Abstract
Objectives
This study used the long-short-term memory (LSTM) artificial intelligence method to model multiple time points of clinical laboratory data, along with demographics and comorbidities, to predict hospital-acquired acute kidney injury (AKI) onset in patients with COVID-19.
Methods
Montefiore Health System data consisted of 1982 AKI and 2857 non-AKI (NAKI) hospitalized patients with COVID-19, and Stony Brook Hospital validation data consisted of 308 AKI and 721 NAKI hospitalized patients with COVID-19. Demographic, comorbidities, and longitudinal (3 days before AKI onset) laboratory tests were analyzed. LSTM was used to predict AKI with fivefold cross-validation (80%/20% for training/validation).
Results
The top predictors of AKI onset were glomerular filtration rate, lactate dehydrogenase, alanine aminotransferase, aspartate aminotransferase, and C-reactive protein. Longitudinal data yielded marked improvement in prediction accuracy over individual time points. The inclusion of comorbidities and demographics further improves prediction accuracy. The best model yielded an area under the curve, accuracy, sensitivity, and specificity to be 0.965 ± 0.003, 89.57 ± 1.64%, 0.95 ± 0.03, and 0.84 ± 0.05, respectively, for the Montefiore validation dataset, and 0.86 ± 0.01, 83.66 ± 2.53%, 0.66 ± 0.10, 0.89 ± 0.03, respectively, for the Stony Brook Hospital validation dataset.
Conclusion
LSTM model of longitudinal clinical data accurately predicted AKI onset in patients with COVID-19. This approach could help heighten awareness of AKI complications and identify patients for early interventions to prevent long-term renal complications.
Keywords: SARS-CoV-2, Long-short-term memory, Artificial intelligence, Acute kidney injury, Multiorgan failure, D-dimer
Introduction
Acute kidney injury (AKI) is common in hospitalized patients with COVID-19 (Huang et al., 2020; Zhu et al., 2020) and is associated with critical illness and mortality (Brienza et al., 2021; Farouk et al., 2020; Nadim et al., 2020; Oliveira et al., 2021; Shao et al., 2020). Contributing factors to hospital-acquired AKI include direct SARS-CoV-2 viral infection of renal cells and indirect effects such as sepsis, host-immune responses (i.e., inflammatory cytotropic and cytokine-mediated immune responses, among others), hemodynamic compromise, acute respiratory distress syndrome, and systemic hypoxia (Adapa et al., 2020; Ahmadian et al., 2021; Khan et al., 2020). Kidney complications in patients with COVID-19 might not receive adequate attention because medical providers need to address more medically urgent issues of SARS-CoV-2 infection. In patients with COVID-19, failure to identify at-risk patients could result in long-term renal damage.
A few studies have used clinical variables at hospital admission to predict AKI (Gabarre et al., 2020; Hectors et al., 2021; Xia et al., 2020), but using only clinical variables at admission to predict the development of AKI is likely inaccurate because patients come into hospitals with different disease severities, or patients might have pre-existing or community-acquired AKI. Although the temporal changes of clinical variables associated with hospital-acquired AKI have been reported (Lu et al., 2021b; Lu et al., 2022), no studies have integrated multiple time points of clinical variables to predict in-hospital AKI onset in COVID-19 to our knowledge. The ability to anticipate which patients will develop AKI would lead to better patient management, such as hemodynamic support, renal replacement therapy, and avoiding nonsteroidal anti-inflammatory drugs, nephrotoxins, and contrast (Chan et al., 2021; Fisher et al., 2020; Hamilton et al., 2020; Hirsch et al., 2020; Ouyang et al., 2021; Trabulus et al., 2020; Wagner et al., 2020), appropriate follow-up care, and timely intervention to prevent long-term kidney damage.
Machine learning (ML) helps tackle complex and multimodal data and is increasingly being used in medicine. ML learns relationships between different data elements to inform outcomes. In contrast to traditional analysis methods such as logistic regression, ML does not require relationships between different input variables and outcomes to be explicitly specified a priori. Time series prediction using ML is possible but with added complexity because there are additional dependencies among input variables. A recurrent neural network is a powerful type of neural network designed to handle sequence dependence. The long short-term memory (LSTM) network (Hochreiter and Schmidhuber, 1997), an artificial recurrent neural network architecture in deep learning, is ideal for processing sequential data. Unlike standard feedforward neural networks, LSTM has feedback connections and is well-suited to classify, process, and make predictions based on time series data.
The goal of this study was to develop an LSTM-ML algorithm to integrate longitudinal clinical data to predict hospital-acquired AKI onset in hospitalized patients with COVID-19. Inputs to the ML model included longitudinal clinical laboratory values, longitudinal vital signs, demographics, and comorbidities. We trained and validated our model on Montefiore Health System data and further cross-validated on Stony Brook Hospital data.
Methods
Study population and data collection
The Montefiore Health System serves a large low-income, racially and ethnically diverse population. Data from the Montefiore Health System consisted of 15 hospitals located in the Bronx and the Lower Hudson Valley and Westchester County. De-identified health data were made available for research after standardization to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) version 6. OMOP CDM represents health care data from diverse sources stored in standard vocabulary concepts (Hripcsak et al., 2015). This approach allows for the systematic analysis of disparate observational databases, including data from the electronic medical record (EMR), administrative claims, and disease classification systems (e.g., ICD-10, SNOWMED, LOINC). ATLAS, a web-based tool developed by the Observational Health Data Sciences and Informatics (OHDSI) community that enables navigation of patient-level, observational data in the CDM format, was used to search vocabulary concepts and facilitate cohort building. Data were subsequently exported and queried as SQLite database files using the DB Browser for SQLite (version 3.12.0).
