Abstract
The edible fungus Pleurotus ostreatus (oyster mushroom) is an industrially produced heterothallic homobasidiomycete whose mating is controlled by a bifactorial tetrapolar genetic system. Two mating loci (matA and matB) control different steps of hyphal fusion, nuclear migration, and nuclear sorting during the onset and progress of the dikaryotic growth. Previous studies have shown that the segregation of the alleles present at the matB locus differs from that expected for a single locus because (i) new nonparental B alleles appeared in the progeny and (ii) there was a distortion in the segregation of the genomic regions close to this mating locus. In this study, we pursued these observations by using a genetic approach based on the identification of molecular markers linked to the matB locus that allowed us to dissect it into two genetically linked subunits (matBα and matBβ) and to correlate the presence of specific matBα and matA alleles with differences in monokaryotic growth rate. The availability of these molecular markers and the mating type dependence of growth rate in monokaryons can be helpful for marker-assisted selection of fast-growing monokaryons to be used in the construction of dikaryons able to colonize the substrate faster than the competitors responsible for reductions in the industrial yield of this fungus.
Incompatibility systems are mechanisms for the creation of variability preventing selfing. The phytopathogenic fungus Ustilago maydis and the mushrooms Coprinus cinereus and Schizophyllum commune have been used as models to study mating incompatibility in basidiomycetes. In these species, mating is controlled by two unlinked multiallelic loci whose independent segregation generates four mating specificities in the progeny of a single individual (these fungi are then called tetrapolar) (for reviews, see references 2, 7, and 11). In tetrapolar basidiomycetes, a single basidiospore produces upon germination a hypha in which all nuclei are identical (homokaryon). Two hyphae with different mating alleles at the two incompatibility loci are able to fuse and give rise to a mycelium in which the two parental nuclei do not fuse throughout vegetative growth. This kind of mycelium is called dikaryotic, and the individual mycelium is called a dikaryon. Vegetative growth is maintained until a set of environmental conditions triggers fruit body formation. Karyogamy occurs within the basidia, and it is immediately followed by meiosis producing four uninucleate spores. The monokaryotic and dikaryotic conditions can be distinguished by the presence of clamp connections in dikaryons and their lack in monokaryons. Clamp connections are hook-shaped structures involved in equal nuclei sorting to the daughter cells produced by mitosis.
Genetic studies carried out in C. cinereus and S. commune have shown that the A incompatibility locus codes for homeodomain-containing transcription factors (2, 13, 14, 18, 21, 24, 27, 28). The b mating-type locus of U. maydis is homologous to the A locus and also codes for homeodomain proteins (6, 20). The B incompatibility locus of C. cinereus and S. commune, homologous to the a locus of U. maydis, codes for pheromones and pheromone receptors (1, 12, 29, 30). Two subloci (Bα and Bβ) form this locus in S. commune, and recombination between them is possible, giving rise to nonparental incompatibility alleles (2). The bipartite structure of the B locus has been described also for other basidiomycetes such as Flammulina velutipes and Pleurotus ostreatus (3, 5, 15).
Larraya et al. (15) previously reported a genetic analysis of the A incompatibility locus in P. ostreatus var. florida using molecular markers; parental and nonparental B genes with distorted segregation were described, but the reasons for their occurrence were not examined. In this study, we examined the genetic bases for this distorted segregation by analyzing the structure of the mat B incompatibility locus and found a relationship between the polygenic trait vegetative growth rate and the mating genes. Moreover, we have developed molecular markers genetically linked to the mat B locus that will provide information allowing one to select in a quick, certain, and easy way monokaryons with allelic combinations suitable to produce compatible crosses for use in breeding programs and to establish the basis for the isolation of genomic clones that either contain the B locus or are adjacent to it.
MATERIALS AND METHODS
Strains, culture conditions, and experimental protocols.
The strains of P. ostreatus used in this work have been previously described (15–17, 19) (Table 1). Strains N001 (Navarra 001, P. ostreatus var. florida), N002, N005, and N006 are commercial, while N003 is a wild isolate from Viana, Spain. The two nuclei present in the dikaryotic strain N001 have been previously separated by dedikaryotization (16), and the corresponding protoclones are deposited in the Spanish Type Culture Collection (PC9 [CECT20311] and PC15 [CECT20312]). For comparisons with other Agaricales, Pleurotus quebecoise and commercial strains of Agaricus bisporus, Lentinus edodes, and Agrocybe aegerita were used.
