Abstract
The randomly amplified polymorphic DNA (RAPD) method was used to investigate the genetic diversity in Xanthomonas cynarae, which causes bacterial bract spot disease of artichoke. This RAPD analysis was also intended to identify molecular markers characteristic of this species, in order to develop PCR-based markers which can be used to detect this pathogenic bacterium in artichoke fields. Among the 340 RAPD primers tested, 40 were selected on their ability to produce reproducible and reliable fingerprints in our genetic background. These 40 primers produced almost similar patterns for the 37 X. cynarae strains studied, different from the fingerprints obtained for other Xanthomonas species and other xanthomonad-like bacteria isolated from artichoke leaves. Therefore, X. cynarae strains form a homogeneous genetic group. However, a little DNA polymorphism within this species was observed and the collection of X. cynarae isolates was divided into two groups (one containing three strains, the second one including all other strains). Out of seven RAPD markers characteristic of X. cynarae that were cloned, four did not hybridize to the genomic DNA of strains belonging to other Xanthomonas species. These four RAPD markers were converted into PCR markers (specific characterized amplified regions [SCARs]); they were sequenced, and a PCR primer pair was designed for each of them. Three derived SCARs are good candidates to develop PCR-based tests to detect X. cynarae in artichoke fields.
Water-soaked and dark green spots on capitulum bracts of artichoke (Cynara scolymus L.) were observed for the first time in 1954 in Brittany and near Angers (France). Warm and humid periods are favorable for development of this disease, which has been observed in numerous artichoke crops in Brittany and has resulted in substantial economic losses for the last decade. The causal agent of this disease was first isolated by Ridé (25) from such bract spots and was identified as a phytopathogenic bacterium belonging to the genus Xanthomonas (25). It was recently classified at the species level as Xanthomonas cynarae (33). An identification test of X. cynarae was described based on biochemical, physiological, and pathogenicity tests (33). Colonies of X. cynarae are yellow and surrounded with a white halo when grown on a Tween medium (17) and produce the typical symptoms of this disease when inoculated to detached scarified bracts of artichoke. A pathogenicity test is required, since other Xanthomonas species look like X. cynarae when grown on Tween medium. This identification procedure, including pathogenicity tests, was used to monitor bacterial populations in artichoke fields and showed that X. cynarae is present on the leaf surface of artichoke before the capitulum development.
A rapid and specific identification test would be very useful to monitor the contamination of artichoke plants in order to develop strategies to control the disease in fields. Since the diagnostic test described above is time consuming and requires, for the pathogenicity test, artichoke bracts which are not available early in the season when the test is required, the aim of this work was to develop a rapid and specific test for the identification of X. cynarae, based on the PCR technique (12). To achieve this goal, we used a two-step strategy. First, the genetic diversity in X. cynarae was assessed by random amplified polymorphic DNA (RAPD) (36) to determine the population structure of X. cynarae, as previously described for many other bacterial species (5, 7, 10, 15, 18, 21, 22, 31). Secondly, we selected RAPD markers specific to X. cynarae strains in order to convert them into specific characterized amplified region (SCAR) markers (20) which would be useful in an identification scheme of X. cynarae; as they have proven to be with numerous other pathogenic bacteria (4, 9, 16, 23, 24, 26).
MATERIALS AND METHODS
Isolates used.
Details on the bacterial strains used in this study are given in Table 1. In addition, 38 strains nonpathogenic to artichoke (C. scolymus L.), isolated either from artichoke leaves (28 strains) or from weed leaves (10 strains; these weeds were either Senebiera sp. or Lunaria annua), were used specifically to assess the specificity to X. cynarae of the SCAR markers obtained in this study. All strains were restreaked for colony uniformity on LPGA medium (yeast extract, 7 g; Bacto Peptone, 7 g; glucose, 7 g; agar-agar, 15 g; H2O, 1,000 ml) and on a Tween medium (17). All strains were grown on Luria-Bertani medium (28) for 24 h at 24°C, except for Xanthomonas populi, which was grown at 19°C.
TABLE 1.