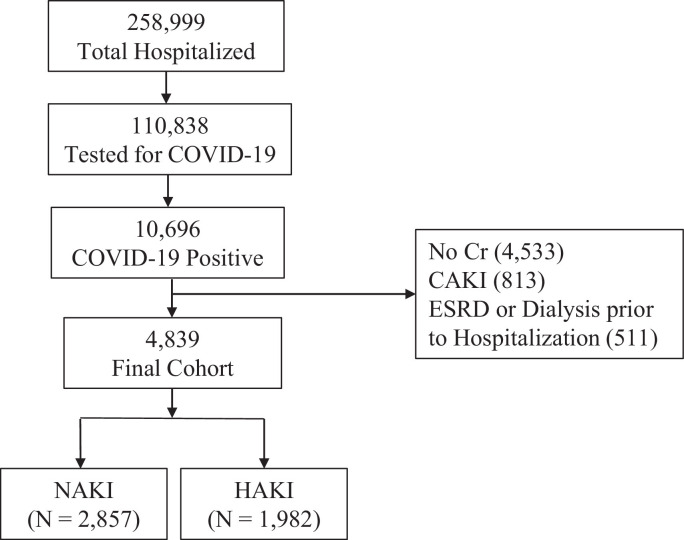
Montefiore Health System data consisted of 258,999 hospitalized patients from March 11, 2020, to January 21, 2021, in the Montefiore Health System. SARS-CoV-2 infection was confirmed by real-time polymerase chain reaction (PCR) test via nasopharyngeal swab specimen. Exclusion criteria included patients who were not tested for COVID-19 or had a negative test result for COVID-19, without creatinine measurements, with community-acquired AKI (CAKI), and with end-stage kidney disease or requiring dialysis before hospitalization (Figure 1 ). The final sample size used in the analysis was 4839 patients with COVID-19, of whom 2857 had NAKI and 1982 had AKI. Montefiore Health System data were used for training and validation with fivefold cross-validations. Subsets of the Montefiore Health system data have been used to address different questions (Hoogenboom et al., 2021a, Hoogenboom et al., 2021b, Iosifescu et al., 2022, Lu et al., 2022, Lu et al., 2021c).
Figure 1.
Flowchart of patient selection of patients with hospital-acquired AKI and those without AKI. AKI, acute kidney injury; CAKI, community-acquired AKI; Cr, creatinine; ESRD, end-stage renal disease; HAKI, hospital-acquired AKI; NAKI, non-AKI; Pts, patients.
Stony Brook Hospital data were used for validation only (not for training). Stony Brook Hospital data consisted of 6678 persons clinically suspected of COVID-19 infection in the emergency department from February 7, 2020, to June 30, 2020, of whom 2892 tested positive using a real-time PCR test for SARS-CoV-2 on a nasopharyngeal swab specimen (Chen et al., 2021; Lu et al., 2021a, 2021b; Zhao et al., 2020). Only hospitalized patients with COVID-19 with creatinine (n = 1029) were used, of whom 308 had AKI and 721 had NAKI. A variance and a subset of Stony Brook Hospital data have been used to address different questions (Chen et al., 2021; Hou et al., 2021; Lam et al., 2020; Li et al., 2020; Shen et al., 2021; Zhao et al., 2020).
AKI definitions
Hospital-acquired AKI was defined using the Kidney Disease: Improving Global Outcomes (KDIGO) criteria (Ad-hoc working group of ERBP et al., 2012; Khwaja, 2012) as either a 0.3 mg/dl increase in serum creatinine within 48 hours or a 1.5-times increase in serum creatinine within a 7-day iterative window. Fifty-seven percent of the patients had pre-existing creatinine baseline values. For patients who did not have creatinine baseline values, the lowest creatinine value during hospitalization was considered the baseline creatinine (Hirsch et al., 2020; Pelayo et al., 2020). CAKI was defined as AKI within 24 hours of admission. Urine output was not used to define AKI because it was not reliably documented. AKI was staged per KDIGO guidelines: stage 1: ≥0.3 mg/dl or to >1.5 to 2-times increase in creatinine; stage 2: >2 to ≤3-times increase in creatinine; and stage 3: >3-times increase in creatinine or rise to ≥4.0 mg/dl, or new initiation of renal replacement therapy (RRT) (Ad-hoc working group of ERBP et al., 2012, Khwaja, 2012).
Demographics, clinical comorbidities, longitudinal vital signs, laboratory blood tests, and blood gases were extracted from EMRs. Demographic data included age, sex, ethnicity, and race. Chronic comorbidities included obesity, diabetes, congestive heart failure, chronic kidney disease, coronary artery disease, chronic obstructive pulmonary disease (COPD), and asthma.
Longitudinal laboratory tests and vital signals included creatinine, albumin, alanine aminotransferase (ALT), aspartate aminotransferase (AST), brain natriuretic peptide, C-reactive protein (CRP), d-dimer, eGFR, ferritin, lactate dehydrogenase (LDH), lymphocytes, troponin-T, white blood cells (WBCs), diastolic blood pressure, systolic blood pressure, temperature, heart rate, and pulse oximetry. An average of daily values was used.
Prediction model
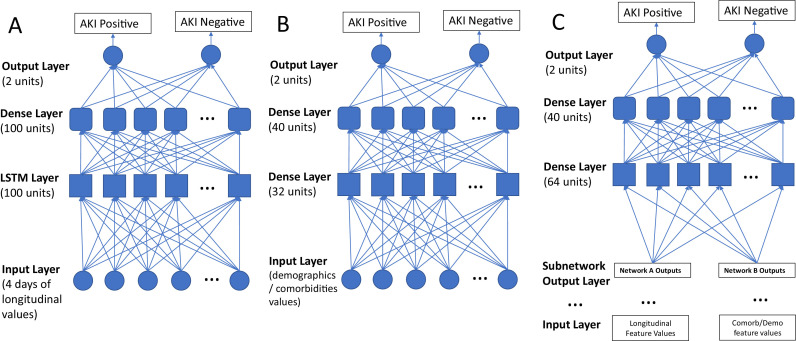
We built a model combining LSTM recurrent neural networks and traditional feedforward neural networks, which take as input a combination of longitudinal vital signs, laboratory blood tests, demographics, and chronic comorbidities to predict AKI outcomes. Prediction was made using values from individual time points and a combination of multiple time points before the onset of AKI, as well as demographics and comorbidities. Figure 2 describes the prediction models. In Figure 2A, the LSTM network takes multiple time points of clinical laboratory values and vital signs as input. Each LSTM unit comprises a cell, an input gate, an output gate, and a forget gate. The cell remembers values over arbitrary time intervals, and the three gates regulate the flow of information in and out of the cell. LSTM is well-suited to classify, process, and predict the given time series of unknown durations. The popular rectified linear function is set as the activation function at each node except for the nodes in the output layer where the Softmax function is used. In Figure 2B, the network takes demographics and comorbidities as input. In Figure 2C, the model combines longitudinal clinical laboratory values, vital signs, demographics, and comorbidities values to predict hospital-acquired AKI onset in patients with COVID-19.
