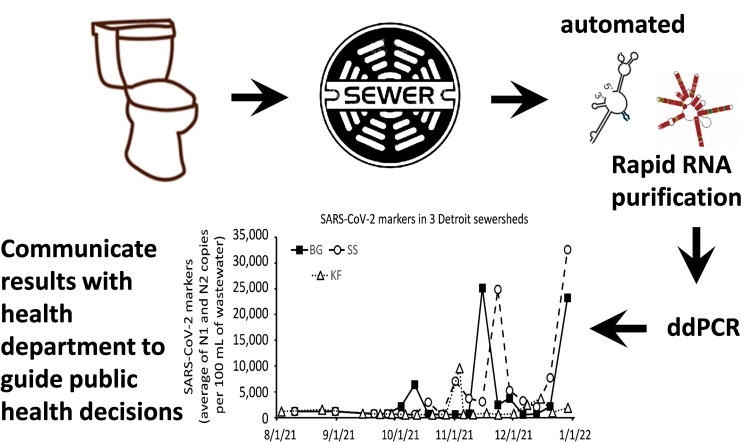
Abstract
Wastewater based epidemiology (WBE) has emerged as a strategy to identify, locate, and manage outbreaks of COVID-19, and thereby possibly prevent surges in cases, which overwhelm local to global health care networks. The WBE process is based on assaying municipal wastewater for molecular markers of the SARS-CoV-2 virus. Standard processes for purifying viral RNA from municipal wastewater are often time-consuming and require the handling of large quantities of wastewater, negatively affecting throughput, timely reporting, and safety. We demonstrate here an automated, faster system to purify viral RNA from smaller volumes of wastewater but with increased sensitivity for detection of SARS-CoV-2 markers. We document the effectiveness of this new approach by way of comparison to the PEG/NaCl/Qiagen method prescribed by the State of Michigan for SARS-CoV-2 wastewater monitoring and show its application to several Detroit sewersheds. Specifically, compared to the PEG/NaCl/Qiagen method, viral RNA purification using the PerkinElmer Chemagic™ 360 lowered handling time, decreased the amount of wastewater required by ten-fold, increased the amount of RNA isolated per μl of final elution product by approximately five-fold, and effectively removed ddPCR inhibitors from most sewershed samples. For detection of markers on the borderline of viral detectability, we found that use of the Chemagic™ 360 enabled the measurement of viral markers in a significant number of samples for which the result with the PEG/NaCl/Qiagen method was below the level of detectability. The improvement in detectability of the viral markers might be particularly important for early warning to public health authorities at the beginning of an outbreak. Applied to sewersheds in Detroit, the technique enabled more sensitive detection of SARS-CoV-2 markers with good correlation between wastewater signals and COVID-19 cases in the sewersheds. We also discuss advantages and disadvantages of several automated RNA purification systems, made by Promega, PerkinElmer, and ThermoFisher.
Keywords: Automated, COVID-19, Detroit, Wastewater, RNA purification, Viral RNA
Graphical abstract
1. Introduction
The rapid global spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the RNA virus that causes coronavirus disease 2019 (COVID-19), has put enormous strain on healthcare institutions and on epidemiological efforts to monitor, track, and predict the spread and evolution of SARS-CoV-2. Early knowledge of the sequences of the SARS-CoV-2 virus (Wu et al., 2020) enabled molecular tests for the presence of the virus to be developed for its presence in both clinical samples and in virus shed by infected individuals into wastewater. Community-wide monitoring efforts in the United States were hampered initially by applying tests only to symptomatic individuals who had traveled from China and by lack of availability of tests (Bendix, 2020; CDC Health Alert Network, 2020). However, even with tests subsequently made more widely available, resistance or low response rates to requests to be tested have occurred in various settings, including college campuses (Gibas et al., 2021). The testing of only symptomatic individuals was especially problematic for controlling the disease because after infections a delay occurs between capability to spread the virus and the occurrence of symptoms. Furthermore, some individuals remain asymptomatic even with observed viral RNA quantities similar to people presenting symptoms (Arons et al., 2020; Oran and Topol, 2021).
For assessing community levels of COVID-19 disease, detection of the virus in wastewater can be a successful strategy. Molecular markers for SARS-CoV-2 virus are shed in feces early in infection and have been observed in raw wastewater collected from locations where infected people are present (Ahmed et al., 2020, Ahmed et al., 2020). Wastewater monitoring has been used to surveil diseases and other public health relevant markers in the past (Castiglioni et al., 2006; Heijnen and Medema, 2011; Brouwer et al., 2018; Xiao et al., 2019). Accordingly, wastewater based epidemiology of SARS-CoV-2 has been underway in multiple locations (Naughton, 2020; Rose et al., 2021). The State of Michigan funded 19 health department and university laboratories across the state to implement wastewater analysis of SARS-CoV-2 for the purposes of providing early warning signs of COVID-19 infections in various communities across the state (State of Michigan, 2021).
Numerous reports have confirmed the value of detecting SARS-CoV-2 markers in wastewater for public health purposes. Researchers have found that wastewater measurements of SARS-CoV-2 markers provide population-level insights including data about disease prevalence and predicted symptomatic cases and hospitalizations (Panchal et al., 2021; Galani et al., 2022). Wastewater monitoring detected SARS-CoV-2 six to eight days before positive tests were reported in New Haven, Connecticut, USA (Peccia et al., 2020), 12–16 days before COVID-19 cases were declared in four municipalities in Spain (Randazzo et al., 2020) and before the exponential growth of the epidemic in Paris, France (Wurtzer et al., 2020). A further advantage of measuring SARS-CoV-2 markers in wastewater as a way of assessing the presence of COVID-19 in the community is that such tests can be accomplished with a comparatively small burden on local healthcare resources compared to frequent and intensive testing of multiple individuals.
In order to successfully detect viral particles from large volumes of wastewater from hundreds to thousands of individuals, the viral RNA in the wastewater samples must first be concentrated and purified. The two steps are usually accomplished sequentially and listed in separate columns of methods review tables (see, for example, Ali et al. (2021) and Alygizakis et al. (2021)). Among previously used methods for concentrating RNA viruses from wastewater are precipitation with polyethylene glycol (PEG) 8000 and high salt followed by centrifugation (Ye et al., 2016; Borchardt et al., 2017), ultracentrifugation (Ye et al., 2016), ultrafiltration (Borchardt et al., 2017; Medema et al., 2020), adsorption-extraction on electronegative membranes (Ahmed et al., 2020, Ahmed et al., 2020; Ahmed et al., 2021), and dead-end filtration on InnovaPrep's Concentrating Pipette device (Ahmed et al., 2021; McMinn et al., 2021). A recent study by Flood et al. (2021) compared the PEG, ultracentrifugation, and ultrafiltration methods using spiked-in Pseudomonas phage Phi6 to compare efficiency of these methods in concentrating enveloped RNA viruses from wastewater. The PEG method significantly increased recovery of Phi6, yielding between 2-times and up to 13-times more Phi6 detected in PEG concentrate than the in the concentrate isolated by ultracentrifugation or ultrafiltration methods. However, as noted above, concentration is usually followed by a subsequent purification step. For example, in the study by Flood et al. (2021), RNA from wastewater was purified from the concentrate on silica-based spin columns (QIAmp Viral RNA Minikit by Qiagen). Other purification methods have been based on binding and selective elution from magnetic silica beads (Biomerieux Nuclisens kit), as implemented by Medema et al. (2020) and many others, as reviewed by Ali et al. (2021).
