Abstract
We released genetically modified Pseudomonas putida WCS358r into the rhizospheres of wheat plants. The two genetically modified derivatives, genetically modified microorganism (GMM) 2 and GMM 8, carried the phz biosynthetic gene locus of strain P. fluorescens 2-79 and constitutively produced the antifungal compound phenazine-1-carboxylic acid (PCA). In the springs of 1997 and 1998 we sowed wheat seeds treated with either GMM 2, GMM 8, or WCS358r (approximately 107 CFU per seed), and measured the numbers, composition, and activities of the rhizosphere microbial populations. During both growing seasons, all three bacterial strains decreased from 107 CFU per g of rhizosphere sample to below the limit of detection (102 CFU per g) 1 month after harvest of the wheat plants. The phz genes were stably maintained, and PCA was detected in rhizosphere extracts of GMM-treated plants. In 1997, but not in 1998, fungal numbers in the rhizosphere, quantified on 2% malt extract agar (total filamentous fungi) and on Komada's medium (mainly Fusarium spp.), were transiently suppressed in GMM 8-treated plants. We also analyzed the effects of the GMMs on the rhizosphere fungi by using amplified ribosomal DNA restriction analysis. Introduction of any of the three bacterial strains transiently changed the composition of the rhizosphere fungal microflora. However, in both 1997 and 1998, GMM-induced effects were distinct from those of WCS358r and lasted for 40 days in 1997 and for 89 days after sowing in 1998, whereas effects induced by WCS358r were detectable for 12 (1997) or 40 (1998) days. None of the strains affected the metabolic activity of the soil microbial population (substrate-induced respiration), soil nitrification potential, cellulose decomposition, plant height, or plant yield. The results indicate that application of GMMs engineered to have improved antifungal activity can exert nontarget effects on the natural fungal microflora.
There is increasing interest in commercial application of genetically modified microorganisms (GMMs) with improved biocontrol properties toward soil-borne plant pathogens. Despite long-term experience with the introduction of nonmodified microorganisms, concern about the ecological impact of large-scale release of GMMs remains. To date, risk assessment studies under field conditions have focused mainly on microorganisms genetically modified with markers, such as antibiotic resistance, lacZY, or xylE, and attention has been given to the potential impact of GMMs on the indigenous soil microflora (8, 29). Biologically mediated processes are central to the ecological functioning of the soil system. Microorganisms play a crucial role in nutrient cycling, and they contribute to suppression of soil-borne plant pathogens (7, 28). The introduction of large numbers of GMMs into the soil could alter microbial populations and disturb microbially driven soil processes. Effects of introduced GMMs on soil ecosystems have been studied mainly in microcosm experiments. Transient perturbations have been observed in indigenous bacterial (26), fungal (30), and protozoal (3) populations, in carbon turnover (38), and in soil enzyme activities (24). However, these microcosms lack the full biotic and abiotic components of a field environment.
Most studies of nontarget effects of GMMs examine only the culturable microflora. This limitation reduces the value of the results, because only a small proportion (0.5 to 2%) of the fungal and bacterial microflora can be cultured using currently available media (37). Techniques such as amplified ribosomal DNA (rDNA) restriction analysis (ARDRA), temperature gradient gel electrophoresis (TGGE), or denaturing gradient gel electrophoresis (DGGE) (23) can be used to more accurately monitor microbial communities without cultivation. Shifts in microbial communities, at the 16S rDNA level, have been studied mainly in soil polluted with heavy metals or with pesticides (12, 13, 31). Reports on the effects of functional genetically modified bacteria on microbial communities, using molecular methods, are scarce. Robleto et al. (29) demonstrated a reduction in the diversity of trifolitoxin-sensitive bacteria after inoculation of field-grown Phaseolus vulgaris with Rhizobium strains differing in their trifolitoxin production. As far as we are aware, no reports have been published on the effects of genetically modified biocontrol bacteria on the composition of the fungal microflora at the 18S rDNA level.
We modified Pseudomonas putida WCS358r to produce the antifungal agent phenazine-1-carboxylic acid (PCA) (35), resulting in improved biocontrol activity toward fungal pathogens, such as Gaeumannomyces graminis var. tritici (D. Glandorf et al., unpublished data). Our objective in this study was to determine if genetically improved biocontrol bacteria could exert effects on nontarget members of the fungal rhizosphere microflora by using both cultivation-dependent and cultivation-independent methods.
MATERIALS AND METHODS
Bacterial strains.
P. putida WCS358r is a rifampin-resistant (15), plant growth-promoting rhizobacterial strain (4) with disease-suppressive properties, based on the production of its fluorescent siderophore (11, 20, 27). We inserted a 6.8-kb BglII-XbaI fragment containing the phzABCDEFG genes from Pseudomonas fluorescens 2-79 (21, 34) under the control of the Ptac promoter into a kanamycin-resistant mini-Tn5 lacZ1 transposon (10). We recovered PCA-producing mutants following mating of WCS358r with Escherichia coli SM10 (λ pir) containing plasmid pUT/Km (10) harboring the transposable phenazine biosynthetic locus. In vitro production of PCA by these GMMs was measured by high-pressure liquid chromatography (HPLC) (6). The presence of a single insertion of the mini-Tn5 in the chromosome of WCS358r and the absence of the transposase gene were confirmed by Southern blotting. Cultures were stored at −80°C in 35% glycerol.
Seed treatment.