Figure 2.
(A) A LSTM network that takes multiple time points of clinical laboratory values and vital signs as input. (B) A network that takes as input the demographics and comorbidities values. (C) An LSTM network that combines both longitudinal feature values and the demographics and comorbidities values to predict hospital-acquired AKI onset in COVID-19 patients. AKI, acute kidney injury; LSTM, long-short-term memory.
The available dataset was split into 80% for training and 20% for testing. Adam optimizer was used with the default learning rate of 0.001. To determine the optimal number of epochs without overfitting, one round of training with a validation split ratio of 20% was first carried out with a large number of epochs (50 in this case). Afterward, both the training loss history and validation loss history were examined. The optimal epoch was 15, at which the validation loss started worsening, as further training should result in model overfitting. Once the optimal epoch of 15 was determined, the model was retrained with this optimal number of epochs. Finally, the trained model was tested on the test dataset. Prediction performance was evaluated by the area under the curve (AUC) of the receiver operating characteristic (ROC) curve, accuracy, sensitivity, specificity, and Brier score.
Ranking of clinical predictors
To provide interpretability of the LSTM model, the importance of different clinical variables in predicting AKI onset was measured based on game theory–based optimal Shapley values. A prediction can be explained by assuming that each feature value of the instance is a “player” in a game where the prediction is the payout. Shapley values—a method from coalitional game theory—explain the fair distribution of the “payout” among the features. The python package SHAP (SHapley Additive exPlanations) package was used to calculate the feature importance values (Lundberg and Lee, 2017). The results of top predictors were plotted as Beeswarm plots. In addition, calibration plots of the predicted vs true probability for predicting AKI onset for individual-day and all-day data with and without comorbidities and demographics were analyzed.
Statistical analysis
All statistical analyses were performed using Python packages (Tensorflow, Sklearn and Statsmodels) and R. Frequencies and percentages for categorical variables between the AKI and NAKI groups were compared using the chi-square test. Group differences in frequencies and percentages for categorical variables were tested using the chi-square test or Fisher exact test. Continuous variables, expressed as median (interquartile range), were compared between groups using nonparametric Mann-Whitney U test. Mortality rates were compared between groups with the chi-square test adjusted with covariates. The mortality odds ratio was obtained using logistic regression with adjustment for sex, age, and major comorbidities. P-values < 0.05 were considered statistically significant unless otherwise specified.
Results
Table 1 (A) summarizes patient demographics and comorbidities of 1982 patients with AKI and 2857 patients with NAKI from the Montefiore Health System. The AKI cohort was older and had fewer females compared with the NAKI cohort (P < 0.001). Race (P = 0.085) was not, but ethnicity (P < 0.05) was, significantly different between groups. Patients with AKI generally had more comorbidities (P < 0.05). Diabetes, congestive heart failure, chronic kidney disease, and COPD (P < 0.05)—but not hypertension, coronary artery disease and liver disease (P > 0.05)—were significantly different between groups. The unadjusted mortality rate was 37.4% for the AKI cohort and 7.1% for the NAKI cohort. The mortality odds ratio of AKI compared with NAKI was 6.32 (95% CI: 5.3, 7.6). Table 1 (B) shows patient profiles for the Stony Brook University Hospital data. Although there were some differences in the two datasets, they were overall quite similar.
Table 1.
Demographic information of (A) Montefiore Health System data and (B) Stony Brook Hospital data.
| (A) Montefiore health system data | NAKI (2857) | AKI (1982) | P-values | |
|---|---|---|---|---|
| Demographics | ||||
| Age, median (IQR) | 61 (47, 75) | 70 (60, 81) | <0.001 | |
| Female gender, n (%) | 1491 (52.2) | 843 (42.5) | <0.001 | |
| Race, n (%) | 0.085 | |||
| White | 246 (16.0) | 177 (14.1) | ||
| Black/African American | 850 (55.4) | 724 (57.6) | ||
| Asian | 92 (6.0) | 59 (4.7) | ||
| Other | 205 (13.3) | 154 (12.3) | ||
| Unknown | 141 (9.3) | 142 (11.3) | ||
| Ethnicity, n (%) | <0.001 | |||
| Hispanic | 1323 (46.3) | 726 (36.7) | ||
| Non-Hispanic | 1534 (53.7) | 1256 (63.3) | ||
| Comorbidities, n (%) | ||||
| Diabetes | 506 (17.7) | 483 (24.4) | <0.001 | |
| Congestive heart failure | 162 (5.7) | 195 (9.8) | <0.001 | |
| Chronic kidney disease | 328 (11.5) | 376 (19.0) | <0.001 | |
| Hypertension | 741 (26.0) | 519 (26.2) | 0.087 | |
| Coronary artery disease | 183 (6.4) | 144 (7.3) | 0.265 | |
| COPD/asthma | 304 (10.6) | 139 (7.0) | <0.001 | |
| Liver disease | 41 (1.4) | 34 (1.7) | 0.511 | |
| Unadjusted mortality, n (%) | 204 (7.1) | 743 (37.4) | <0.001 | |
| Mortality odds ratio | 6.3 [5.3, 7.6] | <0.001 | ||
| (B) Stony Brook Hospital data | NAKI (721) | AKI (308) | P-values | |
|---|---|---|---|---|
| Demographics | ||||
| Age, median (IQR) | 59 (47, 73) | 70 (55, 80) | <0.001 | |
| Female gender, n (%) | 312 (43.3) | 118 (38.3) | 0.139 | |
| Race, n (%) | 0.66 | |||
| White | 384 (53.3) | 177 (57.5) | ||
| Black/African American | 52 (7.2) | 26 (8.4) | ||
| Asian | 22 (3.1) | 10 (3.3) | ||
| Other | 6 (0.8) | 2 (0.6) | ||
| Unknown | 257 (35.6) | 93 (30.2) | ||
| Ethnicity, n (%) | 0.015 | |||
| Hispanic | 197 (27.3) | 62 (20.1) | ||
| Non-Hispanic | 524 (72.7) | 246 (79.9) | ||
| Comorbidities, n (%) | ||||
| Diabetes | 159 (22.1) | 108 (35.0) | <0.001 | |
| Congestive heart failure | 38 (5.3) | 55 (17.9) | <0.001 | |
| Chronic kidney disease | 45 (6.2) | 58 (18.8) | <0.001 | |
| Hypertension | 302 (41.9) | 196 (63.6) | <0.001 | |
| Coronary artery disease | 91 (12.6) | 73 (23.7) | <0.001 | |
| COPD/asthma | 57 (7.9) | 33 (10.7) | 0.144 | |
| Liver disease | 7 (1.0) | 2 (0.7) | 0.612 | |
| Unadjusted mortality, n (%) | 50 (6.9) | 97 (31.5) | <0.001 | |
| Mortality odds ratio | 4.67 [3.1,7.0] | <0.001 | ||
Demographic characteristics and comorbidities of NAKI and AKI patients. Chi-square test or Fisher's exact test was used for group comparison of categorical variables in frequencies and percentages. Mann-Whitney U test was used for group comparison of continuous variables in medians and IQR. Mortality odds ratios were adjusted for demographics and comorbidities (see Methods).