These approaches, (concentration first, then purification), are time-consuming and may be equipment-intensive. For example, the PEG/NaCl/Qiagen protocol recommended for use in the State of Michigan is based on the method described by Flood et al. (2021) and comprises several hours precipitation time and centrifugation of large volumes (100 mL) of wastewater prior to removal of supernatant, transfer of pellets that may vary in size and consistency, and ultimately running the concentrated extract through Qiagen columns. This may also allow PCR inhibitors to accompany the final product resulting in varying analytical efficiency (Monteiro et al., 2022). In the present study, we investigated whether viral RNA could be obtained from the sewershed by adapting an automated method with novel manipulations. This combined concentration and purification of RNA in a single automated, viral RNA concentration and purification process. We describe here our success in achieving improved recovery of RNA with low levels of PCR inhibitors based upon adapting a rapid automated method on the PerkinElmer Chemagic™ 360 platform to our process. We further apply the system to the detection and SARS-CoV-2 in sewersheds in Detroit, MI.
2. Material and methods
2.1. Collection of wastewater
Wastewater was collected once a week from 20 sites in Detroit, MI during most of this study (exceptions occurred during the early weeks of the study). Access to the sewer system at each site was by manhole, with many being located in local roads. One 250 mL grab sample was collected in a 250 mL or 500 ml high density polyethylene bottle from each site between 7 am and 9:30 am at approximately the same time each week. At least one out of every 10 samples was collected as a “field blank” of deionized water transferred in the field into a high-density polyethylene bottle identical to those used for wastewater samples. Sample bottles were placed in a Ziplock bag with a sheet of paper towel and stored on ice at 4–6 °C in coolers until delivery to the laboratory by 10:15 a.m.
Chain-of-custody (CoC) forms were used to document sample collection information (location, date-time of sample collection, sample type [grab vs. composite]) and to record the transfer of sample custody from the field sampling crew to the laboratory. Physical parameter data, including pH, temperature, dissolved oxygen, and specific conductance, were also measured at each sampling location on each day of sampling using a YSI ProDSS with GPS (YSI, Yellow Springs, OH). The data is collected on a physical field sheet that is transferred to the lab after collection. A digital record of each CoC and each YSI data sheet has been preserved and archived with the project records. Weekly numbers of positive COVID-19 tests among residents and staff of long-term care facilities (LTC) in some of the sewersheds was obtained from public data on a Michigan COVID-19 dashboard ((Michigan.gov, 2022), https://www.michigan.gov/coronavirus/0,9753,7-406-98163_98173-526,911--,00.html.
2.2. PEG/NaCl/Qiagen protocol to assay wastewater
This study directly compared the Chemagic™ 360 method to the PEG/NaCl/Qiagen method, as described by Flood et al. (2021) and in protocols provided by the State of Michigan. The PEG/NaCl/Qiagen method described here is required for use by Michigan Department of Health and Human Services (MDHHS)-funded SARS-CoV-2 monitoring laboratories, with exceptions allowed (as with our laboratory) where justified by demonstrated improvements over the state protocol method. The PEG/NaCl/Qiagen method, described in more detail by Flood et al. (2021), is briefly described as follows: 100 mL of wastewater was added to 8 g PEG and 1.7 g NaCl, held at 4 °C for 2 h with continuous agitation, then centrifuged at 4696 ×g at 4 °C for 45 min. Most of the supernatant was removed, leaving a soft pellet of 2 to 4 mL volume (depending on the sample quality and non-fecal matter). RNA in 200 μl of the resuspended pellet was then purified using Qiagen QIAmp Viral RNA mini kit, yielding 80 μL of purified RNA.
Phi6 virus is used as a “spiked-in” internal standard, required in the state protocol, for assessing RNA recovery and potential PCR inhibition. Phi6 virus was added either (a) to all wastewater and field blank controls prior to processing (100 μL of 106 PFU/mL added per 100 mL of the sample, prior to the addition of PEG and NaCl) to test overall recovery and inhibition or (b) to the 200 μl pellet aliquot (10 μL of 105 PFU/mL) of all wastewater and field blank samples prior to purification on the Qiagen column. The higher concentration of the Phi6 spike when put directly into the wastewater is to compensate for losses expected to occur in the precipitation step in comparison to adding the spike directly to the pellet.
2.3. Chemagic™ 360 adapted protocol
We adapted a Chemagic™ 360 instrument (catalog number, 2024-0020) with the 12-rod head (CMG-371, PerkinElmer Health Sciences Inc., Shelton, CT, USA) to process 10 mL wastewater samples without prior concentration. Previous publications have described using the Chemagic™ instrument with a 96-rod head for processing 1 mL samples, for which wastewater RNA must first be concentrated from a larger volume by a time-consuming concentration step such as PEG/NaCl (Laturner et al., 2021) or ultracentrifugation (Hokajärvi et al., 2021). Here we describe application of the Chemagic™ instrument to concentrate and purify PCR-ready viral RNA from 10 mL wastewater samples in a single integrated Chemagic™-based procedure without a preceding concentration step required. The method is described in detail in a published protocol (Vasquez et al., 2021) and briefly here: Prior to placing samples on the automated instrument, 45 mL of the wastewater sample is centrifuged at 4696 ×g for 15 min; 10 mL of the supernatant is transferred and mixed into a tube containing Poly A RNA (7 μL PerkinElmer CMG842), Proteinase K (50 μL PerkinElmer CMG749), and lysis buffer 1 (8 mL PerkinElmer CMG749); incubated for 30 min at 55 °C; and then magnetic beads (50 μL PerkinElmer CMG749) are added and mixed. After placement of the incubated sample into the Chemagic™ 360 instrument and a set of receiving tubes (4 mL Sarstedt®, containing 100 μL elution buffer CMG749), the instrument is run with protocol chemagic™Viral10k 360 H12 prefilling drying VD210119.che for 75 min. The product, in the Sarstedt® tube in a final volume of ~85 μL elution buffer, is then transferred to a 1.5 mL Lo-Bind centrifuge tube for subsequent analysis or long-term storage at −80 °C. Depending on the purpose of the experiment, Phi6 virus was added as an internal standard either to the 10 mL of wastewater (10 μL of 106 PFU/mL added to 10 mL wastewater) after the initial centrifugation or, alternatively, to the eluted sample (10 μL of 106 PFU/mL to the eluate) or to elution buffer that had not undergone processing (reference positive control; 10 μL 106 PFU/mL added to 75 μL elution buffer).