TABLE 1.
P. ostreatus strains studied in this work and B alleles identified in their progeny
| Strain (mating genotype) | Variety | Origin | B allele | Occurrence of B allele (%) | Recombination frequency (%) | Pa |
|---|---|---|---|---|---|---|
| N001 (A1A2 B1B2) | florida | United States | B1 | 63 | 15.8 | <0.05 |
| B2 | 38 | |||||
| B3 | 11 | |||||
| B4 | 8 | |||||
| N017 (A1A2 B3B4) | florida | This study | B3 | 45 | 15.7 | NS |
| B4 | 41 | |||||
| B1 | 8 | |||||
| B2 | 8 | |||||
| N002 (A5A6 B5B6) | ostreatus | Germany | B5 | 39 | 8.2 | NS |
| B6 | 51 | |||||
| B15 | 6 | |||||
| B16 | 2 | |||||
| N018 (A5A6 B15B16) | ostreatus | This study | B15 | 41 | 4.8 | NS |
| B16 | 59 | |||||
| B5 | 3 | |||||
| B6 | 2 | |||||
| N003 (A7A8 B7B8) | ostreatus | Spain | B7 | 86 | 0.6 | NS |
| B8 | 83 | |||||
| B17 | 1 | |||||
| N005 (A11A12 B11B12) | colombinus | Italy | B11 | |||
| B12 | ||||||
| N006 (A13A14 B13B14) | sajor-caju | India | B13 | |||
| B14 |
The significance of any deviation from 1:1 in the segregation ratio of the two parental B alleles was determined by a chi-squared test. NS, not significant.
Molecular techniques, mating, and linkage analysis were performed as described by Larraya et al. (15–17), with the following modifications: (i) for the generation of rapidly amplified polymorphic DNA (RAPD) markers, oligonucleotides belonging to the L, P, R, and S Operon series (Operon Technologies Inc., Alameda, Calif.) were used as primers; and (ii) the PCR amplification program used included a 4-min denaturation at 94°C followed by 39 cycles of 1-min denaturation at 94°C, 1-min annealing at 37°C, and 1.5-min extension at 72°C.
Statistical analysis.
The quantitative trait vegetative mycelium growth rate was measured as the time elapsed from when a 16-mm2 agar plug containing the monokaryon was placed at the center of the plate until it reached the edge of the petri dish (9-cm diameter). Three repetitions for each of the 120 monokaryons derived from strain N001 were performed. The data were subjected to a normality test, and subsequently significant differences in vegetative growth rate among the different mating types were determined following one-way variance analysis using SPSS version 8.0 (SPSS Inc.) with treatment effect fixed.
RESULTS
Determination of B incompatibility alleles present in P. ostreatus.
The mating genotype of P. ostreatus N001 (A1A2 B1B2), as well as those of the other strains, was determined by crossing spore-derived monokaryons against the corresponding testers. In a previous study, Larraya et al. (15) analyzed the segregation of the incompatibility genes present in P. ostreatus N001, examining progeny of 120 monokaryons, and found that in addition to the two expected B mating genotypes (B1, B2), new nonparental B alleles appeared. These new B types were identified because monokaryons harboring them were compatible with two different N001 mating testers having the same A but a different B allele. In all progeny, the frequencies of the four B mating types were 52.5% (B1), 31.6% (B2), 9.2% (B3), and 6.7% (B4) (Table 1). Two alternative hypothesis explaining the generation of these new B alleles were posited: they appeared as the result of an intralocus recombination event; alternatively, alleles B3 and B4 were produced by some kind of instability of the B2 allele. The occurrence of nonparental alleles was studied in two other P. ostreatus strains, N002 and N003 (Table 1). In both cases, new B specificities appeared (B15B16 in strain N002; B17 in strain N003), albeit at a frequency lower than in P. ostreatus N001.