AKI, acute kidney injury; COPD, chronic obstructive pulmonary disease; IQR, interquartile range; NAKI, non-AKI.
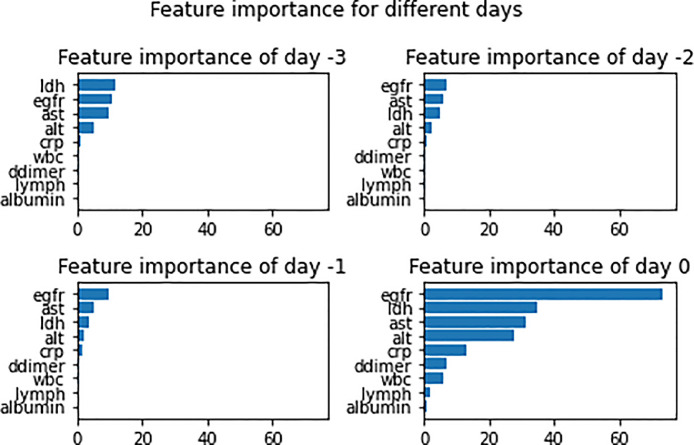
Figure 3 shows the feature importance of clinical variables in predicting AKI at −3, −2, −1, and 0 days before onset. The top four variables at day 0 of AKI onset were eGFR, LDH, AST, and ALT; at −1 day, eGFR, AST, LDH, and ALT; at −2 days, eGFR, AST, LDH, and ALT; and at −3 day, LDH, eGFR, AST, and ALT. The feature importance had higher weighting for days closer to onset. Most of the top variables were consistent across different days.
Figure 3.
Bar graph displays the importance of different clinical variables from a single day in predicting AKI positivity, measured using game theory–based optimal Shapley values. AKI, acute kidney injury; alt, alanine transaminase; ast, aspartate aminotransferase; crp, C-reactive protein; egfr, estimated glomerular filtration rate; ldh, lactate dehydrogenase.
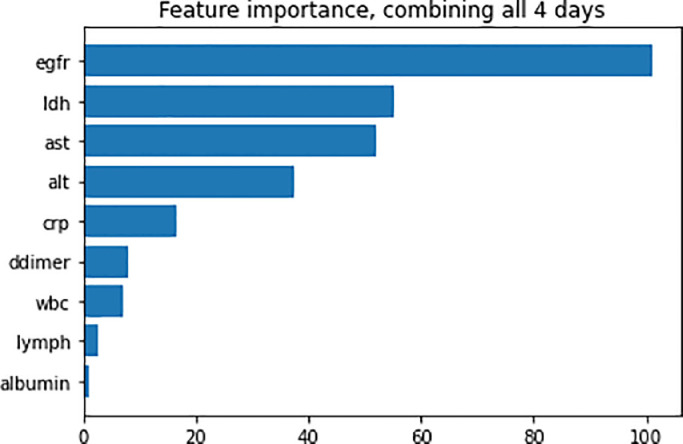
Figure 4 shows the feature importance of clinical variables in predicting AKI integrating all four time points combined. The top five variables that predict AKI onset using all 4 days of data were eGFR, LDH, AST, ALT, and CRP. The importance indices were overall higher for the model integrating four time points combined compared with those using individual time points.
Figure 4.
Bar graph displays the importance of different clinical variables from all 4 days in predicting AKI positivity, measured using game theory–based optimal Shapley values. AKI, acute kidney injury; alt, alanine transaminase; ast, aspartate aminotransferase; crp, C-reactive protein; egfr, estimated glomerular filtration rate; ldh, lactate dehydrogenase.
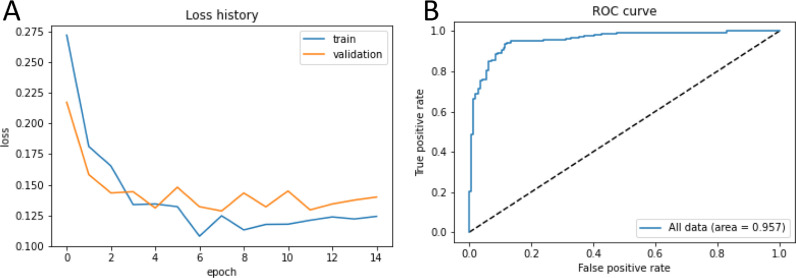
To further evaluate prediction models, we analyzed the loss functions of the training and validation dataset to predict AKI onset for one run that included clinical data, demographics, and comorbidities at four time points (Figure 5 ). Both training and validation models performed well with loss functions rapidly decreasing toward zeros with increasing epochs. A typical ROC curve for AUC of the validation dataset to predict AKI onset for the same model (one of fivefold cross-validation) is shown in Figure 5. The AUC was 0.957.
