Abstract
Nitrogen nutrition in cyanobacteria is regulated by NtcA, a transcriptional activator that is subject to negative control by ammonium. Using Synechococcus sp. strain WH7803 as a model organism, we show that ntcA expression was induced when cells were exposed to nitrogen stress but not when they were subjected to phosphorus or iron deprivation. Transcript levels accumulated in cells grown on a variety of inorganic and organic nitrogen sources, with the sole exception of ammonium. ntcA transcription was induced when ammonium levels dropped below 1 μM and reached maximal levels within 2 h. Furthermore, the addition of more than 1 μM ammonium led to a rapid decline in ntcA mRNA. The negative effect of ammonium was prevented by the addition of l-methionine-d,l-sulfoximine (MSX) and azaserine, inhibitors of ammonium assimilation. Thus, basal ntcA transcript levels are indicative of ammonium utilization. Conversely, the highest ntcA transcript levels were found in cells lacking a nitrogen source capable of supporting growth. Therefore, maximal ntcA expression would indicate nitrogen deprivation. This state of nitrogen deprivation was induced by a 1-h incubation with MSX. The rapid response of ntcA gene expression to the addition of ammonium and MSX was used to design a protocol for assessing relative ntcA transcript levels in field populations of cyanobacteria, from which their nitrogen status can be inferred. ntcA was basally expressed in Synechococcus at a nutrient-enriched site at the northern tip of the Gulf of Aqaba, Red Sea. Therefore, these cyanobacteria were not nitrogen stressed, and their nitrogen requirements were met by regenerated nitrogen in the form of ammonium.
Phytoplankton biomass is thought to be limited by nitrogen availability in many oligotrophic bodies of water (10, 54). Yet the growth of all phytoplankton taxa in such waters is not necessarily rate limited. A significant proportion of phytoplankton biomass and production in oligotrophic seas is contributed by unicellular cyanobacteria of the genera Synechococcus and Prochlorococcus (3, 5, 43). These picophytoplankton taxa grow rapidly despite low ambient nitrogen concentrations in oligotrophic waters (for a review, see reference 15). It has been hypothesized that they acquire the nitrogen they require for growth from nitrogen sources that are rapidly recycled in the photic layer (24, 45). However, no unequivocal evidence for the exclusive utilization of regenerated nitrogen sources (e.g., NH4+ and organic N) by these picophytoplanktonic taxa exists (6, 9, 53). Nor is it apparent whether the use of such nitrogen sources enables these taxa to avoid nitrogen stress. This is mainly because standard oceanographic methods are not conducive to assessing nitrogen deprivation of and nitrogen source utilization by (defined here collectively as the nitrogen status) a certain phytoplankton taxon among the myriad of organisms found in the sea. Therefore, it is necessary to develop techniques capable of assessing the nitrogen status of phytoplankton along taxonomic lines.
Ammonium is the preferred source of inorganic nitrogen in cyanobacteria (13, 18, 34, 35). It may be obtained from the environment by either passive diffusion or active uptake and is assimilated into organic matter via the activities of glutamine synthetase (GS) and glutamate synthase (GOGAT) (13). In the absence of sufficient ammonium, the cyanobacterial cell undergoes a series of adaptive processes in order to obtain the nitrogen required for growth and survival. The initial responses to ammonium deficiency include the induction of higher-affinity ammonium uptake systems and the synthesis of proteins required for the utilization of other nitrogenous compounds such as nitrate, nitrite, urea, and amino acids (13). The utilization of alternative nitrogen sources is energetically more expensive than that of ammonium as, in most cases, it requires both active transport over the cell membrane and conversion to ammonium before assimilation into organic compounds (13, 23). It should be noted that ammonium prevents the utilization of alternative nitrogen sources such as nitrate and nitrite by inhibiting their transport and repressing synthesis of the proteins required for their assimilation at the level of gene transcription (13, 23, 37, 52). Once all external nitrogen sources suitable for growth have been exploited, the cell enters a stage of nitrogen deprivation. During the adaptation of the cell to nitrogen stress, growth may continue transiently as many physiological changes take place, including the specific degradation of phycobiliproteins, which results in chlorosis (22, 56). This process would allow reuse of the nitrogen for the synthesis of proteins required for survival under conditions of nitrogen deprivation (21, 22). Growth is halted once both external and internal nitrogen supplies have been exhausted.
Synthesis of the nitrogen regulatory protein, NtcA, is an essential step in cyanobacterial adaptation to conditions of ammonium depletion. ntcA mutants are incapable of growth on nitrate and nitrite (55; A. Moyal, D. Lindell, and A. F. Post, submitted for publication), and they do not degrade phycobiliproteins in a timely manner under conditions of nitrogen depletion (47; Moyal et al., submitted). This transcriptional activator is subject to negative control by ammonium at the level of gene expression (34, 37). ntcA expression is down-regulated to basal levels in the presence of ammonium. In the absence of ammonium, NtcA enhances the expression of its own gene as well as of those required for the uptake and assimilation of nitrogen sources like nitrate and nitrite (37; Moyal et al., submitted). However, ntcA expression levels appear to be higher under conditions of nitrogen deprivation than in nitrate-grown cells (34). NtcA may also be involved in the expression of genes required for urea utilization (7). The mode of action of NtcA in the process of chlorosis under conditions of nitrogen depletion has yet to be elucidated.
The responsiveness of ntcA to nitrogen availability and the pivotal role it plays in the adaptation of cells to conditions of ammonium and nitrogen depletion suggests that basal and maximal ntcA expression may be useful indicators of ammonium sufficiency and nitrogen deprivation, respectively, in field populations of cyanobacteria. As pointed out by others (31, 41, 50), before a gene or protein can be used in field studies, its pattern of expression must be rigorously studied in relation to appropriate environmental factors under controlled laboratory conditions. In this study we focus on the response of ntcA gene expression to ecologically relevant nitrogen conditions, and we develop a protocol for the investigation of the nitrogen status of cyanobacterial field populations using ntcA gene expression. Our model organism for this study is Synechococcus sp. strain WH7803, a strain type with representatives found in various seas including the Red Sea (4, 36, 44).
MATERIALS AND METHODS
Culture conditions.
Marine Synechococcus sp. strain WH7803 was grown in batch cultures on ASW (56), a defined artificial seawater medium buffered to pH 8, modified as previously described to remove inorganic nitrogen from the trace metal mix (34). The medium was further modified by replacing Tris with HEPES. ASWNO3 and ASWNH4 contained 9 mM NO3− and 2 mM NH4+ as sole nitrogen sources, respectively, whereas ASW0 was devoid of a combined nitrogen source. Organic N sources used in this study were filter sterilized before addition to ASW0 medium. The compositions of organic nitrogen mixes were as follows: for purines, 100 μM (each) adenine, xanthine, guanine, and hypoxanthine; for pyrimidines, 100 μM (each) cytosine, thymine, and uracil; for amides, 100 μM (each) formamide and acetamide; for basal medium Eagle (BME) amino acids, between 50 to 100 μM l isomers of arginine, cysteine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, tyrosine, and valine with the addition of glutamine. The organic N sources used in this study were obtained from Sigma Chemicals except for the BME amino acid mix which was supplied by Biological Industries (Beit HaEmek, Israel) and were of tissue culture grade. NH4+ concentrations were determined during growth experiments on organic sources using a sodium sulfite-adjusted orthophthaldialdehyde method (29) to ensure that ammonium did not accumulate due to spontaneous release from the organic sources.
Cultures were grown at 25°C under continuous white light (provided by cool fluorescent lamps) at an intensity of 40 to 50 μmol of photons · m−2 · s−1 with constant agitation on an orbital shaker at 125 rpm. Growth of cultures was monitored by optical density at 750 nm. Doubling times were in the range of 13 to 16 h. Cells were maintained for a minimum of 5 generations in exponential growth prior to experimentation.