Cells of WCS358r, GMM 2, and GMM 8 were grown on KB agar plates (18) supplemented with rifampin (150 μg/ml) for WCS358 and additional kanamycin (50 μg/ml) for the GMMs. Single colonies were subcultured on KB agar plates (without antibiotics) for 24 h at 28°C. Bacteria were harvested by scraping cells from the agar, suspending them in MgSO4 (10 mM), and washing the suspensions twice by centrifugation (for 20 min at 8,000 × g). Bacterial densities were measured spectrophotometrically at 660 nm and determined using a calibration curve determined for each strain. Commercial wheat seeds (Triticum aestivum cv. Baldus) were incubated for 30 min at room temperature in the washed bacterial suspensions (1.5 × 1010 to 2 × 1010 CFU per ml) in 1% (wt/vol) methylcellulose (Sigma, St. Louis, Mo.) and then placed under a vacuum for 20 min (20 mm Hg) to facilitate bacterial adherence to the seeds. For the control treatment, bacterial suspensions were replaced by 10 mM MgSO4. Treated seeds were allowed to dry overnight on filter paper in a laminar flow cabinet and were sown the next day. Bacterial populations on seeds were about 107 CFU/seed at the time of seeding and consisted virtually exclusively of the strain applied.
Experimental field.
Experiments were conducted in 1997 and 1998 in a site located at De Uithof, Utrecht, The Netherlands. The experimental field had a planting history of grass and consisted of clay soil, with an organic matter content of 4% and a pH (KCl) of 5.0. No fertilizers or chemicals were applied before or during the field trials. Treated seeds were sown on April 23, 1997, and, in an adjacent experimental field, on April 15, 1998, in 1-m2 plots. A randomized block design was used with four treatments, with six replicates each, resulting in a total of 24 plots. The four treatments were seeds treated with WCS358r, GMM 2, GMM 8, and nonbacterized seeds (control). Per plot about 1,750 wheat seeds were sown manually in 11 rows 1 m long at a depth of 2 to 3 cm. Plots were located in three soil strips that were separated from each other by a tile path (width, 60 cm). Within each soil strip, plots were separated by bare soil strips (width, 60 cm) to reduce cross-contamination. Plots were surrounded by an unplanted buffer zone (1.8 m) and straw mats to minimize dispersal of bacteria. The plot was fenced to block rabbits from entering the site, and bird entry was prevented by using bird nets. Plants were harvested on August 8 (1997) and August 4 (1998).
Population dynamics of bacterial inoculants.
Sampling dates were 5, 12, 27, 40, 62, 90, 105 (harvest), and 131 days after sowing. At harvest, wheat plants were cut off above the soil using hedge shears, thereby leaving the roots for postharvest samples. In each plot, three root samples with adhering soil were taken using a 10-cm knife and pooled, resulting in six samples for each treatment. Samples were stored overnight at 4°C. The next day, excess soil was removed from the roots, and 0.5- to 1.0-g (wet weight) samples of roots with tightly adhering soil (rhizosphere samples) were shaken vigorously for 30 s in test tubes containing 5 ml of 10 mM MgSO4 and glass beads (diameter 0.11 mm). Appropriate dilutions were plated on KB+ agar (KB agar containing 13 μg of chloramphenicol, 40 μg of ampicillin, and 100 μg of cycloheximide per ml), amended with rifampin (150 μg/ml) and/or kanamycin (50 μg/ml), using a Spiral Plater (Spiral System model C; Spiral Systems Inc., Cincinnati, Ohio). Plates were incubated at 28°C for 48 h, after which CFU were counted.
PCA extraction from wheat rhizosphere.
To determine whether PCA was produced by the GMMs in the rhizosphere, in 1998 we collected samples from all six field plots for each treatment 12 days after sowing. Samples from three plots were pooled, and roots with tightly adhering soil (25 g) were extracted according to the work of Bonsall et al. (6), resulting in two replicates per treatment. After precipitation of the soil by centrifugation (for 20 min at 8,000 × g), the supernatant was evaporated to dryness in a vacuum centrifuge at room temperature (for 3 h at 100 mtorr). The presence of PCA in the residues was determined by HPLC, by the procedure of Bonsall et al. (6), with some modifications. HPLC fractionation was performed using a Shimadzu (Kiya-Machi, Kyoto, Japan) HPLC gradient system equipped with two LC-9A pumps, a low-pressure mixing chamber, and an SPD-6A UV spectrometric detector operated at 254 nm. The column used was a C18 (5 μm) reversed-phase column (2.1 by 250 mm; Vydac, Hesperia, Calif.), on which the rhizosphere extracts were injected in either 5 μl (for recording chromatograms) or 50 μl (for collecting fractions to obtain mass spectra) of 35% acetonitrile (ACN)–0.1% aqueous trifluoroacetic acid (TFA) and fractionated using an ACN–0.1% TFA gradient as follows: 10% ACN–0.1% TFA for 2 min, followed by a linear gradient over 20 min to 100% ACN, followed by 5 min at 100% ACN. Commercial PCA (Maybridge Chemical Company Ltd., Trevillett, Cornwall, United Kingdom) and PCA produced by strain 2-79 in liquid culture were used as reference samples. To confirm the presence of PCA, 1-min fractions were collected and the fraction corresponding to the Rt of PCA was evaporated to dryness, redissolved in 100 μl of 35% ACN–0.1% TFA, and subjected to mass spectrometric analysis using a hybrid quadrupole orthogonal tandem time-of-flight Q-Tof mass spectrometer (Micromass UK Ltd., Manchester, United Kingdom) fitted with a Z-spray sample introduction system and gold-coated glass capillaries in a nanospray source. The mass spectrometer was operated in the positive-ion mode with a cone voltage of 25 V and a capillary voltage of 1,000 V. Spectra were acquired via the Tof analyzer and were integrated every 2.4 s. Data were recorded and processed using MassLynx software, version 3.1, from Micromass UK Ltd. (http://www.micromass.co.uk). Mass calibration was performed by multiple-ion monitoring of singly charged sodium and cesium iodide signals. Tandem mass spectra were recorded using argon as the collision gas, and spectra were obtained with collision energy settings of 10, 20, and 30 eV.
