Abstract
Regulatory non‐coding RNAs (ncRNAs) including small non‐coding RNAs (sRNAs), long non‐coding RNAs (lncRNAs), and circular RNAs (circRNAs) have gained considerable attention in the last few years. This is mainly due to their condition‐ and tissue‐specific expression and their various modes of action, which suggests them as promising biomarkers and therapeutic targets. One important mechanism of ncRNAs to regulate gene expression is through translation of short open reading frames (sORFs). These sORFs can be located in lncRNAs, in non‐translated regions of mRNAs where upstream ORFs (uORFs) represent the majority, or in circRNAs. Regulation of their translation can function as a quick way to adapt protein production to changing cellular or environmental cues, and can either depend solely on the initiation and elongation of translation, or on the roles of the produced functional peptides. Due to the experimental challenges to pinpoint translation events and to detect the produced peptides, translational regulation through regulatory RNAs is not well studied yet. In the case of circRNAs, they have only recently started to be recognized as regulatory molecules instead of mere artifacts of RNA biosynthesis. Of the many roles described for regulatory ncRNAs, we will focus here on their regulation during inflammation and in immunity.
Keywords: immunology, inflammation, non‐coding RNA, regulation, translation
Abbreviations
- circRNA
circular RNA
- cpuORF
conserved peptide upstream open reading frame
- DC
dendritic cell
- eIF
eukaryotic initiation factor
- IFN
interferon
- IL
interleukin
- IRES
internal ribosome entry site
- lncRNA
long non‐coding RNA
- LPS
lipopolysaccharide
- m6A
N6‐methyladenosine
- MHC
major histocompatibility complex
- miRNA
micro RNA
- mRNA
messenger RNA
- NAT
natural antisense transcript
- ncRNA
non‐coding RNA
- NK
natural killer
- NMD
nonsense‐mediated decay
- ORF
open reading frame
- piRNA
Piwi‐interacting RNA
- RBP
ribosome binding protein
- RP
ribosome profiling
- RPF
ribosome‐protected fragments
- rRNA
ribosomal RNA
- sdRNA
sno‐derived RNA
- siRNA
small interfering RNA
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- SNV
single nucleotide variants
- sORF
short open reading frame
- sRNA
small non‐coding RNA
- SuRE
stem‐loop structured RNA element
- Th2
T helper 2
- tRF
tRNA‐derived fragment
- tRNA
transfer RNA
- uORF
upstream open reading frame
- uTIS
upstream translation initiation site
- UTR
untranslated region
1. INTRODUCTION
Transcription in eukaryotes comprises a major non‐protein‐coding component. The human GENCODE genome assembly version 38 annotates a total number of 60,649 genes. These are comprised of 19,955 (32.9%) protein‐coding genes, 17,944 (29.6%) long non‐coding RNA (lncRNA) genes, 7567 (12.5%) small non‐coding RNA (sRNA) genes, 14,773 (24.4%) pseudogenes, and 645 (1.1%) immunoglobulin/T‐cell receptor gene segments. The total number of annotated transcripts is 237,012, of which 86,757 (36.6%) are protein‐coding transcripts and 48,752 (20.6%) are lncRNA loci transcripts 1 (Figure 1). The annotation of the human genome is constantly refined and expanded. While genomic regions that are annotated constitute only a small proportion of the total genome, it was found that around 75% of the genome is actually transcribed, and 62% of the genome is transcribed resulting in transcripts that are 5′‐capped and 3′‐polyadenylated. 2
FIGURE 1.
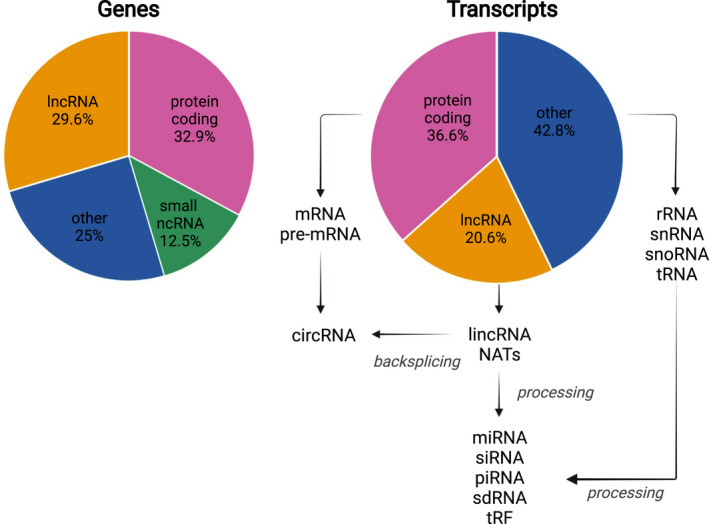
Genes encoding ncRNAs contribute substantially to the total annotated genome and transcriptome, respectively. Transcripts such as pre‐mRNAs or lncRNAs can further be processed by backsplicing, resulting in the generation of circRNAs. The short non‐coding RNAs miRNA, siRNA, piRNA, sno‐derived RNA (sdRNA), and tRNA‐derived fragment (tRF) are cleaved from longer transcripts
ncRNA genes encode housekeeping RNAs including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and regulatory ncRNAs. Regulatory ncRNAs comprise sRNAs that are processed from longer precursors and lncRNAs that are typically longer than 200 nucleotides and function without major processing. Of the annotated long protein‐coding and non‐coding RNAs, around 6% are probably precursors of sRNAs, as part of their sequences were found to overlap with the sequences of sRNAs that comprise micro RNAs (miRNAs), small interfering RNAs (siRNAs), and Piwi‐interacting RNAs (piRNAs). These sRNAs are mostly found in introns, yet exons from lncRNAs often seem to harbor snoRNAs. 2 The separation of ncRNAs into housekeeping and regulatory RNAs is being challenged as more and more noncanonical functions of either group are discovered. 3 , 4 , 5 In addition, fragments derived from ncRNAs such as tRNAs and snoRNAs are conserved between species and involved in the regulation of gene expression, which adds another layer of complexity to the modulation of cellular processes. 3 , 5 , 6 , 7
lncRNAs can be transcribed from both strands from intergenic regions (long intergenic ncRNAs), from introns of protein‐coding genes (long intronic ncRNAs), or from the antisense strand of protein‐coding genes in the case of natural antisense transcripts (NATs). 8 The term lncRNA was initially used to only describe RNAs that were transcribed by RNA polymerase II, are 5′‐capped and 3′‐polyadenylated, and lack known coding capacity or a long open reading frame (ORF). 9 By now, the term ncRNAs is mostly used to describe RNAs without known protein‐coding capacity, meaning that they do not contain ORFs starting with an AUG start codon that code for more than 100 amino acids. 10 The restriction that unknown ORFs needed to code for more than 100 amino acids was required in automated annotation procedures, which else would have resulted in the annotation of many spurious protein‐coding genes. This definition will evidently lead to the under‐annotation of the coding capacity of shorter ORFs or of ORFs with an alternative start codon.
Regulatory ncRNAs are often expressed in response to external cues, during differentiation or in specific stages of development. Their differential expression can modulate transcription or translation of other genes or interfere directly with signaling pathways. lncRNAs can function as target mimics, induce alternative splicing, regulate transcription, modulate chromatin function for example through RNA‐dependent DNA methylation, regulate nuclear bodies, alter the stability of mRNAs or act as scaffolds, and more functions and modes of action continue to get discovered. 4 , 9 , 11 , 12 , 13 , 14 , 15 , 16 , 17 Interestingly, lncRNAs play important roles in inflammatory pathways and in immune reactions in general. 18 , 19 An important factor in determining the function of ncRNAs is their subcellular localization. While ncRNAs localized to the nucleus can modulate epigenetic modifications and transcription, alter splicing or modify RNA, cytoplasmic ncRNAs are rather involved in the regulation of RNA stability and translation. 15 , 20 Further spatial compartmentalization can contribute to the regulation of these processes. 21 Quantifying the expression of protein‐coding and lncRNA genes, it was found that transcripts of protein‐coding genes were present in more copies per cell than transcripts of lncRNAs, respectively. As 29% of all expressed polyadenylated ncRNAs were identified in only one of the studied cell lines, the lower copy numbers are probably the result of a specific expression pattern of ncRNAs. 2 Corresponding with this, it was found that gene expression is complementary between lncRNAs that are often expressed in a tissue‐specific manner or not at all, and protein‐coding RNAs that are generally ubiquitously expressed. 22 lncRNAs therefore constitute an important class of regulatory molecules, and their tissue‐ and condition‐specific expression points to specialized functions, which makes them interesting biomarker candidates or treatment targets. 4 Their identification and the characterization of their different modes of action have therefore gained a lot of attention. Despite their annotation as non‐coding, regulatory RNAs have the propensity to be associated with ribosomes at sORFs located in their sequences, and translational control is a not widely known way to foster their function. Therefore, we will focus here on the translation of short open reading frames (sORFs) in lncRNAs and of upstream ORFs (uORFs) in non‐coding regions of messenger RNAs (mRNAs), on the role of sORF and uORF translation in inflammation and immunity, and on the biogenesis and function of circular RNAs (circRNAs).