2.4. ddPCR analysis
Primers and TaqMan® probes were designed to amplify and detect nucleocapsid gene markers 1 and 2 (N1 and N2) of SARS-CoV-2 and the P8 nucleocapsid gene of bacteriophage Phi6. The primer and probe sequences used in this study, including citations, are shown in Table 1 .
Table 1.
Primer and probe sequences used for ddPCR.
| Target | Primer/probe | Sequence | Reference |
|---|---|---|---|
| SARS-CoV-2 | 2019-nCoV_N1-F | 5′-GACCCCAAAATCAGCGAAAT-3′ | Lu et al. (2020) |
| 2019-nCoV_N1-R | 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | ||
| 2019-nCoV_N1-P | 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1–3′ | ||
| 2019-nCoV_N2-F | 5′-TTACAAACATTGGCCGCAAA-3′ | ||
| 2019-nCoV_N2-R | 5′-GCGCGACATTCCGAAGAA-3′ | ||
| 2019-nCoV_N2-P | 5′-HEX-ACAATTTGCCCCCAGCGCTTCAG-BHQ1-3′ | ||
| Phi6 | Phi6-F | 5′-TGGCGGCGGTCAAGAGC-3′ | Gendron et al. (2010) |
| Phi6-R | 5′-GGATGATTCTCCAGAAGCTGCTG-3′ | ||
| Phi6-P | 5′-6FAM-TCCGCCTGGCACGGTACTCCCT-BHQ1-3′ |
Reactions are assembled in a PCR hood using Bio-Rad's One-step RT-ddPCR Advanced Kit for Probes and their suggested protocol (Bio-Rad, CA, USA). Briefly, a 16.5 μL reaction is prepared to contain a final concentration of 1 x Supermix, 20 U/μL reverse transcriptase, 15 mM DTT, 900 nmol/μL of gene target primers, 250 nmol/μL of gene target probe, and 1.1 μL RNAse-free water. TaqMan® probes are labeled with different fluorescent markers enabling primers and probes for both N1 and N2 to be included in the same reaction with duplex detection. To this reaction 5.5 μL of template nucleic acids purified either by Chemagic™ or PEG/NaCl/Qiagen is added for a final reaction mix of 22 μL, of which 20 μL is processed for each ddPCR reaction. ddPCR reactions for each sampling site and quality controls (positive, no-template, extraction, and processing controls) are prepared in triplicate and loaded onto a 96-well PCR plate. The plate is sealed using the Bio Rad PX1 PCR Plate Sealer at 180 °C for 5 s. The plate is vortexed thoroughly and centrifuged for 1 min at 850 ×g on a microplate centrifuge (Thermo Fisher Scientific, MA, USA) and then transferred to the QX200 Automated Droplet Generator (Bio-Rad, CA, USA). Upon completion of droplet generation according to the manufacturer's instruction, the 96-well plate containing droplets is removed and sealed with the PX1 PCR Plate Sealer at 180 °C for 5 s.
The plate is then transferred to the Bio-Rad C1000 Thermo Cycler and PCR is initiated with the following settings: Hold 25 °C for 3 min., reverse transcription 50 °C for 60 min., enzyme activation 95 °C for 10 min, denaturation 95 °C for 30 s., annealing/extension 55 °C for 1 min, denaturation and annealing/extension steps cycled 40 times, enzyme deactivation 98 °C for 10 min, and hold 4 °C until ready for droplet reading. The 96-well plate is transferred to the Bio-Rad QX200 Droplet Reader which is prepared for analysis by using the Bio-Rad QuantaSoft software package version 1.7.4.0917. Once droplet reading is complete and signal threshold values entered for each reaction the resulting file is exported in comma separated value (.csv) format for subsequent analysis.
Results were calculated in terms of # of copies of PCR targets per 100 mL of the original wastewater (or field blank) sample as described by Flood et al. (2021) for PEG/NaCl/Qiagen. Similarly, adjustment of various volumes in the calculation enabled a comparable calculation of SARS-CoV-2 per 100 mL wastewater for samples processed here by Chemagic™ 360.
Statistics and graphics mainly used the Data Analysis Tools of Excel (Microsoft Office Professional Plus 2019, Excel Version 1808 Build 10,387.20023), supplemented by various on-line calculators using https://mycurvefit.com/ for additional curve-fitting analysis and https://www.statskingdom.com/shapiro-wilk-test-calculator.html for the Shapiro-Wilks normality test and outlier identification.
3. Results
3.1. Recovery of spiked-in Phi6
3.1.1. Amount of spiked-in Phi6 purified from field blank compared to eluate blank
To assess the ability to recover a known amount of RNA, without any interfering factors from wastewater, the recovery of bacteriophage Phi6 RNA was compared by the amount of ddPCR copies detected from Phi6 spiked into 10 mL of pure water (field blank control) to the same amount of Phi6 spiked directly into elution buffer (eluate control) similar in volume to the final eluate from the Chemagic™ process. For sixteen pairs of spiked-in field blank and eluate controls analyzed between 15 November 2021 and 29 December 2021, the Phi6 measured by ddPCR from the pure water field blank averaged 70 ± 9 % (mean ± SD, n = 16) of the spiked eluate control.
3.1.2. Amount of spiked-in Phi6 recovered from wastewater varies from site to site
Over the same period during which the amount of Phi6 measured in the field blank was evaluated (15 November to 29 December 2021), we similarly spiked 10 mL wastewater samples with Phi6 and then purified RNA using the Chemagic™ process. The amount of spiked-in Phi6 measured in wastewater from 21 sewersheds, sampled on 7–10 occasions for each site, averaged 29 ± 20 % of the Phi6 eluate control, significantly less than the 70 ± 9 % that was measured in spiked-in field blank controls (p < 0.001, t-test). The amount of spiked-in Phi6 measured in wastewater samples averaged 39 ± 18 % of the spiked-in Phi6 measured in same day field blanks. For a subset of these wastewater samples, we compared the amount of spiked-in Phi6 measured by ddPCR when the RNA had been purified by the Chemagic™ process versus RNA from the same set of wastewater samples that had been purified by the PEG/NaCl/Qiagen protocol. Chemagic™ samples averaged 25 ± 15 % of the eluate blank (n = 10 wastewater samples); whereas PEG/NaCl/Qiagen samples averaged 6.5 ± 3.3 % of the comparable spiked-in Qiagen blank (p < 0.01, paired t-test, two tailed).