To test if nonparental B alleles were formed by an intralocus recombination event, we generated two hybrid strains, N017 (A1A2 B3B4) and N018 (A5A6 B15B16), carrying the nonparental B alleles appeared in the progeny of N001 and N002, respectively. The analysis of monokaryotic progenies derived from strains N017 and N018 showed that both parental and nonparental B alleles were obtained, which confirmed the complex nature of this locus in P. ostreatus and the recombinational nature of the new formed B alleles.
Recombination frequencies varied among different strains (Table 1). Strains N001 and N017 (both belonging to variety florida) showed recombination frequencies higher than 15%, whereas the strains belonging to variety ostreatus (N002, N018, and N003) had values of 8.2% or lower. This result suggest that the recombination frequency at the B locus is variable.
Additionally, we carried out a statistical analysis to determine if the frequency of parental and recombinant B alleles appeared in monokaryotic progenies of each strain was that expected as a consequence of a single recombinational event. In every case but one, the observed frequencies were as expected. The only exception was strain N001, where a significant bias was detected in the progeny of the B alleles observed (Table 1).
The B alleles differed among the P. ostreatus strains used in this work (Table 1). Nevertheless, it could be possible that the new B mating-type genes that appeared by intralocus recombination in N001 or N002 were functionally identical to those present in another P. ostreatus strain. To test this, monokaryons carrying nonparental B alleles (B3, B4, B15, and B16) were crossed with the mating testers corresponding to strains N001, N002, N003, N005, and N006 to search for incompatibilities revealing common B alleles. No common B genes were found among the strains analyzed in this study. Thus, the allelic compositions of B types B1, B2, B3, and B4 were defined as matBα1 matBβ1, matBα2 matBβ2, matBα1 matBβ2, and matBα2 matBβ1, respectively.
Molecular markers to tag the B incompatibility locus and verification of its bipartite structure.
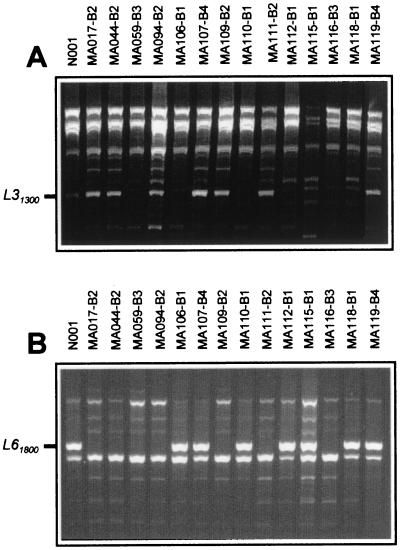
The identification of molecular markers genetically linked to characters of interest was the strategy of choice to facilitate their cloning. To generate molecular markers linked to the B incompatibility locus, an approach combining RAPD and bulk segregant analysis was used in a population of 80 monokaryons derived from dikaryotic strain N001. Two RAPD markers linked to the B locus were found. Marker L31300 (obtained by using as a primer Operon oligonucleotide L3, 1,300 bp long) was present in all monokaryons bearing either B2 or B4, while it was absent in monokaryons carrying B1 or B3 (Fig. 1A). On the other hand, marker L61800 was present in monokaryons with B1 or B4, while it was absent in those with B2 and B3 (Fig. 1B). No recombinants between marker L31300 and B2 or B4 and a single recombinant between L61800 and B1 or B4 were found in the analyzed population. These results indicate that RAPD markers L31300 and L61800 were genetically linked in coupling phase to matBα2 and matBβ1 alleles, respectively (Table 2).
FIG. 1.
RAPD markers found in dikaryon N001 of P. ostreatus and in different monokaryons derived from it, using primers L3 (A) and L6 (B). Markers L31300 and L61800, genetically linked to matBα and matBβ, respectively, are indicated. The incompatibility type of each monokaryon is indicated.
TABLE 2.
Correspondence between RAPD markers, RFLP alleles, and matB allelesa
| RAPD marker
|
RFLP allele
|
matB allele | ||
|---|---|---|---|---|
| Name | Size (bp) | Name | Size (bp) | |
| L31300 | 1,300 | rL313002 | 6,800 | matBα2 |
| rL313001 | 8,600 | matBα1 | ||
| L61800 | 1,800 | rL618003 | 3,900 | matBβ1 |
| rL618002 | 4,300 | matBβ2 | ||
Alleles in the same row are in coupling phase.
