Abstract
Well‐adapted root systems allow plants to grow under resource‐limiting environmental conditions and are important determinants of yield in agricultural systems. Important staple crops such as rice and maize belong to the family of grasses, which develop a complex root system that consists of an embryonic root system that emerges from the seed, and a postembryonic nodal root system that emerges from basal regions of the shoot after germination. While early seedling establishment is dependent on the embryonic root system, the nodal root system, and its associated branches, gains in importance as the plant matures and will ultimately constitute the bulk of below‐ground growth. In this review, we aim to give an overview of the different root types that develop in cereal grass root systems, explore the different physiological roles they play by defining their anatomical features, and outline the genetic networks that control their development. Through this deconstructed view of grass root system function, we provide a parts‐list of elements that function together in an integrated root system to promote survival and crop productivity.
Keywords: embryonic roots, environmental stress, Grasses, Oryza sativa, postembryonic roots, root development, root system architecture, Zea mays
Summary
The review focuses on the root types that make up root systems of members of the grass family such as Zea mays and Oryza sativa. We summarize research that explores the development and genetic networks associated with these root types and describe how root‐type functions are involved in the acclimation to environmental stresses.
1. INTRODUCTION
The evolution of rooting structures has helped plants invade a diverse array of terrestrial environments to access a heterogeneous distribution of water and nutrients. The establishment of a branched architecture in roots increases the absorptive surface area of the organ system and also leads to a differentiation of root types based on the timing of development for each branch and their relative spatial topology (Hetherington & Dolan, 2018, 2019; Hetherington et al., 2020; Rellán‐Álvarez et al., 2016). Such positional differences ultimately lead to root types with distinct growth patterns as exemplified by the unique features of primary roots and their secondary branches (lateral roots) in a typical seed‐plant root system.
Within the flowering plant lineage, substantial differences in the origin of root branches have evolved. Eudicotyledon root systems are typically composed of one embryonic primary root, also called a taproot, and many lateral root branches that form postembryonically (Rellán‐Álvarez et al., 2016). Additionally, postembryonic adventitious roots may form from shoot tissue. Research on roots of the Eudicot model plant Arabidopsis (Arabidopsis thaliana) has helped to establish very detailed knowledge of the molecular mechanisms that pattern root development (Petricka et al., 2012; Van Norman et al., 2013). In contrast, root systems of members of the grass family (Poaceae) show a more complex structure with root types that differ in their time and place of origin as well their anatomical and physiological characteristics (Hochholdinger & Zimmermann, 2008). Understanding the development and function of grass root systems is important since many major crop plants like rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare) belong to this plant family. The following review is meant to provide an overview of the different root types in a grass root system, summarizing research findings on their anatomy, developmental control, and physiological function. For this, we mainly focused on literature of the aforementioned crops species in the grass family. We also attempt to clarify the function of this complex organ system by first deconstructing it into its components. This ‘parts‐level’ view deconvolves the available literature, which often focuses on different parts of the root system and plants studied at different developmental stages. At the end of the review, we attempt to coalesce our understanding into a holistic view of how each root type functions together in a root system that enables survival under environmental stress conditions such as drought, flooding, salinity, changes in nutrient availability, and soil compaction.
2. OVERVIEW OF ROOT TYPES IN GRASS ROOT SYSTEMS
While roots in the model plant A. thaliana can be differentiated into adventitious, lateral, and primary roots based on their site of origin, only a few studies have demonstrated clear physiological differences that distinguish their role in the root system (Figure 1), for example, in gravitropism (Guyomarc'h et al., 2012; Roychoudhry et al., 2013; Waidmann et al., 2019). In stark contrast, grasses establish their root systems through the development of root types that are not only distinguished by their site of initiation but also in their anatomical and physiological roles in supporting the growth of the plant (Atkinson et al., 2014). The initial parts of the root system are established during embryogenesis, leading to root types that are considered embryonic in origin. During embryo development in grasses, a basal pole is specified and forms the coleorhiza, which is a determinate structure that ceases growth before the end of seed development. Within the coleorhiza, the primary root initiates, and like all other root types in grasses, breaks through the outer tissues of the plant to emerge into the environment, post‐germination (Abbe & Stein, 1954; Kiesselbach, 1949). The other components of the embryonic root system are the seminal roots, which initiate at the embryonic root‐shoot junction, called the scutellar node.