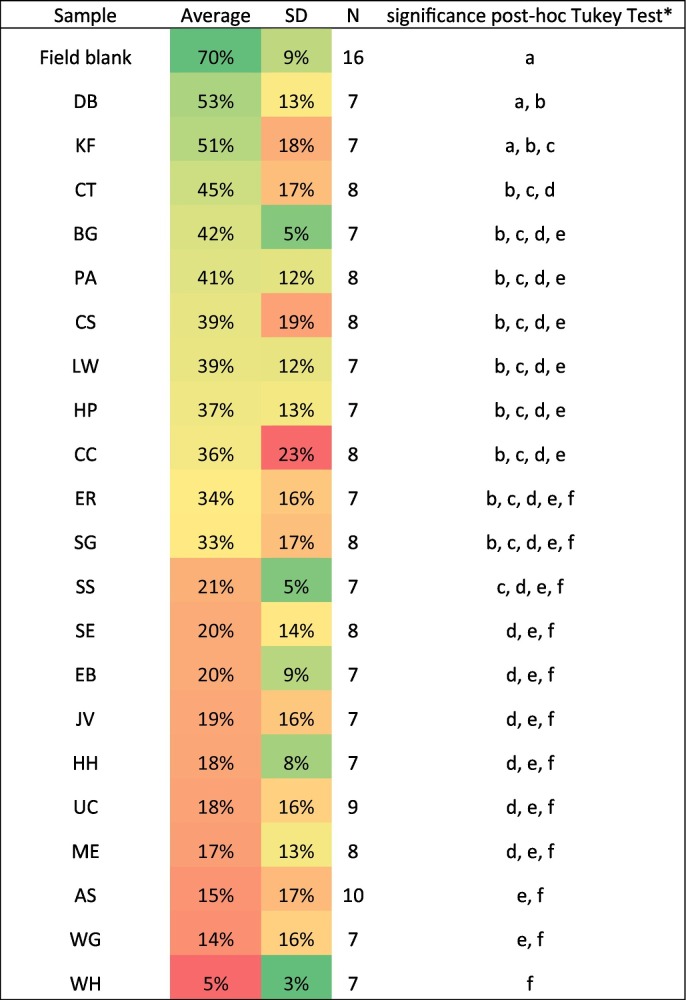
While the Chemagic™ process clearly enabled the measurement of more of the spiked-in Phi6 than the PEG/NaCl/Qiagen method, Table 2 shows that a significant amount of variation occurred in the amount of spiked-in Phi6 measured in wastewater samples collected from different locations (One way ANOVA, p < 0.0001). As percent of the eluate controls, the amount of spiked-in Phi6 in wastewater samples varied from as low as 5 ± 3 % for wastewater samples from site WH to as high as 53 ± 13 % for wastewater samples from site DB (significantly different, post-hoc Tukey, p < 0.05).
Table 2.
Phi6 spiked into wastewater purified by Chemagic™, as percent of eluate control.
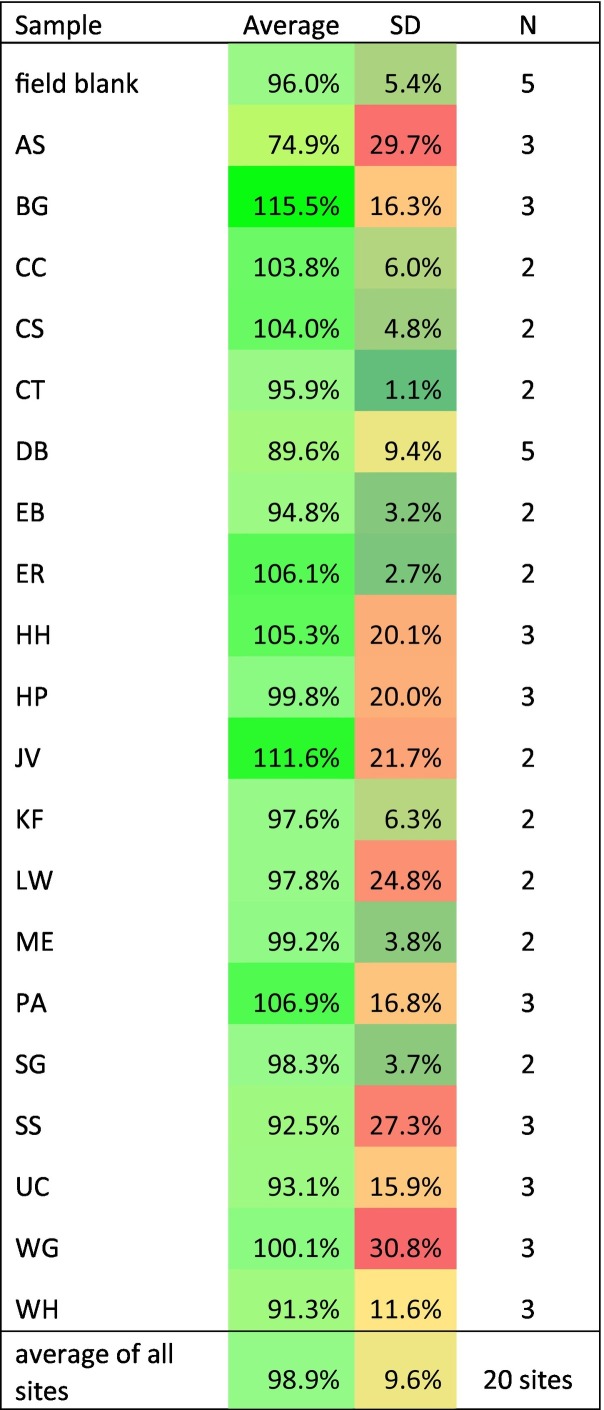
To determine if the variation between Phi6 measured from different sites might be due to varying amounts of PCR inhibitors present in the purified RNA, these results are compared to a set of experiments in which the Phi6 was spiked into all wastewater samples at the elution step and compared to the elution control for the same experiments (Table 3 ).
Table 3.
Phi6 spiked into Chemagic™ wastewater eluate as percent of eluate control.
This procedure was done for 52 wastewater samples and 5 field blanks, yielding an average amount of Phi6, compared to Phi6 spiked into the eluate control, of 98.9 ± 9.6 % for wastewater samples and 96.0 ± 5.4 % for the field blanks. This indicates that, PCR inhibition due to factors in the eluted RNA solution did not usually occur. The Shapiro-Wilks test for normality indicates a normal distribution of values (W = 0.956, p = 0.462), and that only site AS, exhibiting an average recovery of Phi6 of 74.9 %, is a low significant outlier and likely to be partially inhibited. Thus, except for samples from site AS, the differences in Phi6 measurements observed for the various sites when Phi6 was spiked into wastewater before Chemagic™ processing were predominantly due to differences in recovery of RNA in the purification process, rather than the presence of PCR inhibitors in the eluted RNA solutions.