Determination of culturable fungal microflora.
We made dilution plates of processed rhizosphere samples on several agar media specific for fungi. For enumeration of fungal propagules, we chose media that supported the growth of fungi known to be inhibited by the GMMs in vitro. Media included 2% malt amended with Solacol (14) to isolate total filamentous fungi, 2% malt medium amended with benomyl (50 μg/ml) for fast-growing Mucorales (5), and two media selective for Fusarium spp.: Komada's medium (19) and PCNB medium (25). Chlorotetracycline (200 μg/ml; Sigma) was added to prevent bacterial growth. Plates were incubated at 20°C for 6 days, after which CFU were counted.
Determination of composition of fungal microflora by ARDRA.
We examined the composition of the fungal microflora by 18S rDNA analysis of wheat rhizosphere samples by using ARDRA according to the method of Smit et al. (31). For this assay two independent replicate rhizosphere samples were used for each treatment. Each replicate was obtained by pooling samples from three plots. Samples of roots with tightly adhering soil (approximately 3 g) were mixed with 10 ml of sterile phosphate buffer (120 mM; pH 8.0) and 1 g of sterile gravel (diameter, 2 to 3 mm) in 50-ml polypropylene tubes. Tubes were vortexed for 30 s, and the buffer-soil slurry mixture was decanted into a new tube, leaving the gravel and roots behind. After addition of 70 to 90 mg of lysozyme (Merck, Darmstadt, Germany), the slurry was incubated at 37°C for 15 min, followed by a 10-min incubation on ice. The slurry was amended with 15 g of glass beads (diameter, 0.11 mm), after which total DNA was extracted by disrupting cells using a bead beater (31).
One microliter of 0.1× extract was used for PCR amplification. 18S rDNA was amplified by using a primer set that amplifies 18S rDNA sequences from a wide range of fungal taxa: EF3 (5′-TCCTCTAAATGACCAAGTTTG-3′) and EF4 (5′-GGAAGGG[G/A]TGTATTTATTAG-3′) (34). This primer set amplifies a major part of the 18S rDNA, resulting in 1.4-kb DNA fragments. DNA fragments were digested with either TaqI, HinfI, or StyI to generate ARDRA profiles. PCR products (10 μl) were digested for 2 h at 37°C (with StyI or HinfI) or 65°C (with TaqI) after addition of 20 U of each enzyme. Approximately 6 μl per sample was analyzed on precast acrylamide gels (GeneGel Excel 12.5; Amersham Pharmacia Biotech AB, Uppsala, Sweden). Samples from the different treatments obtained at each sample date were run on the same gel in order to exclude differences between ARDRA patterns caused by other factors. As a standard, gels were run for 2 h at 25 mA on a GenePhor horizontal electrophoresis unit (Amersham Pharmacia Biotech), and gels were silver stained using a Hoefer Automated Gel Stainer and a DNA Silver Staining Kit (Amersham Pharmacia Biotech). Diagrams representing percent similarity of resulting banding patterns per sample date were constructed by UPGMA cluster analysis with the Biogene program (version V96.15; Biogene, Vilber, Loumat, France), using the algorithm of Nei and Li (2% confidence) (13a).
SIR, NPA, and cellulose decomposition.
We measured substrate-induced respiration (SIR), soil nitrification potential (NPA), and cellulose decomposition during the growing season. SIR represents the activity of the total metabolically active soil microbial community. NPA is very sensitive to perturbation in the microbial community (36). Cellulose decomposition is a microbial activity of numerous soil fungi. SIR and NPA were determined monthly, starting after 5 days after sowing until 1 month after harvest in 1997. Cellulose decomposition was measured in the 1st, 3rd and 5th months after sowing in 1997. In 1997 the GMM-induced effects appeared to occur primarily in the first part of the growing season, so in 1998 all assays were performed only during the first 2 months of the season.
(i) SIR.
To determine SIR (2), we evaluated 300 g of soil (including roots) from samples taken at depths of 0 to 10 cm from each field plot. Soil samples (without roots) were incubated in glass jars at 22°C for 2 days, after which a surplus of glucose (5 g) was added to each sample to activate the metabolism of the microflora. The accumulation of CO2 over 3 h was determined by taking 5-ml gas samples from the headspace of the glass jars both directly after addition of the glucose and after 3 h of incubation at 22°C. Samples from the headspace were taken with a syringe, through a rubber valve in the lid of the jar. Gas samples were immediately analyzed on a 5890A gas chromatograph (Hewlett-Packard, Pittsburgh, Pa.). The amount of CO2 was determined from a calibration curve. Leakage of CO2 from jars was checked by including a control jar containing a known concentration of CO2. Corrections for the amount of CO2 present in soil moisture and for carbon in the HCO3− form were made as described by Aerts and Toet (1).
(ii) NPA.
Soil was sampled from the field plots as described for SIR. NPA was determined by the procedure of Stienstra et al. (33). Accumulation of nitrate and nitrite was measured for 24 h after addition of 25 ml of 2 mM (NH4)2SO4 to 10 g of soil and incubation of the soil slurries at 25°C on a gyratory shaker (200 rpm). Samples were taken both directly after addition of ammonium sulfate and after 24 h of incubation. Nitrification in samples was stopped by mixing the supernatant of the slurry (centrifuged at 15,700 × g for 1 min) with 2 M KCl (1:1). Samples were stored at −20°C until further analysis. Amounts of nitrate and nitrite were determined colorimetrically on a Skalar (Breda, The Netherlands) SA-40 autoanalyzer.
(iii) Cellulose decomposition.