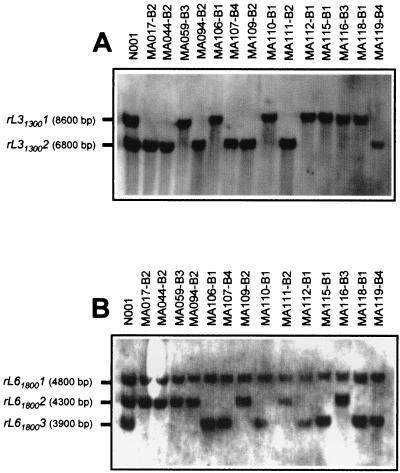
The RAPD markers genetically linked to the B mating-type locus were converted into restriction fragment length polymorphic (RFLP) markers. RFLP analysis using the cloned L31300 and L61800 RAPD markers as probes revealed that both of them corresponded to nonrepetitive DNA sequences (Fig. 2). Two different PstI restriction alleles were found using marker L31300 (Fig. 2A): rL313001 (8,600 bp), present in monokaryons bearing B1 or B3, and rL313002 (6,800 bp), present in monokaryons carrying B2 or B4. Considering the matBα alleles present in each of the B genes, alleles rL313001 and rL313002 are linked in coupling phase to matBα1 and matBα2, respectively, with no recombinants between RFLP alleles and the corresponding matBα alleles. On the other hand, when the cloned RAPD marker L61800 was used as probe, three PstI DNA fragments were identified (Fig. 2B): rL618001, which was a monomorphic 4,800-bp band; rL618002, a 4,300-bp-long band present in B2 or B3 monokaryons; and rL618003, which was 3,900 bp long and detected in monokaryons carrying incompatibility types B1 or B4. Segregation of bands rL618002 and rL618003 indicated that they were alleles of the same locus. Taking into account the allelic composition of the B incompatibility locus, RFLP alleles rL618002 and rL618003 appeared to be genetically linked in coupling phase to matBβ2 and matBβ1, respectively. Finally, RAPD markers L31300 and L61800 cosegregated with RFLP alleles rL313002 and rL618003, respectively (Table 2).
FIG. 2.
RFLP patterns detected in PstI genomic DNA digestions of P. ostreatus dikaryon N001 and of different monokaryons derived from it, using the L31300 (A) and L61800 (B) RAPD markers as probes. The incompatibility type of each monokaryon is indicated.
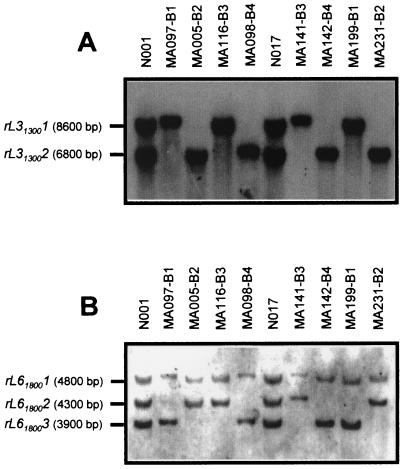
The consistency of the linkage phases found in strain N001 was tested using a monokaryotic progeny derived from dikaryon N017 (A1A2 B3B4). DNA samples from each of the four testers corresponding to N017 were digested using PstI, and the RFLP alleles rL31300 and rL61800 were studied. Figure 3 shows that markers rL313001 and L313002 cosegregated with matBα alleles, and markers rL618002 and rL618003 cosegregated with matBβ alleles, as expected (RFLP markers in monokaryons MA097, MA005, MA116, and MA098). Additionally, the RFLP markers linked to the different mating type genes present in the progeny of dikaryon N017 were also those expected from the previous analysis (markers in monokaryons MA141, MA142, MA199, and MA231). These results indicate that the intralocus recombination event that recovered B1 and B2 alleles in the progeny of N017 also recovered their corresponding rL31300 and rL61800 genotypes, corroborating molecularly the recombinational nature of the newly formed B alleles after meiosis.
FIG. 3.