Figure 5.
(A) The loss functions for the training and validation dataset to predict AKI onset for one run for the model that included clinical data at for four time points along with demographics and comorbidities. (B) ROC curve generated with the AKI onset predictive model, built and trained using clinical variables across all 4 days combined with comorbidities and demographic information. AKI, acute kidney injury; ROC, receiver operating characteristic.
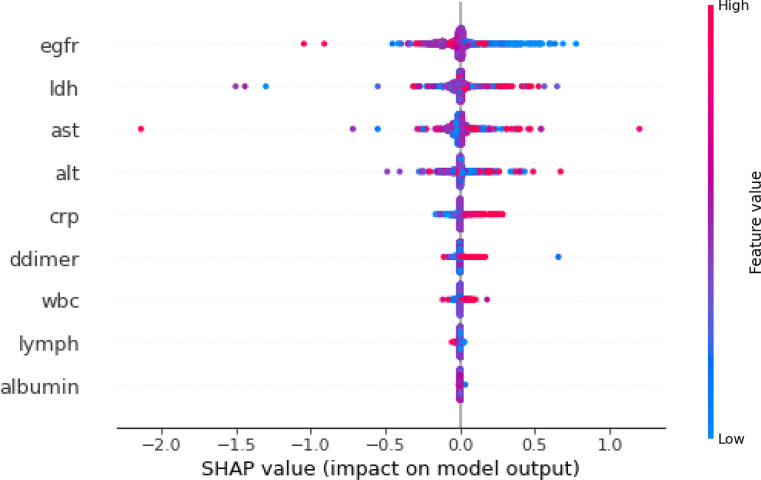
To improve interpretability of the models, Beeswarm plot (Figure 6 ) was used. There was high density of reduced (blue) eGFR that had positive SHAP values, indicative of improved prediction. There was high density of elevated (red) LDH that had positive SHAP values, indicative of improved prediction. Similarly, there were high densities of elevated AST, ALT, and CRP that had positive SHAP values. The remaining variables had relatively lower SHAP, reflecting lower importance for prediction.
Figure 6.
Beeswarm plot for model interpretability for Montefiore data. Importance of variables are ranked from top to bottom. Colors indicate higher (red) or lower (blue) feature values. Positive SHAP values (to the right of the 0 line) indicate data contributing to improved prediction of AKI onset. AKI, acute kidney injury; alt, alanine transaminase; ast, aspartate aminotransferase; crp, C-reactive protein; egfr, estimated glomerular filtration rate; ldh, lactate dehydrogenase; SHAP, SHapley Additive exPlanations; wbc, white blood cell.
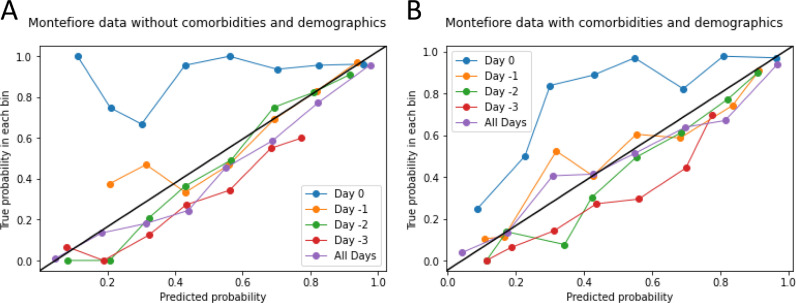
To further improve the interpretability of the models, we generated calibration plots of the predicted versus true probability for predicting AKI onset for individual-day and all-day data with and without comorbidities and demographics (Figure 7 ). Except for day 0 data, all other data points showed good performance. The all-day data showed the best performance. The addition of comorbidities and demographics further improved the performance.
Figure 7.
Predicted vs true probability plots for predicting AKI onset. (A) Montefiore data without comorbidities and demographics. (B) Montefiore data with comorbidities and demographics. The best predictions fall along the y = x line. AKI, acute kidney injury.
Table 2 (A and B) summarizes the performance metrics for LSTM model using individual time points and all four time points with fivefold cross-validation for the Montefiore Health System validation dataset. AUC and other metrics for individual time points were inferior compared with those of all four time points. The AUC for −3, −2, −1, and 0 days and all 4 days were 0.75 ± 0.01, 0.82 ± 0.01, 0.75 ± 0.01, 0.79 ± 0.01, and 0.956 ± 0.005, respectively. The addition of comorbidities and demographics into the LSTM model further improved performance, with the corresponding AUC of 0.77 ± 0.01, 0.84 ± 0.01, 0.80 ± 0.01, 0.83 ± 0.01, and 0.965 ± 0.003, respectively. The best model (all time points along with comorbidities and demographics) yielded AUC of 0.965 ± 0.003, accuracy of 89.57 ± 1.64%, sensitivity of 0.95 ± 0.03, specificity of 0.84 ± 0.05, and Brier score of 0.08 ± 0.01. Low Brier scores indicated good performance.
Table 2.
Area under the curve (AUC), accuracy, sensitivity, specificity, and Brier scores of predicting AKI onset using data from individual days and all 4 days before AKI onset, (A) without and (B) with comorbidities and demographic data from the Montefiore health system test dataset. (C) performance indices of the Stony Brook Hospital testing dataset.