For nitrogen source and nutrient deprivation experiments, cells were collected by centrifugation at 25°C for 10 min at 10,000 × g, washed twice, and then resuspended in the new growth medium. Samples for RNA analysis were taken from cultures in early- to mid-log phase except during nutrient deprivation experiments. Each experiment was repeated independently at least twice.
RNA extraction and analyses from Synechococcus cultures.
Cells were harvested by filtration onto 0.45-μm-pore-size polycarbonate membrane filters (Poretics). RNA was extracted with the Ultraspec RNA reagent (Biotecx) or by using a hot phenol method (48) modified as set out by Lindell et al. (34) and followed by DNase treatment. Total RNA was quantified spectrophotometrically and from ethidium bromide-stained RNA run on nondenaturing agarose gels.
RNase protection assays (RPAs) were carried out on equal amounts of total RNA with a probe internal to the ntcA gene (designated the internal probe). In one experiment a probe that is partially upstream of the ntcA gene (referred to below as the upstream probe) was also used. RPA analysis with the internal probe produces a single protected fragment of 450 bp, whereas the upstream probe produces two protected fragments: a 400- and a 165-bp fragment corresponding to the constitutively expressed and ammonium-regulated ntcA transcripts, respectively (34). Antisense biotinylated RNA probes were transcribed using the Ambion BrightStar BiotinScript kit as previously described (34). The Ambion RPAII kit was used for RPAs as follows. After coprecipitation, the probe and RNA were hybridized for approximately 16 h at 43.5°C in 20 μl of hybridization solution. The denatured RNA and probe were electrotransferred (NovaBlot; Pharmacia) for 30 min at 4 mA · cm−2 to a positively charged nylon membrane (BrightStar Plus membrane; Ambion). Nonisotopic detection was carried out using Ambion's BrightStar BiotinDetect kit followed by exposure on X-ray film. ntcA transcript levels were quantified using a model SL-TRFF scanning densitometer (Biomed Instruments Inc., Falkerton, Calif.).
It should be noted that the experiments presented in Fig. 1, 2, and 6 were carried out using RNA extracted using the Biotecx reagent. RNA extracted in this way produced a low signal-to-noise ratio during the RPA procedure. Thus, banding in these three experiments appears somewhat fainter than in experiments in which the RNA was extracted with hot phenol.
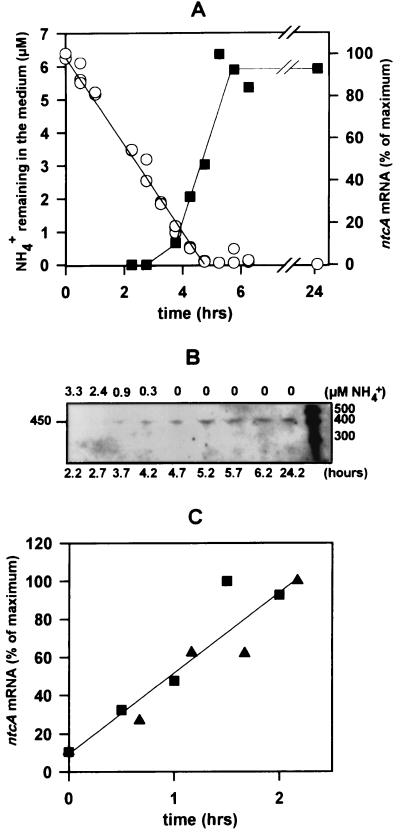
FIG. 1.
ntcA induction and NH4+ uptake. Cells grown on ASWNH4 were transferred to ASW0 plus 6 μM NH4+. (A) NH4+ concentration remaining in the medium (circles) and ntcA transcript levels determined by densitometry from panel B (squares) with time after transfer. (B) ntcA transcript levels determined by RPA analysis of 7 μg of total RNA extracted from cells at different time points after transfer. The scale above the lanes refers to the micromolar concentration of ammonium remaining in the medium at the time the cells were harvested. The scale below the lanes shows the time (in hours) after transfer. The sizes of single-stranded RNA standards are indicated (in bases). (C) Increase in relative ntcA transcript levels with time after they first became apparent. Transcript levels were quantified by densitometry. Squares correspond to the data shown in panels A and B; triangles represent data from an independent experiment. Time zero corresponds to time 3.7 h in panels A and B. The regression line for the combined data from the two experiments is shown.
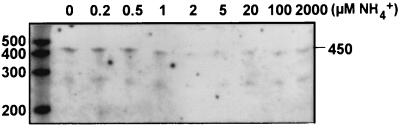
FIG. 2.
Effect of the addition of a range of NH4+ concentrations on ntcA expression. Cells were grown on ASWNO3 and exposed for 5 min to the micromolar concentrations of ammonium indicated above the lanes. ntcA transcript levels were determined by RPA analysis of 13 μg of total cellular RNA. The sizes of single-stranded RNA standards are indicated (in bases).
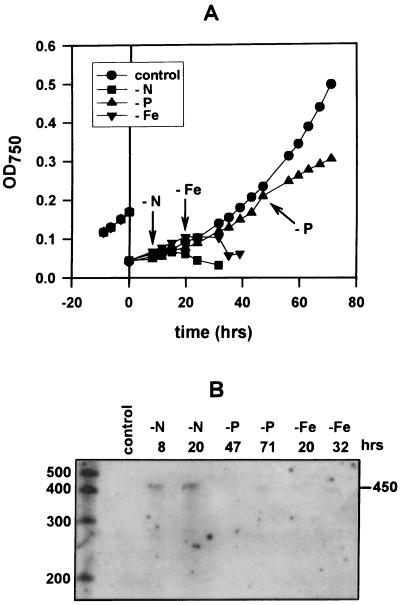
FIG. 6.
The response of Synechococcus sp. strain WH7803 to nutrient deprivation. (A) Growth curves of cultures transferred from ASWNH4 to medium lacking in nitrogen (squares), phosphorus (triangles), or iron (inverted triangles) or to full growth medium (circles). Zero hours indicates the time of transfer from nutrient-replete to nutrient-depleted medium. Growth prior to 0 h is that of the mother culture used for transfers. Arrows indicate the onset of chlorosis and limited growth for each nutrient-deprived culture. (B) ntcA transcript levels of the cells in panel A, as determined by RPA analysis of 7 μg of total RNA extracted from cells subjected to no (control), N, P, or Fe deprivation. Times after transfer to nutrient-deprived medium are indicated above the lanes. The first sample in each pair coincided with the onset of limited growth and chlorosis, as marked by the arrows in panel A, and the second sample in each pair was taken 12 to 24 h after growth limitation set in. The sizes of single-stranded RNA standards are indicated, in bases.
NH4+ uptake assays in Synechococcus cultures.
Cells were grown on ASWNH4, washed twice, and resuspended in filter-sterilized ASW0 plus 6 μM NH4Cl from which HEPES was omitted. The pH remained at 8 to 8.2 throughout the uptake experiments. The concentration of NH4+ remaining in the medium was determined with time using the Spectroquant kit for ammonium determination (Merck), which is based on the indophenol blue reaction. Absorbance was measured at 690 nm using a 5-cm cell. The limit of detection was 100 nM with a precision of ±50 nM. ASW0 medium for standard curves was prepared by adjusting the pH to 13 and bubbling with helium to remove traces of ammonium. This medium was then spiked with different ammonium concentrations and assayed.
Field sampling.