List of bacterial strains used in this study
| Species | CFBPa strain no. | Host | Geographical origin | Yr of isolation |
|---|---|---|---|---|
| Xanthomonas cynarae | ||||
| 18, 19 | C. scolymus L. | France | 1957 | |
| 2044 | C. scolymus L. | France | 1981 | |
| 4182, 4183, 4185, 4186, 4187, 4188Tb, 4189, 4190, 4194, 4195, 4196, 4197, 4198, 4199 | C. scolymus L. | France | 1996 | |
| 4200, 4201, 4926, 4927, 4928, 4929, 4930, 4932, 4934, 4935, 4936, 4937, 4938, 4939, 4940, 4941 | C. scolymus L. | France | 1997 | |
| 4719 | C. scolymus L. | France | 1998 | |
| 4209 | Senebiera sp. | France | 1996 | |
| 4943 | Senebiera sp. | France | 1997 | |
| 4942 | Lunaria annua | France | 1997 | |
| Other Xanthomonas species | ||||
| X. albilineans | 2523T | |||
| X. arboricola pv. juglandis | 2528T | |||
| X. arboricola pv. corylina | K100st | |||
| X. axonopodis pv. phaseoli | 2534PTc | |||
| X. bromi | 3625PT | |||
| X. campestris pv. campestris | 2350T, 1713, 1869 | |||
| X. cassavae | 4642T | |||
| X. codiaei | 4690T | |||
| X. cucurbitae | 2542T | |||
| X. fragariae | 2057T | |||
| X. hortorum pv. pelargonii | 2533PT | |||
| X. hyacinthi | 1156T | |||
| X. melonis | 4644T | |||
| X. oryzae pv. oryzae | 2532T | |||
| X. pisi | 4643T | |||
| X. populi | 1817T | |||
| X. sacchari | 4641T | |||
| X. theicola | 4691T | |||
| X. translucens pv. translucens | 2054T | |||
| X. vasicola pv. holcicola | 2543T | |||
| X. vesicatoria | 4645T | |||
| Pseudomonas syringae (two strains) | ||||
CFBP, Collection Française de Bactéries Phytopathogènes, Station de Pathologie Végétale, Angers, France.
T, type strain.
PT, pathotype strain.
DNA extraction.
For RAPD experiments, genomic DNA was extracted following the standard cetyltrimethylammonium bromide (CTAB) methodology described by Ausubel et al. (2). DNA was approximately quantified by electrophoresis in 1.4% (wt/vol) agarose gels in 0.5× Tris-borate-EDTA (28) in comparison with a few DNA samples previously precisely quantified (by using a spectrophotometer). Each DNA concentration was adjusted to 10 ng/μl in sterile distilled water and stored at −20°C. For PCR analyses, crude bacterial DNAs were obtained by a simple boiling method: 20 μl of bacterial suspension (optical density at 600 nm, 0.1) was boiled for 5 min and stored at −20°C. Phytophthora DNA and artichoke DNA were extracted by a standard CTAB-based procedure.
RAPD analysis.
The bacterial RAPD fingerprints were obtained using the procedure described by Williams et al. (36), with minor modifications (3). Briefly, amplifications were carried out in a final volume of 15 μl containing 1× reaction buffer (Promega, Madison, Wis.), 100 μM concentrations of each deoxynucleotide (Eurobio, Les Ulis, France), 1.5 mM MgCl2, a 0.5 μM concentration of the primer (Operon Technologies, Alameda, Calif.), 0.06 U of Taq DNA polymerase (Promega) per μl, and approximately 10 ng of genomic DNA. RAPD reactions were performed in a 96-well thermal cycler (PTC-100; MJ Research Inc., Watertown, Mass.), with one step at 93°C for 2 min 30 s and 45 cycles at 93°C for 30 s, 35°C for 45 s, and 72°C for 90 s, followed by one step at 72°C for 5 min. Following agarose gel electrophoresis and ethidium bromide staining, amplified patterns were visualized under UV light (300 nm). RAPD amplified bands were scored visually according to their presence or absence for the bacteria studied. Only clear, unambiguous, and reproducible RAPD markers were taken into account. The reproducibility of each scored marker was checked by at least two RAPD experiments. Genetic distances between X. cynarae strains were calculated according to the formula established by Nei and Li (19).
Development of specific SCAR markers for X. cynarae, from characteristic RAPD markers. (i) Cloning of DNA fragments from RAPD fingerprints.
Several RAPD fragments common to all X. cynarae strains were recovered from agarose gels with a scalpel and purified by using the Ultrafree-DA kit (Millipore, Bedford, Mass.). The extracted DNA fragments were used as templates for another round of amplification using the same RAPD primers with a slightly increased annealing temperature. When such reamplifications resulted in amplicons of the expected sizes, they were cleaned using the Centrifugal Filter Services kit (Millipore) and cloned into the pCR4-TOPO vector using the TOPO TA Cloning kit (Invitrogen, Carlsbad, Calif.), as recommended by the manufacturer.
(ii) DNA probes.
DNA probes were made from recombinant plasmids (one for each cloned DNA marker) by PCR amplifications using oligonucleotide primers T7 and T3 (Invitrogen) flanking the cloning site of the pCR4-TOPO vector and by incorporating digoxigenin-labeled nucleotide (DIG-dUTP) as described by Hoisington (14). Probes were stored at −20°C before hybridization tests.
(iii) Southern blots and membrane hybridization.
DNAs from three strains of X. cynarae (from the Collection Française de Bactéries Phytopathogènes [CFBP], CFBP 19, CFBP 4188T, and CFBP 4199) and from type strains of 19 of the 20 Xanthomonas species described by Vauterin et al. (34) were amplified with the RAPD primer which produced the cloned fragment to be tested. After electrophoresis, DNA patterns were transferred onto a Hybond-N+ nylon membrane (Amersham, Arlington Heights, Ill.) by capillary blotting (30). Moreover, Xanthomonas strains were grown on nylon membranes according to the method of Sambrook et al. (28) before a colony hybridization step. Prehybridization, hybridization, and DNA hybrid detection were performed as described by Hoisington (14). Autoradiographies were obtained on Kodak X-Omat films after up to 15 min of exposure.