We measured cellulose decomposition by determining loss of tensile strength of cotton strips (Shirley Dyeing and Finishing Ltd., Hyde, United Kingdom) after soil incubation (17). At each of the six replicate plots for each treatment, a cotton strip 12 cm wide was inserted into the soil to a depth of 10 cm and incubated for 3 to 5 weeks. At the beginning of each incubation, another strip was inserted in the soil and immediately removed to serve as a control for strength loss by insertion, wetting, and drying of the strip. After incubation, the strips were washed with tap water, air dried, and stored until further analysis. Each cotton strip was cut into substrips of 3.5 cm corresponding to different distances from the soil surface (0 to 3.5, 4 to 7.5, and 8 to 10.5 cm). The tensile strength of each cotton strip was determined at 70% relative humidity using an M1000e tensiometer (Mecmesin Ltd., Horsham, United Kingdom) at speed 3. Tensile strength loss was calculated as the difference between the tensile strengths of the control strips and the strips that had been in the soil for 3 to 5 weeks. Six replicates per treatment were used.
Determination of plant growth.
We evaluated plant growth by determining the height, fresh weight, and dry weight of each of 15 plants per plot at each sampling date. In addition, seed weight (100 seeds per replicate plot) was measured after harvest of the plants. Plant dry weight was determined after incubation of the plant material for 5 days at 70°C.
Statistics.
All data obtained for the culturable microflora, microbial activities, and plant growth (six replicates per treatment) were, if necessary after log10 transformation, statistically analyzed with repeated-measures analysis of variance (ANOVA) using SAS/STAT software, version 6.04 (SAS Institute, Cary, N.C.) by assessing the interaction between time (5 to 31 days) and treatment (bacterial treatment). A significant interaction between time and treatment was determined after Bonferroni's correction of P values. Results for populations of WCS358r and the GMMs, effects on microbial activities in 1998, and kernel weight were analyzed, for each sample date, by one-way ANOVA, followed by a t test (least significant difference [LSD]) using the same SAS software. For the dendrograms obtained from the ARDRA data, similarity indexes were calculated. The similarity index represents the difference between the two values for each treatment at each time point, relative to the mean similarity of all treatments. Subsequently these similarity indexes were analyzed over different periods of time by regression analysis using SAS software. The predictor variable was time (in days) since the start of the experiment, and the dependent variable was the difference between the mean similarities of pairs within clusters and pairs among clusters.
RESULTS
GMMs.
PCA-producing, kanamycin-resistant derivatives of P. putida WCS358r were obtained by transposition of the phz biosynthetic gene locus into the chromosome of WCS358r using the mini-Tn5 transposon delivery system (16). Two GMMs were selected for study, GMM 2 and GMM 8. GMM 8 produced three times as much PCA as GMM 2. The presence of a single insert of the mini-Tn5 in both GMMs was confirmed by Southern blotting, and, as expected, the transposase gene was absent.
Population dynamics of WCS358r and the GMMs and stability of the phz genes in the rhizosphere.
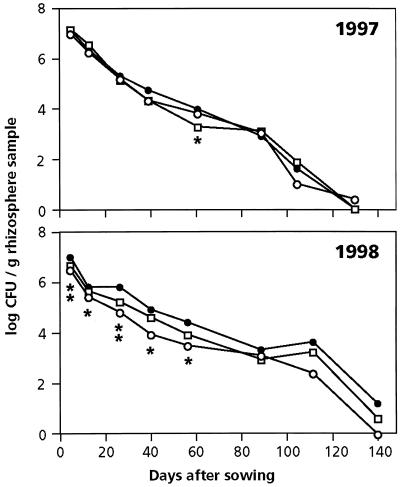
In both years populations of WCS358r and the GMMs decreased from about 107 CFU per g of rhizosphere sample to 102 to 104 CFU per g at harvest, and to near the detection limit (102 to 103 CFU/g of rhizosphere sample) 1 month after harvesting (131 or 139 days after sowing) (Fig. 1). No kanamycin and rifampin-resistant CFU were detected in the WCS358r-treated plots. Rifampin-resistant (but not rifampin- and kanamycin-resistant) CFU were detected in one of the six replicate control plots 40 days after sowing in 1997 at a density of 8 × 102 CFU per g of rhizosphere sample and only after 5 days in 1998 (2 × 105 CFU/g). Mean numbers of rifampin-resistant CFU (in log CFU per gram of rhizosphere sample) in the control plots at these sampling dates were 0.48 (in 1997) and 0.88 (in 1998). No statistically significant differences in the number of cells of WCS358r and the GMMs were detected during the 1997 season, except for GMM 8 at 62 days after sowing. During the 1998 season, however, populations of the GMMs declined within 5 days of sowing and remained lower than that of WCS358r for the first 60 days. Neither in 1997 nor in 1998 were statistically significant differences observed between numbers of GMMs on rifampin-containing KB+ with or without kanamycin.
FIG. 1.
Populations of P. putida WCS358r (●) and its phenazine-producing derivatives GMM 2 (○) and 8 (□) in wheat rhizosphere samples during the field trials of 1997 and 1998. Bacteria were introduced into the soil by treatment of wheat seeds (approximately 107 CFU/seed). Plants were harvested 105 days (1997) and 111 days (1998) after sowing. Statistically significant differences (P = 0.05) are indicated by asterisks: one asterisk indicates a difference between WCS358r and either GMM 2 or GMM 8, and two asterisks indicate a difference between WCS358r and both GMMs. P values were lower than 0.05 at day 62 (P = 0.016) in 1997 and at days 5, 12, 27, 40, and 56 (P = 0.000, 0.048, 0.017, 0.033, and 0.027, respectively) in 1998.
Detection of PCA in rhizosphere extracts using HPLC and mass spectrometry.