Twelve liters of seawater was collected with Niskin bottles from a depth of 5 m at a nutrient-enriched site (the Ardag offshore floating commercial fish farm) at the northern tip of the Gulf of Aqaba (Red Sea) on 11 April 2000 at 8:45 am and again at 9:15 am and transported to the laboratory. This site has nutrient concentrations higher than those in the surrounding oligotrophic waters, but they are similar to those found in many coastal regions (see Results). Subsamples were taken for the determination of Synechococcus cell abundance enumerated by epifluorescence microscopy as outlined by Lindell and Post (33), nitrate and nitrite concentrations were determined according to the work of Parsons et al. (42) with a Quick Chem 8000 autosampler (LACHAT Instruments), and the ammonium concentration was estimated using the orthophthaldialdehyde method (29). The remaining water was split into 3 equal volumes of 3.4 liters and kept at 25°C for 60 min, illuminated with 200 μmol of photons · m−2 · s−1 with the following additions: 100 μM NH4Cl was added to one aliquot, 100 μM l-methionine-d,l-sulfoximine (MSX) was added to a second aliquot, and the third aliquot remained untreated. Each subsample was then filtered onto a 47-mm-diameter, 0.45-μm-pore-size Supor-450 filter (Gelman Sciences) under a vacuum of 25 in of Hg while illuminated. The filter was immersed in storage buffer (20 mM EDTA, 400 mM NaCl, 0.75 M sucrose, 50 mM Tris [pH 9]) according to the method of Gordon and Giovannoni (20), frozen immediately in liquid nitrogen, and stored at −70°C until nucleic acid extraction.
RNA extraction from field samples.
Samples were thawed on ice and incubated with lysozyme (1 mg · ml−1) at 37°C for 15 min. The pH of the sample was brought down to 7.5 with HCl. Sodium dodecyl sulfate (SDS) was added to a final concentration of 1%, and the sample was heated in a microwave to near-boiling. An equal volume of phenol (pH 7.8) preheated to 65°C was added, and the sample was mixed vigorously and incubated at 65°C for 5 min. An equal volume of chloroform-isoamyl alcohol (24:1) was added and mixed vigorously before centrifugation at 1,700 × g for 5 min. The sample was extracted again with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), followed by extraction with an equal volume of chloroform-isoamyl alcohol (24:1). Nucleic acids were precipitated with 0.4 volume of 7.5 M ammonium acetate and 1 volume of isopropanol at −20°C for 1 h and centrifuged at 14,000 × g for 30 min at 4°C. Nucleic acids were resuspended in TE2 (10 mM Tris–0.1 mM EDTA [pH 8]) prior to the removal of DNA using Ambion's DNA-free. The absence of DNA was verified by nested PCR for the maximal number of cycles as described below.
Reverse transcription.
Reverse transcription was carried out with 500 ng of total RNA denatured for 10 min at 70°C in 20-μl reaction mixtures containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 0.5 mM deoxynucleoside triphosphates (dNTPs), 10 μM dithiothreitol (DTT), 40 U of RNasin (Promega), and 2 pmol of primer 4AR [5′ AT GGC (C/T)TC GGC (G/T)AT GGC (C/T)TG (A/G)T 3′] at 42°C with 65 U of SuperScript II (Gibco-BRL) reverse transcriptase. The reverse primer 4AR is located at bp positions 551 to 530 relative to the first nucleotide of the ntcA initiation codon found in Synechococcus sp. strain WH7803.
Nested PCR.
Two microliters of the reverse transcription reaction product was used in the first 20-μl PCR (PCR1) with primers 1AF [5′ AT(A/T/C) TT(C/T) TT(C/T) CC(G/T/C) GGG GA(C/T) CC(G/A/T) GC 3′], which anneals to bp positions 103 to 125 relative to the first nucleotide of the initiation codon, and 4AR. These primers amplify a 449-bp ntcA fragment from all marine Synechococcus and Prochlorococcus strains tested (data not shown). PCR1 mixtures contained 10 mM Tris-HCl (pH 9), 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2, 200 μM dNTPs, 2.4 μM 1AF, 0.8 μM 4AR, and 1 U of Taq polymerase (Promega). PCRs were run on an MJ Research thermocycler for 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and elongation for 2 min at 68°C following an initial 4-min denaturation step at 94°C. Amplification was still in the exponential phase after 30 cycles of PCR1. One microliter of the PCR1 product was used as a template for each of the six or seven second PCRs (PCR2) with primer set G15–16F [a 1:1.3 ratio of primers G15F, 5′ GA(A/G) TC(A/C/G/T) GG(G/T/C) GAA GAG ATC AC(C/T) GT 3′, and G16F, 5′ GA(A/G) TC(A/T) GG(A/T) GAA GA(A/G) AT(A/T) AC(A/T) GT 3′] and primer S50R [5′ G CAG (A/G)TC (A/G)AT (G/C)GT GAT (G/C)CC (G/C)(A/T/C)G 3′]. PCR2 mixtures were 20 μl in volume and contained 10 mM Tris-HCl (pH 9), 50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, 200 μM dNTPs, 0.8 μM G15–16F, 0.8 μM S50R, and 1 U of Taq polymerase (Promega). PCR2 cycling conditions were identical to those for PCR1 except that reaction tubes were removed manually at 2-cycle intervals between 11 and 25 cycles. G15–16F and S50R are located, respectively, at bp positions 178 to 200 and 521 to 500 relative to the first nucleotide of the ntcA initiation codon and produce a fragment of 344 bp. While primer set G15–16F anneals to the ntcA gene from all marine Synechococcus and Prochlorococcus strains tested, primer S50R anneals specifically to Synechococcus strains (data not shown) such that amplification with this primer pair yields an ntcA fragment specifically from marine Synechococcus. Reaction mixtures were overlaid with 2 drops of mineral oil (Sigma). PCR products were quantified densitometrically using one-dimensional (1D) image analysis software (Kodak Digital Science).
RESULTS
Response of ntcA to ecologically relevant ammonium concentrations.
Ammonium concentrations found in the marine environment are generally below 0.5 μM in open ocean environments (39, 51) but can reach >5 μM in particle-associated microhabitats and in coastal waters (17, 32, 51). In order to determine whether ntcA expression is modulated within this range of ammonium concentrations, we monitored ntcA transcript levels as NH4+ was exhausted from the growth medium (Fig. 1). Cells grown on ASWNH4 and transferred to ASW0 plus 6 μM NH4+ took up ammonium at a rate of 0.41 ± 0.04 fmol · cell−1 · h−1 (n = 3) in a linear fashion down to our detection limit of 100 nM NH4+ (Fig. 1A). Enhanced ntcA expression was first apparent when the ammonium concentration dropped below 1 μM (Fig. 1A and B). Transcript levels increased from that point on at a constant rate of approximately 45% per h (Fig. 1C) and reached a maximum within 2 h of induction. ntcA transcript levels remained high for at least 20 h. Figure 2 shows the response of ntcA-expressing cells to the addition of a range of ammonium concentrations. Cultures were grown on ASWNO3 at low densities (typically 8.5 × 106 cells · ml−1) so that in treatments with low ammonium concentrations, no more than 0.3 μM ammonium would be utilized during the 5-min incubations prior to harvesting of the cells (assuming an uptake rate of 0.41 fmol · cell−1 · h−1; see above). Rapid filtration ensured that cells were harvested and transferred to lysis buffer at −70°C within 3 min. The addition of ammonium concentrations greater than 1 μM led to a significant reduction in ntcA transcript levels, whereas no decline in ntcA mRNA was apparent when concentrations below 1 μM were added.
Nature of ammonium inhibition.
To determine whether the negative effect of ammonium on ntcA expression is direct or requires its incorporation into the cell, we made use of MSX and azaserine, inhibitors of GS and GOGAT, respectively. The addition of 100 μM NH4+ to ASWNO3-grown cells led to a drastic decline in ntcA transcript levels after 5 min, as expected (Fig. 3A). However, incubation of cells for 1 h with 100 μM MSX prevented the ammonium-mediated decline in ntcA mRNA. The same results were obtained when the cells were incubated with azaserine (data not shown). These data indicate that ammonium must be incorporated into cell material via the activities of GS and GOGAT for its negative effect on ntcA transcription to occur.