2. sORFS, THE HIDDEN CODING POTENTIAL IN lncRNAs
As lncRNAs were often annotated as non‐coding if they do not contain AUG start codon ORFs coding for more than 100 amino acids, some lncRNAs can still harbor peptide‐coding ORFs. Sequencing ribosome‐associated RNAs in murine macrophages revealed that about 10% were annotated as non‐coding, and that more than half of the ncRNAs that showed similar features as translated protein‐coding RNAs used the noncanonical start codons CUG, UUG, or GUG. 23 In addition, a few examples of sORFs have already been known to be translated into peptides or to have a regulatory function in the 5′ untranslated region (UTR). 10 , 24 With the establishment of ribosome profiling (RP), it became possible to directly determine the RNA regions that actually get translated, and to analyze different aspects of translation. 25 , 26 , 27 , 28 In RP, the RNA stretches that are located inside elongating ribosomes are protected from RNase treatment in the cell extract, and the ribosome‐protected fragments (RPF) are afterward isolated and sequenced. 29 , 30 Recently, the establishment of single‐cell RP has expanded the toolkit for research of translation even further, making it possible to distinguish between cell state‐specific translation events. 31 These technical advances led to the identification of actively translated sORFs in lncRNAs including NATs and circRNAs of which some started with non‐AUG start codons, as well as of translation of ORFs in 3′UTRs and 5′UTRs of mRNAs. 14 , 29 , 32 , 33 , 34 , 35 For some of the sORF peptides, the in vivo accumulation could be confirmed with the use of mass spectrometry, 14 and with systematic CRISPR‐based screening that can precisely disrupt protein‐coding regions, the functional roles of sORF‐encoded peptides could be validated on a larger scale. 33 While more and more lncRNAs with coding potential and translated sORFs are discovered and shown to be differentially translated under specific conditions, more examples of sORF peptides that are involved in various cellular functions are found. 36 , 37 , 38 , 39 , 40 , 41 , 42 Their mode of action includes the regulation of larger proteins or protein complexes, while some are secreted and act as signal peptides 37 , 39 , 43 , 44 , 45 (Figure 2). However, in some cases, the mere association of ribosomes with sORFs in lncRNAs or mRNAs is sufficient to have a biological impact, for example as explained below in more detail through the regulation of translation initiation efficiency at the start codon of the main ORF (mORF) or of transcript stability through ribosome stalling (Figure 3) (Box 1).
FIGURE 2.
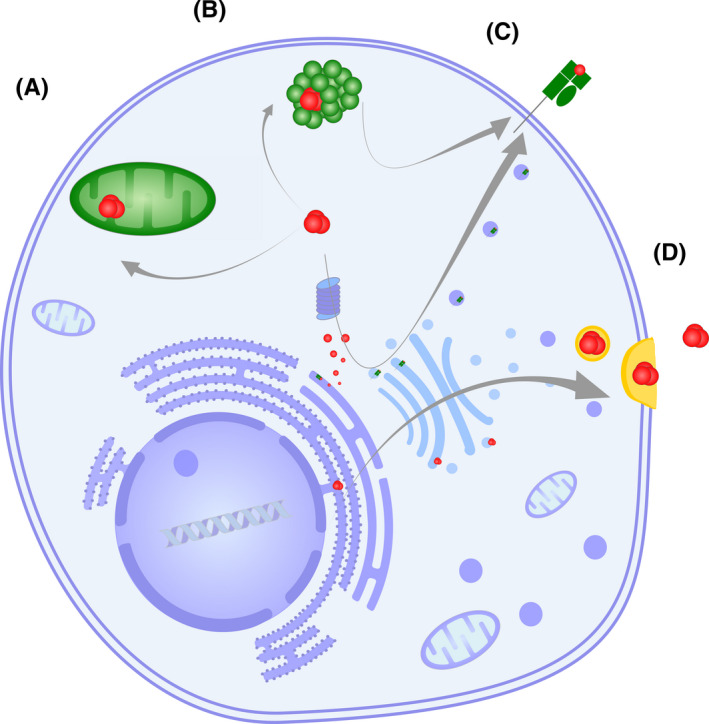
sORF‐encoded peptides (in red) can regulate cellular behavior in various ways. (A) sORF‐encoded peptides can localize to different cellular compartments including mitochondria; an example is Mm47, which plays a role in inflammasome activation. (B) sORF‐encoded peptides can bind to protein complexes and affect their function; an example is miPEP155, which is associated with a chaperone and thereby influences antigen presentation. (C) The presentation of peptides on MHC receptors contributes to cellular immunosurveillance. (D) sORF‐encoded peptides can be secreted and influence neighboring cells
FIGURE 3.
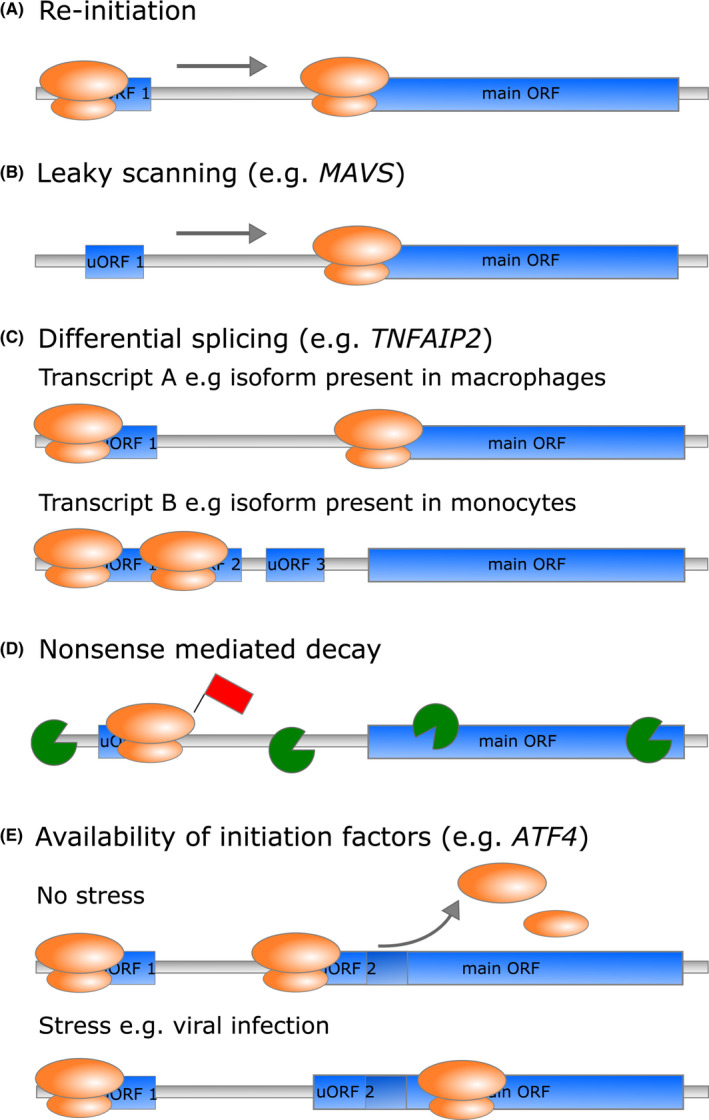
uORFs can regulate translation of the mORF in various ways. The presence of uORFs in the 5′UTR mainly represses translation of the mORF. (A) When translation initiation occurs on the start codon of the uORF, translation needs to be re‐initiated at the mORF after translation of the uORF is completed. (B) In leaky scanning, the pre‐initiation complex does not initiate translation at the start codon of the uORF, but keeps scanning until it initiates translation at the start codon of the mORF. (C) Translational repression of the mORF can be regulated through the expression of transcript isoforms containing different numbers of uORFs. (D) Ribosomes that are stalled on uORFs during elongation or termination of translation can lead to the induction of NMD and degradation of the mRNA. (E) The repressive properties of uORFs can change through internal or external cues such as stress induced through viral infection
BOX 1. Major milestone discoveries.