(iv) DNA sequencing and primer design.
Cloned DNA fragments of interest were sequenced (around 300 nucleotides from both ends of each DNA fragment) by Cybergene (Evry, France). Nucleotide sequence data were compared against the Genbank nucleotide sequence database (BLAST search) and were analyzed with the Primer 3 software in order to design pairs of PCR primers (20-mers) for obtaining SCAR markers characteristic of X. cynarae. Oligonucleotides were synthesized by Cybergene.
PCR conditions for the amplification of X. cynarae-specific markers (SCARs).
PCR amplifications were carried out in a final volume of 40 μl containing 1× buffer (Promega), 1.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, 0.2 μM concentrations of each primer, 0.02 U of Taq DNA polymerase (Promega) per μl, and 2 μl of a boiled bacterial suspension. The reaction mixtures were subjected to 35 cycles of the following incubations: 45 s at 93°C (except for the first one, 3 min 45 s), 45 s at the appropriate annealing temperature (52°C for primer pair XC800, 55°C for XC420, and 60°C for primer pairs XC1000 and XC850), and 60 s at 72°C (except for the last cycle, 10 min).
RESULTS
RAPD analysis.
In a first trial, 340 RAPD primers (10-mer oligonucleotides with arbitrary sequences) were tested on a few bacterial strains (four strains of X. cynarae, two strains of other Xanthomonas species, and two xanthomonad-like strains isolated from artichoke leaves but nonpathogenic to this plant) in order to select a set of RAPD primers which produce reliable and reproducible fingerprints for the bacteria studied. Forty primers were retained for assessment of the genetic diversity within the X. cynarae species.
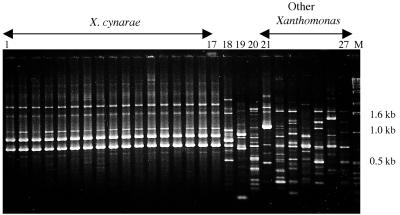
The 40 selected RAPD primers were used to analyze genomic DNAs of all 37 X. cynarae strains, 7 other Xanthomonas bacteria (K 100st, CFBP 2534, CFBP 1713, CFBP 1869, CFBP 2542, CFBP 2532, and CFBP 2054), and 4 epiphytic xanthomonad-like strains isolated from the leaves of artichokes (or weeds [Senebiera sp. or Lunaria annua]) but nonpathogenic to artichoke plants (Table 1). With each primer, we obtained RAPD patterns which were similar for all X. cynarae strains but distinct from those observed for the other bacteria studied (Fig. 1 and 2). Since all X. cynarae strains shared one or several common RAPD markers characteristic of this species with each primer used, RAPD results showed that these 37 strains form a single genetic group. Overall, we obtained 112 RAPD markers shared among all X. cynarae strains. The reproducibility of all these markers was checked by performing at least two experiments per RAPD primer.
FIG. 1.
RAPD fingerprints characteristic of X. cynarae species, epiphytic strains nonpathogenic to artichoke, and other Xanthomonas species. Lanes 1 to 17, X. cynarae strains (CFBP no. 4182, 4183, 4185, 4186, 4188, 4190, 4199, 4200, 4942, 4943, 2044, 4719, 4209, 4930, 4931, 18, and 19) belonging to the three genetic subgroups A1, A2, and B (see text); lanes 18 to 20, epiphytic strains nonpathogenic to artichoke (CFBP no. 4184, 4191, and 4192); lanes 21 to 27, strains belonging to 6 other Xanthomonas species (CFBP no. 1713, 1869, 2542, 2532, 2054, K100st, and 2534: Table 1); lane M, molecular weight marker (1-kb ladder; Gibco BRL).
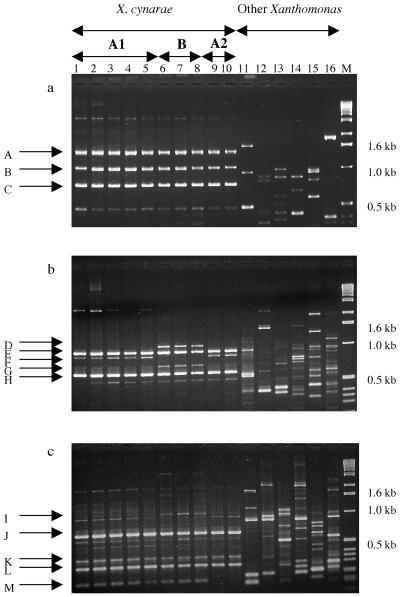
FIG. 2.
