Abstract
It has frequently been reported that chitinolytic soil bacteria, in particular biocontrol strains, can lyse living fungal hyphae, thereby releasing potential growth substrate. However, the conditions used in such assays (high bacterial density, rich media, fragmented hyphae) make it difficult to determine whether mycolytic activity is actually of importance for the growth and survival of chitinolytic bacteria in soils. An unidentified group of β-subclass Proteobacteria (CβPs) was most dominant among the culturable nonfilamentous chitinolytic bacteria isolated from Dutch sand dune soils. Here we demonstrate that the CβPs grew at the expense of extending fungal mycelium of three dune soil fungi (Chaetomium globosum, Fusarium culmorum, and Mucor hiemalis) under nutrient-limiting, soil-like conditions. Aggregates of CβPs were also often found attached to fungal hyphae. The growth of a control group of dominant nonchitinolytic dune soil bacteria (β- and γ-subclass Proteobacteria) was not stimulated in the mycelial zone, indicating that growth-supporting materials were not independently released in appreciable amounts by the extending hyphae. Therefore, mycolytic activities of CβPs have apparently been involved in allowing them to grow after exposure to living hyphae. The chitinase inhibitor allosamidin did not, in the case of Mucor, or only partially, in the cases of Chaetomium and Fusarium, repress mycolytic growth of the CβPs, indicating that chitinase activity alone could not explain the extent of bacterial proliferation. Chitinolytic Stenotrophomonas-like and Cytophaga-like bacteria, isolated from the same dune soils, were only slightly stimulated by exposure to fungal hyphae.
Possession of chitinase genes is widespread among many taxa of nonfilamentous soil bacteria (11). Yet several of these bacteria have been shown to be poor degraders of chitin in soil or soil-like model systems, due to their inability to penetrate chitin particles (7, 12). Filamentous microorganisms, such as fungi and actinomycetes, and gliding bacteria are much more efficient in degradation of chitin particles (7, 10, 12). Indeed, the decomposition of chitin in terrestrial soils appears to be attributable mainly to fungi and actinomycetes (7, 10). The continued maintenance of chitinase genes by many nonfilamentous bacteria, however, strongly suggests that they confer some selective advantage on the cells harboring them.
Chitin is an important constituent of the cell walls of most fungi (3), and chitinolytic bacteria have received considerable attention as potential biocontrol agents due to their ability to lyse hyphae of fungal crop pathogens (4, 14, 23). Chitinases of some mycolytic bacterial strains have been shown to destabilize cell walls of fungal pathogens (4, 15). Hyphal tips appear to be especially susceptible to the lytic activities of chitinolytic bacteria.
The ability to lyse the tips of fungal hyphae may allow nonfilamentous chitinolytic soil bacteria to utilize living fungal hyphae as an additional growth substrate (5). This could be an important selection pressure for the maintenance of chitinase genes, given the limited availability of growth substrates in soils (25). There is, however, only limited information on the population dynamics of chitinolytic bacteria during mycolytic activities. This is because most studies have been done using either high densities of chitinolytic bacteria or media that, in addition to hyphae, contain other nutrients supporting bacterial growth (2, 15, 19). In the few cases in which minimal liquid media were used, pretreatment (homogenization or partial fragmentation during washing steps) and age (autolysis) of precultured hyphae may have resulted in leakage of hyphal contents and, consequently, stimulation of bacterial growth and mycolytic activity (13, 16, 23). Hence, it is not clear whether mycolytic growth can be initiated by chitinolytic soil bacteria under natural conditions, i.e., when they are exposed to intact growing fungal hyphae in soil in the absence of other nutrients.