RFLP patterns detected in PstI genomic DNA digestions of P. ostreatus dikaryons N001 and N017 and of four monokaryons (bearing different B alleles) derived from each of them, using the L31300 (A) and L61800 (B) RAPD markers as probes. The incompatibility type of each monokaryon is indicated.
Molecular markers linked to the B mating-type locus are species specific.
To determine whether loci rL31300 and rL61800 were also associated with the B locus in other P. ostreatus strains, the corresponding probes were hybridized to membranes containing genomic DNA from dikaryon N001, N002, N003, N005, or N006 and from some monokaryons derived from them, digested with EcoRI, PstI, or XhoI. Both probes gave clear signals in each case and revealed a high level of polymorphism (Table 3). To test the presence of RFLP markers homologous to those revealed by L31300 and L61800 in other mushrooms, PstI digestions of genomic DNA purified from P. quebecoise, A. bisporus, Agrocybe aegerita, and L. edodes were probed. P. quebecoise gave a weak hybridization signal when L31300 was used as probe, whereas no homologous sequences could be detected in the other species (data not shown).
TABLE 3.
Restriction length polymorphism of loci L31300 and L61800 in different P. ostreatus strains
| RFLP probe | Enzyme | Size (kbp) of corresponding restriction fragments
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B1 | B2 | B5 | B6 | B7 | B8 | B11 | B12 | B13 | B14 | ||
| L31300 | EcoRI | 4.5 | 11.5 | 17.0 | 17.0 | 17.0 | 17.0 | 17.0 | 17.0 | 11.5 | 11.5 |
| PstI | 8.6 | 6.8 | 8.6 | 8.6 | 11.2 | 8.6 | 8.6 | 8.6 | 8.6 | 8.6 | |
| XhoI | 3.4 | 9.0 | 12.5 | 4.2 | 3.7 | 9.0 | 4.2 | 4.2 | 14.0 | 3.2 | |
| L61800 | EcoRI | 13.0 | 13.0 | 20.0 | NDa | 21.5 | 21.5 | 15.3 | 15.3 | 15.3 | 18.5 |
| 11.0 | 11.0 | 4.5 | 10.0 | 14.0 | 3.7 | 3.1 | |||||
| 3.5 | |||||||||||
| PstI | 4.8 | 4.8 | 8.6 | 8.6 | 8.6 | 8.6 | 8.6 | 8.6 | 7.4 | 7.4 | |
| 3.9 | 4.3 | ||||||||||
| XhoI | 12.0 | 4.5 | 10.2 | 12.5 | 12.5 | 7.8 | 12.0 | 14.3 | 16.5 | 16.5 | |
| 4.5 | 4.2 | 3.3 | 10.2 | 3.8 | 4.2 | 4.2 | |||||
| 7.8 | |||||||||||
ND, not determined.
Analysis of distorted segregation in parental B alleles.
The monokaryotic progeny derived from strain N001 carried four different B alleles, two parental (B1 and B2) and two nonparental B types (B3 and B4), as a result of recombination between the two subunits (matBα and matBβ) on the B locus. Because the parental B alleles did not segregate 1:1 as expected (Table 1), we investigated the reason for such a discrepancy. To determine whether this bias was explained by differences in either growth rate of the vegetative mycelium or spore germination, we measured the vegetative growth rate of each of the 80 monokaryons forming the sample population. A one-way variance analysis test was applied to look for differences in the quantitative trait vegetative mycelium growth rate for the different mating genotypes. No significant differences in growth rate were found between matBα1 and matBα2 alleles (P = 0.8) or between matBβ1 and matBβ2 alleles (P = 0.9) in monokaryons bearing the A1 mating allele, whereas significant differences (P = 0.04) were observed between matBα1 and matBα2 alleles in monokaryons whose genomes bore the A2 mating allele. Interestingly, no significant differences appeared between matBβ1 and matBβ2 alleles (P = 0.5) in an A2 genome context. Monokaryons with mating genotype B1 or B3 (matA2 matBα1matBβ−) grew faster than those with the genotype B2 or B4 (matA2 matBα2matBβ−). Finally, significant differences (P = 0.01) in growth rate were observed for both alleles of the A locus. Monokaryons carrying the A2 allele grew faster than those with A1 specificity.
