various miRNAs in HCC. According to the bioinformatics analysis, miR-139 was predicted to regulate CBX3 in HCC. Then, the interplay between miR-139 and CBX3 was verified by dual-luciferase reporter assays. The qRT-PCR and western blot results showed that miR-139 expression was negatively correlated with CBX3 expression. Confirmatory experiments showed that CBX3 repression could reverse the proliferation and invasion induced by miR-139 downregulation. These data revealed that miR-139 functioned as a tumor suppressor involved in HCC progression, which is consistent with the results of previous reports [12,30,31]. Wong Carmen Chak-Lui et al. demonstrated that miR-139 is downregulated in HCC and plays a tumor suppressive role [12]. Moreover, Zan Ying et al. reported that miR-139 overexpression led to decreased c-Myc levels in HCC [32]. Our results also showed that miR-139 expression was negatively related to the expression of E2F, Myc, CDK1 and Cyclin B1, as demonstrated by western blots. Rescue experiments showed that miR-139 inhibited HCC progression through direct inhibition of CBX3 expression.
Conclusion
Collectively, the present study demonstrated that CBX3 has a tumor-promoting role in HCC development and is a marker of poor prognosis in HCC patients. We found that miR-139 may have an anticancer effect by inhibiting the expression of CBX3 in HCC. Our results demonstrated that the miR-139/CBX3 axis may be involved in HCC development by regulating cell cycle progression and may be a promising target in the treatment of HCC.
Supplementary Material
Funding Statement
This work was supported by the Henan Medical Science and Technology Public Relations Project (Grant No. 2018020914), GL provided this funding and considered as corresponding author in our paper. And the Foundation of Luoyang Central Hospital (Grant No. LHGJ20191222), PZ provided this funding and considered as first author in our paper.
Authors’ contributions
GL conceived the experiments; XY gave an experimental guidance in the lab; ZZ and YZ performed the experiments; GZ designed the experiments, PZ analyzed the data and wrote the paper. All authors have read and approved the manuscript and ensure that this is the case.
Availability of data and materials
The data used to support the findings of this study and related materials are available from the corresponding author upon request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Declarations
Ethics approval and consent to participate:
This study obtained the approval of the Ethics Committee of Luoyang Central Hospital Affiliated to Zhengzhou University. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All of the patients were consent to participate in this study.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Poon D, Anderson BO, Chen LT, et al. Management of hepatocellular carcinoma in Asia: consensus statement from the asian oncology summit 2009. Lancet Oncol. 2009;10(11):1111–1118. [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. [DOI] [PubMed] [Google Scholar]
- [3].Dhanasekaran R, Nault JC, Roberts LR, et al. Genomic medicine and implications for hepatocellular carcinoma prevention and therapy. Gastroenterology. 2019;156(2):492–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Galle PR PR , Forner A , Llovet MJ. et al. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. [DOI] [PubMed] [Google Scholar]
- [5].Smallwood A, Hon GC, Jin F, et al. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22(8):1426–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Akaike Y, Kuwano Y, Nishida K, et al. Homeodomain-interacting protein kinase 2 regulates DNA damage response through interacting with heterochromatin protein 1γ. Oncogene. 2014;34(26):3463–3473. [DOI] [PubMed] [Google Scholar]
- [7].Rupaimoole R, Slack FJ.. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. [DOI] [PubMed] [Google Scholar]
- [8].Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. [DOI] [PubMed] [Google Scholar]