The first aim of this study was to monitor the dynamics of nonfilamentous chitinolytic soil bacteria during exposure to growing fungal hyphae under nutrient-limited conditions in a soil-like system. The second aim was to address the role of chitinase in the response of bacterial populations to the presence of fungal hyphae.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study were selected from a collection of chitinolytic bacteria isolated from sandy coastal dune soils in the Netherlands (5). Analysis of whole-cell fatty acid methyl ester profiles (MIDI-FAME) and general cell and colony morphology had revealed that Pseudomonas spp., Stenotrophomonas spp., and Cytophaga spp. were the most commonly isolated nonfilamentous, chitinolytic bacteria in these soils (4), and representatives of each of these bacterial groups were chosen for experimental use. Amplified ribosomal DNA (rDNA) restriction analysis (ARDRA), as well as denaturing gradient gel electrophoresis (DGGE) of 16S rDNA fragments (17), revealed a high level of relatedness between the chitinolytic pseudomonads chosen for this study (W. De Boer, P. J. A. Klein Gunnewiek, and G. A. Kowalchuk, unpublished data). Nearly full-length 16S rDNA sequences (±1,450 bp) were determined for two of these strains (EMBL accession numbers AJ310394 and AJ310395). Sequence comparison against known 16S rDNA sequences (1) revealed that these bacteria were not true pseudomonads but rather comprised a group within the β-subclass of the Proteobacteria, with 94 to 95% sequence identity to isolates within the genera Herbaspillum, Matsuebacter, Janthinobacter, and Ultramicrobacterium. The sequences of the two strains examined showed 98% sequence identity with each other. Therefore, in this article we will refer to this group as unidentified chitinolytic β-subclass Proteobacteria, or CβPs. The number of chitinolytic strains selected were 9, 8, and 3 for the CβPs, stenotrophomonads, and cytophageles, respectively. Preliminary experiments had shown that the CβPs and stenotrophomonads were slow and inefficient degraders of chitin particles in soil microcosms, whereas the Cytophaga-like strains were fast and efficient chitin degraders (7; W. De Boer and P. J. A. Klein Gunnewiek, unpublished data). In addition, eight common strains of nonchitinolytic dune soil bacteria, belonging to the genera Pseudomonas, Burkholderia, and Comamonas, were selected for control experiments.
Fungal strains.
The study was conducted using three filamentous fungal species that are abundant in coastal dune soils, namely, the ascomycete Chaetomium globosum, the hyphomycete Fusarium culmorum, and the zygomycete Mucor hiemalis (8).
Bacterial dynamics during mycelial development in sand.
Chitinolytic bacterial isolates were grown on chitin-yeast agar (CYA) at 20°C for 14 days. Medium composition was as described by De Boer et al. (7) but with the addition of 0.01 g of yeast extract (Difco, Detroit, Mich.) liter−1. Nonchitinolytic bacteria were grown on 10-fold-diluted tryptic soy broth agar (TSBA) at 20°C for 7 days (7). Fungi were grown on potato-dextrose agar (PDA) at 20°C for 2 (Mucor and Fusarium) or 7 (Chaetomium) days (6).
For each bacterial group, i.e., CβPs, stenotrophomonads, cytophagales, and nonchitinolytic bacteria, a mixture was made by adding equal numbers of cells of each strain to P buffer (KH2PO4 at 1 g liter−1 [pH 6.5]). The suspensions were mixed into autoclaved, acid-purified beach sand to give a moisture content of 5% (wt/wt) and a total microbial density of 104 CFU g−1 of sand. Portions (50 g) of sand were transferred to petri dishes (diameter, 8.5 cm) and spread evenly in an 8-mm layer. The petri dishes were sealed, placed at 20°C, and preincubated for 1 week to allow the bacteria to adapt to conditions in sand, as preliminary experiments showed some bacterial growth due to the introduction of organic compounds during bacterial inoculation.
After the preincubation, an agar disk (PDA; diameter, 1 cm) from the growing margins of the fungi was inverted and placed centrally on an autoclaved glass slide that had been put on top of the inoculated sand. The glass slide was used to prevent the diffusion of nutrients from PDA into the soil. PDA disks without fungi served as a control for potential diffusion effects. The petri dishes were again sealed and incubated at 20°C. Hyphal extension was inspected regularly using a binocular microscope and was marked on the lid of the petri dishes. Growth of Fusarium was strongly inhibited by the glass slides, and hyphae of this fungus did not reach the sand in most cases. The other two fungal species reached their maximum extension between the 1st and 2nd weeks of incubation.