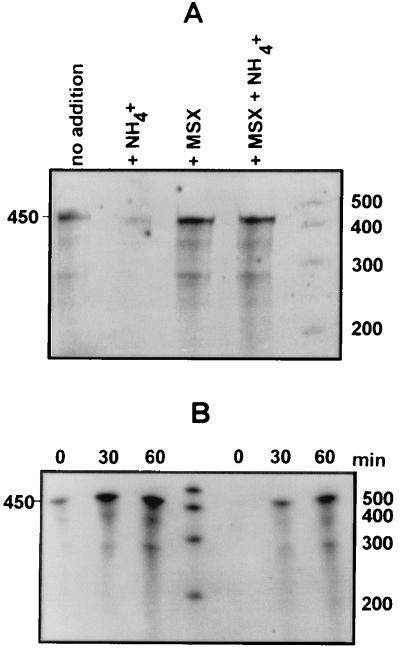
FIG. 3.
(A) Effect of MSX on NH4+-promoted decline in ntcA expression. ntcA transcript levels were determined by RPA analysis of 7 μg of total RNA extracted from cells grown on ASWNO3, either with no addition, incubated for 5 min with 100 μM NH4+, incubated with the GS inhibitor MSX (100 μM) for 60 min, or incubated for 60 min with MSX prior to a 5-min incubation with 100 μM NH4+. (B) Time series of ntcA transcript levels, determined by RPA on 5 μg of RNA, after the addition of 100 μM MSX to cells grown on ASWNO3 (lanes left of the marker) or ASWNH4 (lanes right of the marker). Time after MSX addition is given above the lanes. The sizes of single-stranded RNA standards are indicated (in bases).
The addition of MSX to both ammonium- and nitrate-grown cells led to enhanced ntcA expression within 60 min (Fig. 3B). This suggests that the prevention of ammonium assimilation via GS served to induce a nitrogen starvation response in Synechococcus sp. strain WH7803. Furthermore, it confirms that ntcA expression in nitrate-grown cells is less than maximal (see also Fig. 5C).
FIG. 5.
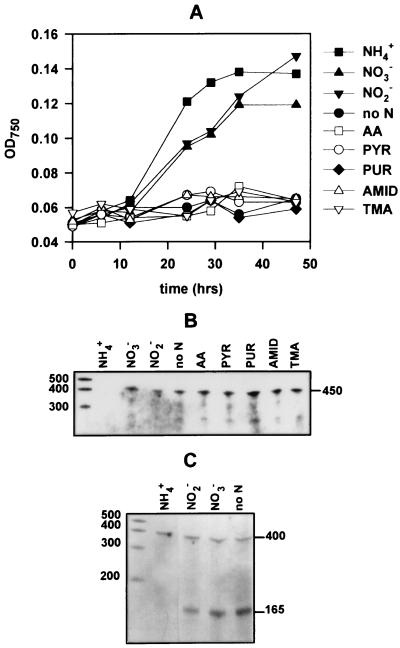
(A) Growth curves of cells transferred from ASWNO3 to ASW0 supplemented with a variety of nitrogenous compounds. (B) ntcA transcript levels as determined by RPA analysis using the internal ntcA probe with 5 μg of RNA extracted from cells 4 h after transfer to the different nitrogen sources. Abbreviations: no N, lacking a nitrogenous source; AA, amino acids; PYR, pyrimidines; PUR, purines; AMID, amides; TMA, trimethylamine. (C) RPA analysis with the upstream ntcA probe of 5 μg of RNA extracted from cells grown on inorganic nitrogen sources or deprived of nitrogen (nitrogen sources are shown above the lanes). The sizes of single-stranded RNA standards are indicated (in bases).
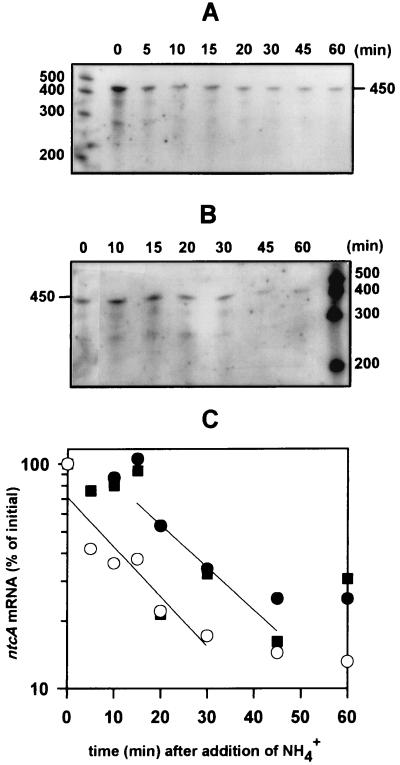
An important aspect of the applicability of a molecular probe is the capability to monitor processes as they occur. Figure 4 shows the time course response of ntcA transcription to NH4+ addition. The addition of 100 μM NH4+ to ASWNO3-grown cells led to an immediate decline in ntcA transcript levels (Fig. 4A). Cells that had been starved of a nitrogen source for 20 h responded to NH4+ addition. However, in contrast to ASWNO3-grown cells, there was a 15-min delay in response (Fig. 4B and C). After this short lag period, ntcA mRNA dropped rapidly to basal levels at a similar rate to ASWNO3-grown cells, with a half-life of approximately 6 min under the growth conditions used here (Fig. 4C).
FIG. 4.
Decline of ntcA transcript levels upon the addition of NH4+ to cells grown on ASWNO3 (A) or deprived of a nitrogen source for 20 h (B). Five micrograms of total RNA, extracted from cells at the indicated times after NH4+ addition, was subjected to RPA analysis. Times (in minutes) after the addition of NH4+ (100 μM) are shown above the lanes. The sizes of single-stranded RNA standards are indicated (in bases). (C) Relative ntcA mRNA levels determined by densitometry and plotted on a semilogarithmic graph. Cells were grown on ASWNO3 (open symbols) or deprived of a nitrogen source for 20 h (solid symbols). Circles correspond to the experiments presented in panels A and B. Squares are from an independent experiment. Regression lines for the linear portion of each treatment are shown.
Response of ntcA to nitrogen sources.
Studies so far have tested the expression of ntcA in the presence of ammonium, in the presence of nitrate, and in the absence of a combined nitrogen source. In order to determine the response of ntcA to other potential nitrogen sources found in the sea (e.g., nitrite and a variety of organic nitrogen compounds [1, 51]), we transferred ASWNO3-grown cells to an ASW0 medium supplemented with a variety of inorganic and organic nitrogen sources. Urea was not tested, as Synechococcus sp. strain WH7803 lacks the genes required for its assimilation (7). Cell growth was supported by the inorganic nitrogen sources only (until the source became exhausted after approximately 24 to 34 h) (Fig. 5A). RPA analysis showed that the transfer of cells for 4 h to all nitrogen sources tested, other than ammonium, led to enhanced ntcA transcript levels irrespective of whether they supported growth or not (Fig. 5B). Use of the upstream ntcA probe in RPA analysis showed that the ammonium-regulated transcript (denoted by the 165-bp protected fragment), while absent from ammonium-grown cells, was most greatly enhanced in cells deprived of a nitrogen source (Fig. 5C). Interestingly, expression of this transcript was considerably lower in cells transferred to nitrite than in cells transferred to nitrate. Note that the expression of the constitutively expressed transcript (denoted by the 400-bp protected fragment) remained at the same low level under all nitrogen conditions.
Response of ntcA to other nutrients.