Mapping of the B incompatibility locus.
The progeny of 80 monokaryons described above were used to map the B locus. Fourteen RAPD markers (L141525, P161525, P12950, P31375, P11600, P22650, R21600, L31300, P22100, L61800, R15675, P2725, P19525, and R32275) were assigned to the linkage group to which the B locus belongs (17). The matBα and matBβ subunits were 19.0 centimorgans (Kosambi units [10]) apart, easily tagged by the tightly linked markers L31300 and L61800. The linkage group to which the B locus maps corresponds to the physically separated chromosome IX (16). Four molecular markers which were linked to the matBα sublocus (P11600, P22650, R21600, and L31300) showed distorted segregation. This was not the case for markers P22100, L61800, R15675, P2725, P19525, and R32275, which are close to the matBβ sublocus at the end of the chromosome.
DISCUSSION
The control of hyphal fusion and dikaryon formation is essential for filamentous fungi, as their mycelia form intricate mats in which the chance for contacts between sister branches is high. In P. ostreatus, as in other higher basidiomycetes, this control is based on two unlinked loci (A and B) responsible for different steps involved in the fusion process and in sorting of nuclei during dikaryotic hyphal growth. These two genes have been called either incompatibility loci or mating genes throughout the literature, and these two terms were considered here to be synonyms. In a previous paper, Larraya et al. (15) analyzed the A locus in five different P. ostreatus strains, isolated molecular markers genetically linked to it, and concluded that the A gene is controlled by a multiallelic single locus for which nine functionally different members were identified. In the present study, genetic experiments have allowed the identification of molecular markers genetically linked to the B mating-type gene, which confirmed that new B alleles can be formed as a consequence of intralocus recombination between the two subloci (matBα and matBβ) of the B gene. Considering the B incompatibility locus as a complex unit, an allelic series similar to that described for the A gene (15) has been found. Fifteen functionally different B mating alleles were distinguished, some of which resulted from intralocus recombination (Table 1).
The frequency of intralocus recombination yielding new (i.e., nonparental) B types is an estimate of the intergenic linkage distances between loci matBα and matBβ. However, it is known that the recombination frequency in S. commune does not depend exclusively on the physical distances between the two subunits of the B locus but also is under genetic control of a different locus where alleles for low recombination frequency are dominant over those for high recombination rate (9, 22, 26). Mating genes and recombination-controlling genes are genetically linked, although they can be physically separated by recombination (25, 26). It could be possible that similar mechanisms account for differences in recombination frequencies in the different P. ostreatus strains.
In a previous study (17), a distorted segregation was observed for all molecular markers surrounding matA and matBα genes, whereas no bias in the segregation was found in molecular markers surrounding the matBβ gene. Three hypotheses were put forward to explain this observation: (i) a nonrandom segregation of mating types that would drive a skewed segregation of markers linked to them, (ii) differences in viability, germination, or vegetative growth rate associated with different mating haplotypes that may cause preferred selection for some phenotypes in the population, and (iii) the occurrence of balancing selection on mating types that could counteract some negative selection on loci linked to the mating type. The results presented here indicate that there exists a relationship (linkage) between mating genes matA and matBα and the quantitative trait vegetative mycelium growth rate which could explain the distortion observed. The statistical analysis carried out here shows that monokaryons bearing the A2 mating allele grew faster than those bearing the A1 allele. The same is true for monokaryons with the matBα1 allele (B1 and B3) with respect to those carrying the matBα2 allele (B2 and B4). It is conceivable that slow-growing monokaryons need more time than fast-growing ones to develop a colony after germination, and these differences could have promoted a preferred selection for some genotypes when the population analyzed here was established. This skewed selection produced an increase in the frequency of alleles derived from the leading genotype in relation to those derived from the lagging one. The effect of negative selection against slowly germinating spores has been previously discussed by Eger (4) with respect to P. ostreatus var. florida and by Kerrigan et al. (8) with respect to A. bisporus. When the monokaryotic growth rate was studied in crossing programs using S. commune as the model system, it was also seen that faster-growing monokaryons belong to a certain mating type (23). Taken together, these data suggest that evolution has conserved genome regions in Agaricales, where genetic determinants affecting growth rate and mating type genes are kept together. In this way, those monokaryons whose mating alleles and polygenic traits related to growth rate display a cis configuration would be preferentially selected over those with a trans genetic organization.