- [9].Zhu W, Li H, Yu Y, et al. Enolase-1 serves as a biomarker of diagnosis and prognosis in hepatocellular carcinoma patients. Cancer Manag Res. 2018;10:5735–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bao J, Yu Y, Chen J, et al. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9(10):1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang L, Yin D, Wang Y, et al. Inhibition of the growth of hepatocellular carcinoma cells through fibroblast growth factor 18 suppressed by miR-139. Oncol Rep. 2017;38(4):2565–2571. [DOI] [PubMed] [Google Scholar]
- [12].Wong CC, Wong CM, Tung EK, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140(1):322–331. [DOI] [PubMed] [Google Scholar]
- [13].Gu W, Li X, Wang J. miR-139 regulates the proliferation and invasion of hepatocellular carcinoma through the WNT/TCF-4 pathway. Oncol Rep. 2014;31(1):397–404. [DOI] [PubMed] [Google Scholar]
- [14].Sridharan R, Gonzales-Cope M, Chronis C, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1 gamma in reprogramming to pluripotency. Nat Cell Biol. 2013;15(7):872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- [16].Dialynas GK, Vitalini MW, Wallrath LL. Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat Res. 2008;647(1–2):13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chang C, Liu J, He W, et al. A regulatory circuit HP1gamma/miR-451a/c-Myc promotes prostate cancer progression. Oncogene. 2018;37(4):415–426. [DOI] [PubMed] [Google Scholar]
- [18].Chang SC, Lai YC, Chen YC, et al. CBX3/heterochromatin protein 1 gamma is significantly upregulated in patients with non-small cell lung cancer. Asia Pac J Clin Oncol. 2018;14(5):e283–e288. [DOI] [PubMed] [Google Scholar]
- [19].Ma C, Nie XG, Wang YL, et al. CBX3 predicts an unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Mol Med Rep. 2019;19(5):4205–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang H, Chen W, Fu X, et al. CBX3 promotes tumor proliferation by regulating G1/S phase via p21 downregulation and associates with poor prognosis in tongue squamous cell carcinoma. Gene. 2018;654:49–56. [DOI] [PubMed] [Google Scholar]
- [21].Liu M, Huang F, Zhang D, et al. Heterochromatin protein HP1gamma promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75(21):4593–4604. [DOI] [PubMed] [Google Scholar]
- [22].Chen LY, Cheng CS, Qu C, et al. CBX3 promotes proliferation and regulates glycolysis via suppressing FBP1 in pancreatic cancer. Biochem Biophys Res Commun. 2018;500(3):691–697. [DOI] [PubMed] [Google Scholar]
- [23].Zhang H, Fu X, Su X, et al. CBX3/HP1gamma is upregulated in tongue squamous cell carcinoma and is associated with an unfavorable prognosis. Exp Ther Med. 2018;15(5):4271–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhong X, Kan A, Zhang W, et al. CBX3/HP1gamma promotes tumor proliferation and predicts poor survival in hepatocellular carcinoma. Aging (Albany NY). 2019;11(15):5483–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Klungboonkrong V, Das D, McLennan G. Molecular mechanisms and targets of therapy for hepatocellular carcinoma. J Vasc Interv Radiol. 2017;28(7):949–955. [DOI] [PubMed] [Google Scholar]
- [26].Shen S, Dean DC, Yu Z, et al. Role of cyclin-dependent kinases (CDKs) in hepatocellular carcinoma: therapeutic potential of targeting the CDK signaling pathway. Hepatol Res. 2019. DOI: 10.1111/hepr.13353 [DOI] [PubMed] [Google Scholar]
- [27].Chen LY, Cheng CS, Qu C, et al. Overexpression of CBX3 in pancreatic adenocarcinoma promotes cell cycle transition-associated tumor progression. Int J Mol Sci. 2018;19(6):1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rossi JJ. New hope for a microRNA therapy for liver cancer. Cell. 2009;137(6):990–992. [DOI] [PubMed] [Google Scholar]
- [29].Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen J-A, Yu Y, Xue C, et al. Low microRNA-139 expression associates with poor prognosis in patients with tumors: a meta-analysis. Hepatobiliary Pancreat Dis Int. 2019;18(4):321–331. [DOI] [PubMed] [Google Scholar]
- [31].Li P, Xiao Z, Luo J, et al. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23(4):2475–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zan Y, Wang B, Liang L, et al. MicroRNA-139 inhibits hepatocellular carcinoma cell growth through down-regulating karyopherin alpha 2. J Exp Clin Cancer Res. 2019;38(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study and related materials are available from the corresponding author upon request.