Gene expression regulation through changes in translation is a relatively rare, but important mode of action of ncRNAs
Translation of some sORFs and uORFs results in the production of functional peptides of which some were shown to accumulate in vivo
Translational regulation plays an important role in fine‐tuning gene expression during inflammation or in immunity
CircRNAs are regulatory ncRNAs rather than mere biosynthesis artifacts
The special properties of circRNAs make them promising candidates for RNA‐based therapeutic strategies
2.1. The role of sORF translation in inflammation and immunity
While a comprehensive picture of the regulatory role of sORF peptides in the immune system is still missing, some interesting cases have already emerged (Figure 2). In murine macrophages, a sORF peptide encoded in the lncRNA Aw112010 was identified, which plays a proinflammatory role in the mucosa. Through mutations in the RNA sequence that changed the RNA folding, but not the peptide sequence, it could be shown that the actual translated peptide promotes the defense response against Salmonella enterica and increases the susceptibility to induce colitis. 23 Another murine peptide that is connected to inflammatory responses of macrophages is Mitochondrial micropeptide‐47 (Mm47), which is translated from the lncRNA 1819958I24Rik and localized to mitochondria. Lower levels of Mm47 were associated with lower levels of interleukin (IL)‐1β and decreased NOD‐, LRR‐, and pyrin domain‐containing protein 3 (NLRP3) inflammasome activation, which is closely connected to mitochondrial function 46 (Figure 2A). Several other sORF peptides including mitochondrial elongation factor 1 microprotein (MIEF1‐MP), micropeptide regulator of β‐oxidation (MOXI), and mitoregulin have been identified in different contexts to play a role in mitochondrial processes such as mitochondrial translation, fatty acid oxidation, and mitochondrial supercomplex formation. 38 , 47 , 48 Whether these peptides are involved in immunologic processes is not known yet. However, metabolic reprogramming through translational changes is a central aspect of immune cell function. 49 , 50 , 51 The MIR155HG transcript and the miRNA that is processed from this transcript have known functions in inflammatory diseases and cancer. 52 , 53 In addition, the 17 amino acid peptide termed miPEP155 that is encoded by the transcript MIR155HG was recently described to be involved in antigen presentation in human and murine dendritic cells (DCs) in an anti‐inflammatory context. Through the interaction of miPEP155 with Heat shock cognate protein 70 (HSC70) that is required for antigen presentation, the HSC70‐Heat shock protein 90 (HSP90) machinery is disrupted and antigen presentation on major histocompatibility complex (MHC) class II molecules is modulated 54 (Figure 2B). The discovery of the translated peptide adds another layer of regulatory function to this multifunctional lncRNA (Box 1).
RP experiments in murine DCs identified several new translated sORF and uORF peptides on ncRNA, which mirrored the known early, intermediate, and late response to stimulation with lipopolysaccharide (LPS). 55 The detected translated peptides included Solute carrier 35a4 (SLC35a4), MIEF1‐MP and a 68 amino acid peptide, which was later shown to be involved in RNA decapping and called Nobody. 55 , 56 Interestingly, another sORF peptide is encoded in the same transcript as Nobody. However, this peptide does not get translated in DCs, but has been associated with T helper 2 (Th2) differentiation and aggravation of allergic airway inflammation in a murine model system. 57 As RP studies on immune cells such as T or B cells have so far not aimed at the detection of unconventional translation events, 58 , 59 , 60 , 61 the question of the involvement of sORF peptides in adaptive immunity still remains largely unanswered (Box 2).
BOX 2. Future research perspectives.
Due to the environment‐ and tissue‐specific expression pattern of regulatory ncRNAs, their number is expected to increase in future studies
The characterization of the functional roles and location of sORF and cpuORF peptides promises to be an interesting avenue of research
Owing also to the recent inclusion of RNA vaccines into the clinical practice, further studies on circRNA immunogenicity and translation are highly indicated
3. uORFs AS CELLULAR TOOLS FOR THE FINE ADJUSTMENT OF GENE EXPRESSION
Computational sequence analyses have revealed that uORFs are present in the 5`UTRs of about 50% of human transcripts. 62 This finding has been validated with results from RP experiments with which upstream translation initiation sites (uTISs) were identified in more than 50% of human transcripts. Interestingly, the majority of translation at uTISs initiated at a near‐cognate start codons differing in one base from the AUG start codon. 63 As uORFs can function as response elements that rapidly adapt protein production to altered environmental conditions through translational regulation, their properties and mode of action have attracted quite some research interest (Figure 3) (Box 1).
3.1. uORF mode of action
uORFs regulate the expression of the downstream mORF by different mechanisms. In most cases, the presence of uORFs inhibits translation of the mORF under homeostatic conditions, as the presence of upstream start codons in general decreases the efficiency of the rate‐limiting initiation step at the mORF 62 , 64 (Figure 3A). In translation initiation according to the scanning model, the small 40S ribosomal subunit is in a pre‐initiation complex loaded with the Eukaryotic initiation factor 2 (eIF2)‐GTP‐initiator methionyl tRNA (tRNAi Met) ternary complex, and it scans the 5′UTR of the mRNA in the 5′ to 3′ direction until it encounters a start codon. Here, the scanning is arrested and the GTP in the ternary complex is hydrolyzed, which leads to the release of eIF2‐GDP and other initiation factors. This allows the binding of the large 60S ribosomal subunit and the formation of the 80S ribosome that can now start peptide elongation. 65 For translation of the mORF in the presence of uORFs, the ribosome either has to re‐initiate translation at the start codon of the mORF or to bypass the uORF via leaky scanning of the transcript 65 , 66 (Figure 3A,B). The efficiency of these processes depends on different factors such as the sequence context of the uORF and mORF start codons, that is, the presence of a favorable Kozak consensus sequence, 67 the presence of cognate or near‐cognate start codons, uORF termination efficiency, intercistronic distance, availability of initiation factors, mRNA secondary structure and sequence, and in some cases the peptide sequence encoded by the uORF. 16 , 64 , 68 Adding to the complexity of uORF‐mediated translation control, certain arrangements or structural features of these factors can lead to enhanced instead of repressed translation. One way of dynamically modulating mORF translation efficiency is therefore to vary these features through the creation of transcript variants, for example, by changing the number of uORFs through alternative splicing or alternative promoter usage 16 (Figure 3C). Accordingly, many of these properties are under pressure of negative selection and are often conserved between different species. 68 , 69
Another mode of action of translational repression by uORFs is through stalling of ribosomes. Ribosome stalling of the elongating or terminating ribosome is associated with mRNA secondary structures, interactions with trans‐acting factors or the nascent peptide, and usage of nonsense codons. Ribosomes stalled in translating uORFs both inhibit progression of the ribosomes to the mORF and induce nonsense‐mediated decay (NMD). NMD is a cellular surveillance pathway that degrades translationally abnormal RNA, which includes mRNAs that prematurely terminate translation. As the uORF stop codon can be recognized as a premature termination signal when ribosomes stall at the uORF stop codon, the mRNA can be subjected to degradation leading to reduced translation efficiency of the mORF 16 , 66 (Figure 3D).