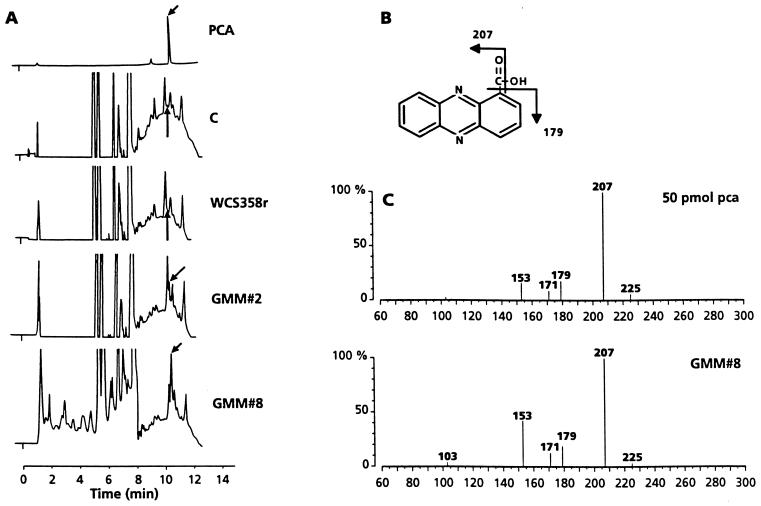
Rhizosphere extracts obtained in the 1998 field trial 12 days after sowing were fractionated using reversed-phase HPLC. The chromatogram obtained from standard PCA (Fig. 2A) has a clear peak with a retention time of 10.48 min. When the component in this fraction was analyzed using nanoelectrospray tandem mass spectrometry, the fragment ions were identical to those obtained from a sample of standard PCA (Fig. 2C). In extracts of control- and WCS358r-treated wheat rhizospheres a very minor peak with a retention time similar to that of standard PCA appears, but, based on mass spectrometric analysis of these fractions, no PCA is present. HPLC chromatograms of rhizosphere extracts of wheat plants treated with GMM 2 and GMM 8 have peaks with the same retention time as standard PCA (Fig. 2A), and the presence of PCA was confirmed by spectrometric analysis of these peaks (Fig. 2C). We could not accurately measure the amount of PCA in the samples, but comparison of the heights of the peaks corresponding to PCA suggests that PCA production by GMM 8 is higher than that of GMM 2. This conclusion is consistent with the greater intensity of PCA-specific ions in the mass spectra compared to the intensities of the background ions.
FIG. 2.
HPLC and collision-induced dissociation nanoelectrospray mass spectrometric analyses of PCA extracted from rhizosphere samples of wheat plants treated with P. putida WCS358r, its phenazine-producing derivatives GMM 2 and GMM 8, and control plants from the 1998 field trial. (A) HPLC chromatograms. Arrows indicate the position of the PCA peak. PCA, standard PCA (50 pmol). Results for extracts from rhizosphere samples of control plants (C), WCS358r-treated plants (WCS358r), and GMM 2- and GMM 8-treated plants (GMM 2 and GMM 8, respectively) are shown. (B) Mass spectrum fragmentation scheme of PCA. (C) Fragmentation tandem mass spectra of standard PCA and PCA collected following HPLC of the rhizosphere extract from wheat plants treated with GMM 8 (fraction indicated with an arrow in panel A) obtained using a collision energy setting of 20 eV. Mass assignments are given as nominal masses.
Effects on indigenous culturable fungal microflora.
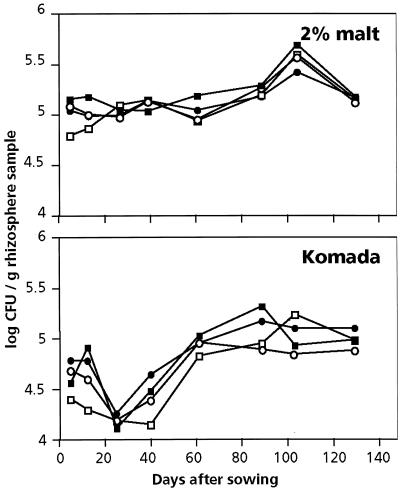
Rhizosphere populations were analyzed using a repeated-measurements ANOVA that determines whether the development of fungal populations during the growing season is affected by the bacterial treatment (interaction between time and treatment; a P value of 0.05 was considered significant). On the media used, only the development of rhizosphere populations quantified during the growing season on 2% malt (total filamentous fungi) and Komada's medium (mainly Fusarium spp.) for GMM 8-treated plants was significantly affected (P = 0.05) compared to the control treatment (Fig. 3). The development of fungal populations in the GMM 8-treated plants seemed to be different from that for the other treatments only in the beginning of the season. Although the effects are minor, it seems that GMM 8 exerts a transient suppressive effect on certain fungal populations in the rhizosphere. This effect was only observed in the beginning of the season of 1997 and only on Komada's medium and malt medium. In 1998 none of the fungal populations quantified on the different agar media was significantly affected by the introduction of the GMMs (data not shown).
FIG. 3.
Population dynamics of culturable fungi in rhizospheres of wheat plants treated with either P. putida WCS358r (●) or its phenazine-producing derivative GMM 2 (○) or GMM 8 (□) and in rhizospheres of control plants (■) during the field trial of 1997. Fungal propagules in rhizosphere samples were quantified on 2% malt medium and Komada's medium. For both media, fungal population dynamics of GMM 8-treated plants differ in time from fungal population dynamics of all other treatments (interaction time × treatment, P = 0.0001).
Effects on the composition of indigenous fungal microflora.