Examples of RAPD fingerprints revealing the genetic diversity within the X. cynarae species. Lanes 1 to 10, X. cynarae strains (CFBP no. 2044, 4188, 4199, 4719, 4200, 4209, 4929, 4930, 18, and 19) belonging to RAPD groups A1, A2, or B, as indicated above panel a; lanes 11 to 16, strains belonging to other Xanthomonas species (CFBP no. 1713, 2542, 2532, 2054, K100st, and 2534; Table 1); lane M, molecular weight marker (1-kb ladder; Gibco BRL). (a) A, B, and C are RAPD markers characteristic of X. cynarae. (b) E and H are RAPD markers shared by all X. cynarae strains (A1, A2, and B); D and G are RAPD markers characteristic of group B; F is a RAPD marker characteristic of group A (A1 and A2). (c) I is a faint marker characteristic of group B; J, K, and L are RAPD markers shared by all X. cynarae strains (A1, A2, and B); M is a RAPD marker shared by subgroup A1 and group B.
More precisely, 20 RAPD primers provided identical fingerprints for all 37 X. cynarae strains (Fig. 1 and 2a). However, the 20 remaining primers revealed a genetic polymorphism within this species (Fig. 2b and c). Therefore, we identified 3 RAPD subgroups for X. cynarae, named A1, A2, and B. Table 2 recapitulates these results, in terms of the number of markers that distinguish each subgroup and in terms of genetic distances between subgroups. Subgroups A1 and A2 are very similar, since the distance between them is only 0.007. Both of them are much more distant to group B, since the distances observed between A1 and B and between A2 and B are 0.177 and 0.185, respectively. These results explain our subgroup names. The distribution of all 37 X. cynarae strains within the three identified RAPD subgroups is presented in Table 3. Most of them belong to subgroup A1, which contains strains isolated in Brittany and in the Loire valley mainly from artichoke leaves, but also from weed leaves (2 strains out of 32). Subgroup A2 contains two strains isolated from artichoke leaves in Brittany in 1957. The three strains which belong to group B were recently isolated in Brittany, either from artichoke leaves or from a weed (Senebiera sp.) present in an artichoke field.
TABLE 2.
Genetic distances between the 3 RAPD subgroups identified within the 37 X. cynarae strains studied with 40 RAPD primers
| RAPD subgroup | No. of RAPD fragments or genetic distancea
|
||
|---|---|---|---|
| A1 | A2 | B | |
| A1 | 2 | 49 | |
| A2 | 0.007 | 51 | |
| B | 0.177 | 0.185 | |
Numbers of RAPD fragments distinguishing RAPD subgroups are indicated above the diagonal; genetic distances, calculated according to the method of Nei and Li (19), are given below the diagonal.
TABLE 3.
Distribution in the three RAPD subgroups and characteristics of the 37 X. cynarae strains
| RAPD subgroup | No. of strains | Isolated on: | Geographic location | Yr of isolation |
|---|---|---|---|---|
| A1 | 28 | Artichoke | Brittany, France | 1996–97 |
| 2 | Weeds | Brittany, France | 1997 | |
| 2 | Artichoke | Loire valley, France | 1981–98 | |
| A2 | 2 | Artichoke | Brittany, France | 1957 |
| B | 1 | Senebiera sp. | Brittany, France | 1996 |
| 2 | Artichoke | Brittany, France | 1997 |
Selection of RAPD markers characteristic of X. cynarae for development of SCARs.
We detected at least one RAPD band common to all X. cynarae strains which was absent in all other studied bacteria for each of the 40 RAPD primers used. In order to obtain SCARs specific to X. cynarae, and therefore usable for the identification of this pathogen, we selected seven of these RAPD fragments for cloning. Our selection criteria were a sufficient DNA length (>400 bp) in order to maximize the availability of convenient sites for designing PCR primers and a strong intensity of the DNA band associated with its location as far as possible from other bands, to increase the chance of cloning the targeted marker. Figure 2a displays three examples (markers A, B, and C) of such selectable RAPD markers.
The lack of DNA sequences homologous to the seven cloned RAPD bands in other Xanthomonas genomes was tested by using them as probes in two hybridization experiments: (i) on the RAPD patterns of a representative subset of bacterial strains previously used and obtained with the primer which produced the cloned marker (after Southern blotting) and (ii) on the genomic DNA of 3 X. cynarae strains (CFBP 19, CFBP 4188T, CFBP 4199) and 19 other Xanthomonas strains (after dot blotting). In the first experiment, for each of the seven probes no hybridization signal was obtained in the RAPD patterns of non-X. cynarae strains, whereas a very strong signal was obtained at the location of the cloned marker in X. cynarae RAPD patterns (data not shown). In the second analysis, only four of the seven probes hybridized exclusively to the genomic DNA of the three X. cynarae strains (no signal was obtained for other Xanthomonas strains). The three other probes produced a strong signal with the three X. cynarae strains (high DNA homology) but also a faint one with at least one other Xanthomonas strain. Little DNA homology was observed with Xanthomonas fragariae DNA in the three cases, with Xanthomonas bromi DNA with one probe, and with Xanthomonas cucurbitae DNA with another probe. We therefore retained the first four probes (cloned RAPD markers) as good candidates to design PCR primers specific to the X. cynarae species.