The molecular markers isolated and described in this report constitute a first step toward the cloning and characterization by chromosome walking of the B mating-type genes and their flanking regions and useful tools for identifying in a quick and easy way monokaryons used as parentals in breeding programs. The RFLP profiles revealed by probe L61800 and restriction enzymes EcoRI and XhoI allow the identification of each B allele present in our collection. The genomic sequences detected by these RFLP probes are present in all of the P. ostreatus strains tested, although they bear different mating alleles, but they cannot be detected in P. quebecoise or in other Agaricales, suggesting that these sequences can be considered bonafide species-specific molecular markers. This was also the case of molecular markers S11900 and S181300, linked to the A locus (15). The availability of RFLP markers linked to the mating-type genes can be useful in marker-assisted selection for fast-growing monokaryons eligible for construction of dikaryons presumably able to colonize the substrate quicker than other competitors responsible for the reduction of yield in the industrial production of oyster mushrooms.
ACKNOWLEDGMENTS
This work was supported by research project BIO99-0278 of the Comisión Nacional de Ciencia y Tecnología and by Funds of the Universidad Pública of Navarra (Pamplona, Spain). M.A. holds a grant from the Departamento de Industria del Gobierno de Navarra.
We acknowledge the technical assistance of Gabriel Pardo, Amaya Iriarte, and Nerea Olaberría.
REFERENCES
- 1.Bölker M, Kahmann R. Sexual pheromones and mating responses in fungi. Plant Cell. 1993;5:1461–1469. doi: 10.1105/tpc.5.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casselton L A, Olesnicky N S. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S-T, Miles P G. Edible mushrooms and their cultivation. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 96–111. [Google Scholar]
- 4.Eger G. Pleurotus ostreatus-breeding potential of a new cultivated mushroom. Theor Appl Genet. 1976;47:155–163. doi: 10.1007/BF00278373. [DOI] [PubMed] [Google Scholar]
- 5.Eugenio C P, Anderson N A. The genetics and cultivation of Pleurotus ostreatus. Mycology. 1968;60:627–634. [Google Scholar]
- 6.Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell. 1992;68:647–657. doi: 10.1016/0092-8674(92)90141-x. [DOI] [PubMed] [Google Scholar]
- 7.Hiscock S J, Kües U. Cellular and molecular mechanisms of sexual incompatibility in plants and fungi. Int Rev Cytol. 1999;193:165–295. doi: 10.1016/s0074-7696(08)61781-7. [DOI] [PubMed] [Google Scholar]
- 8.Kerrigan R W, Royer J C, Baller L M, Kohli Y, Horgen P A, Anderson J B. Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics. 1993;133:225–236. doi: 10.1093/genetics/133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koltin Y, Raper J R, Simchen G. The genetic structure of the incompatibility factors of Schizophyllum commune: the B factor. Proc Natl Acad Sci USA. 1967;57:55–62. doi: 10.1073/pnas.57.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosambi D D. The estimation of map distance from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- 11.Kothe E. Mating types and pheromone recognition in the Homobasidiomycete Schizophyllum commune. Fungal Genet Biol. 1999;27:146–152. doi: 10.1006/fgbi.1999.1129. [DOI] [PubMed] [Google Scholar]
- 12.Kronstad J V, Staben C. Mating type in filamentous fungi. Annu Rev Genet. 1997;31:245–276. doi: 10.1146/annurev.genet.31.1.245. [DOI] [PubMed] [Google Scholar]
- 13.Kües U, Casselton L A. The origin of multiple mating types in mushrooms. J Cell Sci. 1993;104:227–230. [Google Scholar]
- 14.Kües U, Richardson W V, Tymon A M, Mutasa E S, Gottgens B, Gaubatz S, Gregoriades A, Casselton L A. The combination of dissimilar alleles of the A alpha and A beta gene complexes, whose proteins contain homeo domain motifs, determines sexual development in the mushroom Coprinus cinereus. Genes Dev. 1992;6:568–577. doi: 10.1101/gad.6.4.568. [DOI] [PubMed] [Google Scholar]