After 2 and 6 weeks of incubation, 6 replicate petri dishes per treatment (bacterial group × fungal species) were removed for enumeration of bacteria. Bacterial CFU were determined in sand taken from the mycelial extension zone using diluted TSBA as described in reference 6. In the controls, i.e., PDA disks without fungi, sand was collected directly adjacent to the slide glasses, because that is the zone where any possible stimulation of bacterial growth by nutrient diffusion was expected to occur.
Since the CβPs were by far the most stimulated by the presence of fungi, a second, more detailed experiment was performed to monitor the temporal dynamics of this bacterial group in the mycelial zones of Mucor and Chaetomium. The experimental conditions were the same as those given above except that bacterial counts (four replicates) were made 0, 1, 2, 3, 4, and 6 weeks after introduction of the fungus.
Effect of allosamidin on bacterial dynamics.
Allosamidin is a powerful inhibitor of endochitinases, previously detected in the mycelial extract of a Streptomyces sp. (21). A stock solution of allosamidin (Eli Lilly and Company, Indianapolis, Ind.) was made in 5 mM acetic acid (22). Preliminary tests showed that allosamidin (10 μM) was an effective inhibitor of chitinase activity for the bacteria used in this study, both in sand and on agar media. Mycelial extension of the three fungi used was not affected (De Boer and Klein Gunnewiek, unpublished results).
The effect of allosamidin (10 μM in soil solution) on the dynamics of bacteria in sand within the mycelial zone was studied for the CβPs. Bacteria were inoculated and preincubated in sand as described in the preceding subsection with and without allosamidin. PDA disks containing the fungi C. globosum, F. culmorum, or M. hiemalis inocula were placed on stainless steel disks (diameter, 2 cm; thickness, 1 mm) as opposed to glass slides. The choice for stainless steel was made after an additional experiment indicated that it did not inhibit the extension of Fusarium hyphae. After 4 weeks of incubation, bacteria from the mycelial and control zones were enumerated as described in the previous subsection, and four replicates were counted per treatment (fungal species with or without allosamidin) for a total of 32 samples.
Microscopic observations.
Hyphae were picked aseptically from sand containing a mixture of the CβPs and were fixed by heat on glass slides. A drop of sterile, demineralized water containing 2 μg of 4′,6-diamidino-2-phenylindole (DAPI) (Sigma Chemical Co., St. Louis, Mo) liter−1 was put on top of the hyphae. After 5 min of incubation in the dark, the slides were rinsed with sterile demineralized water. Excess water was removed with filter paper. A cover glass was mounted with a drop of antifade solution (24) and sealed with clear nail polish. Microscopic examination of the hyphae for the presence of bacteria was done under UV excitation using a Leitz epifluorescence microscope.
Data analysis.
Data were analyzed by means of analysis of variance (ANOVA). Where necessary, log transformations were applied to data sets in order to establish homogeneity of variances. Differences between means were inspected using Tukey's honestly significant difference at the 5% level.
Nucleotide sequence accession numbers.
The 16S rDNA sequences determined in this study have been deposited in EMBL under accession numbers AJ310394 and AJ310395.
RESULTS
Response of bacterial groups to mycelial development in sand.
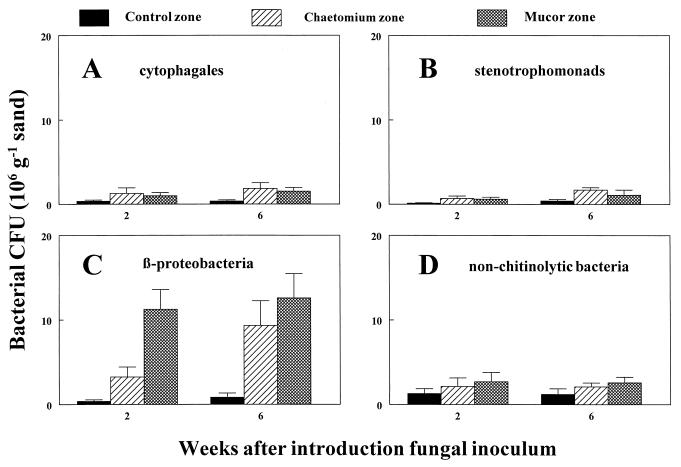
During the 1-week preincubation in sand, bacterial numbers had increased from 104 to about 105 CFU g of sand−1 for all bacterial groups tested. The introduction of fungi resulted in a strong increase in the number of CβPs in the mycelial zone, whereas the numbers of nonchitinolytic bacteria were not significantly affected (Fig. 1C and D). Two weeks after introduction of the fungi, the growth stimulation of the CβPs was already detectable, especially in the Mucor zone. FAME-gas chromatography (GC) analysis and morphological inspection of colonies revealed that this growth stimulation was not restricted to a single strain. The other two chitinolytic groups, namely, cytophageles and stenotrophomonads, were only slightly, albeit significantly, stimulated by the presence of fungi (Fig. 1A and B).