Figure 1.
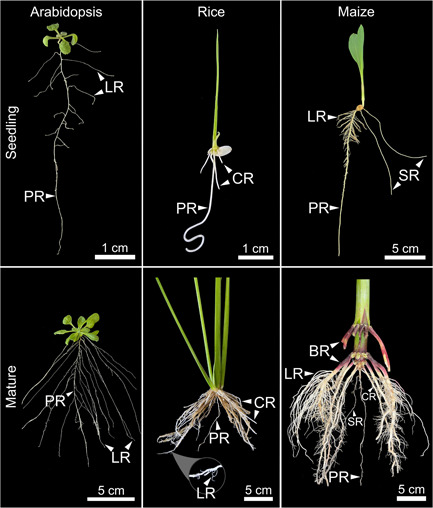
Root system architecture in the Eudicot model (Arabidopsis) and two members of the grass family (rice and maize) at early and late developmental stages. BR, brace root; CR, crown root; LR, lateral root; PR, primary root; SR, seminal root
Post‐germination development of the plant leads to the establishment of the bulk of the root system. At positions where leaves join the stem, nodal roots develop. Nodal roots that initiate from stem tissues below the soil surface are called crown roots since this basal region of the shoot is termed the crown. In maize and other panicoid grasses, nodes above the soil surface originate branches called brace roots (Hochholdinger et al., 2004). Both embryonic and postembryonic roots are able to form secondary root branches called lateral roots. Together, the diverse developmental origins of root types in grasses create a multi‐axial system that is complex, highly responsive to environmental cues and performs diverse functions from anchoring the plant to the ground, to providing mechanical support to the stem, to acquiring resources from the soil. In the proceeding sections of the review, we will provide a detailed description of the origin of these different root types, their anatomical properties, and the developmental mechanisms that affect their formation.
2.1. Primary and seminal roots: Overview
In grasses, the embryonic root system consists of one primary root, also called the radicle, and varying numbers of seminal roots (Figure 1). Both anecdotal and empirical evidence suggest that the embryonic root system is frequently ephemeral in grasses and senesces after a period of growth, after germination (Fusseder, 1987). Significant genotypic variance for seminal root numbers has been observed in grasses, though the molecular basis for this variation has not been determined (Tai et al., 2016). Seminal root numbers have increased in domesticated grasses such as maize and barley compared to their wild relatives, which may improve nutrient acquisition. It was shown that seminal roots contributed to approximately 35% of total phosphorus and nitrogen uptake in the first 25 days of seedling growth in maize landraces (Perkins & Lynch, 2021). The physiological basis for the senescence of the embryonic root system has not been well investigated and may result as a consequence of a regulated developmental program or from being starved of resources due to the rapid growth of the postembryonic root system.
2.1.1. Primary and seminal roots: Anatomy and function
Root anatomy can be conceptualized as concentric cylinders of tissue made of different cell types. The outermost tissue layer of the root is the epidermis (Figure 2a,b), which forms root hairs that function in nutrient and water acquisition. Internal to the epidermis, the ground tissue is composed of one to several layers of cortical cells followed by the endodermis. While seedling roots of Arabidopsis have only one cortical cell layer, the primary roots of grasses typically have multiple cortical cells layers, ranging from 8 to 9 layers in maize, 5 to 6 layers in rice, and 4 to 5 layers in Setaria viridis (Clark & Harris, 1981; Ortiz‐Ramírez et al., 2021). The endodermis serves as a semipermeable barrier between the outer ground tissues and the inner stele where the vasculature is housed. Endodermal cell walls are impregnated with lignin patterned into bands termed Casparian strips, which prevent the apoplastic movement of water and ions into the inner root cylinder. Depending on environmental conditions and age of the root, an exodermis may form as well, which is a subepidermal tissue layer (hypodermis) with Casparian bands. The inner stele consists of pericycle, the vascular system, and pith. In roots of Eudicots such as Arabidopsis and tomato, the diarch vascular system shows a bilateral symmetry with two xylem and two phloem poles (Figure 2a). In contrast, roots of grasses have a polyarch symmetry with multiple xylem and phloem poles that are arranged along the root's circumference (Figure 2b). This basic anatomy applies to all root types in the grass root system. It is thought that the higher number of cortical layers in grasses may allow for additional functions to be performed by this tissue (Ortiz‐Ramírez et al., 2021). In the section on environmental regulation, we discuss how cortical cells are modified in response to abiotic stressors.