While the function of most uORFs is sequence‐independent, meaning that the amino acid sequence encoded by the uORFs is not important for their regulatory function, a small fraction of uORFs relies on the encoded peptide sequence. For the sequence‐specific uORFs, the encoded peptide sequences have been conserved in evolution and have therefore been named conserved peptide uORFs (cpuORFs). 17 In humans, around 1.7% of uORFs were initially found to be cpuORFs with likely conservation at the amino acid level. 68 By using a novel pipeline to detect cpuORFs conserved in evolutionary divergent animal genomes, additional cpuORFs encoded in the human genome could be identified of which several were confirmed to repress mORF translation in a peptide sequence‐dependent manner. 70 Interestingly, for those uORF peptides for which we found evidence for in vivo accumulation by mass spectrometry in Arabidopsis, 70.8% had a homologous uORF peptide sequence in other species. 14 As mass spectrometry has a bias for the identification of more abundant peptides or proteins and usually follows experimental procedures that will de‐enrich small proteins, the identification of uORF peptides is a sign of their pronounced accumulation. This might indicate that sequence‐specific cpuORF peptides have longer half‐lives than sequence‐independent uORF peptides, which might be linked to their functional roles (Box 1). In general, the function of the cpuORF peptides is not necessarily connected to the functions of the proteins encoded by the respective mORFs. 33 , 71 , 72 The functional roles of cpuORF peptides therefore remain largely unknown and may hold interesting surprises (Box 2).
3.2. uORF translational control in inflammation and immune response pathways
During the differentiation of monocytes into macrophages, leukocyte‐specific transcript 1 (LST1) protein levels are upregulated. Protein upregulation is at least partly controlled through differential splicing, as the amount of LST1 transcripts containing exon 1C is increased, while the transcript variant including exon 1B, which contains a long repressive uORF, displayed only a moderate increase. 73 Similarly, the 5´UTR of the TNF alpha‐induced protein 2 (TNFAIP2) mRNA contains multiple uORFs that inhibit translation of the mORF in monocytes. uORF‐mediated repression of translation is relieved in mature macrophages, leading to increased TNFAIP2 protein expression 74 (Figure 3C). Further examples of proteins whose expression or function is translationally modulated through the presence of uORFs in the context of inflammation and immunity include signaling lymphocytic activation molecule family member 1 (SLAMF1) (CD150), CD36, suppressor of cytokine signaling 1 (SOCS1), 75 , 76 , 77 and mitochondrial antiviral signaling (MAVS). MAVS is involved in the retinoic acid‐inducible gene I (RIG‐I)/melanoma differentiation‐associated protein 5 (MDA5)‐dependent sensing of viral nucleic acids in the cytoplasm. Under homeostatic conditions, uORFs in the MAVS mRNA initiate leaky scanning of the full‐length MAVS start codon of ORF1. By working together to inhibit translation of the MAVS mORF, three uORFs control production of either the full‐length MAVS from ORF1 or the truncated version from downstream ORF2. They thereby maintain immune homeostasis through prevention of MAVS spontaneous aggregation and stimulation of Interferon (IFN)‐β production 78 (Figure 3B).
Responses to cellular stress such as viral infections lead to phosphorylation of Eukaryotic initiation factor 2 (eIF2α), which causes inhibition of global translation. However, increased eIF2α phosphorylation can also lead to preferential translation of transcripts involved in the stress response by mechanisms involving uORFs. 79 One well‐characterized example is the transcription factor activating transcription factor 4 (ATF4) whose translation is regulated by two uORFs of which one overlaps with the mORF. Under homeostatic conditions, translation of the ATF4 transcript is inhibited, as the first and the second uORF get translated through the quick acquisition of the necessary initiation factors, which enable re‐initiation at the start codon of the second uORF. This inhibits initiation of translation at the start codon of the overlapping mORF. Upon cell stress, ribosome re‐initiation at the start codon of the second uORF is less efficient due to reduced availability of functional ternary complex caused by the phosphorylation of eIF2α. The reduced re‐initiation at the start codon of the second uORF increases the probability of leaky scanning, and thereby promotes re‐initiation at the start codon of the mORF of ATF4 instead 80 , 81 (Figure 3E).
In a systematic study investigating mutations in the UTR that affect uORF start or stop codons or uORF peptide sequences, a set of single nucleotide variants (SNVs) in 296 genes that were associated with human diseases was identified. These include the previously characterized UTR variants of IFN regulatory factor 6 (IRF6) and Neurofibromin (NF1). 82 In addition, expression of Alpha‐1‐antitrypsin (SERPINA1) that is involved in inflammatory conditions is associated with the transcription of isoforms that differ in their 5′UTRs including the presence of uORFs. 83 The properties of uORFs to regulate and fine‐tune the expression of specific genes therefore play an important role in health and disease (Box 1).
4. circRNAs ACT ON SEVERAL LEVELS IN THE CONTROL OF GENE TRANSCRIPTION AND TRANSLATION
circRNAs are circular single‐stranded RNA molecules created by covalently joining the 5′ and 3′ free ends of a linear transcript 84 (Figure 4A). The existence of circular forms of RNA has been long acknowledged however, they were considered to be biosynthesis artifacts or splicing by‐products with no or little biological effect. Owing also to the advent of next generation sequencing techniques, circRNAs have been finally recognized as a common feature in eukaryotes, revealing a distinct biogenesis and diverse cellular functions. 85 , 86 , 87
FIGURE 4.
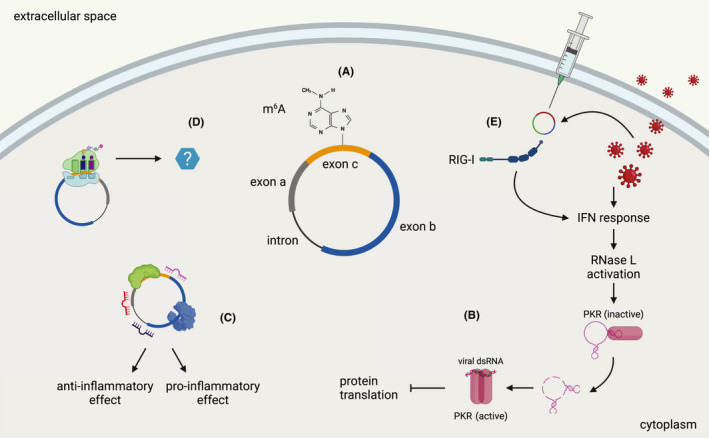
circRNA functions in inflammation and immunity. (A) The schematic drawing depicts the simplified structure of an eukaryotic circRNA, which may contain different combinations of exons and introns. In humans, RNA modifications such as m6A and the presence of introns are key to avoid the development of an immune response against circular RNA structures. (B) Endogenous circRNAs can participate in the response to viral infections. Upon viral infection and interferon (IFN) response, RNase L is activated and degrades a circRNA that is bound to PKR and keeps it in an inactive state. Activated PKR can recognize viral dsRNA and dimerize, which ultimately results in inhibition of overall protein translation. (C) circRNAs with classical functions of protein and miRNA sponging can regulate inflammatory responses, both with pro‐ and anti‐inflammatory properties. (D) Accumulating evidence points to the production of circRNA‐derived peptides, yet their functions in inflammation and immunity have to be investigated and clarified. (E) circRNAs that are produced by viruses or engineered and exogenously administered and do not contain m6A modifications or introns can elicit an immune response by activating pattern recognition receptors such as RIG‐I
4.1. circRNA biogenesis and regulation
circRNAs can be generated from the circularization of several precursors, including pre‐mRNA, housekeeping RNA, and regulatory RNA. 88 The mechanism of circularization is usually referred to as backsplicing, with two main models being proposed: the direct backsplicing and the lariat models, which differ in the order of occurrence of the splicing event. 88 Several signals and factors, both in cis and in trans, are involved in circRNA biogenesis, 89 and the presence of long flanking introns and repetitive elements was shown to strongly favor RNA circularization. 90 , 91 , 92 Both main backsplicing models could explain how expression of the linear counterpart is regulated, and there is evidence of both co‐regulation and competition between linear and circRNA biogenesis. 93 , 94
Several RNA‐binding proteins (RBPs) have been demonstrated to regulate the biogenesis of circRNAs. 93 , 95 , 96 Of note, the splice factor encoded by the Nudix hydrolase 21 (NUDT21) gene is one of the earliest factors intervening in the 3′ end maturation and polyadenylation of pre‐mRNAs. Its reduction in hepatocellular carcinoma was associated with overall lower circRNA levels, 97 suggesting an important role in circRNA biogenesis in close association with canonical pre‐mRNA processing. Other RBPs have also been associated with specific circRNAs, which are linked with a role in various disease (Table 1).