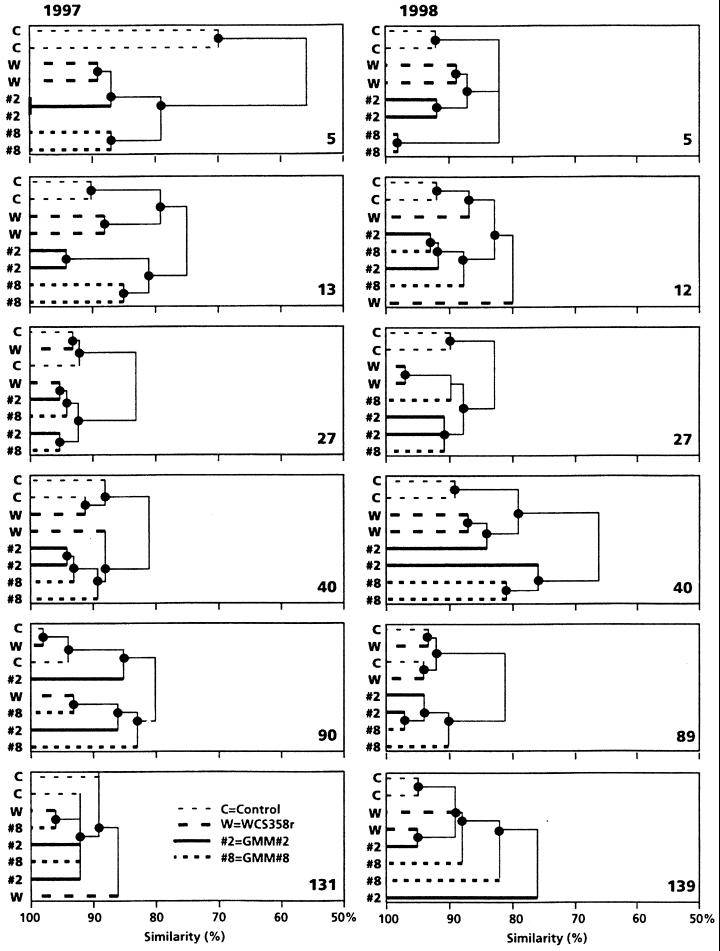
Application of WCS358r or either of the PCA-producing GMMs to seeds caused a shift in the fungal population of wheat roots, as indicated by cluster analysis of replicate ARDRA-generated profiles of rhizosphere samples (Fig. 4). Treatments are considered to be different if both the replicate ARDRA patterns for one treatment cluster together, apart from patterns of other treatments. In this case the replicate ARDRA patterns for each treatment are more similar to each other than to other patterns. For both years similarity indexes were calculated from the dendrograms. No significant interaction between the year of the experiment and the treatments were observed, allowing us to pool the data of the two experiments. Regression analysis of the similarity indexes demonstrated significant differences between the treatments when they were analyzed over a period as long as 40 days. After this time there were no significant differences between the treatments. Effects on the fungal microflora as a result of bacterization with WCS358r or the GMMs seemed differential, since the ARDRA profiles from the GMM-treated samples clustered separately from those from the WCS358r-treated samples and those from the control treatment. In addition, GMM 2 and 8 appeared to differ in their initial effects on fungal microflora composition (days 5 and 13 in 1997 and day 5 in 1998), as the ARDRA profiles of the two GMMs were not similar during this initial period. GMM-induced effects tended to last longer than WCS358r effects, as demonstrated by the prolonged clustering of profiles from the GMM-treated plants. On the other hand, replicates of the WCS358r treatment clustered together early in the season, but this clustering disappeared as the season proceeded. Effects of the GMMs could be observed up to 40 days (1997) and 89 days (1998) after sowing, whereas WCS358r-induced effects were detectable up to 12 and 40 days, respectively. In both years all treatments cluster together 1 month after harvest, indicating that the effects induced by the bacterial treatments were transient.
FIG. 4.
Effects of P. putida WCS358r (W) and its phenazine-producing derivatives GMM 2 (#2), and GMM 8 (#8) on the composition of the fungal rhizosphere community during the field trials of 1997 and 1998 as determined by ARDRA. C, control. Dendrograms represent percent similarity of fungal communities of roots of wheat plants grown from bacterized seeds (107 CFU/seed) during the growing seasons of 1997 and 1998. Samples were taken 5 to 139 days (as indicated to the right of each dendrogram) after sowing of bacterized seeds. Similarities are based on ARDRA patterns generated from TaqI-digested, amplified 18S rDNA obtained from wheat rhizosphere DNA extracts. Two independent replicates per treatment were used, each representing a pooled sample of three field plots.
Effects on soil microbial activities.
In 1997 values obtained for SIR, NPA, and cellulose decomposition fluctuated throughout the growing season for all treatments (data not shown). There were no statistically significant effects of the introduction of either WCS358r or the GMMs on these microbial activities.
Effects on plant growth.
The introduction of WCS358r and the GMMs had no effect on plant height, plant fresh weight, or plant dry weight (data not shown) in either 1997 or 1998. Kernel weight (100-seed weight) of plants at harvest did not differ between treatments and ranged from 3.9 g (±0.1) to 4.3 g (±0.1) in 1997 and from 3.8 g (±0.3) to 3.9 g (±0.3) in 1998.
DISCUSSION
We introduced the ability to produce PCA into a plant growth-promoting bacterial strain, P. putida WCS358r, using the mini-Tn5 transposon system as a delivery vector. When the GMMs were introduced into the field, no effects of the genetic modification on population densities of the two selected GMMs were observed in the wheat rhizosphere in 1997, indicating that the extra metabolic load did not affect the ecological fitness of these PCA-producing derivatives. In 1998, however, cell numbers of the GMMs dropped during the first 5 days after sowing in comparison to numbers of the parental strain, WCS358r, and these differences were maintained throughout the growing season. The weather conditions, rainfall (in millimeters), and mean temperature (in degrees Centigrade), during these first 5 days in 1998 were different (14.8 mm and 7.6°C) from those in 1997 (4.7 mm and 9.5°C) (Dutch Meteorological Institute [KNMI], de Bilt, The Netherlands). De Leij et al. (9) also found indications that changes in the ecological fitness of genetically modified variants of P. fluorescens in soil depend upon the environmental conditions that the GMMs encounter. Mazzola et al. (22) demonstrated that the production of PCA contributes to the ecological fitness of P. fluorescens 2-79. The PCA-producing WCS358r strains, however, do not appear to be more fit then their PCA− parent.