Figure 2.
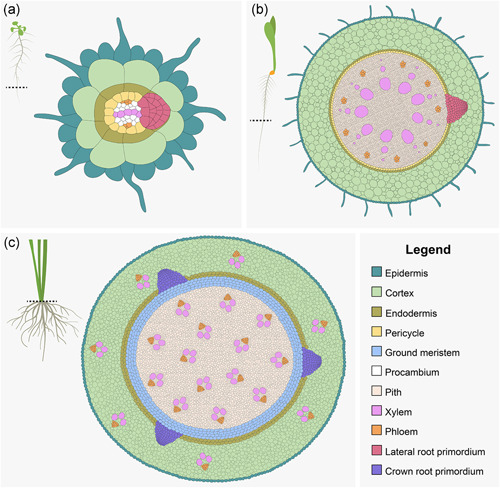
Schematics showing the emerging lateral root primordium in Arabidopsis and maize (a, b) and crown root primordium in rice (c). The dotted line represents the transverse section through the primary roots of Arabidopsis and maize, and the crown (base of the stem) of rice
Seminal roots are often significantly smaller in diameter compared to primary roots. In maize, this is primarily due to a reduction in cortical cell layer number, while the proportion of xylem area to total stele area is greater in seminal roots (Tai et al., 2016). This anatomical difference may give seminal roots a greater capacity for nutrient and water transport at a reduced energy cost. Consistent with this hypothesis, a correlation was found between larger cortical cell diameters and energy cost for root growth in a set of diverse wheat genotypes (Colombi et al., 2019).
While primary and seminal roots might have different characteristics for water and nutrient uptake, research has shown that most water is likely taken up by lateral roots that emerge from primary and seminal roots, as shown in young seedlings of maize (Ahmed et al., 2016). These lateral roots have a significantly higher conductivity for water uptake. Primary and seminal roots are most likely mainly involved in the axial transport of water from their lateral roots to the shoot.
2.1.2. Primary and seminal roots: Developmental regulation
In maize embryos, it was observed that ZmPIN1‐mediated auxin transport was significantly correlated with the establishment of the shoot and root apical meristems in a similar way to Arabidopsis (Forestan et al., 2010). A maize mutant called rootless with undetectable meristems 1 (rum1) showed a total lack of seminal and lateral roots in maize (Woll et al., 2005). The causal locus was identified as a monocot‐specific AUX/IAA protein (GRMZM2G037368). AUX/IAAs are repressor proteins that interact with AUXIN RESPONSE FACTOR (ARF) proteins and inhibit the expression of genes regulated by ARFs. Auxin causes the degradation of the AUX/IAA proteins, which then leads to the expression of auxin‐responsive genes. In Arabidopsis, the AUX/IAA protein BODENLOS has been identified as being necessary for primary root formation during early embryogenesis (Hamann et al., 1999; Herud et al., 2016). This points at a sequence of events, where precise control of auxin distribution and perception is needed to define the localization of the root apical meristem and initiate its establishment in the developing embryo.
Once the apical and basal poles of the embryo are established, the division and differentiation of cells from the quiescent center into the distinct root cell layers occurs. In rice, it was found that OsWOX4, a transcription factor from the WUSCHEL‐RELATED HOMEOBOX (WOX) protein family, is required for the maintenance of meristem size and control of cell length in the maturation zone (Chen et al., 2020). Similar to Arabidopsis, the expression of WOX transcription factors in rice may be confined to the quiescent center through the action of CLAVATA3 (CLV3)/ENDOSPERM SURROUNDING REGION (ESR)‐related (CLE) proteins (Chu et al., 2013).