3.2. Recovery and detection of SARS-CoV-2 markers
3.2.1. More sensitive detection of SARS-CoV-2 with Chemagic™, compared to PEG/NaCl/Qiagen
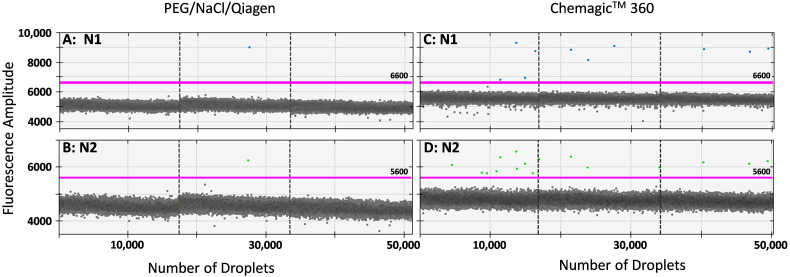
The performance of the Chemagic™ process for detection of SARS-CoV-2 markers in wastewater was compared to the PEG/NaCl/Qiagen protocol. On several occasions during August, October, and November 2021 wastewater samples from various sewersheds in Detroit were processed with both the Chemagic™ process and the PEG/NaCl/Qiagen protocol, spanning a period in which the markers were rarely detected by either method (August), only detected in about half the samples (October), and detected in most samples (November). Out of a total of 20 samples analyzed on the dates for which both processes were used, 9 samples occurred for which SARS-CoV-2 was detected in wastewater by one method and not the other. For all 9 samples, SARS-CoV-2 was detected in samples processed via the Chemagic™ process and not after processing by PEG/NaCl/Qiagen (significantly different, Fisher exact test, p < 0.0001). Fig. 1 illustrates a representative example of the greater number of positive droplets detected after Chemagic™ purification than after PEG/NaCl/Qiagen purification.
Fig. 1.
Representative example of detection of N1 and N2 from the same wastewater sample processed by Chemagic™ compared with PEG/NaCl/Qiagen, out of nine for which detection occurred with one method but not the other (all nine were positive for the Chemagic™ purified sample but not for PEG/NaCl/Qiagen; see text). ddPCR droplet results are shown for the same wastewater sample processed by PEG/NaCl/Qiagen (A: N1 and B: N2) and Chemagic™ (C: N1 and D: N2). All ddPCR results are technical triplicates with dashed vertical line separating the droplet results of the three reactions. Positive droplets are the points above the horizontal threshold line; the broad band of droplets below the threshold is negative droplets, approximately 17,000 per reaction. Bio-Rad considers three or more positive droplets to yield reliable Poisson statistics, which was achieved for this sample using Chemagic™ purification but not for PEG/NaCl/Qiagen.
3.2.2. Higher amount of SARS-CoV-2 measured in wastewater samples processed via Chemagic™ compared to the PEG/NaCl/Qiagen protocol
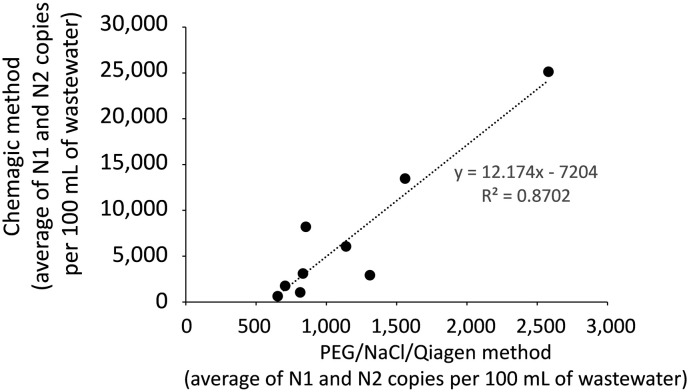
For wastewater samples processed by both PEG/NaCl/Qiagen and Chemagic™ protocols, the amounts of SARS-CoV-2 molecular markers that were measured by ddPCR was usually greater with the Chemagic™ process. In our data set, 10 sites processed by both protocols had detectable signals from at least 1 of the 2 gene markers. The N1 gene target was detected in all 10 samples processed via Chemagic™ but only 8 of the samples processed by PEG/NaCl/Qiagen. The N2 gene target was detected in all 10 samples processed by Chemagic™ while only 9 samples processed by PEG/NaCl/Qiagen were detectable. The quantity of viral copies per 100 mL of wastewater for all samples that were above the limit of detection for each method was correlated, as illustrated in the linear regression in Fig. 2 . In all but 2 samples the Chemagic™ purification produced more signal than its PEG/NaCl/Qiagen counterpart (significantly different for both markers: N1: Paired t-test, two-tailed p = 0.007; N2: Paired t-test, two-tailed p = 0.013). On average, the number of copies of SARS-CoV-2 detected per 100 mL in the Chemagic™-purified samples was 4.9 ± 3.4 times the number of copies measured in the PEG/NaCl/Qiagen-purified samples.
Fig. 2.
Comparison of the amounts of N1 and N2 markers measured in samples purified by the Chemagic™ process versus the PEG/NaCl/Qiagen method, for samples in which both processes yielded a detectable measurement. Note the much higher values on the Chemagic™ axis versus the PEG/NaCl/Qiagen axis.
3.3. SARS-CoV-2 markers in wastewater from three sites in Detroit
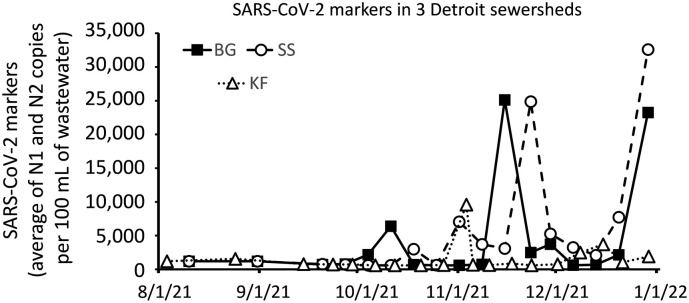
Here we illustrate application of these processes to analyze SARS-CoV-2 molecular markers in wastewater collected from three sites in Detroit from the second week of August through the last week of December. Initially, the PEG/NaCl/Qiagen protocol was used for sample processing and continued until the last week of September, after which samples were processed on the Chemagic™ platform. During August and September, the majority of samples had no detectable signal and this trend continued after switching to Chemagic™. Beginning in November we observed increases in samples from all three sites, with two of them rising simultaneously to a high level (and the third one moderately) on the last illustrated sampling day, the beginning of the “omicron surge.”
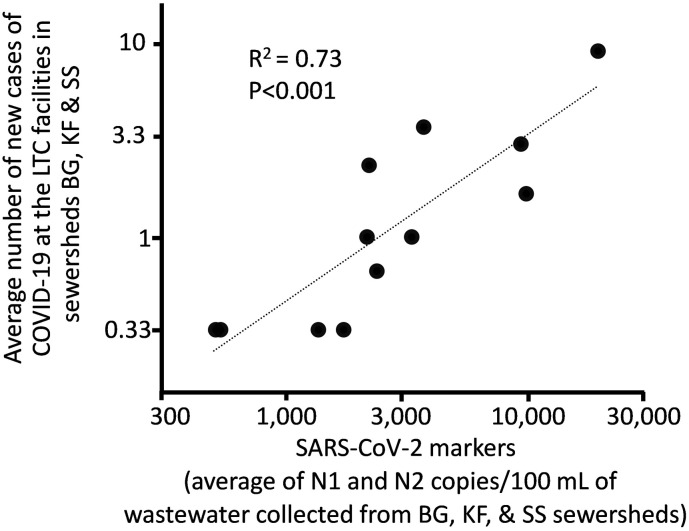
Each of the sewersheds for which the data are illustrated in Fig. 3 contained a long-term care facility (nursing home or similar) accounting for a part of the input to that wastewater stream. Comparing the levels of SARS-CoV-2 markers in wastewater in the three sewersheds to the number of COVID-19 cases (positive tests occurring the first time the same week that the sewage was collected) in the LTC facilities by linear regression yielded the significant relationship illustrated in Fig. 4 (R2 = 0.73, p < 0.001).