The introduction of GMM 8 resulted in a relatively minor and transient effect on the number of fungi that grew on malt medium and Komada's medium. These findings suggest that PCA production by GMM 8 can suppress some culturable fungi. The lack of effect of the introduction of GMM 2 on numbers of culturable fungi may be explained by its lower level of PCA production, as determined in vitro (16).
Both introduction of WCS358r and introduction of the GMMs resulted in a transient effect on the composition of the rhizosphere fungal microflora, as determined by 18S rDNA analysis. The distinct effects of WCS358r and the GMMs were most prominent at the beginning of the field trials in both 1997 and 1998 (Fig. 4), when the numbers of bacteria introduced were relatively high. The WCS358r-induced effect on the fungal microflora is probably caused by the production of pseudobactin 358, the fluorescent siderophore of WCS358 (4). Siderophore production by WCS358r is a prerequisite for suppression of fusarium wilt in carnations and radishes by this strain (11, 20, 27). The GMM-induced impact on the composition of the fungal microflora lasted longer than the WCS358r-induced impact. The observation that the GMM-induced shift in the fungal microflora was longer-lasting and differed qualitatively from the shift caused by the parental strain probably indicates that the PCA produced by the GMMs also affected the composition of the fungal microflora. The detection of PCA in the rhizospheres of GMM-treated plants and not in rhizosphere samples of WCS358-treated plants or of control plants supports the role of PCA in these shifts in the fungal microflora.
Individual species involved in the observed shift in composition of the fungal microflora have not yet been identified. Our future research will focus on identification of fungal species that are affected by the introduced GMMs by sequencing the discriminatory amplified 18S rDNA fragments after their separation by TGGE or DGGE and by analyzing the cloned 18S rDNA sequences as described by Smit et al. (32).
We found that introduction of PCA-producing GMMs can transiently affect the composition of the rhizosphere fungal microflora of field-grown wheat. This field experiment was set up at the request of the Coordination Commission for Risk Assessment in the Netherlands (CCRO), and a permit for this small-scale field release was granted by the Ministry of Housing, Spatial Planning, and the Environment. For future studies this field release has provided the development of tools to detect effects on microbial communities. Comparative studies of the impact of common agricultural practices, such as plowing or crop rotation, on microbial communities are needed to evaluate the importance of the transient shifts we observed. We expect that the impact of introduced (genetically modified) microorganisms is only minor compared to that of these practices. Further research also will focus on the nature of the induced effects and on the question of whether repeated subsequent applications of GMMs in the same field intensifly the observed GMM-induced effects.
ACKNOWLEDGMENTS
This study was initiated by the CCRO, which is financed by the Dutch Ministry of Economic Affairs.
We thank W. Gams of the Central Bureau of Fungal Cultures (CBS), Baarn, The Netherlands, for identification of soil fungi, R. van Logtenstein (Section of Landscape Ecology, Utrecht University) for help with measuring soil microbial activities, and Bas Valstar, Jeroen van Schaik, and Fred Siesling (Botanical Gardens, Utrecht University) for construction of the experimental field and care of the field site.
REFERENCES
- 1.Aerts R, Toet S. Nutritional controls on carbon dioxide and methane emission from Carex-dominated peat soils. Soil Biol Biochem. 1997;29:1683–1690. [Google Scholar]
- 2.Anderson J P E, Domsch K H. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol Biochem. 1978;10:215–221. [Google Scholar]
- 3.Austin H K, Hartel P G, Coleman D C. Effects of genetically altered Pseudomonas solanacearum on predatory protozoa. Soil Biol Biochem. 1990;22:115–117. [Google Scholar]
- 4.Bakker P A H M, Lamers J G, Bakker A W, Marugg J D, Weisbeek P J, Schippers B. The role of siderophores in potato tuber yield increase by Pseudomonas putida in a short rotation of potato. Neth J Plant Pathol. 1986;92:249–256. [Google Scholar]
- 5.Bollen G J. A comparison of the in vitro antifungal spectra of thiophanates and benomyl. Neth J Plant Pathol. 1972;78:55–64. [Google Scholar]
- 6.Bonsall R F, Weller D M, Thomashow L S. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl Environ Microbiol. 1997;63:951–955. doi: 10.1128/aem.63.3.951-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook R J, Baker K F. The nature and practice of biological control of plant pathogens. St. Paul, Minn: The American Phytopathological Society; 1983. [Google Scholar]
- 8.De Leij F A A M, Sutton E J, Whipps J M, Fenlon J S, Lynch J M. Impact of field release of a genetically modified Pseudomonas fluorescens on indigenous microbial populations of wheat. Appl Environ Microbiol. 1995;61:3443–3453. doi: 10.1128/aem.61.9.3443-3453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Leij F A A M, Thomas C E, Bailey M J, Whipps J M, Lynch J M. Effect of insertion site and metabolic load on the environmental fitness of a genetically modified Pseudomonas fluorescens isolate. Appl Environ Microbiol. 1998;64:2634–2638. doi: 10.1128/aem.64.7.2634-2638.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative Eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duijff B J, Bakker P A H M, Schippers B. Suppression of Fusarium wilt of carnation by Pseudomonas putida WCS358 at different levels of disease incidence and iron availability. Biocontrol Sci Technol. 1994;4:279–288. [Google Scholar]