FIG. 1.
Response of chitinolytic and nonchitinolytic dune soil bacteria to invading hyphae of the fungi C. globosum and M. hiemalis in purified sand. CFU of the different bacterial groups in the mycelial zone and in a comparable zone without fungal inoculum were determined 2 and 6 weeks after introduction of the fungal inoculum (PDA disks on glass slide). Data represent the means and standard deviations for six replicates that were harvested at the indicated times.
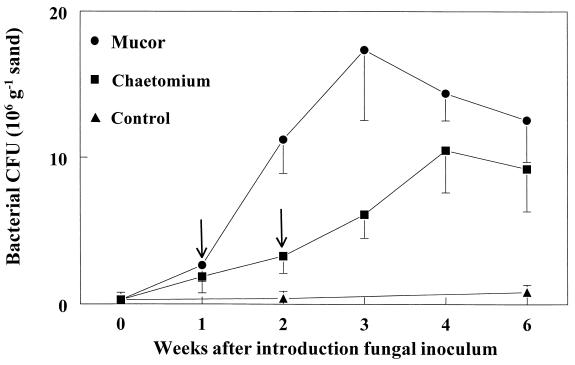
The more detailed analysis of the dynamics of the CβPs in the mycelial zones of Mucor and Chaetomium confirmed that both fungi stimulated bacterial growth and that stimulation by Mucor was more rapid (Fig. 2). This experiment also showed that the strongest stimulation occurred about 2 weeks after the mycelium had reached its maximum extension.
FIG. 2.
Temporal dynamics of chitinolytic β-subclass Proteobacteria in purified sand in the mycelial zones of C. globosum and M. hiemalis. Arrows indicate the times when the mycelial extension stopped. Data represent means and standard deviations for four replicates that were harvested at the indicated times.
Effect of allosamidin on dynamics of CβPs in mycelial zones.
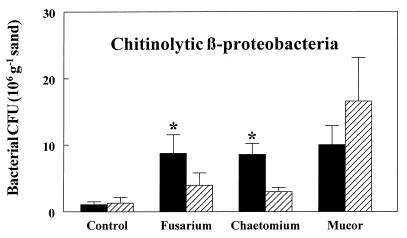
In the sand containing allosamidin, the CβPs still increased in number in response to the presence of each of the three fungi tested (Fig. 3). However, without addition of allosamidin, this bacterial increase was significantly greater in the mycelial zones of Chaetomium and Fusarium. In contrast, allosamidin had no significant effect on the CβPs in the mycelial zone of Mucor.
FIG. 3.
Effect of the chitinase inhibitor allosamidin (10 μM in soil solution) on proliferation of chitinolytic β-subclass Proteobacteria in the mycelial zones of three fungi in purified sand. Data represent means for four replicates that were harvested 4 weeks after introduction of the fungal inoculum (PDA disks on stainless steel disks). Striped bars, allosamidin treatments; solid bars, controls. Statistical significant differences (P < 0.05) between allosamidin and control treatments are indicated by asterisks.
Microscopic observations.