Phytoplankton may be subject to nitrogen, phosphorus, or iron stress in different regions of the world's oceans (see reference 50 and references therein). To test whether enhanced ntcA expression is a general nutrient stress response, we transferred ASWNH4-grown cells to a medium lacking either nitrogen (N), phosphorus (P), or iron (Fe), or back to the nonlimiting medium. Synechococcus sp. strain WH7803 responded rapidly to N deprivation, showing reduced pigmentation (Fig. 6A) followed by a cessation of growth within 12 h after transfer. Fe-deprived cells also responded by a reduction in cellular pigmentation prior to an arrest in growth, but this took longer to occur than in N-deprived cultures. Cells deprived of P grew longest prior to chlorosis. In contrast to the response of N- and Fe-deprived cells, the growth rate declined to about half of nonlimited growth following chlorosis, but did not cease, prior to phosphorus readdition. Growth recommenced after the addition of the missing nutrient (data not shown), verifying that indeed the limitation observed was due to deprivation of the nutrient in question. Figure 6B shows ntcA transcript levels of cells deprived of the different nutrients at time intervals chosen to correspond to chlorosis and the end of nonlimited growth (the first sample in the pair for each nutrient) and to 12 to 24 h after growth had become limited (the second sample in each pair). Elevated ntcA transcript levels were detected at both time points, but only in cells deprived of nitrogen. Enhanced ntcA expression was also not evident in cells deprived of P or Fe prior to chlorosis and growth limitation (data not shown). Thus, enhanced ntcA expression is a specific nitrogen stress response.
Determining the nitrogen status of field populations.
We have used the findings presented above to design a sampling protocol capable of gauging ntcA transcript levels in field populations of cyanobacteria relative to basal and maximal ntcA expression. While basal expression is indicative of ammonium sufficiency, maximal expression suggests nitrogen deprivation, and intermediate levels of ntcA expression suggest that an alternative nitrogen source, like nitrate or nitrite, is being utilized. The protocol involves comparing ntcA transcript levels in an untreated subsample to those in subsamples incubated with either 100 μM NH4+Cl or 100 μM MSX for 60 min in the light at 25°C. The ammonium addition serves to reduce ntcA expression down to basal levels even in cells deprived of a nitrogen source for 20 h (Fig. 4), whereas the MSX addition serves to enhance ntcA expression to maximal levels even in the presence of sufficient ammonium (Fig. 3C).
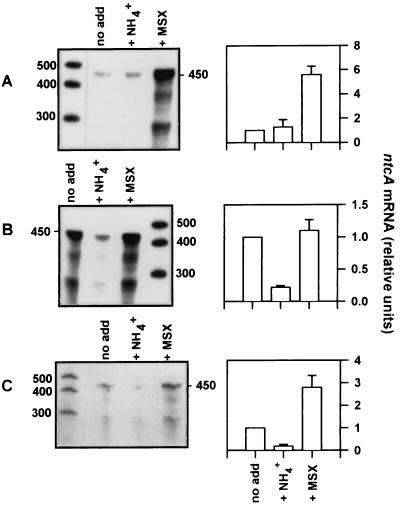
This protocol was tested under controlled laboratory conditions on Synechococcus sp. strain WH7803 grown either on ammonium or nitrate or deprived of a nitrogen source for 20 h (Fig. 7). ntcA expression was assayed by RPA analysis using the internal probe, and representative experiments are shown in the left panels. Results from three independent experiments were quantified by densitometry and are presented graphically in the right panels. Transcript levels from the “+NH4+” and the “+MSX” treatments are presented relative to the “no add” treatment, which was normalized to the value of 1 in all experiments regardless of growth conditions. ntcA transcript levels in ammonium-grown cells were significantly enhanced (five to sevenfold) by the addition of MSX (Fig. 7A), whereas they remained low upon further addition of ammonium. In contrast, ntcA expression declined fivefold upon addition of ammonium to nitrogen-deprived cells but did not change upon addition of MSX (Fig. 7B). However, ntcA expression levels in nitrate-grown cells (Fig. 7C) were both enhanced by the addition of MSX (two to fourfold) and reduced upon ammonium addition (declined approximately fivefold). These results conformed with those expected and suggested that the nitrogen status of field populations of marine Synechococcus can be determined using this protocol.
FIG. 7.
Testing of protocol on cultures of Synechococcus sp. strain WH7803. Cells grown on ASWNH4 (A), deprived of a nitrogen source for 20 h (B), or grown on ASWNO3 (C) were subjected to the protocol described in the text. One-third of each culture was incubated for 90 min either untreated (no add), with 100 μM NH4Cl (+NH4+), or with 100 μM MSX (+MSX). ntcA transcript levels were analyzed using RPAs with the internal ntcA probe on 7 μg of total RNA. The left panels show the autoradiograms of representative experiments. ntcA transcript levels were determined densitometrically from three independent experiments under each growth condition and are shown graphically in the right panels. Transcript levels from the “+NH4+” and the “+MSX” treatments are presented relative to the “no add” treatment, which was normalized to the value of 1 in all experiments (and thus has no error bars) regardless of growth conditions. Error bars, standard errors. The sizes of single-stranded-RNA standards are indicated (in bases).
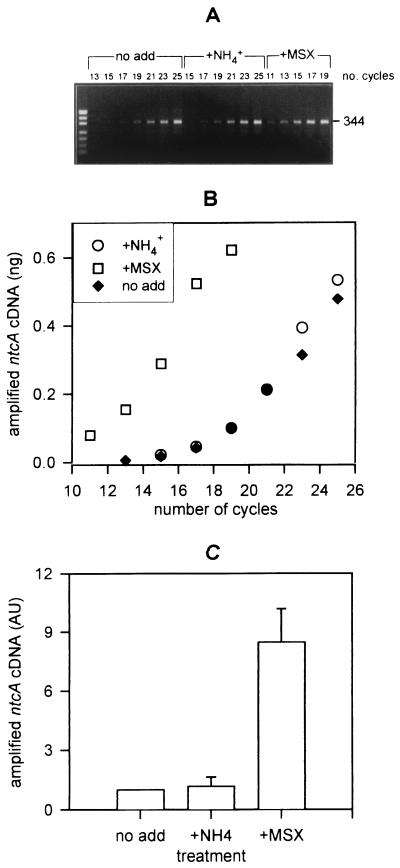
The ntcA expression method was then used to determine the nitrogen status of field populations of Synechococcus (54,000 cells · ml−) found at the nutrient-enriched site in the Gulf of Aqaba during the spring bloom of the year 2000. Nitrogen concentrations in these waters were 0.22 μM nitrite, 0.63 μM nitrate, and approximately 0.6 μM ammonium. A maximal error of ±0.2 μM ammonium resulted from an unexpected matrix effect probably caused by residual fish feed. Relative ntcA transcript levels for the three subsamples (untreated, ammonium addition, and MSX addition) were determined using reverse transcription followed by nested PCR (nested RT-PCR). Figure 8A shows the amounts of amplified Synechococcus-specific ntcA DNA in the different treatments after increasing numbers of cycles of PCR2. The amounts of ntcA DNA were determined densitometrically and used to plot a graph of ntcA DNA as a function of the number of PCR2 cycles (Fig. 8B). These data were then used to determine at which cycles ntcA DNA was apparent, yet still in the exponential phase of amplification. The amounts of ntcA DNA from such cycles were used to plot the bar graph (Fig. 8C) of the relative amount of ntcA DNA determined from two independent nested RT-PCR procedures for each of the two separate field samplings taken at the same site 30 min apart. Figures 8A and B show that an ntcA fragment from the MSX-treated subsample was already visible after 11 cycles of PCR2 and that the amount of ntcA DNA increased exponentially for a further 6 cycles. An ntcA fragment from the ammonium-treated subsample was apparent from cycle 15 and increased exponentially through to cycle 25. The untreated subsample produced a similar amount of ntcA DNA to that from the ammonium-treated subsample after the same number of cycles. Therefore, the results from cycles 15 and 17 in this field sample were used to plot Fig. 8C along with the results in analogous cycles of the replicate analysis and in the second field sample. Figure 8C shows that ntcA transcript levels in the untreated subsample were not significantly different from those in the ammonium-added subsample, but were eightfold lower than those in the MSX-added subsample. These data indicate that ntcA in the untreated subsamples was basally expressed and infer that the Synechococcus populations close to the fish farm were utilizing ammonium.