Design of X. cynarae-specific primers.
The DNA sequences of the first 300 bases from both terminal ends of the four selected RAPD fragments were determined. None of these sequences showed a highly significant DNA homology with gene sequences stored in the Genbank database. The analysis of these sequences, using the software Primer 3, allowed us to design a PCR primer pair usable for amplifying each of the four markers. The designed primers are 20-mers with melting temperatures (Tm) of around 60°C (usual values for PCR primers; Table 4). We named the four primer pairs XC1000, XC850, XC800, and XC420 based on the expected sizes of the fragments to be amplified.
TABLE 4.
Characteristics of the four primer pairs designed for amplifying PCR markers specific to the X. cynarae species
| Primer pair | Sequence | %G-C | Tm (°C) |
|---|---|---|---|
| XC800 | TAA GGG AAA TGA TCG GCA TC | 45 | 55.3 |
| CAA GGA ATT GAG CTT CTC CG | 50 | 57.3 | |
| XCS6 | TGA TAG ATC CCA AGA TGG GC | 50 | 57.3 |
| AAT GTC TCG CCT TCG TAT GG | 50 | 57.3 | |
| XC420 | GGC AGG CTG TTC GGT AGG TG | 65 | 63.5 |
| GGC AGG CTG TAG TTG GTT GA | 55 | 59.4 | |
| XC1000 | ACT CGC TCG GAG ACT CAT GT | 55 | 59.4 |
| AGG ATG TCG TGT TGT TGC CC | 55 | 59.4 |
Only the primer pair XC420 was expected to produce the whole RAPD fragment (full length), since both PCR primers were designed with the RAPD primer sequence at their 5′ end (the 10 additional nucleotides of each primer are the ones which directly followed the RAPD primer sequence on both ends). In the three other cases, expected SCARs are shorter than the initial RAPD fragments, since the DNA sequences upon which the primers were designed were internal to the RAPD fragments.
Specificity of the four SCARs for X. cynarae.
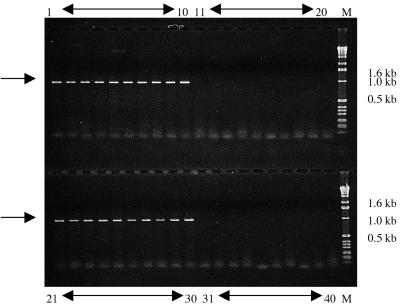
Each of the four designed PCR primer pairs was then tested for its ability to produce an amplicon specific to the X. cynarae species, which is a requirement in order to use such PCR primers for identifying this pathogenic species on artichoke leaves. For that purpose, each primer pair was used on a large panel of microorganisms. Each primer pair produced the same PCR fragment for all 37 X. cynarae strains, with the expected DNA length. Primer pairs XC1000, XC850, and XC800, when used with a sufficiently high annealing temperature, did not produce any PCR fragment for all other microorganisms studied (including Phytophtora and Pseudomonas strains) or from artichoke genomic DNA. Figure 3 illustrates the specificity to X. cynarae of the SCAR marker obtained using the primer pair XC1000. However, the primer pair XC420 amplified from the DNA of one nonpathogenic xanthomonad-like strain a DNA fragment similar in size and intensity to the one obtained from X. cynarae strains, even when the most stringent annealing conditions were applied.
FIG. 3.
SCAR marker characteristic of X. cynarae obtained by using the primer pair XC1000. Lanes 1 to 10, X. cynarae strains (CFBP no. 4182, 4183, 4188, 4190, 4192, 4194 4199, 4200, 18, and 19); lanes 11 to 20, strains belonging to other Xanthomonas species (CFBP no. 2523, 2528, 2534, 3625, 2350, 4642, 4690, 2542, 2057, and 2533; Table 1); lanes 21 to 30, X. cynarae strains (CFBP no. 4926, 4927, 4928, 4942, 4943, 2044, 4719, 4209, 4929, and 4930); lanes 31 to 40, epiphytic strains nonpathogenic to artichoke; lane M, molecular weight marker (1-kb ladder; Gibco BRL). The SCAR marker locations are indicated by arrows.
DISCUSSION
The RAPD analysis we performed was intended to assess the genetic diversity within the X. cynarae species. RAPD fingerprints allow intraspecific genetic diversity studies but are not very good tools for such analyses at the interspecific level (8). Therefore, we did not use the RAPD technique for a phylogenic study in the genus Xanthomonas. More appropriate technical tools (phenotypical and biochemical tests, DNA-DNA hybridization, and 16S ribosomal DNA sequencing) were used to classify X. cynarae in the genus Xanthomonas (33).