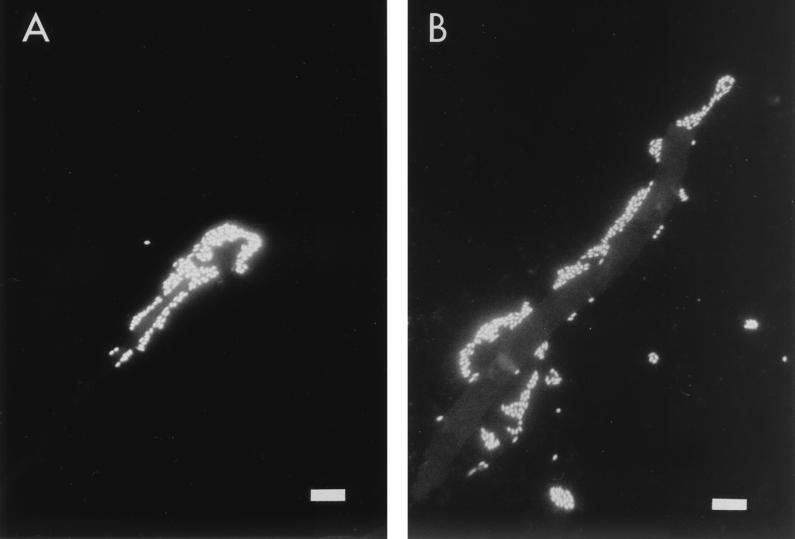
Bacterial aggregates were attached to many hyphae picked from sand containing the CβPs, as observed by fluorescent microscopy (Fig. 4), and this was true for all three fungi tested. In several cases, hyphal tips were completely covered by bacteria (Fig. 4A). Occasionally, crimping of such tips was apparent. Bacteria were, however, also found to be attached to other parts of hyphae (Fig. 4B). Addition of allosamidin reduced the numbers of attached CβPs, but this was not quantified.
FIG. 4.
Microscopic observations of DAPI-stained fungal hyphae that were picked from sand containing chitinolytic β-subclass Proteobacteria. (A) A hyphal tip of F. culmorum covered by bacteria. (B) Separate clusters of bacteria along a hypha of F. culmorum. Scale bar, 10 μm.
DISCUSSION
Several studies have dealt with the effects of chitinolytic soil bacteria on plant-pathogenic fungi, in the context of demonstrating their potential as biocontrol agents. These studies have involved applications of high densities of bacteria, use of nutrient-rich liquid or agar media, or addition of hyphal fragments (15, 16, 19). Under such conditions, many chitinolytic soil bacteria were observed to lyse fungal hyhae. In addition, several studies have demonstrated that purified chitinase of potential biocontrol strains can cause deformation of living hyphae (18, 26). However, from these studies it is not clear whether chitinolytic bacteria are also able to attack living fungi under natural conditions, i.e., at low cell densities in the soil.
This study showed that chitinolytic β-subclass Proteobacteria (CβPs) isolated from coastal dune soils were able to grow at the expense of living fungal hyphae in sand without the presence of other carbon sources. They could do so at cell densities that are common for dune soils (5, 7). Both pathogenic and saprophytic dune soil fungi stimulated the growth of the CβPs, suggesting that their growth on living fungi is a general phenomenon in dune soils. The actual increase of the CβPs is probably even higher than that indicated by the plate counts. Microscopic observations showed that many bacteria were attached to hyphae, and dispersion of such cells for plate counts may not have been complete.
Unlike those of the CβPs, numbers of nonchitinolytic bacteria did not increase in the mycelial zone. This indicates that the amount of hypha-derived materials supporting bacterial growth was small. Therefore, the proliferation of the CβPs was probably due to direct mycolytic interaction with the living fungal hyphae. Furthermore, this finding suggests that chitinase was required for growth on living hyphae. The role of chitinase, however, is not completely clear. Proliferation of the CβPs in the mycelial zone was inhibited only partly (for Chaetomium and Fusarium) or not at all (for Mucor) by the chitinase inhibitor allosamidin. Additional experiments showed that this was not attributed to ineffectiveness of allosamidin in the soil-like system. Apparently, other factors, in addition to chitinase, contributed to the growth response of the CβPs. Bacterial chitinase production has previously been observed in combination with other antifungal factors, such as other lytic enzymes and antibiotics, which may also have been involved in the observed growth response (4, 9).