FIG. 8.
ntcA expression in field populations of Synechococcus from a nutrient-enriched site (0.22 μM NO2−, 0.63 μM NO3−, and approximately 0.6 μM NH4+) in the Gulf of Aqaba, Red Sea, determined from nested RT-PCR of untreated subsamples (no add) or subsamples following a 1-h incubation with ammonium (+NH4+) or MSX (+MSX). (A) Amounts of amplified ntcA cDNA from the three treatments after increasing numbers of cycles in the nested PCR (PCR2) run on a 3% agarose gel. Both treatment type and number of PCR2 cycles are shown above the lanes. The pUC19-MspI DNA standard (501 + 489, 404, 331, 242, 190, 147, 111 + 110 bp) is shown in the far-left lane. (B) The amount of amplified ntcA cDNA from panel A was quantified densitometrically and is presented graphically as a function of the number of PCR2 cycles. Both panels A and B show results from a representative experiment. (C) Cycles at which ntcA DNA was above the detection limit in all treatments, yet still in the exponential phase of amplification, were used to determine the average amount of ntcA cDNA amplified from two independent nested RT-PCR procedures from each of the two seawater samples. Amplified ntcA cDNA from the +NH4+ and +MSX treatments are presented relative to that from the untreated subsamples (no add), which was normalized to the value of 1 in each nested RT-PCR procedure. Error bars, standard errors.
DISCUSSION
Nitrogen metabolism and ntcA expression.
Synechococcus sp. strain WH7803 grew only on inorganic nitrogen sources despite evidence for its capacity to assimilate organic compounds such as leucine, methionine, uracil, and adenine into proteins and nucleic acids (30, 8, 48; personal observations). In addition, cells supplied with leucine as the sole nitrogen source retained their pigmentation for several days (personal observations). Therefore, organic nitrogen sources may play a role in cell maintenance and survival during periods of severe inorganic nitrogen limitation in this cyanobacterium. Even though the growth rate of Synechococcus sp. strain WH7803 did not differ when it was grown on the various inorganic nitrogen sources, ntcA transcript levels were substantially different. Cells grown on ammonium expressed ntcA at basal levels (i.e., expressed only the constitutive transcript), whereas ntcA expression was greater in nitrate-grown than in nitrite-grown cells (Fig. 5C). Furthermore, ntcA expression was greatest in cells deprived of a nitrogen source (34) (Fig. 5C). While we have no experimental information to help explain these findings, one feasible explanation may be that ntcA transcription is controlled by feedback inhibition that depends on the rate of supply of nitrogen, if nitrogen derived from nitrite is incorporated more rapidly than that derived from nitrate. Nitrite assimilation may occur faster than nitrate assimilation because only one reduction step is required and there may be more than one nitrite transport system (13). Furthermore, nitrite assimilation is energetically cheaper than nitrate assimilation (23).
The ammonium-promoted repression of nitrate and nitrite uptake as well as nirA transcription requires the assimilation of ammonium into carbon skeletons via the GS/GOGAT pathway (13, 52). Our findings show that the negative effect of ammonium on ntcA expression also requires its prior assimilation via the activities of GS and GOGAT (Fig. 3). The direct effector molecule acting downstream of ammonium assimilation has remained elusive. It is feasible that the ammonium-promoted down-regulation of transcription acts separately on ntcA and the nirA operon. Alternatively, the effector molecule may directly affect ntcA transcription, whereas the effect on ntcA-regulated genes like nirA may be due to the absence of NtcA.
The prerequisite of ammonium assimilation for the decline in ntcA transcript levels could explain the observed delay in the response of nitrogen-deprived cells following ammonium addition. Ammonium uptake or assimilation may take time to commence following nitrogen deprivation. It is also possible that internal ammonium supplies or the direct effector molecule leading to ammonium-promoted down-regulation takes time to accumulate subsequent to starvation.
The inhibition of GS activity by the addition of MSX led to enhanced ntcA expression (Fig. 3B). This mimics the response for nitrogen deprivation despite the presence of suitable nitrogen sources, as shown by halted growth after the addition of MSX (36). This suggests that the major, if not the sole, pathway for the assimilation of ammonium is via the activity of GS in Synechococcus sp. strain WH7803. The addition of MSX to Anacystis nidulans growing on nitrate or ammonium also invokes a nitrogen starvation response. In these cells MSX served to stimulate nitrate and nitrite uptake and reduction rates (11, 12, 27, 28). Furthermore the addition of MSX to Synechococcus sp. strain PCC 7942 led to enhanced expression of the nirA operon (52).
The induction of ntcA gene expression in Synechococcus sp. strain WH7803 occurred when NH4+ dropped below ca. 1 μM. In the same organism Scanlan et al. (49) found that the expression of a phosphate-binding protein (PstS) was induced when PO4− dropped below ca. 50 nM. The 20-fold-higher concentration required for induction of expression of ntcA relative to pstS is interesting when one considers that the cellular requirement of nitrogen and phosphorus for unlimited growth is generally considered to be at a ratio of 16:1 (46). It therefore appears that the adaptive responses to low concentrations of nitrogen and phosphorus are induced at a similar level relative to the biochemical requirements of the cells. However, it should be noted that Cuhel and Waterbury (8) reported much higher cellular phosphorus levels for this organism than expected from the Redfield ratio.
ntcA expression as an indicator of the nitrogen status of Synechococcus spp.
For ntcA expression to be a suitable indicator of the nitrogen status of field populations of unicellular cyanobacteria, a number of requirements must be met. Previous reports have shown that the ntcA gene is present in a single copy in a wide range of cyanobacteria (14, 34). In a separate study we have shown that in the absence of ammonium, ntcA expression was enhanced over a range of photon fluxes as well as over the entire diel cycle, but that maximal differences between the constitutive and regulated ntcA transcripts occurred in the morning hours (D. Lindell and A. Post, unpublished data). Therefore, for highest resolution, field sampling should be carried out in the morning. In addition, ntcA expression was similarly enhanced in cells grown at 18 and 25°C (Lindell and Post, unpublished), which is within the range of temperatures usually found in tropical and subtropical waters. Here we have shown that ntcA expression responded specifically to nitrogen availability and that ntcA gene expression responded rapidly to ammonium addition and nitrogen deprivation.
In the discussion below, we assume that ntcA expression and NtcA-regulated nitrogen acquisition in field populations of marine Synechococcus are similar to those for the model organism Synechococcus sp. strain WH7803. This is a fair assumption considering that these functions are very similar in the evolutionarily more distant marine and freshwater model Synechococcus strains, WH7803 and PCC 7942 (compare references 37 and 55 to reference 34 and Moyal et al., submitted). Moreover, the nucleotide sequences of the ntcA gene are more similar among marine Synechococcus strains than between Synechococcus sp. strains WH7803 and PCC 7942 (36). We further assume that urea utilization is indeed regulated by NtcA (7).
The rapid response of ntcA expression to both ammonium and MSX enables us to compare actual ntcA transcript levels to basal and maximal levels. Basal levels of ntcA expression can be used to infer ammonium sufficiency. The use of inhibitors of the assimilation of ammonium showed that ntcA transcript levels declined only after ammonium was incorporated into cellular organic matter. Thus, basal ntcA expression is indicative not only of the presence of ammonium but of its utilization by the cell. Furthermore, when Synechococcus sp. strain WH7803 displays basal ntcA expression, it is incapable of utilizing nitrate or nitrite (34, 35; Moyal et al., submitted). Therefore, basal ntcA expression is indicative of exclusive ammonium utilization. In contrast to basal expression, maximal ntcA expression is indicative of the absence of a nitrogen source capable of supporting growth. Intermediate transcript levels will demonstrate that while ammonium supplies are inadequate to prevent enhanced ntcA expression, a nitrogen source is being obtained. However, the identity of these nitrogen sources cannot be unequivocally ascertained using the present protocol. The discovery of genes or proteins that are specifically induced when alternative nitrogen sources are available (e.g., urea, nitrate, or nitrite) would enable their use in conjuction with ntcA expression analysis to further determine nitrogen source utilization by Synechococcus.