Both the endodermal and cortical cell layers are clonally related since they originate from a cortical‐endodermal stem cell. In Arabidopsis, differentiation of the endodermal and cortical layers is controlled by the SHORT‐ROOT (SHR) protein, which travels from the stele to the next‐most outer tissue layer and acts as a transcription factor establishing the fate of the endodermis (Cui et al., 2007; Nakajima et al., 2001; Petricka et al., 2012). Through heterologous expression in Arabidopsis, it was shown that SHR proteins from grasses, such as Brachypodium and rice, have greater mobility across tissues and induce multiple cortex layers instead of multiple endodermal layers (Wu et al., 2014). This finding is supported by a recent publication showing higher mobility of SHR protein in maize, as well (Ortiz‐Ramírez et al., 2021). Interestingly, the expression of SHR in maize is localized to the endodermis and not the stele, as observed in Arabidopsis.
Taken together, these findings show that regulation of root development in grasses shows many parallels to programs that were discovered in Arabidopsis. However, due to differences in anatomy, for example, multiple cortical layers, and additional root types like seminal roots, there is much to learn that will be exclusive to grasses.
2.2. Lateral roots: Overview
Root branching, or the formation of lateral roots, is an important aspect of root system architecture that leads to a significant increase in overall root surface area. In winter rye, for example, plants that were excavated just before flowering had a more than 1000‐fold increase in root surface area due to the formation of lateral root branches (Dittmer, 1937). This increase in root surface area through root branching is necessary for plants to access resources such as water and nutrients, which are non‐uniformly distributed in the soil. Both in natural and agricultural ecosystems it was determined that the variability in nutrient distribution across an entire field site can be of a similar scale to the variance that can be observed within the root zone of a single plant at the same field site, which can be several orders of magnitude (Jackson & Caldwell, 1993; Robertson et al., 1997). This variance is likely shaped by biotic and abiotic factors. For example, nutrient availability depends on microbial activity, which varies with the seasons. Precipitation or irrigation patterns and local soil texture, which refers to the relative proportions of different soil particle sizes, also contribute to differences in nutrient fluxes. It has been shown that plants, including maize and rice, are able to tune lateral root branching in response to spatial differences in water availability, a process called hydropatterning (Bao et al., 2014; Robbins & Dinneny, 2018). It is likely that this response also leads to more effective nutrient utilization.
Both their importance for resource acquisition and their high degree of developmental plasticity in response to the environment make lateral root branching an interesting breeding target.
2.2.1. Lateral roots: Anatomy and function
Lateral roots originate from other roots and are formed postembryonically. Patterning of lateral roots occurs in the elongation and early maturation zone of their parent root, as revealed by studies that have examined their induction in response to moisture cues (Babé et al., 2012; Robbins & Dinneny, 2018). While the parent root is growing, new lateral roots are initiated continuously, albeit depending on environmental conditions, and they emerge and elongate as their parent root matures.
Lateral root anatomy and development have been extensively studied in the eudicot model plant Arabidopsis (Péret et al., 2009; Van Norman et al., 2013), and the genetic pathways involved in lateral root initiation and emergence have been described in detail. Both in eudicots and grasses, lateral roots are initiated in the pericycle. In contrast to other cell types generated by the root meristem, pericycle cells maintain their competence for cell divisions (Dubrovsky et al., 2000). In Arabidopsis, this is particularly true for pericycle cells located at the xylem poles, though pericycle cells neighboring the xylem poles also contribute to primordium formation (Figure 2a) (Torres‐Martínez et al., 2020). In contrast, lateral root initiation in grasses was observed in pericycle cells close to the phloem poles in crown roots of rice (Kawata & Shibayama, 1965) and in the primary root of maize (Figure 2b) (Casero et al., 1995).
While lateral root patterning in Arabidopsis is controlled by oscillatory gene expression, also called the root‐clock, which can be tuned by cues such as the plant hormone auxin and retinals, it is not known whether this is also the case in grasses (Dickinson et al., 2021; Perianez‐Rodriguez et al., 2021; Van Norman et al., 2013). In maize, anticlinal (i.e., perpendicular to the longitudinal axis of the root) and periclinal (i.e., parallel to the longitudinal axis of the root) cell divisions are observed in the phloem pole pericycle, which is similar to the cell divisions observed in the xylem pole pericycle of Arabidopsis leading to lateral root initiation (Figure 2a,b) (Jansen et al., 2012). While in Arabidopsis, cell divisions for lateral root initiation are exclusively observed in the pericycle, in maize, cell divisions are also observed in the endodermis (Hochholdinger & Zimmermann, 2008). Moreover, the emerging lateral root has to break through one to two layers of cortex cells and the epidermis in Arabidopsis, while in grasses, such as maize, 10–15 cortical cells layers have to be traversed.