Fig. 3.
Levels of SARS-CoV-2 markers in wastewater collected from 3 sewersheds in Detroit located in southeast, central, and northwest areas of the City. These are small sewersheds with estimated populations ranging from 400 to 1000 people and therefore their exact locations are restricted. The number of people in the LTC facilities in these sewersheds are estimated to account for 20 % to 70 % of the people in the sewershed. On October 4, the method of viral RNA purification was changed from PEG/NaCl/Qiagen to Chemagic™ 360.
Fig. 4.
SARS-CoV-2 marker levels in sewershed wastewater compared to COVID-19 cases in long term care facilities in the sewersheds. Data points represent the average N1 and N2 marker levels for sewersheds BG, KF, and SS, the same sites described in the caption and illustrated in Fig. 3, versus the average of the staff and resident cases for the facilities as reported by Michigan.gov (2022). Data were transformed and analyzed on a logarithmic (base 10) scale and do not include weeks when no COVID-19 cases were reported for the facilities as log (0 cases) cannot be plotted.
4. Discussion
Application of an improved viral purification procedure based on the 12-Rod PerkinElmer Chemagic™ 360 platform enabled a more sensitive detection of SARS-CoV-2 in wastewater. Compared to the PEG/NaCl/Qiagen protocol described by Flood et al. (2021) and used by many laboratories in Michigan, the Chemagic™ system enabled a more sensitive detection of SARS-CoV-2 at the lowest detectable limit, as exemplified by 9 wastewater samples in which SARS-CoV-2 markers was detected after purification with Chemagic™ but not after purification by PEG/NaCl/Qiagen (Section 3.2.1).
The increased sensitivity of detection using the Chemagic system is evident in the measurement of N1 and N2 markers in wastewater from three small sewersheds in Detroit over a 5-month period, illustrated in Fig. 3. Most detections in these sewersheds occurred after October 4, 2022, when the RNA purification method was switched to the Chemagic automated system although that result also largely reflects the overall increase in COVID-19 prevalence in the community. Indeed, the N1, N2 marker measurements as a whole correlated well with “same week” COVID-19 case rates of nursing home facilities in the sewersheds (Fig. 4. Not shown: much worse R2 levels for these sewersheds when the comparison was made to case data in later weeks). While some reports (summarized recently by Shah et al. (2022)) have shown wastewater to provide an “early warning” of outbreaks a week or two later, a question for the future is whether rapid mitigation efforts taken in response to new infections detected by symptoms, clinical tests, or wastewater can prevent correlation with a later surge. The illustrated data are part of a city-wide project to monitor 20 sewersheds in Detroit (https://www.ramlabwsu.org/public-data-page.html) in which sewersheds range from closely monitored facilities (e.g. long-term care institutions, as illustrated here, in small sewersheds) to larger sewersheds sampling more people in less supervised communities. Future comparisons of these sewersheds and associated case data may enable examination of the effects of sewershed size on detectability and variability of the SARS-CoV-2 markers in wastewater, as has been studied by others comparing detectability in a range of sizes of much larger sewersheds (Weidhaas et al., 2021). Regardless of the size of the sewershed, the increased speed and greater sensitivity of the automated method described here is more likely to yield detections that could trigger public health actions than the less sensitive PEG/NaCl/Qiagen method.
Application of a PEG/NaCl/Qiagen method similar to the one used here by Flood et al. (2021) reported obtaining a Phi6 recovery from the initial amount spiked into wastewater of about 20 %, which is 3-fold higher than we report here for the PEG/NaCl/Qiagen protocol. The recovery calculated by Flood et al. (2021) is for the PEG/NaCl concentration step and does not include losses from the Qiagen step. Using a comparable PEG/NaCl/RNA purification method, Torii et al. (2022) reported RNA virus recoveries of 0.07 % - 2.6 % and values from other recent PEG/NaCl/RNA purification, publications ranging from <1 % to 50 %. Our recovery of 4-times as much Phi6 spiked-in RNA (25 + 15 % of the eluate control for Chemagic™ v 6.5 + 3.3 % for the PEG/NaCl/Qiagen protocol) and the 4.9:1 fold ratio of the amount of N1 and N2 measured (3.1.2, 3.2.2 and Fig. 2) could potentially provide better data for management of public health responses.
The higher amounts measured for both SARS-CoV-2 markers and Phi6 using the Chemagic™ method v the PEG/NaCl/Qiagen method might be related to various differences in the methods. Among those differences are: (a) no preliminary “solids removal step” (the 15 minute centrifugation step at the beginning of the Chemagic™ method) is done prior to adding PEG/NaCl for precipitation in the PEG/NaCl/Qiagen procedure. We included centrifugation in the Chemagic™ protocol because ddPCR droplet generation in preliminary experiments without centrifugation often had unacceptably low droplet counts (<10,000 droplets); low droplet counts were not a problem for the state-prescribed PEG/NaCl/Qiagen procedure, (b) the Qiagen purification step uses only a part of the pellet after PEG/NaCl precipitation; the Chemagic™ method final product is from the entire 10 mL starting material, and (c) PCR inhibitors may be present after the Qiagen purification step that the magnetic bead separation method seems to have effectively removed. The effective removal of inhibitors from the Chemagic™ eluates is demonstrated by the >95 % measurement of Phi6 spiked into the wastewater eluates compared to spikes directly into the elution medium control (Table 3).
Besides Chemagic™, alternative systems for automated recovery of nucleic acids from wastewater include the Promega Maxwell® RSC and the Thermo Fisher Scientific Kingfisher™. Table 4 provides a comparison of these systems and the PEG/NaCl/Qiagen process based on hands-on testing (PEG/NaCl/Qiagen, Chemagic™ and Maxwell® platforms) and analysis of published protocols (Kingfisher™) (Karthikeyan and Humphrey, 2020; Appliedbiosystems, 2021; Karthikeyan et al., 2021).
Table 4.