We have exploited the fact that maximal ntcA expression can be chemically induced by the addition of MSX and that expression is reduced to basal levels after the addition of ammonium to develop a protocol capable of differentiating between basal, maximal, and intermediate levels of ntcA expression. It should be noted that this assay relies on the ability of cyanobacteria to transport MSX into the cell, and it will not be informative for those cells incapable of such transport. The coupling of this protocol to nested RT-PCR expression analysis has enabled us to determine the nitrogen status of Synechococcus field populations. RT-PCR rather than RPAs was used for determining ntcA expression levels in field populations for two major reasons. First, the low abundance of Synechococcus in the sea (relative to yields achieved in laboratory cultures) requires the more-sensitive RT-PCR method for the detection of ntcA mRNA. Second, the use of ntcA primers with differing taxonomic specificity in RT-PCR enables the assessment of the nitrogen status of cyanobacteria at various taxonomic levels. Such flexibility can not be afforded by RPAs, which are sensitive to even minor mismatches between probe and template sequences. Furthermore, this RT-PCR assay could easily be adapted for use in a quantitative PCR machine.
At the northern tip of the Gulf of Aqaba, at a site with measurable levels of nitrate, nitrite, and ammonium (yet below 1 μM in all cases), Synechococcus populations were expressing ntcA basally (Fig. 8). Therefore these Synechococcus populations were not nitrogen deprived. Moreover, from these results we can infer that they were exclusively utilizing regenerated nitrogen in the form of ammonium despite relatively high concentrations of both nitrate and nitrite. The nitrogen concentrations at this site are similar to those found in coastal and estuarine waters (32, 51) as well as in association with micro- and macroaggregates and Trichodesmium blooms in oligotrophic seas (17, 19). It is therefore likely that ntcA would be basally expressed by Synechococcus populations inhabiting such environments. Thus ammonium would be the major, if not the sole, nitrogen source utilized by Synechococcus under such conditions. Inherent to this situation is the fact that the action of the transcriptional activator, NtcA, would not be required for the cell's nitrogen demands to be met in these waters.
At this stage we do not know whether ntcA expression is induced, and the action of NtcA required, for nitrogen acquisition by Synechococcus spp. in the vast regions of the open ocean where ammonium is found at nanomolar concentrations. Reports of nitrogen preference in the field have shown that at ambient ammonium concentrations of 1 μM or higher, planktonic assemblages preferentially utilized ammonium irrespective of the concentrations of other nitrogen sources, whereas at ammonium concentrations below 0.5 to 1 μM, nitrogen sources were used according to availability (16, 38). Other reports, however, show a clear preference for ammonium even in oligotrophic waters, where ammonium concentrations are well below 1 μM (26, 40). In these waters, ammonium is rapidly regenerated by bacteria and zooplankton such that ammonium utilization is often balanced by its regeneration (2, 17, 25). Therefore, the flux of ammonium rather than ambient concentrations is likely to be a better indication of ammonium availability. Whether ntcA gene expression responds to ambient ammonium concentrations or to the flux of ammonium is unclear at this stage. Regardless of this current uncertainty, by allowing Synechococcus to report on its own nitrogen status through ntcA expression levels, we will be able to assess whether field populations of marine Synechococcus in waters with differing nitrogen regimes are deprived of nitrogen, are utilizing a suitable nitrogen source, or are thriving solely on ammonium.
ACKNOWLEDGMENTS
This work was supported by grants from the European Union Mast III program PROMOLEC (MAS3-CT97-0128), the Ecological Foundation of the Keren Kayemet Le'Israel (190/1/702/6), and the Moshe Shilo Center for Marine Biogeochemistry, Minerva Stiftung-Gesellschaft fuer die Forschung, Munich, Germany.
We thank Nick Fuller and Efrat David for the OPA ammonium measurements during organic N experiments and field sampling respectively, Aliza Moyal for help with ammonium uptake experiments, and Tanya Korpal and Boaz Lazar for the nitrate and nitrite field measurements. We thank the Ardag fish farm for permission to sample from their farm and Dror Angel and Noa Eden for logistic help with the field sampling. We also thank Gitai Yahel, Nir Peleg, and two anonymous reviewers for constructive comments on an earlier version of the manuscript.
REFERENCES
- 1.Antia N J, Harrison P J, Oliveira L. The role of dissolved organic nitrogen in phytoplankton nutrition, cell biology and ecology. Phycologia. 1991;30:1–89. [Google Scholar]
- 2.Bronk D A, Ward B B. Gross and net nitrogen uptake and DON release in the euphotic zone of Monterey Bay, California. Limnol Oceanogr. 1999;44:573–585. [Google Scholar]
- 3.Burkhill P H, Leakey R J G, Owens N J P, Mantoura R F C. Synechococcus and its importance to the microbial food web of the northwestern Indian Ocean. Deep-Sea Res. 1993;40:773–782. [Google Scholar]
- 4.Campbell L, Carpenter E J. Characterization of phycoerythrin-containing Synechococcus spp. by immunofluorescence. J Plank Res. 1987;9:1167–1181. [Google Scholar]
- 5.Campbell L, Liu H, Nolla H A, Vaulot D. Annual variability of phytoplankton and bacteria in the subtropical North Pacific ocean at station ALOHA during the 1991–1994 ENSO event. Deep-Sea Res. 1997;44:167–192. [Google Scholar]
- 6.Chisholm S W. Phytoplankton size. In: Falkowski P G, Woodhead A D, editors. Primary productivity and biogeochemical cycles in the sea. New York, N.Y: Plenum Press; 1992. pp. 213–237. [Google Scholar]
- 7.Collier J C, Brahamsha B, Palenik B. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology. 1999;145:447–459. doi: 10.1099/13500872-145-2-447. [DOI] [PubMed] [Google Scholar]
- 8.Cuhel R L, Waterbury J B. Biochemical composition and short-term nutrient incorporation patterns in a unicellular marine cyanobacterium, Synechococcus sp. Limnol Oceanogr. 1984;29:370–374. [Google Scholar]
- 9.Dauchez S, Legendre L, Fortier L, Levasseur M. Nitrate uptake by size-fractionated phytoplankton on the Scotian Shelf (Northwest Atlantic): spatial and temporal variability. J Plank Res. 1996;18:577–595. [Google Scholar]
- 10.Fanning K A. Nutrient provinces in the sea: concentration ratios, reaction rate ratios, and ideal covariation. J Geophys Res. 1992;97:5693–5712. [Google Scholar]
- 11.Flores E, Guerrero M G, Losada M. Short-term ammonium inhibition of nitrate utilization by Anacystis nidulans and other cyanobacteria. Arch Microbiol. 1980;128:137–144. [Google Scholar]
- 12.Flores E, Herrero A, Guerrero M G. Nitrite uptake and its regulation in the cyanobacterium Anacystis nidulans. Biochim Biophys Acta. 1987;896:103–108. [Google Scholar]
- 13.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- 14.Frias J E, Merida A, Herrero A, Martin-Nieto J, Flores E. General distribution of the nitrogen control gene ntcA in cyanobacteria. J Bacteriol. 1993;175:5710–5713. doi: 10.1128/jb.175.17.5710-5713.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnas M, Crosbie N D. In situ growth dynamics of the photosynthetic prokaryotic picoplankters Synechococcus and Prochlorococcus. Bull Inst Oceanogr. 1999;19:387–417. [Google Scholar]