TABLE 1.
Regulation of circRNA biogenesis by specific RBPs and their role in disease
| circRNA | Gene | RBP | RBP function | Disease and effect | Ref |
|---|---|---|---|---|---|
| circ0006916 | HOMER1 | TNRC6A | Binding to flanked intron regions, promotion of circ0006916 production | Use of TNRC6A as a possible strategy to increase circ0006916 levels in lung cancer cells, where the expression of this circRNA is downregulated. Regulation of cell cycle and proliferation. | 142 |
| SCD‐circRNA 2 | SCD | RBM3 | Binding to flanking regions, promotion of SCD‐circRNA 2 biogenesis | In hepatocellular carcinoma, upregulation of both SCD‐circRNA 2 and RBM3. Regulation of cell cycle. | 143 |
| circAMOTL1L | AMOTL1 | RBM25 | Promotion of circAMOTL1L biogenesis | In prostatic cancer, downregulation of both circAMOTL1L and RBM25 due to lower p53 activity. Regulation of EMT. | 144 |
| TTN‐derived circRNAs | TTN | RBM20 | Promotion of biogenesis of a specific circRNAs subset from the titin gene I‐band. Alternative backsplicing. | In dilated cardiomyopathy, RBM20 activity is lost, and a complete set of circRNAs together with it. | 145 |
As circRNAs are longer‐lived molecules compared to other RNA types due to their lack of free ends enabling exonucleolytic digestion, the control of their degradation is essential. Some mechanisms have been proposed, such as structure recognition and decay mediated by RBPs, 98 Argonaute 2 (Ago2)‐directed splicing by miRNA targeting, 99 , 100 and endolytic cleavage upon recognition of m6A modifications or imperfect duplex regions. 99 It could also be shown that circRNAs can form imperfect RNA duplexes under homeostatic conditions, which inhibit double‐stranded RNA‐activated protein kinase (PKR) that is related to innate immunity. Upon stimulation or viral infection, the circRNAs get degraded by RNase L, which is required for the activation of PKR 101 (Figure 4B). In addition to circRNA stability, transport of circRNAs is also critical for their function. This process is still poorly studied, but circRNA size might be decisive on how nuclear export is regulated. 102
4.2. circRNA functions
circRNAs regulate several cellular functions, including cell cycle progression, 103 , 104 ribosomal RNA transcription, 105 and maturation. 106 circRNAs act as competing endogenous RNAs as they retain multiple miRNA binding sites in their sequence and were suggested to act as sponges to limit the ability of miRNAs to reach their actual targets (Figure 4C). For example, circular Cerebellar degeneration‐related protein 1 antisense (CircCDR1as), also known as cIRS‐7, possesses more than 70 conserved binding sites for miR‐7 and other miRNAs, thereby regulating a large variety of pathways including immune cell functions. 107 , 108 , 109 , 110 , 111 Similarly, circular Sex‐determining region on the Y chromosome (Sry) has been shown to sponge miR‐138. 112 In an analogous fashion, circRNAs can bind to and sponge RBPs in competition with the linear transcripts for access to the RBPs. 113 , 114
circRNAs also regulate pre‐mRNA splicing. It is suggested that linear and circRNA are often processed co‐transcriptionally and therefore, one form may regulate how the other is expressed. 93 This probably holds true for many, but not all circRNAs. 94 Some circRNAs have also been shown to actively regulate splicing. 115 , 116 In glioblastoma, circular SWI/SNF related, matrix associated, actin‐dependent regulator of chromatin, subfamily A, member 5 (circSMARCA5) regulates alternative splicing of Serine and arginine‐rich splicing factor 1 (SRSF3), Polypyrimidine tract binding protein 1 (PTBP1), and Vascular endothelial growth factor A (VEGFA) by tethering the splicing factor SRSF1 and ultimately inhibiting cell migration and angiogenic activity (Box 1). 117 , 118
Although generally classified as non‐coding RNAs, there is also evidence for the association of circRNAs with the translation machinery, 119 and for the expression of peptides that are encoded by sORFs on circRNAs 120 , 121 , 122 , 123 , 124 (Figure 4D). In a study of translational events in human hearts, 40 translated circRNAs were identified using RP, and for 9 of them, the expression of the encoded peptides was confirmed by mass spectrometry. 32 Even though hundreds of peptides are predicted to be translated from sORFs on circRNAs, only a few of them have been characterized so far. 120 , 121 , 125 , 126 This includes the expression of circular Muscleblind (circMbl)‐derived peptides in Drosophila and of a circZNF609 peptide in human and murine cells. 120 , 121 At least two circRNA‐derived peptides were shown to regulate the Wnt pathway in human cells with oncogenic effects, 126 , 127 and a novel 198‐aa peptide from a Collagen type VI alpha 3 chain (COL6A3)‐derived circRNA was found to regulate the aggressiveness of colorectal cancer cells by regulating the stability of the COL6A3 mRNA. 128 In contrast, for a selection of 1000 highly expressed circRNAs no evidence of translation could be identified in a study when multiple publicly available datasets of different experimental conditions were analyzed. 129 With covalently closed ends and devoid of the characteristics of a classically translated mRNA, circRNAs are depending on cap‐independent mechanisms for translation. Those involve internal ribosome entry sites (IRES) or IRES‐like structures containing N6‐methyladenosine (m6A) modifications, which have been shown to be able to recruit the pre‐initiation complex and start translation. 124 , 130 Most importantly, two specific features of the IRES have recently been recognized to play a crucial role in driving circRNA translation, which are 18S rRNA complementarity and a structured RNA element. This peculiar RNA secondary structure is a stem‐loop structured RNA element (SuRE) that is located 40–60 nucleotides from the first nucleotide of the IRES. 131 Future studies are needed to elucidate the regulation and role of circRNA translation, as well as the function of their encoded peptides in cellular pathways beyond tumor biology (Box 2).
4.3. circRNAs play a role in inflammation and immune regulation
In the last few years, there was an almost exponential growth in the number of papers reporting novel circRNAs and their functions. In many of these papers, a role of circRNAs in a variety of disease models and in the regulation of inflammation and immune responses was suggested, especially for circRNAs with miRNA‐sponging functions, but the validity and biomedical relevance of these findings will need to be further substantiated. Notwithstanding, there is increasing evidence on the important regulatory role of circRNAs in CD4+ and CD8+ lymphocytes, macrophages, and natural killer (NK) cells, which has an impact on tumor and antiviral immunity and on autoimmune disorders. 132
Besides circRNAs that act through miRNA sponging, at least two circRNAs with a protein‐binding function were identified that play a role in inflammation (Figure 4C). Circular antisense non‐coding RNA in the INK4 locus (circANRIL) shows pro‐apoptotic and atheroprotective functions as it binds to a protein complex that assembles with pre‐ribosomes and precursor rRNA, which affects rRNA maturation and ribosome biogenesis in human peripheral blood mononuclear cells and vascular smooth muscle cells. 106 circ_0075932 directly binds to and increases the expression of the RBP Pumilio 2 (PUM2), which positively regulates AuroraA kinase and thus activates the nuclear factor‐kappaB (NF‐κB) pathway. Exosomes derived from circ_0075932‐overexpressing human adipocytes induce inflammation and apoptosis in dermal keratinocytes. 133 The abovementioned mechanism of circRNAs to regulate the activity of PKR through the formation of intramolecular imperfect duplex regions and the activity of RNase L links the action of circRNAs to the recognition of foreign nucleic acids and the direct antiviral activity of the innate immune system 101 (Figure 4B). The mechanism of PKR activation through the degradation of PKR‐bound circRNA by RNase L is also proposed to be dysregulated in some autoimmune disorders. 101
4.4. Implications for circRNAs as a biotechnological tool
The discovery of circRNAs and their peculiar characteristics and diverse functions quickly led to the exploration of their potential as therapeutic agents. However, circRNAs are not an exclusive feature of eukaryotes, but they can also be encoded by viruses, and circular RNA structures can be recognized by the immune system as part of the antiviral response. 134 , 135 Therefore, the question of how circRNA are recognized by the immune system was addressed in several studies. 136 , 137 , 138 , 139 Engineered circRNAs have been shown to elicit a response from pattern recognition receptors, in particular RIG‐I, while endogenous circRNAs appear to be protected from immune activation (Figure 4E). Whether a specific circRNA elicits an immune response depends on the type of biogenesis, the specific sequence, and how the RNA is delivered into cells. The presence of human introns 136 and of the m6A modification 138 seem to play an important role in suppressing innate immunity (Figure 4A) (Box 1).