The ineffectiveness of allosamidin in reducing growth of the CβPs in the mycelial zone of Mucor suggests that chitinase is not required by the bacteria in the acquisition of nutrients from this fungus. Unlike the other fungi tested, Mucor incorporates chitosan rather than chitin as a major component of the cell wall (3), and it has been shown that chitosanases are required for protoplast formation in Mucor species (20). The possession of both chitinase and chitosanase is not uncommon for soil bacteria (11). Therefore, chitosanase may well be involved in the response of the CβPs to hyphae of Mucor.
The impact of chitinolytic bacteria on fungal development is not yet known, as the actual amount of fungal material converted by bacteria was not determined. However, only partial degradation of the fungal mycelium was observed, as mycelial networks remained visible throughout the incubation period. The growth response of the CβPs appeared to be restricted to young hyphae, as their numbers did not increase in the mycelial zones containing only old (>4 weeks) hyphae, suggesting that degradation of mature or empty fungal hyphae in soils is not a niche for them.
The growth response of CβPs to invading fungal hyphae supports our hypothesis that nonfilamentous soil bacteria with low chitin-degrading ability use their chitinase genes in interactions with fungi rather than for degradation of chitinous material. This hypothesis, however, should also apply to chitinolytic stenotrophomonads. Yet growth of these bacteria was not much stimulated by fungi. Unlike the CβPs, stenotrophomonads isolated from dune soils were not able to degrade colloidal chitin in minimal media but required additional growth factors which could be supplied by adding yeast extract (De Boer and Klein Gunnewiek, unpublished results). Perhaps the lack of these growth factors in the sand suppressed their proliferation in the mycelial zone.
Chitinolytic Cytophaga-like bacteria also showed little response to the presence of living fungal hyphae. In this case, limitation due to lack of additional growth factors is probably not the cause of this poor stimulation, as these bacteria are strong degraders of both colloidal and particulate chitin in minimal media and soil microcosms (11; De Boer and Klein Gunnewiek, unpublished results). Chitinolytic Cytophaga-like bacteria may, therefore, be better equipped to degrade dead chitinous materials than to attack living fungal hyphae.
In conclusion, we present strong evidence for mycolytic growth of chitinolytic β-subclass Proteobacteria, which are dominant among the culturable nonfilamentous chitinolytic bacteria in Dutch coastal dune soils. Future research will be directed to the mechanisms and regulation of this type of growth and to the fitness consequences for both bacteria and fungi.
ACKNOWLEDGMENTS
We are indebted to James H. Prather of Eli Lilly and Company for a generous gift of allosamidin.
Footnotes
This is publication number 2810 of the Netherlands Institute of Ecology.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrows-Broaddus J, Kerr T J. Inhibition of Fusarium moniliforme var. subglutinans, the causal agent of pine pitch canker, by the soil bacterium Athrobacter sp. Can J Microbiol. 1981;27:20–27. doi: 10.1139/m81-004. [DOI] [PubMed] [Google Scholar]
- 3.Carlile M J, Watkinson S C. The fungi. London, United Kingdom: Academic Press; 1994. [Google Scholar]
- 4.Chernin L, Ismailov Z, Haran S, Chet I. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl Environ Microbiol. 1995;61:1720–1726. doi: 10.1128/aem.61.5.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Boer W, Klein Gunnewiek P J A, Lafeber P, Janse J D, Spit B E, Woldendorp J W. Antifungal properties of chitinolytic dune soil bacteria. Soil Biol Biochem. 1998;30:193–203. [Google Scholar]
- 6.De Boer W, Klein Gunnewiek P J A, Woldendorp J W. Suppression of hyphal growth of soil-borne fungi by dune soils from vigorous and declining stands of Ammophila arenaria. New Phytol. 1998;138:107–116. [Google Scholar]