As mentioned in the section on primary and seminal roots, lateral roots have the highest conductivity for water and nutrient uptake. This is not only true for young seedlings but also for more mature plants as shown in maize and barley (Ahmed et al., 2018; Schneider et al., 2020). Since lateral roots emerge first on the proximal segments in young seedlings, most water is taken up closer to the soil surface. Therefore, a good supply of water in the top soil layers is needed for early seedling establishment. Once the nodal root system is established, water uptake shifts predominantly to laterals of crown roots (Ahmed et al., 2018). The authors hypothesized that this might be due to the higher axial conductance of crown roots, compared to the embryonic root system, and may lead to a more direct vascular connection to the shoot allowing for better propagation of xylem tension.
While the tissues involved in lateral root initiation are known both in Arabidopsis and grasses, detailed knowledge of the molecular mechanisms that lead to lateral root initiation in grasses is still lacking. Since lateral and nodal roots are both postembryonically derived root types, they may share some common features. Interestingly, some mutants that have defects in lateral root initiation also show defects in crown root development, as described in the next section.
Since lateral root branching‐dependent soil exploration is pivotal for water and nutrient acquisition, much effort has been made towards understanding the anatomy of lateral root initiation. While we have a comprehensive knowledge of the different stages of lateral root development in Arabidopsis, there are still open questions that need to be answered in grasses.
2.2.2. Lateral roots: Developmental regulation
In maize, the mutant rootless with undetectable meristems 1 (rum1) shows a complete lack of lateral and seminal root initiation, a strong inhibition of crown roots, and also a delayed gravitropic response (Woll et al., 2005). The responsible mutation was identified as a truncated Mutator transposon element inserted in the RUM1 gene, which encodes a monocot‐specific AUX/IAA protein (von Behrens et al., 2011). This protein is most closely related to IAA8 and IAA27 in Arabidopsis, which are involved in lateral root development. Moreover, the RUM1 protein interacts with the AUXIN RESPONSE FACTORS 25 and 34 (ZmARF25 and ZmARF34) in vitro and in vivo in Arabidopsis protoplasts. RNA‐Seq analysis also uncovered that AUXIN RESPONSE FACTORS 8 and 37 (ZmARF8 and ZmARF37) genes were downregulated in the rum1 mutant (Y. Zhang et al., 2014). The authors argue that regulation of the ARF transcription factors may be involved in vascular development, since rum1 showed a strong lack of organization of xylem elements and pith cells in primary roots.
Another interaction partner of RUM1 was identified as LATERAL ROOT PRIMORDIA 1 (LRP1), a transcriptional activator that acts in the root meristems of lateral and crown root primordia (Y. Zhang, von Behrens, et al., 2015). RUM1 directly binds to the promoter of LRP1, indicating a potential function in lateral root initiation. A homeolog of rum1 called rum1‐like1 (rul1) has been identified that shares many of the features of rum1 (Y. Zhang et al., 2016). However, protein‐protein interaction assays also showed that some unique interactions exist for each of the two proteins, which indicate differences in their molecular networks despite their interwoven nature.
In rice, another AUX/IAA gene has been identified to be involved in lateral root initiation. Similar to rum1 in maize, a mutant of OsIAA13 also showed a defect in its gravitropic response and reduced numbers of lateral roots (Kitomi et al., 2012). Analysis of DR5 reporter activity, which is auxin‐responsive and active in lateral root initiation sites (Jansen et al., 2012), showed an absence in roots of the Osiaa13 mutants indicating altered auxin distribution or perception. Further research using the mutant of OsIAA13 showed that the gene is also involved in lysigenous aerenchyma formation in rice (Yamauchi et al., 2019). Through transcriptome analysis and biochemical characterization, it was found that the OsIAA13 protein interacts with OsARF19, which in turn binds to the promoter of a lateral organ boundary domain (LBD)‐encoding gene (OsLBD1‐8).
Through studies in Arabidopsis, it has been shown that auxin efflux carriers PIN‐FORMED (PIN) proteins are important for proper auxin distribution, which is needed for induction of lateral roots. In rice, a mutant for the auxin efflux carrier OsPIN2 has been isolated (Inahashi et al., 2018). Plants showed a basipetal shift of the lateral root formation zone, which was dependent on the growing environment of the mutant plants. These findings confirm that similar to Arabidopsis, auxin efflux carriers control auxin distribution patterns that regulate the localization of lateral root initiation.