Overall, more rigorous research on circRNA immunogenicity and translation is strongly recommended to understand how to exploit their properties at best. Indeed, an unprecedented advance in RNA vaccine development has been elicited very recently by the SARS‐CoV‐2 pandemic. Besides the obvious benefits for the management and control of the pandemic itself, these advances also laid the foundation for the development of other vaccines and therapeutics. In summary, circRNAs are promising for the development of novel RNA‐based treatment strategies, with some approaches already under investigation, 140 , 141 and a raising number of pre‐print papers and biotech companies’ outlets mentioning this technology (Box 2).
5. CONCLUSIONS
Regulatory RNAs including lncRNAs and circRNAs and translation events in ncRNAs or in non‐coding parts of mRNAs provide an interesting mechanism to modulate gene expression and cellular functions. The translation of sORFs located in lncRNAs or circRNAs can result in stable and functional peptides with specialized roles. While the translation of uORFs mainly serves to regulate the efficiency of translation initiation at the corresponding mORFs, some uORF peptides are actually conserved at amino acid level and were shown to accumulate in vivo, which indicates that they might have functional roles that are not necessarily associated with the role of the protein encoded by the mORF. As shown here, regulatory ncRNAs do play important roles in the context of inflammation and immunity. Also the various modes of action of circRNAs point to important roles of these exciting molecules in immune responses. Through their circular nature, their ability to form intramolecular duplex regions, and to interact with DNA, other RNAs, and RBPs, they represent a promising and versatile class of novel RNA‐based therapeutic agents. Further studies with the aim to unravel the mode of action of regulatory ncRNAs, and to characterize the roles of sORF and uORF peptides that accumulate in vivo under specific conditions are expected to provide novel information on a class of molecules that has so far mainly been hiding in plain sight.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGEMENTS
JK was funded through “Stiftung vormals Bündner Heilstätte Arosa.” EDB is funded by AO Foundation and AO Trauma. Figures 1 and 4 were created with Biorender. Open Access Funding provided by Universitat Zurich.
Della Bella E, Koch J, Baerenfaller K. Translation and emerging functions of non‐coding RNAs in inflammation and immunity. Allergy. 2022;77:2025–2037. doi: 10.1111/all.15234
Elena Della Bella and Jana Koch contributed equally to this study.
REFERENCES
- 1. Frankish A, Diekhans M, Ferreira A‐M, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766‐D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wajahat M, Bracken CP, Orang A. Emerging functions for snoRNAs and snoRNA‐Derived Fragments. Int J Mol Sci. 2021;22:10193. doi: 10.3390/IJMS221910193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Statello L, Guo C‐J, Chen L‐L, Huarte M. Gene regulation by long non‐coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22(2):96‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Su Z, Wilson B, Kumar P, Dutta A. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu Rev Genet. 2020;54:47‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alves CS, Nogueira FTS. Plant small RNA world growing bigger: tRNA‐derived fragments, longstanding players in regulatory processes. Front Mol Biosci. 2021;8:638911. doi: 10.3389/FMOLB.2021.638911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z, Stanton BA. Transfer RNA‐derived fragments, the underappreciated regulatory small RNAs in microbial pathogenesis. Front Microbiol. 2021;12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma L, Bajic VB, Zhang Z. On the classification of long non‐coding RNAs. RNA Biol. 2013;10:924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11‐42. [DOI] [PubMed] [Google Scholar]
- 10. Basrai MA, Hieter P, Boeke JD. Small open reading frames: Beautiful needles in the haystack. Genome Res. 1997;7:768‐771. [DOI] [PubMed] [Google Scholar]
- 11. Wirth S, Crespi M. Non‐protein coding RNAs, a diverse class of gene regulators, and their action in plants. RNA Biology. 2009;6(2):161‐164. doi: 10.4161/rna.6.2.8048 [DOI] [PubMed] [Google Scholar]
- 12. Chekanova JA. Long non‐coding RNAs and their functions in plants. Curr Opin Plant Biol. 2015;27:207‐216. [DOI] [PubMed] [Google Scholar]
- 13. Nakano RT, Piślewska‐Bednarek M, Yamada K, et al. PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana . Plant J. 2017;89:204‐220. [DOI] [PubMed] [Google Scholar]
- 14. Bazin J, Baerenfaller K, Gosaid SJ, Gregory BD, Crespi M, Bailey‐Serres J. Global analysis of ribosome‐associated non‐coding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA. 2017;114(46):E10018‐E10027. doi: 10.1073/pnas.1708433114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cech TR, Steitz JA. The noncoding RNA revolution‐trashing old rules to forge new ones. Cell. 2014;157:77‐94. [DOI] [PubMed] [Google Scholar]
- 16. Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2014;5:765‐778. [DOI] [PubMed] [Google Scholar]
- 17. von Arnim AG, Jia Q, Vaughn JN. Regulation of plant translation by upstream open reading frames. Plant Sci. 2014;214:1‐12. [DOI] [PubMed] [Google Scholar]
- 18. Bocchetti M, Scrima M, Melisi F, et al. LncRNAs and immunity: coding the immune system with noncoding oligonucleotides. Int J Mol Sci. 2021;22:1‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walther K, Schulte LN. The role of lncRNAs in innate immunity and inflammation. RNA Biol. 2021;18:587‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong C, Maquat LE. lncRNAs transactivate STAU1‐mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhat P, Honson D, Guttman M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat Rev Mol Cell Biol. 2021;22:653‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melé M, Ferreira PG, Reverter F, et al. The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson R, Kroehling L, Khitun A, et al. The translation of non‐canonical open reading frames controls mucosal immunity. Nature. 2018;564:434‐438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Odermatt A, Becker S, Khanna VK, et al. Sarcolipin regulates the activity of SERCA1, the fast‐twitch skeletal muscle sarcoplasmic reticulum Ca 2‐ATPase. J Biol Chem. 1998;273(20):12360–12369. doi: 10.1074/jbc.273.20.12360 [DOI] [PubMed] [Google Scholar]
- 25. Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arpat AB, Liechti A, De Matos M, Dreos R, Janich P, Gatfield D. Transcriptome‐wide sites of collided ribosomes reveal principles of translational pausing. Genome Res. 2020;30:985‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhatt PR, Scaiola A, Loughran G, et al. Structural basis of ribosomal frameshifting during translation of the SARS‐CoV‐2 RNA genome. Science. 2021;372:1306‐1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma P, Wu J, Nilges BS, Leidel SA. Humans and other commonly used model organisms are resistant to cycloheximide‐mediated biases in ribosome profiling experiments. Nat Commun. 2021;12(1):1–13. doi: 10.1038/S41467-021-25411-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ingolia NT, Brar GA, Stern‐Ginossar N, et al. Ribosome profiling reveals pervasive translation outside of annotated protein‐coding genes. Cell Rep. 2014;8:1365‐1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ingolia NT. Genome‐wide translational profiling by ribosome footprinting. Methods Enzym. 2010;470:119‐142. [DOI] [PubMed] [Google Scholar]
- 31. VanInsberghe M, van den Berg J, Andersson‐Rolf A, Clevers H, van Oudenaarden A. Single‐cell Ribo‐seq reveals cell cycle‐dependent translational pausing. Nature. 2021;597(7877):561‐565. [DOI] [PubMed] [Google Scholar]
- 32. van Heesch S, Witte F, Schneider‐Lunitz V, et al. The translational landscape of the human heart. Cell. 2019;178:242‐260.e29. [DOI] [PubMed] [Google Scholar]
- 33. Chen J, Brunner AD, Cogan JZ, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367:140‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193‐204. [DOI] [PubMed] [Google Scholar]
- 37. Pauli A, Norris ML, Valen E, et al. Toddler: An embryonic signal that promotes cell movement via apelin receptors. Science. 2014;343(6172):1248636. doi: 10.1126/science.1248636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stein CS, Jadiya P, Zhang X, et al. Mitoregulin: a lncRNA‐encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23:3710‐3720.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makarewich CA, Munir AZ, Schiattarella GG, et al. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. Elife. 2018;7:1‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arnoult N, Correia A, Ma J, et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature. 2017;549:548‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo B, Zhai D, Cabezas E, et al. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456‐461. [DOI] [PubMed] [Google Scholar]