Comparison of manual and automated wastewater processing systems.
| Instrument or process |
PEG/NaCl/Qiagen | Promega Maxwell® RSC | Thermo Fisher Kingfisher | PerkinElmer Chemagic |
|---|---|---|---|---|
| Feature being compared | ||||
| Sample preparation | Agitation: 100 mL, 2 h | Centrifugation: 40 mL, 10 min; Lysis incubation: 30 min | N/A | Centrifugation: 45 mL, 15 min; Lysis incubation: 30 min |
| Machine preparation | N/A | Load 1 cartridge and 1 tip per sample | 1st run: Load 2 sample plates and 1 lysis plate per sample. 2nd run: Load 5 plates per sample with reagents as indicated by protocol | Load 1 tip and 3 tubes per sample |
| Sample concentration | Centrifugation: 100 mL, 45 min Volume of resuspended pellet: 2–5 mL (typically 2.5 mL, varies by sample composition) | Vacuum Filtration: 10–60 min, time varies by sample composition | First automated run with 24-well deep well plate, 2 h, up to 24 samples | Combined with purification |
| Sample purification | Manual, Qiagen kit, 40 min - 1.5 h depending on sample number | Automated, Maxwell Cartridge, 45 min per run, up to 48 samples | Manual transfer of lysate from 24-well first run to 2nd automated run on 96-well plate, 1.5 h, up to 24 samples | Automated, 75 min per run, up to 12 samples; can prepare a second run while first run is operating |
| Automated instrument cost | N/A | +++++ | ++++++ | +++++++ |
| Supplies cost/sample (to nearest $5) | $20 | $40 | $40 | $30 |
| Total time | 5–7 h | 2–4 h | 3–4 h | 2.5–3 h |
| Labor | +++++++ | ++ | ++++ | + |
| Reference | Flood et al. (2021), Method 3 | Promega (2021) | Appliedbiosystems (2021); Karthikeyan et al. (2021) | Vasquez et al. (2021) |
All three automated platforms utilize magnetic beads to isolate and purify nucleic acids, yet each system uses different approaches with their own unique benefits and drawbacks. The Promega Maxwell® system utilizes a vacuum-based sample filtration concentration step prior to application of 1 mL concentrated samples to the Maxwell® RSC. The filtration system can be applied to up to 20 samples simultaneously but functioned variably from sample to sample; the use of cartridges prefilled with reagents for the Maxwell® RSC was convenient; and the Maxwell® RSC can purify nucleic acids from up to 48 sample concentrates simultaneously.
The Kingfisher™ does not utilize prefilled cartridges and requires the reagents to be manually aliquoted before the samples can be processed. Although this manual prefilling step is time consuming it does allow the greatest flexibility and control over reaction conditions among the automated platforms. The Chemagic™ instrument is the most expensive of the three; however, it utilizes a large reservoir for its reagents and unlike the other automated systems, Chemagic™ combines the sample concentration and purification steps into a single process, greatly reducing the amount of sample handling and pipetting required. Of the automated systems, the cost of the reagents and supplies per sample was lowest for the Chemagic™. Supplies for the PEG/NaCl/Qiagen method were less expensive than for the automated systems but processing required more time and labor.
Despite the higher instrument cost, the improvement in sensitivity of detection with the Chemagic™ instrument, its lower labor requirements, its fast processing time, and the cost-savings for Chemagic™ supplies compared to the other automated instruments can justify the use of this instrument to process wastewater samples. The more sensitive detection of SARS-CoV-2 markers and the rapid reporting of wastewater data to public health partners, made more possible by this automated system, may be especially important to obtain the earliest warning of resurgences of infections in the community and for considering the application of mitigation and other public health policies that may prevent a larger outbreak.
CRediT authorship contribution statement
William Shuster, Jeffrey L. Ram: Conceptualization; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, Mohammed F. Khan, James Hartrick: Data curation; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, Mohammed F. Khan, James Hartrick, Jeffrey L. Ram: Formal analysis; William Shuster, Jeffrey L. Ram: Funding acquisition; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, Mohammed F. Khan, James Hartrick, Investigation; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, James Hartrick, William Shuster, Jeffrey L. Ram: Methodology; Azadeh Bahmani, Jeffrey L. Ram: Project administration; William Shuster, Jeffrey L. Ram: Resources; William Shuster, Jeffrey L. Ram: Supervision; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, James Hartrick, William Shuster, Jeffrey L. Ram: Validation; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, Jeffrey L. Ram: Visualization; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, Jeffrey L. Ram: Roles/Writing - original draft; Nicholas W. West, Adrian A. Vasquez, Azadeh Bahmani, William Shuster, Jeffrey L. Ram: Writing - review & editing.
Declaration of competing interest
The authors have no known competing personal relationships or financial interests that could have influenced the work reported in this paper.
Acknowledgements
Funding for this work was provided by Project AY of the WSU-MDHHS Master contract MA-2021, entitled “SARS-CoV-2 Epidemiology – Wastewater Evaluation and Reporting (SEWER) Network” (Principal Investigators: Jeffrey L. Ram and William Shuster) from the Michigan Department of Health and Human Services, using Federal Financial Assistance from the U.S. Department of Treasury under the Epidemiology and Laboratory Capacity: Enhancing Detection Expansion through Coronavirus Response and Relief (CRR) Supplemental Appropriations Act of 2021 (P.L. 116-260). Wastewater samples were collected with cooperation and guidance from the Detroit Water and Sewerage Department, the Detroit Health Department, the Wayne State University Campus Health Center, and the Great Lakes Water Authority.
Editor: Warish Ahmed
Data availability
Data will be made available on request.
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:9. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Simpson S.L., Smith W.J.M., Metcalfe S., McMinn B., Symonds E.M., Korajkic A. Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali W., Zhang H., Wang Z., Chang C., Javed A., Ali K., Du W., Niazi N.K., Mao K., Yang Z. Occurrence of various viruses and recent evidence of SARS-CoV-2 in wastewater systems. J. Hazard. Mater. 2021;414 doi: 10.1016/j.jhazmat.2021.125439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alygizakis N., Markou A.N., Rousis N.I., Galani A., Avgeris M., Adamopoulos P.G., Scorilas A., Lianidou E.S., Paraskevis D., Tsiodras S., Tsakris A., Dimopoulos M.-A., Thomaidis N.S. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: protocols and future perspectives. TrAC Trends Anal. Chem. 2021;134 doi: 10.1016/j.trac.2020.116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appliedbiosystems Perform SARS-CoV-2 (or other viral targets) nucleic acid purification from 10 mL wastewater samples using Dynabeads Intact Virus Enrichment beads and KingFisher Flex. MagMAXTM Microbiome Ultra Nucleic Acid Isolation Kit User Guide. 2021. https://laboshop.ae/storage/products/August2021/3OT5z6LZdgazqIJzbme7.pdf 7-8.
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382(22):2081–2090. doi: 10.1056/nejmoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendix A. The US decided to make its own coronavirus test, but the process was plagued by errors and delays. Here's a timeline of what went wrong. Insider Mar 11, 2020, 8:18 PM. 2020. https://www.businessinsider.com/us-coronavirus-testing-problems-timeline-2020-3
- Borchardt M.A., Spencer S.K., Hubbard L.E., Firnstahl A.D., Stokdyk J.P., Kolpin D.W. Avian influenza virus RNA in groundwater wells supplying poultry farms affected by the 2015 influenza outbreak. Environ.Sci.Technol.Lett. 2017;4(7):268–272. doi: 10.1021/acs.estlett.7b00128. [DOI] [Google Scholar]
- Brouwer A.F., Eisenberg J.N.S., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. 2018;115(45):E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni S., Zuccato E., Crisci E., Chiabrando C., Fanelli R., Bagnati R. Identification and measurement of illicit drugs and their metabolites in urban wastewater by liquid chromatography−tandem mass spectrometry. Anal. Chem. 2006;78(24):8421–8429. doi: 10.1021/ac061095b. [DOI] [PubMed] [Google Scholar]
- CDC Health Alert Network Update and interim guidance on outbreak of 2019 novel coronavirus (2019-nCoV) 2020. https://emergency.cdc.gov/han/HAN00427.asp issued 1 February 2020.