Before starting the RAPD study within the X. cynarae species, we performed a primer-screening step, in order to select a set of RAPD primers which produces reliable fingerprints. Indeed, a bacterial genome is small and therefore may not possess enough sites homologous to a given RAPD primer to amplify only genomic DNA fragments based on good matches of the primer on both ends of each fragment. Nevertheless, in such a case, a RAPD fingerprint is produced from the bacterial DNA, but this pattern is usually nonreproducible, since the selected genomic fragments to be amplified may change from one experiment to another, resulting in different RAPD fingerprints (29). Therefore, by using a set of eight different bacteria (four X. cynarae, two xanthomonad-like bacteria isolated from artichoke but nonpathogenic to this plant, and two other Xanthomonas species), we selected 40 RAPD primers (out of 340 tested) for their ability to produce reproducible fingerprints as in other studies (7, 18, 21, 31, 32). Moreover, we took into account only the intensive bands that are highly reproducible (6, 9, 13, 21) when analyzing RAPD fingerprints.
With the 40 selected RAPD primers, we obtained many species-specific markers for X. cynarae, in agreement with the fact that X. cynarae is a genomic species in the genus Xanthomonas (33). Moreover, 112 of 163 RAPD markers were common to all X. cynarae strains. However, a genetic diversity was observed within X. cynarae, since we obtained 51 polymorphic DNA markers within the X. cynarae species, which revealed two main genetic groups, A and B. Furthermore, 2 markers of the 163 which were scored distinguished two subgroups (A1 and A2) in group A. Subgroup A1 seems widespread in Brittany, since 30 strains of the 35 isolated in this area in 1996 and 1997 belong to this subgroup (Table 3). It is also present in the Loire valley, as indicated by the two studied strains isolated there in 1981 and 1998. Subgroup A2 includes only two X. cynarae strains isolated 40 years ago (Table 3). This subgroup A2 is very similar to subgroup A1. If we assume that strains belonging to subgroup A2 are representative of the bacterial population present in Brittany 40 years ago, we can conclude that X. cynarae appears to be stable in this region and that group A forms the core of a clonal population.
We only found three strains belonging to group B, all of them recently isolated in Brittany. This group B, although unambiguously composed of X. cynarae strains, is quite different from group A (Table 2). We were not able to identify any particular characteristic for the artichoke fields where group B strains were found (random geographical distribution, no rain or wind exposure specificity, no correlation with the artichoke cultivar grown or with the previous crop in the field). However, it may be possible that X. cynarae strains belonging to group B were introduced in Brittany by growing artichokes coming from Italy or south-east France. Indeed, Italian artichokes were grown in Brittany during a few years because of an unusual long period of deep frost which destroyed a large part of the local artichoke crop during the winter of 1963-64. Moreover, cultivars of little artichokes, very popular in south-east France, have been grown in Brittany since 1993. Even if Italian artichokes and French little artichokes are not known to be damaged by the bacterial bract spot disease, they may carry a X. cynarae population. In order to confirm these hypotheses, it would be interesting to check whether X. cynarae is present in south-east France and/or in Italy. In such a case, we would then determine whether those strains belong to group B only.
Three bacterial strains (CFBP 4209, CFBP 4942, and CFBP 4943; Table 1) isolated from the leaf surfaces of weeds which were present in artichoke fields were identified as X. cynarae strains based on their phenotypical characteristics and their ability to produce typical symptoms on detached bracts. Two of them belong to subgroup A1, and the third one belongs to group B (Table 3). Therefore, it would be interesting to determine whether X. cynarae is able to multiply on such weeds, which would then be identified as sources of contamination in artichoke crops. The three strains we studied may also have been simply washed off from infected artichoke plants.
In the absence of a simple and fast identification procedure of X. cynarae strains, our aim was to develop a PCR-based test to identify X. cynarae. Indeed, such tests are reported to be sensitive, reliable, and fast, which explains why PCR-based techniques have become more and more widespread for detecting plant pathogens (12). In order to develop PCR markers characteristic of X. cynarae, we first identified RAPD markers shared by all X. cynarae strains (37 isolates) and not present in the RAPD fingerprints of other Xanthomonas species and xanthomonad-like bacteria isolated from artichoke leaves but nonpathogenic to this plant. Such RAPD markers cannot be used to detect X. cynarae in a mixture of different bacteria (for example, after a wash of artichoke leaves), since RAPD primers are nonspecific and can amplify DNA fragments from any genome (11, 27, 35). This is the reason why we had to convert RAPD markers characteristic of X. cynarae into SCAR markers (i.e., PCR markers). Nevertheless, the RAPD analysis was useful for studying the population structure of X. cynarae.
Of the seven RAPD markers characteristic of X. cynarae that were cloned, the four which were homologous only to X. cynarae genomic DNA were sequenced. Of the four PCR primer pairs designed, three produced an X. cynarae-specific amplicon (XC1000, XC850, and XC800) that is amplified for all three X. cynarae genetic subgroups (A1, A2, and B). Therefore, the low genetic diversity observed in the X. cynarae species facilitated the development of species-specific SCARs. For a species (or a pathovar) in which the genetic diversity is high, it may be more difficult to develop a single SCAR characteristic of the whole species (or pathovar), and several SCARs may be required to detect all isolates of the targeted bacterial group (1).