- 42. Pueyo JI, Magny EG, Sampson CJ, et al. Hemotin, a regulator of phagocytosis encoded by a small ORF and conserved across metazoans. PLoS Biol. 2016;14:1‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koh M, Ahmad I, Ko Y, et al. A short ORF‐encoded transcriptional regulator. Proc Natl Acad Sci USA. 2021;118:e2021943118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chu Q, Martinez TF, Novak SW, et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat Commun. 2019;10:4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsumoto A, Pasut A, Matsumoto M, et al. MTORC1 and muscle regeneration are regulated by the LINC00961‐encoded SPAR polypeptide. Nature. 2017;541:228‐232. [DOI] [PubMed] [Google Scholar]
- 46. Bhatta A, Atianand M, Jiang Z, Crabtree J, Blin J, Fitzgerald KA. A mitochondrial micropeptide is required for activation of the Nlrp3 inflammasome. J Immunol. 2020;204:428‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rathore A, Chu Q, Tan D, et al. MIEF1 microprotein regulates mitochondrial translation. Biochemistry. 2018;57:5564‐5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Makarewich CA, Baskin KK, Munir AZ, et al. MOXI is a mitochondrial micropeptide that enhances fatty acid β‐oxidation. Cell Rep. 2018;23:3701‐3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanin DE, Matsushita M, Klein Geltink RI, et al. Mitochondrial membrane potential regulates nuclear gene expression in macrophages exposed to prostaglandin E2. Immunity. 2018;49:1021‐1033.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolf T, Jin W, Zoppi G, et al. Dynamics in protein translation sustaining T cell preparedness. Nat Immunol. 2020;21:927‐937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geltink RIK, Kyle RL, Pearce EL. Unraveling the complex interplay between T cell metabolism and function. Annu Rev Immunol. 2018;36:461‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang C, Li J, Li H, et al. lncRNA MIR155HG accelerates the progression of sepsis via upregulating MEF2A by sponging miR‐194‐5p. DNA Cell Biol. 2021;40:811‐820. [DOI] [PubMed] [Google Scholar]
- 53. Vargova K, Curik N, Burda P, et al. MYB transcriptionally regulates the miR‐155 host gene in chronic lymphocytic leukemia. Blood. 2011;117:3816‐3825. [DOI] [PubMed] [Google Scholar]
- 54. Niu L, Lou F, Sun Y, et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci Adv. 2020;6:eaaz2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fields AP, Rodriguez EH, Jovanovic M, et al. A regression‐based analysis of ribosome‐profiling data reveals a conserved complexity to mammalian translation. Mol Cell. 2015;60:816‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. D’Lima NG, Ma J, Winkler L, et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol. 2017;13:174‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Khan MM, Chatterjee S, Dwivedi VP, et al. CD4 + T cell‐derived novel peptide Thp5 induces interleukin‐4 production in CD4 + T cells to direct T helper 2 cell differentiation. J Biol Chem. 2012;287:2830‐2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moore MJ, Blachere NE, Fak JJ, et al. ZFP36 RNA‐binding proteins restrain T cell activation and anti‐viral immunity. Elife. 2018;7:33057. doi: 10.7554/eLife.33057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Myers DR, Norlin E, Vercoulen Y, Roose JP. Active tonic mTORC1 signals shape baseline translation in Naive T cells. Cell Rep. 2019;27:1858‐1874.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Manfrini N, Ricciardi S, Alfieri R, et al. Ribosome profiling unveils translational regulation of metabolic enzymes in primary CD4+ Th1 cells. Dev Comp Immunol. 2020;109:103697. [DOI] [PubMed] [Google Scholar]
- 61. Jin HY, Oda H, Chen P, et al. Differential sensitivity of target genes to translational repression by miR‐17~92. PLoS Genet. 2017;13:e1006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA. 2009;106(18):7507‐7512. doi: 10.1073/pnas.0810916106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single‐nucleotide resolution. Proc Natl Acad Sci USA. 2012;109(37):E2424‐E2432. doi: 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andreev DE, O'Connor PBF, Fahey C, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4(4):1–21. doi: 10.7554/elife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731‐745. doi: 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Silva J, Fernandes R, Romão L. Translational regulation by upstream open reading frames and human diseases. In: Romão L, ed. Advances in Experimental Medicine and Biology. Springer LLC; 2019:99‐116. [DOI] [PubMed] [Google Scholar]
- 67. Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Johnstone TG, Bazzini AA, Giraldez AJ. Upstream ORF s are prevalent translational repressors in vertebrates. EMBO J. 2016;35:706‐723. doi: 10.15252/embj.201592759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chew GL, Pauli A, Schier AF. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun. 2016;7(1):11663. doi: 10.1038/ncomms11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Takahashi H, Miyaki S, Onouchi H, et al. Exhaustive identification of conserved upstream open reading frames with potential translational regulatory functions from animal genomes. Sci Reports 2020;10(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Akimoto C, Sakashita E, Kasashima K, et al. Translational repression of the McKusick‐Kaufman syndrome transcript by unique upstream open reading frames encoding mitochondrial proteins with alternative polyadenylation sites. Biochim Biophys Acta – Gen Subj. 2013;1830:2728‐2738. [DOI] [PubMed] [Google Scholar]
- 72. Ebina I, Takemoto‐Tsutsumi M, Watanabe S, et al. Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence‐dependent manner. Nucleic Acids Res. 2015;43:1562‐1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schiller C, Nowak C, Diakopoulos KN, Weidle UH, Weiss EH. An upstream open reading frame regulates LST1 expression during monocyte differentiation. PLoS One. 2014;9:e96245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Scholz A, Rappl P, Böffinger N, Mota AC, Brüne B, Schmid T. Translation of TNFAIP2 is tightly controlled by upstream open reading frames. Cell Mol Life Sci. 2020;77:2017‐2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Putlyaeva LV, Schwartz AM, Korneev KV, et al. Upstream open reading frames regulate translation of the long isoform of SLAMF1 mRNA that encodes costimulatory receptor CD150. Biochemistry. 2014;79:1405‐1411. [DOI] [PubMed] [Google Scholar]
- 76. Griffin E, Re A, Hamel N, et al. A link between diabetes and atherosclerosis: glucose regulates expression of CD36 at the level of translation. Nat Med. 2001;7:840‐846. [DOI] [PubMed] [Google Scholar]
- 77. Gregorieff A, Pyronnet S, Sonenberg N, Veillette A. Regulation of SOCS‐1 expression by translational repression. J Biol Chem. 2000;275:21596‐21604. [DOI] [PubMed] [Google Scholar]
- 78. Shi Y, Wu J, Zhong T, et al. Upstream ORFs prevent MAVS spontaneous aggregation and regulate innate immune homeostasis. iScience. 2020;23(5):101059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wek RC. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol. 2018;10: a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269‐11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Blais JD, Filipenko V, Bi M, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24:7469‐7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Whiffin N, Karczewski KJ, Zhang X, et al. Characterising the loss‐of‐function impact of 5′ untranslated region variants in 15,708 individuals. Nat Commun. 2020;11:2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Corley M, Solem A, Phillips G, et al. An RNA structure‐mediated, posttranscriptional model of human α‐1‐antitrypsin expression. Proc Natl Acad Sci USA. 2017;114:E10244‐E10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035‐1045. doi: 10.1080/15476286.2016.1271524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell‐type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Barrett SP, Salzman J. Circular RNAs: Analysis, expression and potential functions. Development. 2016;143(11):1838‐1847. doi: 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA splicing. In: Xiao J, ed. Advances in Experimental Medicine and Biology. Springer LLC; 2018:41‐52. [DOI] [PubMed] [Google Scholar]
- 89. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205‐211. [DOI] [PubMed] [Google Scholar]
- 90. Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168‐2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wilusz JE. Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements 2015;5(3):39‐45. doi: 10.1080/2159256X.2015.1045682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170‐177. [DOI] [PubMed] [Google Scholar]