| (A) Without comorbidities and demographic data | |||||
|---|---|---|---|---|---|
| AUC | Accuracy | Sensitivity | Specificity | Brier Score | |
| Day −3 | 0.75 ± 0.01 | 65.05 ± 1.92% | 0.75 ± 0.03 | 0.61 ± 0.03 | 0.21 ± 0.01 |
| Day −2 | 0.82 ± 0.01 | 73.94 ± 1.22% | 0.57 ± 0.05 | 0.87 ± 0.01 | 0.17 ± 0.01 |
| Day −1 | 0.75 ± 0.01 | 69.47 ± 0.33% | 0.64 ± 0.02 | 0.78 ± 0.03 | 0.20 ± 0.01 |
| Day 0 | 0.79 ± 0.01 | 66.61 ± 0.03% | 0.64 ± 0.03 | 0.81 ± 0.03 | 0.19 ± 0.01 |
| All time points | 0.956 ± 0.005 | 87.98 ± 1.75% | 0.88 ± 0.05 | 0.87 ± 0.05 | 0.09 ± 0.01 |
| (B) With comorbidities and demographic data | |||||
|---|---|---|---|---|---|
| Day −3 | 0.77 ± 0.01 | 69.10 ± 1.24% | 0.68 ± 0.03 | 0.70 ± 0.02 | 0.20 ± 0.01 |
| Day −2 | 0.84 ± 0.01 | 76.46 ± 0.43% | 0.68 ± 0.03 | 0.82 ± 0.03 | 0.17 ± 0.01 |
| Day −1 | 0.80 ± 0.01 | 70.53 ± 1.29% | 0.65 ± 0.04 | 0.79 ± 0.04 | 0.19 ± 0.01 |
| Day 0 | 0.83 ± 0.01 | 65.45 ± 1.05% | 0.63 ± 0.02 | 0.82 ± 0.01 | 0.19 ± 0.01 |
| All time points | 0.965 ± 0.003 | 89.57 ± 1.64% | 0.95 ± 0.03 | 0.84 ± 0.05 | 0.08 ± 0.01 |
| (C) Stony Brook Hospital data prediction performance | |||||
|---|---|---|---|---|---|
| All time points | 0.83 ± 0.03 | 78.93 ± 2.74% | 0.73 ± 0.09 | 0.80 ± 0.05 | 0.16 ± 0.01 |
| All time points + comorbidities and demographics | 0.86 ± 0.01 | 83.66 ± 2.53% | 0.66 ± 0.10 | 0.89 ± 0.03 | 0.12 ± 0.01 |
Cross-validation with Stony Brook Hospital data
For cross-validation using an independent dataset, we tested our predictive model on the Stony Brook Hospital dataset. The AKI and NAKI cohorts were 70 and 59 years old, and consisted of 38.3% and 43.3% female, 20.1% and 27.3% Hispanics, 57.5% and 53.3% Caucasian, 8.4% and 7.2% Blacks, 3.9% and 3.9% other races, 30.2% and 35.6% unknown/not reported, with 31.5% and 6.9% unadjusted mortality rate, respectively. The LSTM predictive model with all four data time points yielded an AUC of 0.83 ± 0.03, accuracy 78.93 ± 2.74%, sensitivity 0.73 ± 0.09, and specificity 0.80 ± 0.05 (Table 2C). The addition of comorbidities and demographics data yielded an AUC of 0.86 ± 0.01, accuracy of 83.66 ± 2.53%, sensitivity of 0.66 ± 0.10, specificity of 0.89 ± 0.03, and Brier score of 0.12 ± 0.01.
Discussion
This study applied LSTM to integrate longitudinal clinical data to predict hospital-acquired AKI onset in patients with COVID-19. The inputs to the LSTM model were longitudinal clinical laboratory values, longitudinal vital signs, demographics, and comorbidities. LSTM using longitudinal data markedly improves prediction accuracy over individual time points, with the top predictors of AKI onset to be eGFR, LDH, AST, ALT, and CRP. Inclusion of comorbidities and demographics further improved prediction performance. The best model was the one that used all time points along with comorbidities and demographics, yielding an AUC of 0.965 ± 0.003, accuracy of 89.57 ± 1.64%, sensitivity of 0.95 ± 0.03, specificity of 0.84 ± 0.05, and Brier score of 0.08 ± 0.01 for the Montefiore validation dataset. When tested on an external validation dataset from Stony Brook Hospital, the corresponding performance indices were 0.86 ± 0.01, 83.66 ± 2.53%, 0.66 ± 0.10, 0.89 ± 0.03, and 0.12 ± 0.01, respectively. In addition, analysis was performed to improve interpretability of LSTM models. This approach could help frontline physicians to raise awareness for AKI complications and identify patients who might need early interventions to prevent AKI and long-term renal complications.
LSTM networks are well-suited to make predictions with time series data. The importance indices and prediction performance indices were overall much higher for the model integrating all four time points compared with those using individual time points. The top predictors of AKI onset were eGFR, LDH, AST, ALT, and CRP. The SHAP Beeswarm plots were helpful in assessing the direction of changes and variability of prediction performance of top predictors, providing some interpretability of LSTM results. LDH is a marker of cell death and multiorgan failure (Lim et al., 2020; Mokhtari et al., 2020; Thierry and Roch, 2020). Reduced eGFR, an indicator of kidney dysfunction, has been associated with AKI in COVID-19 (Mirijello et al., 2021). Elevated CRP is indicative of inflammation and cytokine storm, among others (Lorenz et al., 2020). Elevated ALT and AST are indicative of liver enzyme dysfunction (Zhang et al., 2020).
A few studies have used clinical variables at hospital admission to predict AKI (Gabarre et al., 2020; Hectors et al., 2021; Xia et al., 2020), but predicting AKI development using only clinical variables at admission is likely inaccurate because patients came into hospitals with different disease severities or might have community-acquired AKI. The temporal characteristics of clinical variables leading up to in-hospital AKI development have been reported (Lu et al., 2021b; Lu et al., 2022). Lu et al. previously reported abnormal creatinine, procalcitonin, WBC count, LDH, and lymphocyte count at admission to be associated with a higher likelihood of AKI development (Lu et al., 2022). They applied logistic regression models to determine how accurate clinical variables could also predict hospital-acquired AKI at different, but individual, time points before AKI onset. To our knowledge, there has been no study that integrated multiple time points of clinical variables using machine learning or other methods to predict in-hospital AKI onset in COVID-19. Our study is novel because it used LSTM to integrate multiple time point data to predict in-hospital development of AKI with two health system data, along with analysis to enable interpretability of LSTM models (such as SHAP Beeswarm plots and calibration plots). Our cohort consisted of a diverse population that included Black and Hispanic patients. The top predictors of AKI are similar to those reported previously. The slight differences in predictors from the two studies could be because of difference in analysis methods, sample sizes, patient cohorts, among others.
To improve generalization, we tested our model on an independent dataset that mainly consisted of Caucasians. The Stony Brook Hospital cohort had a 31% AKI incidence rate, whereas Montefiore cohort had a 41% AKI incidence rate. The mortality rates in the AKI and NAKI groups in the Stony Brook Hospital data were slightly lower. The performance indices were excellent for this independent dataset, suggesting that this predictive model has significant generalizability.