- 12.Engelen B, Meinken K, von Wintzintgerode F, Heuer H, Malkomes H P, Backhaus H. Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl Environ Microbiol. 1998;64:2814–2821. doi: 10.1128/aem.64.8.2814-2821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fantroussi E S, Verschuere L, Verstraete W, Top E M. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol. 1999;65:982–988. doi: 10.1128/aem.65.3.982-988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Ferguson A. Biochemical systematics and evolution. Glasgow, United Kingdom: Blackie; 1980. [Google Scholar]
- 14.Gams W, van Laar W. The use of Solacol® (validamycin) as a growth retardant in the isolation of soil fungi. Neth J Plant Pathol. 1982;88:39–45. [Google Scholar]
- 15.Glandorf D C M, Brand I, Bakker P A H M, Schippers B. Stability of rifampin resistance as a marker for root colonization studies of Pseudomonas putida in the field. Plant Soil. 1992;147:135–142. [Google Scholar]
- 16.Glandorf D C M, Thomashow L S, Smit E, Wernars K, Bakker P A H M, van Loon L C. Field release of genetically-modified Pseudomonas putida WCS358r to study effects on the indigenous soil microflora: preliminary experiments. In: Ogoshi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S, editors. Plant growth-promoting rhizobacteria—present status and future prospects. Sapporo, Japan: Faculty of Agriculture, Hokkaido University; 1997. pp. 428–430. [Google Scholar]
- 17.Harrison A F, Latter P M, Walton D W H. Cotton strip assay: an index of decomposition in soils. ITE Symposium no. 24. Cumbria, United Kingdom: Institute of Terrestrial Ecology; 1988. [Google Scholar]
- 18.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 19.Komada H. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soils. Rev Plant Prot Res. 1975;8:114–125. [Google Scholar]
- 19a.Kowalchuk G A, Gerards S, Woldendorp J. Detection and characterization of fungal infections of Ammophila arenaria (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl Environ Microbiol. 1997;63:3858–3865. doi: 10.1128/aem.63.10.3858-3865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leeman M, van Pelt J A, Hendrickx M J, Scheffer R J, Bakker P A H M, Schippers B. Biocontrol of Fusarium wilt of radish in commercial greenhouse trials by seed treatment with Pseudomonas fluorescens WCS374. Phytopathology. 1995;85:1301–1305. [Google Scholar]
- 21.Mavrodi D V, Ksenzenko V N, Bonsall R F, Cook R J, Boronin A M, Thomashow L S. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzola M, Cook R J, Thomashow L S, Weller D M, Pierson L S. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 24.Naseby D C, Lynch J M. Impact of wild type and genetically-modified Pseudomonas fluorescens on soil enzyme activities and microbial population structure in the rhizosphere of pea. Mol Ecol. 1998;7:617–625. [Google Scholar]
- 25.Nash S M, Snyder W C. Quantitative estimations by plate counts of propagules of the bean root rot Fusarium in field soils. Phytopathology. 1962;52:567–572. [Google Scholar]
- 26.Natch A, Keel C, Hebecker N, Laasik E, Défago G. Influence of the biocontrol strain Pseudomonas fluorescens CHAO and its antibiotic overproducing derivative on the diversity of resident root colonising pseudomonads. FEMS Microbiol Ecol. 1997;23:341–352. [Google Scholar]
- 27.Raaijmakers J M, Leeman M, Van Oorschot M M P, van der Sluis I, Schippers B, Bakker P A H M. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology. 1995;85:1075–1081. [Google Scholar]
- 28.Raaijmakers J M, Weller D M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant-Microbe Interact. 1998;11:144–152. [Google Scholar]
- 29.Robleto E A, Borneman J, Triplett E W. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl Environ Microbiol. 1998;64:5020–5022. doi: 10.1128/aem.64.12.5020-5022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Short K A, Seidler R J, Olsen R H. Survival and degradative capacity of Pseudomonas putida induced or constitutively expressing plasmid-mediated degradation of 2,4-dichlorophenoxyacetate (TFD) in soil. Can J Microbiol. 1990;36:821–826. [Google Scholar]
- 31.Smit E, Leeflang P, Wernars K. Detection of shifts in microbial community structure and diversity in soil caused by copper contamination using amplified ribosomal DNA restriction analysis. FEMS Microbiol Ecol. 1997;23:249–261. [Google Scholar]
- 32.Smit E, Leeflang P, Glandorf B, van Elsas J D, Wernars K. Analysis of fungal diversity in soil by sequencing of cloned PCR-amplified 18S rDNA and temperature gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2614–2621. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stienstra A W, Klein Gunnewiek P, Laanbroek H J. Repression of nitrification in soils under a climax grassland vegetation. FEMS Microbiol Ecol. 1994;14:45–52. [Google Scholar]
- 34.Thomashow L S, Weller D M. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomashow L S, Weller D M, Bonsall R F, Pierson L S. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56:908–912. doi: 10.1128/aem.56.4.908-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiedje J M, Colwell R K, Grossman Y L, Hodson R E, Lenski R E, Mack R E, Regal P J. The planned introduction of genetically-modified organisms: ecological considerations and recommendations. Ecology. 1989;70:298–315. [Google Scholar]
- 37.Torsvik V, Daae F L, Sandaa R A, Ovreas L. Novel techniques for analyzing microbial diversity in natural and perturbed environments. J Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z M, Crawford D L, Magnuson T S, Bleakley B H, Hertel G. Effects of bacterial lignin peroxidase on organic-carbon mineralization in soil, using recombinant Streptomyces strains. Can J Microbiol. 1991;37:287–294. [Google Scholar]