- 93. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. CircRNA biogenesis competes with Pre‐mRNA splicing. Mol Cell. 2014;56:55‐66. [DOI] [PubMed] [Google Scholar]
- 94. Zhang Y, Xue W, Li X, et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611‐624. [DOI] [PubMed] [Google Scholar]
- 95. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125‐1134. [DOI] [PubMed] [Google Scholar]
- 96. Fei T, Chen Y, Xiao T, et al. Genome‐wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci USA. 2017;114:E5207‐E5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li X, Ding J, Wang X, Cheng Z, Zhu Q. NUDT21 regulates circRNA cyclization and ceRNA crosstalk in hepatocellular carcinoma. Oncogene. 2020;39:891‐904. [DOI] [PubMed] [Google Scholar]
- 98. Fischer JW, Busa VF, Shao Y, Leung AKL. Structure‐mediated RNA decay by UPF1 and G3BP1. Mol Cell. 2020;78:70‐84.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guo Y, Wei X, Peng Y. Structure‐mediated degradation of CircRNAs. Trends Cell Biol. 2020;30:501‐503. [DOI] [PubMed] [Google Scholar]
- 100. Hansen TB, Wiklund ED, Bramsen JB, et al. MiRNA‐dependent gene silencing involving Ago2‐mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414‐4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865‐880.e21. [DOI] [PubMed] [Google Scholar]
- 102. Huang C, Liang D, Tatomer DC, Wilusz JE. A length‐dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846‐2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chaudhary R, Muys BR, Grammatikakis I, et al. A circular RNA from the MDM2 locus controls cell cycle progression by suppressing p53 levels. Mol Cell Biol. 2020;40:e00473‐19. doi: 10.1128/mcb.00473-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huang X, He M, Huang S, et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4‐dependent rDNA transcription. Mol Cancer. 2019;18(1):166. doi: 10.1186/s12943-019-1098-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Holdt LM, Stahringer A, Sass K, et al. Circular non‐coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7(1):12429. doi: 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Barrett SP, Parker KR, Horn C, Mata M, Salzman J. ciRS‐7 exonic sequence is embedded in a long non‐coding RNA locus. PLoS Genet. 2017;13(12):e1007114. doi: 10.1371/journal.pgen.1007114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhou X, Jiang L, Fan G, et al. Role of the ciRS‐7/miR‐7 axis in the regulation of proliferation, apoptosis and inflammation of chondrocytes induced by IL‐1β. Int Immunopharmacol. 2019;71:233‐240. [DOI] [PubMed] [Google Scholar]
- 109. Wang F, Chen X, Han Y, Xi S, Wu G. CircRNA CDR1as regulated the proliferation of human periodontal ligament stem cells under a lipopolysaccharide‐induced inflammatory condition. Mediators Inflamm. 2019;2019:1‐9. doi: 10.1155/2019/1625381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang W, Zhang C, Hu C, Luo C, Zhong B, Yu X. Circular RNA‐CDR1as acts as the sponge of microRNA‐641 to promote osteoarthritis progression. J Inflamm. 2020;17(1):8. doi: 10.1186/s12950-020-0234-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhao J, Chu F, Xu H, et al. C/EBPα/miR‐7 controls CD4+ T‐cell activation and function and orchestrates experimental autoimmune hepatitis in mice. Hepatology. 2021;74:379‐396. [DOI] [PubMed] [Google Scholar]
- 112. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 113. Ji Q, Zhang C, Sun X, Li Q. Circular RNAs function as competing endogenous RNAs in multiple types of cancer. Oncol Lett. 2018;15:23‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Schneider T, Hung LH, Schreiner S, et al. CircRNA‐protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313. doi: 10.1038/srep31313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R‐loop formation. Nat Plants. 2017;3:53. doi: 10.1038/nplants.2017.53 [DOI] [PubMed] [Google Scholar]
- 116. Qin M, Wei G, Sun X. Circ‐UBR5: an exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem Biophys Res Commun. 2018;503:1027‐1034. [DOI] [PubMed] [Google Scholar]
- 117. Barbagallo D, Caponnetto A, Brex D, et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers. 2019;11(2):194. doi: 10.3390/cancers11020194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Barbagallo D, Caponnetto A, Cirnigliaro M, et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19:480. doi: 10.3390/ijms19020480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bartsch D, Zirkel A, Kurian L. Characterization of circular RNAs (circRNA) associated with the translation machinery. In: Dieterich C, Papantonis A, eds. Methods in Molecular Biology. Humana Press Inc.; 2018:159‐166. [DOI] [PubMed] [Google Scholar]
- 120. Legnini I, Di Timoteo G, Rossi F, et al. Circ‐ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22‐37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66:9‐21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Welden JR, van Doorn J, Nelson PT, Stamm S. The human MAPT locus generates circular RNAs. Biochim Biophys Acta Mol Basis Dis. 2018;1864:2753‐2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. doi: 10.1186/S12943-020-1135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304‐315. doi: 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circβ‐catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li Y, Wang Z, Su P, et al. circ‐EIF6 encodes EIF6‐224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta‐catenin pathway. Mol Ther. 2022;30(1):415‐430. doi: 10.1016/J.YMTHE.2021.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhang C, Zhou X, Geng X, et al. Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death Dis. 2021;12(5):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Stagsted LVW, Nielsen KM, Daugaard I, Hansen TB. Noncoding AUG circRNAs constitute an abundant and conserved subclass of circles. Life Sci Alliance. 2019;2:e201900398. doi: 10.26508/lsa.201900398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N 6 ‐methyladenosine. Cell Res. 2017;27:626‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chen C‐K, Cheng R, Demeter J, et al. Structured elements drive extensive circular RNA translation. Mol Cell. 2021;81:4300‐4318.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Li Z, Cheng Y, Wu F, et al. The emerging landscape of circular RNAs in immunity: breakthroughs and challenges. Biomark Res. 2020;8:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhang X, Chen L, Xiao B, Liu H, Su Y. Circ_0075932 in adipocyte‐derived exosomes induces inflammation and apoptosis in human dermal keratinocytes by directly binding with PUM2 and promoting PUM2‐mediated activation of AuroraA/NF‐κB pathway. Biochem Biophys Res Commun. 2019;511:551‐558. [DOI] [PubMed] [Google Scholar]
- 134. Choudhary A, Madbhagat P, Sreepadmanabh M, Bhardwaj V, Chande A. Circular RNA as an additional player in the conflicts between the host and the virus. Front Immunol. 2021;12:602006. doi: 10.3389/FIMMU.2021.602006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Tan KE, Lim YY. Viruses join the circular RNA world. FEBS J. 2021;288:4488‐4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chen YG, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67:228‐238.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wesselhoeft RA, Kowalski PS, Parker‐Hale FC, Huang Y, Bisaria N, Anderson DG. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell. 2019;74:508‐520.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chen YG, Chen R, Ahmad S, et al. N6‐methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96‐109.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Basavappa MG, Cherry S. Going in circles: the black box of circular RNA immunogenicity. Mol Cell. 2019;76:3‐5. [DOI] [PubMed] [Google Scholar]
- 140. He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Awan FM, Yang BB, Naz A, et al. The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: possible implication as theranostic agents. RNA Biol. 2021;18(1):1‐15. doi: 10.1080/15476286.2020.1790198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Dai X, Zhang N, Cheng Y, et al. RNA‐binding protein trinucleotide repeat‐containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39:981‐992. [DOI] [PubMed] [Google Scholar]
- 143. Dong W, Dai ZH, Liu FC, et al. The RNA‐binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD‐circRNA 2 production. EBioMedicine. 2019;45:155‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yang Z, Qu CB, Zhang Y, et al. Dysregulation of p53‐RBM25‐mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L‐miR‐193a‐5p‐Pcdha pathway. Oncogene. 2019;38:2516‐2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Khan MAF, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996‐1003. [DOI] [PubMed] [Google Scholar]