A PCR-based test to detect X. cynarae on artichoke leaves is now available in our laboratory. Now, we intend to use the primers developed in this study in a quantitative PCR test with a fluorimetric procedure (i.e., the TaqMan technology developed by Perkin-Elmer, Foster City, Calif.). Indeed, a quantitative detection of X. cynarae would be necessary to determine whether outbreaks of the disease are correlated to the level of the bacterial population on artichoke leaves. We are also interested in developing SCAR markers characteristic of X. cynarae groups A and B, in order to develop epidemiological markers which may be useful for studying the spread of the inoculum in Brittany.
ACKNOWLEDGEMENTS
We thank G. Barbeyron and P. Rouault for technical assistance, L. Gardan for providing the type and pathotype strains of the 20 Xanthomonas species described by Vauterin et al. (34), and T. Lunn for reading the manuscript.
Financial support from the Conseil Régional de Bretagne is gratefully acknowledged.
REFERENCES
- 1.Arnold D L, Athey-Pollard A, Gibbon M J, Taylor J D, Vivian A. Specific oligonucleotide primers for the identification of Pseudomonas syringae pv. pisi yield one of two possible DNA fragments by PCR amplification: evidence for phylogenetic divergence. Physiol Mol Plant Pathol. 1996;49:233–245. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingdom R E, Moore D D, Smith J A, Sideman S G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates, Wiley Interscience; 1992. [Google Scholar]
- 3.Barbeyron G, Boury S. Marker assisted selection in cauliflower (Brassica oleracea var. botrytis): molecular mapping of a nuclear recessive male sterility gene. Acta Hortic. 1998;459:149–156. [Google Scholar]
- 4.Berthier F, Ehrlich S D. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int J Syst Bacteriol. 1999;49:997–1007. doi: 10.1099/00207713-49-3-997. [DOI] [PubMed] [Google Scholar]
- 5.Birch P R J, Hyman L J, Taylor R, Opio A F, Bragard C, Toth I K. RAPD PCR-based differentiation of Xanthomonas campestris pv. phaseoli and Xanthomonas campestris pv. phaseoli var. fuscans. Eur J Plant Pathol. 1997;103:809–814. [Google Scholar]
- 6.Boury S, Lutz I, Gavalda M C, Guidet F, Schlesser A. Empreintes génétiques du chou-fleur par RAPD et vérification de la pureté hybride F1 d'un lot de semences. Agronomie. 1992;12:669–681. [Google Scholar]
- 7.Clerc A, Manceau C, Nesme X. Comparison of randomly amplified polymorphic DNA with amplified length polymorphism to assess genetic diversity and genetic relatedness within genospecies III of Pseudomonas syringae. Appl Environ Microbiol. 1998;64:1180–1187. doi: 10.1128/aem.64.4.1180-1187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke D E L, Forster J W, Jenkins P D, Jones D G, Lewis D M. Analysis of intraspecific and interspecific variation in the genus Alternaria by the use of RAPD-PCR. Ann Appl Biol. 1998;132:197–209. [Google Scholar]
- 9.Day W A, Pepper I L, Joens L A. Use of an arbitrarily primed PCR product in the development of a Campylobacter jejuni-specific PCR. Appl Environ Microbiol. 1997;63:1019–1023. doi: 10.1128/aem.63.3.1019-1023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farber J M, Addison C J. RAPD typing for distinguishing species and strains in the genus Listeria. J Appl Bacteriol. 1994;77:242–250. doi: 10.1111/j.1365-2672.1994.tb03070.x. [DOI] [PubMed] [Google Scholar]
- 11.Fegan M, Croft B J, Teakle D S, Hayward A C, Smith G R. Sensitive and specific detection of Clavibacter xyli subsp. xyli, causal agent of ratoon stunting disease of sugarcane, with a polymerase chain reaction-based assay. Plant Pathol. 1998;47:495–504. [Google Scholar]
- 12.Henson J M, French R. The polymerase chain reaction and plant disease diagnosis. Annu Rev Phytopathol. 1993;31:82–109. doi: 10.1146/annurev.py.31.090193.000501. [DOI] [PubMed] [Google Scholar]
- 13.Heun M, Helentjaris T. Inheritance of RAPDs in F1 hybrids of corn. Theor Appl Genet. 1993;85:961–968. doi: 10.1007/BF00215035. [DOI] [PubMed] [Google Scholar]
- 14.Hoisington D. Laboratory protocols: CIMMYT applied molecular genetics laboratory. Mexico, D.F., Mexico: CIMMYT; 1992. [Google Scholar]