- Flood M.T., D’Souza N., Rose J.B., Aw T.G. Methods evaluation for rapid concentration and quantification of SARS-CoV-2 in raw wastewater using droplet digital and quantitative RT-PCR. FoodEnviron.Virol. 2021;13(3):303–315. doi: 10.1007/s12560-021-09488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani A., Aalizadeh R., Kostakis M., Markou A., Alygizakis N., Lytras T., Adamopoulos P.G., Peccia J., Thompson D.C., Kontou A., Karagiannidis A., Lianidou E.S., Avgeris M., Paraskevis D., Tsiodras S., Scorilas A., Vasiliou V., Dimopoulos M.-A., Thomaidis N.S. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron L., Verreault D., Veillette M., Moineau S., Duchaine C. Evaluation of filters for the sampling and quantification of RNA phage aerosols. Aerosol Sci. Technol. 2010;44(10):893–901. doi: 10.1080/02786826.2010.501351. [DOI] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Brazell L.R., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W.W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Medema G. Surveillance of influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in the Netherlands. J. Water Health. 2011;9(3):434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki,Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Humphrey G. Automated high throughput viral concentration from wastewater using the KingFisher Flex platform v1 (protocols.io.bptemnje) Protocols.io. 2020 doi: 10.17504/protocols.io.bptemnje. https://www.protocols.io/view/automated-high-throughput-viral-concentration-from-bptemnje [DOI] [Google Scholar]
- Karthikeyan S., Nguyen A., McDonald D., Zong Y.J., Ronquillo N., Ren J.T., Zou J.J., Farmer S., Humphrey G., Henderson D., Javidi T., Messer K., Anderson C., Schooley R., Martin N.K., Knight R. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems. 2021;6(4) doi: 10.1128/mSystems.00793-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laturner Z.W., Zong D.M., Kalvapalle P., Gamas K.R., Terwilliger A., Crosby T., Ali P., Avadhanula V., Santos H.H., Weesner K., Hopkins L., Piedra P.A., Maresso A.W., Stadler L.B. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S., Harcourt J., Tamin A., Thornburg N., Villanueva J., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg.Infect.Dis.J. 2020;26(8):1654. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn B.R., Korajkic A., Kelleher J., Herrmann M.P., Pemberton A.C., Ahmed W., Villegas E.N., Oshima K. Development of a large volume concentration method for recovery of coronavirus from wastewater. Sci. Total Environ. 2021;774:8. doi: 10.1016/j.scitotenv.2021.145727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ.Sci.Technol.Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michigan.gov Coronavirus/Michigan Data. 2022. https://www.michigan.gov/coronavirus/0,9753,7-406-98163_98173-526911--,00.html Long Term Care Data.
- Monteiro S., Rente D., Cunha M.V., Marques T.A., Cardoso E., Álvaro P., Vilaça J., Ribeiro J., Silva M., Coelho N., Brôco N., Carvalho M., Santos R. Recovery of SARS-CoV-2 from large volumes of raw wastewater is enhanced with the inuvai R180 system. J. Environ. Manag. 2022;304 doi: 10.1016/j.jenvman.2021.114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.C. COVIDPoops19 summary of global SARS-CoV-2 wastewater monitoring efforts by UCuc Merced researchers. 2020. https://ucmerced.maps.arcgis.com/apps/dashboards/c778145ea5bb4daeb58d31afee389082
- Oran D.P., Topol E.J. The proportion of SARS-CoV-2 infections that are asymptomatic: a systematic review. Ann. Intern. Med. 2021;174(5):655–662. doi: 10.7326/M20-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal D., Prakash O., Bobde P., Pal S. SARS-CoV-2: sewage surveillance as an early warning system and challenges in developing countries. Environ. Sci. Pollut. Res. 2021;28(18):22221–22240. doi: 10.1007/s11356-021-13170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Omer S.B. 2020. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv, 2020.2005.2019.20105999. [Google Scholar]
- Promega Maxwell® RSC Enviro Total Nucleic Acid Kit. 2021. https://www.promega.com/-/media/files/resources/protocols/technical-manuals/500/maxwell-rsc-enviro-total-nucleic-acid-protocol-tm663.pdf?rev=268d9ed5379e44c6bd412799a6608308&sc_lang=en 17 pages.
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. 115942-115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J.P., Medema G., Naughton C.C., Katsivelis P. WASTEWATER SPHERE (SARS Public Health Environmental REsponse. 2021. https://sphere.waterpathogens.org/about
- Shah S., Gwee S.X.W., Ng J.Q.X., Lau N., Koh J.Y., Pang J.X. Wastewater surveillance to infer COVID-19 transmission: a systematic review. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State of Michigan . Michigan.gov; 2021. New funding announced for continued COVID-19 wastewater monitoring.https://www.michigan.gov/som/0,4669,7-192-29942_34762-562538--,00.html (posted June 24, 2021) [Google Scholar]
- Torii S., Oishi W., Zhu Y., Thakali O., Malla B., Yu Z., Zhao B., Arakawa C., Kitajima M., Hata A., Ihara M., Kyuwa S., Sano D., Haramoto E., Katayama H. Comparison of five polyethylene glycol precipitation procedures for the RT-qPCR based recovery of murine hepatitis virus, bacteriophage phi6, and pepper mild mottle virus as a surrogate for SARS-CoV-2 from wastewater. Sci. Total Environ. 2022;807 doi: 10.1016/j.scitotenv.2021.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez A.A., West N.W., Bahmani A., Ram J.L. 2021. Rapid and direct method to extract SARS-CoV-2 RNA from municipal wastewater using the Chemagic 360™ 12-rod head platform.10.17504/protocols.io.b2reqd3e [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Vander Laan J., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775:12. doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Hu Y., Song Z.-G., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. Direct submission: severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. GenBank: MN908947. 2020. https://www.ncbi.nlm.nih.gov/nuccore/MN908947.3 (direct submission 5 January 2020)
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25(50):38–44. doi: 10.2807/1560-7917.Es.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Shao X.T., Tan D.Q., Yan J.H., Pei W., Wang Z., Yang M., Wang D.G. Assessing the trend of diabetes mellitus by analyzing metformin as a biomarker in wastewater. Sci. Total Environ. 2019;688:281–287. doi: 10.1016/j.scitotenv.2019.06.117. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ.Sci.Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.