This study had several limitations. These findings need to be replicated on additional data from other hospitals to expand generalizability. As with all observational studies, other residual confounders may exist that were not accounted for in our analysis. Urine analysis data, such as proteinuria and hematuria, were not used in our predictive modeling owing to their small sample sizes. Prospective studies validating our predictive models are warranted.
Conclusions
This study employed a sophisticated ML method to integrate longitudinal clinical data to predict in-hospital onset a few days prior. This approach has the potential to provide frontline physicians with an objective quantitative tool to stratify patients with COVID-19 who are at risk of developing AKI in time-sensitive and potentially resource-constrained environments.
Acknowledgments
Conflict of interest
The authors have no competing interests to declare.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical approval statement
This study was approved by the Einstein-Montefiore Institutional Review Board (#2021-13658) and Stony Brook University Institutional Review Board (#2020-00207) with an exemption for informed consent and a HIPAA waiver.
Authors’ contributions
J.Y. Lu collected and verified the data and created tables and figures. Joanna Zhu collected and analyzed the data. J.Y. Lu and Joanna Zhu drafted the manuscript. Jocelyn Zhu and J.Y. Lu analyzed the data. T.Q. Duong supervised and verified the data, and edited the manuscript. All authors read and approved the final version of the manuscript and contributed to the conceptualization and design of the study.
Data availability
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.07.034.
Appendix. Supplementary materials
References
- Adapa S, Chenna A, Balla M, Merugu GP, Koduri NM, Daggubati SR, et al. COVID-19 pandemic causing acute kidney injury and impact on patients with chronic kidney disease and renal transplantation. J Clin Med Res. 2020;12:352–361. doi: 10.14740/jocmr4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ad-hoc working group of ERBP, Fliser D, Laville M, Covic A, Fouque D, Vanholder R, Juillard L, Van Biesen WA. European Renal Best Practice (ERBP) position statement on the Kidney Disease Improving Global Outcomes (KDIGO) clinical practice guidelines on acute kidney injury: part 1: definitions, conservative management and contrast-induced nephropathy. Nephrol Dial Transplant. 2012;27:4263–4272. doi: 10.1093/ndt/gfs375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian E, Hosseiniyan Khatibi SM, Razi Soofiyani S, Abediazar S, Shoja MM, Ardalan M, et al. Covid-19 and kidney injury: pathophysiology and molecular mechanisms. Rev Med Virol. 2021;31:e2176. doi: 10.1002/rmv.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brienza N, Puntillo F, Romagnoli S, Tritapepe L. Acute kidney injury in coronavirus disease 2019 infected patients: a meta-analytic study. Blood Purif. 2021;50:35–41. doi: 10.1159/000509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L, Chaudhary K, Saha A, Chauhan K, Vaid A, Zhao S, et al. AKI in hospitalized patients with COVID-19. J Am Soc Nephrol. 2021;32:151–160. doi: 10.1681/ASN.2020050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Zhao Z, Hou W, Singer AJ, Li H, Duong TQ. Time-to-death longitudinal characterization of clinical variables and longitudinal prediction of mortality in COVID-19 patients: a two-center study. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.661940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farouk SS, Fiaccadori E, Cravedi P, Campbell KN. COVID-19 and the kidney: what we think we know so far and what we don't. J Nephrol. 2020;33:1213–1218. doi: 10.1007/s40620-020-00789-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M, Neugarten J, Bellin E, Yunes M, Stahl L, Johns TS, et al. AKI in hospitalized patients with and without COVID-19: a comparison study. J Am Soc Nephrol. 2020;31:2145–2157. doi: 10.1681/ASN.2020040509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabarre P, Dumas G, Dupont T, Darmon M, Azoulay E, Zafrani L. Acute kidney injury in critically ill patients with COVID-19. Intensive Care Med. 2020;46:1339–1348. doi: 10.1007/s00134-020-06153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton P, Hanumapura P, Castelino L, Henney R, Parker K, Kumar M, et al. Characteristics and outcomes of hospitalised patients with acute kidney injury and COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hectors SJ, Riyahi S, Dev H, Krishnan K, Margolis DJA, Prince MR. Multivariate analysis of CT imaging, laboratory, and demographical features for prediction of acute kidney injury in COVID-19 patients: a bi-centric analysis. Abdom Radiol (NY) 2021;46:1651–1658. doi: 10.1007/s00261-020-02823-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JS, Ng JH, Ross DW, Sharma P, Shah HH, Barnett RL, et al. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int. 2020;98:209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Hoogenboom W., Fleysher R., Soby S., Mirhaji P., Mitchell W., Morone K., et al. Individuals with sickle cell disease and sickle cell trait demonstrate no increase in mortality or critical illness from COVID-19 - A fifteen hospital observational study in the Bronx, New York. Haematologica. 2021;106(11):3014–3016. doi: 10.3324/haematol.2021.279222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom W., Pham A., Anand H., Fleysher R., Buczek A., Soby S., et al. Clinical characteristics of the first and second COVID-19 waves in the Bronx, New York: A retrospective cohort study. Lancet Regional Health Americas. 2021;3:100041. doi: 10.1016/j.lana.2021.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Zhao Z, Chen A, Li H, Duong TQ. Machining learning predicts the need for escalated care and mortality in COVID-19 patients from clinical variables. Int J Med Sci. 2021;18:1739–1745. doi: 10.7150/ijms.51235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iosifescu AL, Hoogenboom WS, Buczek AJ, Fleysher R, Duong TQ, et al. New-onset and persistent neurological and psychiatric sequelae of COVID-19 compared to influenza: A retrospective cohort study in a large New York City healthcare network. Int J Methods Psychiatr Res. 2022:e1914. doi: 10.1002/mpr.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Chen L, Yang CR, Raghuram V, Khundmiri SJ, Knepper MA. Does SARS-CoV-2 Infect the kidney? J Am Soc Nephrol. 2020;31:2746–2748. doi: 10.1681/ASN.2020081229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract. 2012;120:c179–c184. doi: 10.1159/000339789. [DOI] [PubMed] [Google Scholar]