- 15.Lawrence L M, Harvey J, Gilmore A. Development of a random amplification of polymorphic DNA typing method for Listeria monocytogenes. Appl Environ Microbiol. 1993;59:3117–3119. doi: 10.1128/aem.59.9.3117-3119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manulis S, Valinsky L, Lichter A, Gabriel D W. Sensitive and specific detection of Xanthomonas campestris pv. pelargonii with DNA primers and probes identified by random amplified polymorphic DNA analysis. Appl Environ Microbiol. 1994;60:4094–4099. doi: 10.1128/aem.60.11.4094-4099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuire R G, Jones J B, Sasser M. Tween media for semi-selective isolation of Xanthomonas campestris pv. vesicatoria from soil and plant material. Plant Dis. 1986;70:887–891. [Google Scholar]
- 18.Momol M T, Momol E A, Lamboy W F, Norelli J L, Beer S V, Aldwinckle H S. Characterization of Erwinia amylovora strains using random amplified polymorphic DNA fragments (RAPDs) J Appl Microbiol. 1997;82:389–398. doi: 10.1046/j.1365-2672.1997.00377.x. [DOI] [PubMed] [Google Scholar]
- 19.Nei M, Li W H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paran I, Michelmore R W. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet. 1993;85:985–993. doi: 10.1007/BF00215038. [DOI] [PubMed] [Google Scholar]
- 21.Parent J G, Lacroix M, Pagé D, Vézina L, Végiard S. Identification of Erwinia carotovora from soft rot diseased plants by random amplified polymorphic DNA (RAPD) analysis. Plant Dis. 1996;80:494–499. [Google Scholar]
- 22.Pooler M R, Hartung J S. Genetic relationships among strains of Xylella fastidiosa from RAPD-PCR data. Curr Microbiol. 1995;31:134–137. doi: 10.1007/BF00294290. [DOI] [PubMed] [Google Scholar]
- 23.Pooler M R, Hartung J S. Specific PCR detection and identification of Xylella fastidiosa strains causing citrus variegated chlorosis. Curr Microbiol. 1995;31:377–381. doi: 10.1007/BF00294703. [DOI] [PubMed] [Google Scholar]
- 24.Pooler M R, Ritchie D F, Hartung J S. Genetic relationships among strains of Xanthomonas fragariae based on random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus PCR data and generation of multiplexed PCR primers useful for the identification of this phytopathogen. Appl Environ Microbiol. 1996;62:3121–3127. doi: 10.1128/aem.62.9.3121-3127.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridé M. Sur une maladie nouvelle de l'artichaut (Cynara scolymus) C R Acad Sci. 1956;243:174–177. [Google Scholar]
- 26.Roeckel-Drevet P, Tourvieille J, Drevet J R, Says-Lesage V, Nicolas P, Tourvieille de Labrouhe D. Development of a polymerase chain reaction diagnostic test for the detection of the biotrophic pathogen Plasmopara halstedii in sunflower. Can J Microbiol. 1999;45:797–803. doi: 10.1139/w99-068. [DOI] [PubMed] [Google Scholar]
- 27.Samac D A, Nix R J, Oleson A E. Transmission frequency of Clavibacter michiganensis subsp. insidiosus to alfalfa seed and identification of the bacterium by PCR. Plant Dis. 1998;82:1362–1367. doi: 10.1094/PDIS.1998.82.12.1362. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Smith J J, Scott-Craig J S, Leadbetter J R, Bush G L, Roberts D L, Fulbright D W. Characterization of random amplified polymorphic DNA (RAPD) products from Xanthomonas campestris and some comments on the use of RAPD products in phylogenetic analysis. Mol Phylogenet Evol. 1994;3:135–145. doi: 10.1006/mpev.1994.1016. [DOI] [PubMed] [Google Scholar]
- 30.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 31.Tailliez P, Tremblay J, Ehrlich S D, Chopin A. Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD) Syst Appl Microbiol. 1998;21:530–538. doi: 10.1016/S0723-2020(98)80065-9. [DOI] [PubMed] [Google Scholar]
- 32.Thwaites R, Mansfield J, Eden-Green S, Seal S. RAPD and rep PCR-based fingerprinting of vascular bacterial pathogens of Musa spp. Plant Pathol. 1999;48:121–128. [Google Scholar]
- 33.Trébaol G, Gardan L, Manceau C, Tanguy J L, Tirilly Y, Boury S. Genomic and phenotypic characterization of Xanthomonas cynarae: a new species causing bacterial bract spot of artichoke (Cynara scolymus L.) Int J Syst Evol Microbiol. 2000;50:1471–1478. doi: 10.1099/00207713-50-4-1471. [DOI] [PubMed] [Google Scholar]
- 34.Vauterin L, Hoste B, Kersters K, Swings J. Reclassification of Xanthomonas. Int J Syst Bacteriol. 1995;45:472–489. [Google Scholar]
- 35.Verdier V, Mosquera G, Assigbétsé K. Detection of the cassava bacterial blight pathogen, Xanthomonas axonopodis pv. manihotis, by polymerase chain reaction. Plant Dis. 1998;82:79–83. doi: 10.1094/PDIS.1998.82.1.79. [DOI] [PubMed] [Google Scholar]
- 36.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]