- 7.De Boer W, Gerards S, Klein Gunnewiek P J A, Modderman R. Response of the chitinolytic microbial community to chitin amendments of dune soils. Biol Fertil Soils. 1999;29:170–177. [Google Scholar]
- 8.De Rooij-Van der Goes P C E M. The role of plant-parasitic nematodes and soil-borne fungi in the decline of Ammophila arenaria (L.) Link. New Phytol. 1995;129:661–669. [Google Scholar]
- 9.Gaffney T D, Lam S T, Ligon J, Gates K, Frazelle A, Di Maio J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, Kempf H-J, Becker J O. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 10.Gooday G W. Physiology of microbial degradation of chitin and chitosan. Biodegradation. 1990;1:177–190. [Google Scholar]
- 11.Gooday G W. The ecology of chitin degradation. Adv Microb Ecol. 1990;11:387–430. [Google Scholar]
- 12.Gould W D, Bryant R J, Trofymow J A, Anderson R V, Elliott E T, Coleman D C. Chitin decomposition in a model soil system. Soil Biol Biochem. 1981;13:487–492. [Google Scholar]
- 13.Inbar J, Chet I. Evidence that chitinase produced by Aeromonas caviae is involved in the biological control of soil-borne pathogens by this bacterium. Soil Biol Biochem. 1991;23:973–978. [Google Scholar]
- 14.Kobayashi D Y, Guglielmoni M, Clarke B B. Isolation of the chitinolytic bacteria Xanthomonas maltophilia and Serratia marcescens as biological control agents for summer patch disease of turfgrass. Soil Biol Biochem. 1995;27:1479–1487. [Google Scholar]
- 15.Lim H-S, Kim Y-S, Kim S-D. Pseudomonas stutzeri YPL-1 genetic transformation and antifungal mechanism against Fusarium solani, an agent of plant root rot. Appl Environ Microbiol. 1991;57:510–516. doi: 10.1128/aem.57.2.510-516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell R, Alexander M. Lysis of soil fungi by bacteria. Can J Microbiol. 1962;9:169–177. [Google Scholar]
- 17.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ordentlich A, Elad Y, Chet I. The role of chitinase of Serratia marcescens for the control of Sclerotium rolfsii. Phytopathology. 1988;78:84–88. [Google Scholar]
- 19.Podile A R, Prakash A P. Lysis and biological control of Aspergillus niger by Bacillus subtilis AF1. Can J Microbiol. 1996;42:533–538. doi: 10.1139/m96-072. [DOI] [PubMed] [Google Scholar]
- 20.Reyes F, Lahoz R, Martinez M J, Alfonso C. Chitosanases in the autolysis of Mucor rouxii. Mycopathology. 1985;89:181–187. [Google Scholar]
- 21.Sakuda S, Isogai A, Matsumoto A, Suzuki A, Koseki K. The structure of allosamidin, a novel insect chitinase inhibitor, produced by Streptomyces spec. Tetrahedron Lett. 1986;27:2475–2478. [Google Scholar]
- 22.Sándor E, Pusztahelyi T, Karaffa L, Karányi Z, Pócsi I, Biró S, Szentirmai A, Pócsi I. Allosamidin inhibits the fragmentation of Acremonium chrysogenum but does not influence the cephalosporin C production of the fungus. FEMS Microbiol Lett. 1998;164:231–236. doi: 10.1111/j.1574-6968.1998.tb13091.x. [DOI] [PubMed] [Google Scholar]
- 23.Sneh B. Use of rhizosphere chitinolytic bacteria for biological control of Fusarium oxysporum f. sp. dianthi in carnation. Phytopath Z. 1981;100:251–256. [Google Scholar]
- 24.Weinbauer M G, Beckman C, Höfle M G. Utility of green fluorescent nucleic acid dyes and aluminium oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl Environ Microbiol. 1998;64:5000–5003. doi: 10.1128/aem.64.12.5000-5003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams S T. Oligotrophy in soil: fact or fiction. In: Fletcher M, Floodgate G D, editors. Bacteria in their natural environments. London, United Kingdom: Academic Press; 1985. pp. 81–110. [Google Scholar]
- 26.Zhang Z, Yuen G Y. The role of chitinase production by Stenotrophomonas maltophilia strain C3 in biological control of Bipolaris sorokiniana. Phytopathology. 2000;90:384–389. doi: 10.1094/PHYTO.2000.90.4.384. [DOI] [PubMed] [Google Scholar]