- 16.Glibert P M, Biggs D C, McCarthy J J. Utilization of ammonium and nitrate during austral summer in the Scotia Sea. Deep-Sea Res. 1982;29:837–850. [Google Scholar]
- 17.Glibert P M, Dennett M R, Caron D A. Nitrogen uptake and NH4+ regeneration by pelagic microplankton and marine snow from the North Atlantic. J Mar Res. 1988;46:837–852. [Google Scholar]
- 18.Glibert P M, Ray R T. Different patterns of growth and nitogen uptake in two clones of marine Synechococcus spp. Mar Biol (Berlin) 1990;107:273–280. [Google Scholar]
- 19.Glibert P M, O'Neil J M. Dissolved organic nitrogen release and amino acid oxidase activity by Trichodesmium spp. Bull Inst Oceanogr. 1999;19:265–272. [Google Scholar]
- 20.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorl M, Sauer J, Baier T, Forchhammer K. Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology. 1998;144:2449–2458. doi: 10.1099/00221287-144-9-2449. [DOI] [PubMed] [Google Scholar]
- 22.Grossman A R, Schaefer M R, Chiang G G, Collier J L. The responses of cyanobacteria to environmental conditions: light and nutrients. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 641–675. [Google Scholar]
- 23.Guerrero M G, Lara C. Assimilation of inorganic nitrogen. In: Fay P, Van Baalen C, editors. The cyanobacteria. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1987. pp. 163–186. [Google Scholar]
- 24.Harrison W G, Wood L J E. Inorganic nitrogen uptake by marine picoplankton—evidence for size partitioning. Limnol Oceanogr. 1988;33:468–475. [Google Scholar]
- 25.Harrison W G. Regeneration of nutrients. In: Falkowski P G, Woodhead A D, editors. Primary productivity and biogeochemical cycles in the sea. New York, N.Y: Plenum Press; 1992. pp. 385–407. [Google Scholar]
- 26.Harrison W G, Harris L R, Irwin B D. The kinetics of nitrogen utilization in the oceanic mixed layer: nitrate and ammonium interactions at nanomolar concentrations. Limnol Oceanogr. 1996;41:16–32. [Google Scholar]
- 27.Herrero A, Flores E, Guerrero M G. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J Bacteriol. 1981;145:175–180. doi: 10.1128/jb.145.1.175-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrero A, Guerrero M G. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J Gen Microbiol. 1986;132:2463–2468. [Google Scholar]
- 29.Holmes R M, Aminot A, Kerouel R, Hooker B A, Peterson B J. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- 30.Kramer J G. The effect of irradiance and specific inhibitors on protein and nucleic acid synthesis in the marine cyanobacterium Synechococcus sp. WH 7803. Arch Microbiol. 1990;154:280–285. [Google Scholar]
- 31.LaRoche J, McKay R M L, Boyd P. Immunological and molecular probes to detect phytoplankton responses to environmental stress in nature. Hydrobiologia. 1999;401:177–198. [Google Scholar]
- 32.L'Helguen S, Madec C, Le Corre P. Nitrogen uptake in permanently well-mixed temperate coastal waters. Est Coast Shelf Sci. 1996;42:803–818. [Google Scholar]
- 33.Lindell D, Post A F. Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol Oceanogr. 1995;40:1130–1141. [Google Scholar]
- 34.Lindell D, Padan E, Post A F. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J Bacteriol. 1998;180:1878–1886. doi: 10.1128/jb.180.7.1878-1886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindell D, Padan E, Post A F. Effect of ammonium on nitrate/nitrite uptake and ntcA expression in Synechococcus sp. strain WH 7803. Bull Inst Oceanogr. 1999;19:273–278. [Google Scholar]
- 36.Lindell D. Assessing the nitrogen status of marine prokaryotic phytoplankton using molecular methods. Ph.D. thesis. Jerusalem, Israel: Hebrew University of Jerusalem; 2000. [Google Scholar]
- 37.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarthy J J, Taylor W R, Taft J L. Nitrogenous nutrition of the plankton in the Chesapeake Bay. 1. Nutrient availability and phytoplankton preference. Limnol Oceanogr. 1977;22:996–1011. [Google Scholar]
- 39.McCarthy J J, Garside C, Nevins J L. Nitrogen dynamics during the Arabian Sea northeast monsoon. Deep-Sea Res II. 1999;46:1623–1664. [Google Scholar]
- 40.Metzler P M, Glibert P M, Gaeta S A, Ludlam J M. New and regenerated production in the South Atlantic off Brazil. Deep-Sea Res. 1997;44:363–384. [Google Scholar]
- 41.Palenik B, Wood A M. Molecular markers of phytoplankton physiological status and their application at the level of individual cells. In: Cooksey K E, editor. Molecular approaches to the study of the ocean. London, United Kingdom: Chapman and Hall; 1997. pp. 187–205. [Google Scholar]
- 42.Parsons T R, Maita Y, Lalli C M. A manual of chemical and biological methods for seawater analysis. Oxford, United Kingdom: Pergamon Press; 1984. [Google Scholar]
- 43.Partensky F, Hess W R, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomar M L C A, Caruso G, Maugeri T L, Scarfo R, Zaccone R. Distribution of Synechococcus spp. determined by immunofluorescent assay. J Appl Microbiol. 1998;84:493–500. [Google Scholar]
- 45.Probyn T A, Waldron H N, James A G. Size-fractionated measurements of nitrogen uptake in aged upwelled waters—implications for pelagic food webs. Limnol Oceanogr. 1990;35:202–210. [Google Scholar]
- 46.Redfield A C. The biological control of chemical factors in the environment. Am Sci. 1958;46:205–222. [PubMed] [Google Scholar]
- 47.Sauer J, Margit G, Forchhammer K. Nitrogen starvation in Synechococcus PCC 7942: involvement of glutamine synthetase and NtcA in phycobiliprotein degradation and survival. Arch Microbiol. 1999;172:247–255. doi: 10.1007/s002030050767. [DOI] [PubMed] [Google Scholar]
- 48.Scanlan D J, Mann N H, Carr N G. The response of the picoplanktonic marine cyanobacterium Synechococcus species WH7803 to phosphate starvation involves a protein homologous to the periplasmic phosphate-binding protein of Escherichia coli. Mol Microbiol. 1993;101:181–191. doi: 10.1111/j.1365-2958.1993.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 49.Scanlan D J, Silman N J, Donald K M, Wilson W H, Carr N G, Joint I, Mann N H. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol. 1997;63:2411–2420. doi: 10.1128/aem.63.6.2411-2420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scanlan D J, Wilson W H. Application of molecular techniques to addressing the role of P as a key effector in marine ecosystems. Hydrobiologia. 1999;401:149–175. [Google Scholar]
- 51.Sharp J H. The distribution of inorganic nitrogen and dissolved and particulate organic nitrogen in the sea. In: Carpenter E J, Capone D G, editors. Nitrogen in the marine environment. New York, N.Y: Academic Press; 1983. pp. 1–35. [Google Scholar]
- 52.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 53.Tremblay J E, Legendre L, Klein B, Therriault J C. Size-differential uptake of nitrogen and carbon in a marginal sea (Gulf of Lawrence, Canada): significance of diel periodicity and urea uptake. Deep-Sea Res II. 2000;47:489–518. [Google Scholar]
- 54.Tyrrell T, Law C S. Low nitrate:phosphate ratios in the global ocean. Nature. 1997;387:793–796. [Google Scholar]
- 55.Vega-Palas M A, Madueno F, Herrero A, Flores E. Identification and cloning of a regulatory gene for nitrogen assimilation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1990;172:643–647. doi: 10.1128/jb.172.2.643-647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyman M, Gregory R P F, Carr N G. Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science. 1985;230:818–820. doi: 10.1126/science.230.4727.818. [DOI] [PubMed] [Google Scholar]