- Lam KW, Chow KW, Vo J, Hou W, Li H, Richman PS, et al. Continued in-hospital angiotensin-converting enzyme inhibitor and angiotensin II receptor blocker use in hypertensive COVID-19 patients is associated with positive clinical outcomes. J Infect Dis. 2020;222:1256–1264. doi: 10.1093/infdis/jiaa447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ge P, Zhu J, Li H, Graham J, Singer A, et al. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ. 2020;8:e10337. doi: 10.7717/peerj.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MA, Pranata R, Huang I, Yonas E, Soeroto AY, Supriyadi R. Multiorgan failure with emphasis on acute kidney injury and severity of COVID-19: systematic review and meta-analysis. Can J Kidney Health Dis. 2020;7 doi: 10.1177/2054358120938573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz G, Moog P, Bachmann Q, La Rosée P, Schneider H, Schlegl M, et al. Cytokine release syndrome is not usually caused by secondary hemophagocytic lymphohistiocytosis in a cohort of 19 critically ill COVID-19 patients. Sci Rep. 2020;10:18277. doi: 10.1038/s41598-020-75260-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JQ, Lu JY, Wang W, Liu Y, Buczek A, Fleysher R, et al. Clinical predictors of acute cardiac injury and normalization of troponin after hospital discharge from COVID-19. EBioMedicine. 2022;76:103821. doi: 10.1016/j.ebiom.2022.103821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Anand H, Frager SZ, Hou W, Duong TQ. Longitudinal progression of clinical variables associated with graded liver injury in COVID-19 patients. Hepatol Int. 2021;15:1018–1026. doi: 10.1007/s12072-021-10228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Babatsikos I, Fisher MC, Hou W, Duong TQ. Longitudinal clinical profiles of hospital vs. community-acquired acute kidney injury in COVID-19. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.647023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Buczek A, Fleysher R, Hoogenboom WS, Hou W, Rodriguez CJ, et al. Outcomes of Hospitalized Patients With COVID-19 With Acute Kidney Injury and Acute Cardiac Injury. Frontiers in Cardiovascular Medicine. 2021;8:798897. doi: 10.3389/fcvm.2021.798897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Hou W, Duong TQ. Longitudinal prediction of hospital-acquired acute kidney injury in COVID-19: a two-center study. Infection. 2022;50:109–119. doi: 10.1007/s15010-021-01646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg SM, Lee SI. A unified approach to interpreting model predictions. (Nips 2017) Adv Neural Inf Process Syst. 2017;30 [Google Scholar]
- Mirijello A, Piscitelli P, de Matthaeis A, Inglese M, D'Errico MM, Massa V, et al. Low eGFR is a strong predictor of worse outcome in hospitalized COVID-19 patients. J Clin Med. 2021;10 doi: 10.3390/jcm10225224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhtari T, Hassani F, Ghaffari N, Ebrahimi B, Yarahmadi A, Hassanzadeh G. COVID-19 and multiorgan failure: a narrative review on potential mechanisms. J Mol Histol. 2020;51:613–628. doi: 10.1007/s10735-020-09915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadim MK, Forni LG, Mehta RL, Connor MJ, Jr Liu KD, Ostermann M, et al. COVID-19-associated acute kidney injury: consensus report of the 25th Acute Disease Quality Initiative (ADQI) Workgroup. Nat Rev Nephrol. 2020;16:747–764. doi: 10.1038/s41581-020-00356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CB, Lima CAD, Vajgel G, Campos Coelho AV, Sandrin-Garcia P. High burden of acute kidney injury in COVID-19 pandemic: systematic review and meta-analysis. J Clin Pathol. 2021;74:796–803. doi: 10.1136/jclinpath-2020-207023. [DOI] [PubMed] [Google Scholar]
- Ouyang L, Gong Y, Zhu Y, Gong J. Association of acute kidney injury with the severity and mortality of SARS-CoV-2 infection: a meta-analysis. Am J Emerg Med. 2021;43:149–157. doi: 10.1016/j.ajem.2020.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelayo J, Lo KB, Bhargav R, Gul F, Peterson E, DeJoy Iii R, et al. Clinical characteristics and outcomes of community- and hospital-acquired acute kidney injury with COVID-19 in a US inner city hospital system. Cardiorenal Med. 2020;10:223–231. doi: 10.1159/000509182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Li X, Liu F, Tian T, Luo J, Yang Y. Acute kidney injury is associated with severe infection and fatality in patients with COVID-19: a systematic review and meta-analysis of 40 studies and 24,527 patients. Pharmacol Res. 2020;161 doi: 10.1016/j.phrs.2020.105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Hoshmand-Kochi M, Abbasi A, Glass S, Jiang Z, Singer AJ, et al. Initial chest radiograph scores inform COVID-19 status, intensive care unit admission and need for mechanical ventilation. Clin Radiol. 2021;76:473. doi: 10.1016/j.crad.2021.02.005. e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry AR, Roch B. SARS-CoV2 may evade innate immune response, causing uncontrolled neutrophil extracellular traps formation and multi-organ failure. Clin Sci (Lond) 2020;134:1295–1300. doi: 10.1042/CS20200531. [DOI] [PubMed] [Google Scholar]
- Trabulus S, Karaca C, Balkan II, Dincer MT, Murt A, Ozcan SG, et al. Kidney function on admission predicts in-hospital mortality in COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Garcia-Rodriguez V, Yu A, Dutra B, DuPont A, Cash B, et al. Elevated D-dimer is associated with multiple clinical outcomes in hospitalized Covid-19 patients: a retrospective cohort study. SN Compr Clin Med. 2020;2:2561–2567. doi: 10.1007/s42399-020-00627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wen Y, Duan Y, Su H, Cao W, Xiao M, et al. Clinicopathological features and outcomes of acute kidney injury in critically ill COVID-19 with prolonged disease course: a retrospective cohort. J Am Soc Nephrol. 2020;31:2205–2221. doi: 10.1681/ASN.2020040426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Chen A, Hou W, Graham JM, Li H, Richman PS, et al. Prediction model and risk scores of ICU admission and mortality in COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0236618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.