- 15.Larraya L, Peñas M M, Pérez G, Santos C, Ritter E, Pisabarro A G, Ramírez L. Identification of incompatibility alleles and characterisation of molecular markers genetically linked to the A incompatibility locus in the white rot fungus Pleurotus ostreatus. Curr Genet. 1999;34:486–493. doi: 10.1007/s002940050424. [DOI] [PubMed] [Google Scholar]
- 16.Larraya L M, Pérez G, Peñas M M, Baars J P, Mikosch T S P, Pisabarro A G, Ramirez L. Molecular karyotype of the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol. 1999;65:3413–3417. doi: 10.1128/aem.65.8.3413-3417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larraya L M, Pérez G, Ritter E, Pisabarro A G, Ramírez L. A genetic linkage map of the edible basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 2000;66:5290–5300. doi: 10.1128/aem.66.12.5290-5300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magae Y, Novotny C, Ullrich R. Interaction of the Aα Y and Z mating-type homeodomain proteins of Schizophyllum commune detected by the two-hybrid system. Biochem Biophys Res Commun. 1995;211:1071–1076. doi: 10.1006/bbrc.1995.1920. [DOI] [PubMed] [Google Scholar]
- 19.Peñas M M, Asgeirsdóttir S A, Lasa I, Culiañez-Macià F A, Pisabarro A G, Wessels J G H, Ramírez L. Identification, characterization, and in situ detection of a fruit-body-specific hydrophobin of Pleurotus ostreatus. Appl Environ Microbiol. 1998;64:4028–4034. doi: 10.1128/aem.64.10.4028-4034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 1990;60:295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- 21.Shen G P, Park D C, Ullrich R C, Novotny C P. Cloning and characterization of a Schizophyllum gene with A beta 6 mating-type activity. Curr Genet. 1996;29:136–142. doi: 10.1007/BF02221577. [DOI] [PubMed] [Google Scholar]
- 22.Simchen G. Genetic control of recombination and the incompatibility system in Schizophyllum commune. Genet Res (Cambridge) 1967;9:195–210. [Google Scholar]
- 23.Simchen G. Monokaryotic variation and haploid selection in Schizophyllum commune. Heredity. 1966;21:241–263. doi: 10.1038/hdy.1966.21. [DOI] [PubMed] [Google Scholar]
- 24.Specht C, Stankis M M, Giasson L, Novotny C P, Ullrich R C. Functional analysis of the homeodomain-related proteins of the Aα locus of Schizophyllum commune. Proc Natl Acad Sci USA. 1992;89:7174–7178. doi: 10.1073/pnas.89.15.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamberg J. Genetic control of recombination in Schizophyllum commune: separation of the controlled and controlling loci. Heredity. 1969;24:306–309. doi: 10.1038/hdy.1969.35. [DOI] [PubMed] [Google Scholar]
- 26.Stamberg J. Two independent gene systems controlling recombination in Schizophyllum commune. Mol Gen Genet. 1968;102:221–228. doi: 10.1007/BF00385977. [DOI] [PubMed] [Google Scholar]
- 27.Stankis M M, Specht C P, Yang H, Giasson L, C. U R, Novotny C P. The Aα mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc Natl Acad Sci USA. 1992;89:7169–7173. doi: 10.1073/pnas.89.15.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tymon A M, Kües U, Richarson W V J, Casselton L A. A fungal mating type protein that regulates sexual and asexual development contains a POU-related domain. EMBO J. 1992;11:1805–1813. doi: 10.1002/j.1460-2075.1992.tb05232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaillancourt L, Raper C A. Pheromones and pheromone receptors as mating-type determinants in basidiomycetes. In: Setlow J K, editor. Genetic engineering, principles and methods. Vol. 18. New York, N.Y: Plenum; 1996. pp. 219–247. [DOI] [PubMed] [Google Scholar]
- 30.Wendland J, Kothe E. Allelic divergence at the BαI pheromone receptor genes of Schizophyllum commune. FEMS Microbiol Lett. 1996;145:451–455. doi: 10.1111/j.1574-6968.1996.tb08615.x. [DOI] [PubMed] [Google Scholar]