In wheat, a transcription factor called LATERAL ROOT DENSITY (LRD) has been identified as a negative regulator of lateral root growth under water‐limited conditions (Placido et al., 2020). LRD is a homolog of AtKNAT3 in Arabidopsis. AtKNAT3 is thought to be involved in root development, and it has been shown that AtKNAT3 expression is controlled by abscisic acid. This might explain why LRD in wheat is upregulated under water‐limited conditions, but it is not known whether LRD is controlled by abscisic acid as well.
Other mutants with defects in lateral root initiation and growth have been found in maize and rice, but the molecular mechanisms are mostly unknown. A lateral root mutant identified in maize from an EMS mutant screen is lateral rootless 1 (lrt1), which shows a lack of or aberrant growth of lateral, primary, and seminal roots depending on growth conditions (Hochholdinger & Feix, 1998; Husakova et al., 2013). While it is not known what the molecular basis for the mutant phenotype is, these data suggest there is a common genetic mechanism that acts in both embryonic and postembryonic roots to control their development.
These findings in maize and rice strongly support the hypothesis that lateral root initiation happens through similar pathways in the eudicot model species Arabidopsis and grasses. However, it will be necessary to understand which homologs are involved in these pathways to be able to manipulate them in the future. It also remains unclear what determines the spatial pattern of lateral root initiation at the phloem pole pericycle in grasses, compared to the xylem poles in Arabidopsis.
2.3. Crown roots: Overview
Crown roots are adventitious roots that are unique to grasses. These adventitious roots initiate from non‐root tissues, at the base of the stem close to the soil surface, in a region called the crown, hence the name ‘crown roots’ (Sebastian et al., 2016). In mature plants, they make up the bulk of the root system and are essential for anchorage and nutrient and water absorption (Ahmed et al., 2018; Reneau et al., 2020).
2.3.1. Anatomy and function
Unlike the primary root, which develops during embryogenesis, crown roots develop postembryonically from differentiated cells. Crown root development in rice has been extensively characterized and can be divided into seven stages (Itoh et al., 2005). First, founder cells from the innermost ground meristem cell layers, which are next to the peripheral cylinder of vascular bundles in the stem, divide periclinally to form a crown root primordium (Figure 2c). Subsequently, cells in the crown root primordium divide anticlinally and periclinally to form the endodermis‐epidermis, central cylinder, and root cap initials. At the next stage, the endodermis‐epidermis initial undergoes periclinal divisions to form separate endodermal and epidermal cell layers. The endodermal initials then divide periclinally to generate the cortical tissue layers. The central cylinder initial undergoes anticlinal and periclinal divisions that increase their size to develop a dome shape, while the root cap initials divide periclinally to form the columella. After the fundamental organization of the root has been established, cells from all the different layers become vacuolated and elongate. Finally, the crown root primordium becomes vascularly connected to the stem and emerges.
While important for the establishment of the seedling, the contribution of the primary and seminal roots to resource acquisition is ephemeral. The transition between embryonic and postembryonic root systems occurs 2–3 weeks after germination in maize with the formation of whorls of crown roots that take over the role of water and nutrient acquisition (Hoppe et al., 1986). This was further supported by findings showing that the main source of water uptake in 5‐week‐old maize seedlings was from laterals of crown roots, while the seminal and primary roots made a smaller contribution (Ahmed et al., 2018).
2.3.2. Crown roots: Developmental regulation
The regulation of crown root development has been extensively studied in rice (Figure 3). The phytohormone auxin plays an essential role in crown root formation, controlling development by promoting cell division, differentiation, and expansion. Auxin transport is necessary for the establishment of an auxin gradient and accumulation at the vascular bundles and ground meristem cells adjacent to the peripheral vascular cylinder and induces cell dedifferentiation and crown root primordium formation (Kitomi et al., 2008). Identified by two independent groups, the crown rootless 4 (crl4) and gnom1 mutants show reduced numbers of crown roots and lateral roots. The CRL4 and OsGNOM1 genes encode a membrane‐associated guanine‐nucleotide exchange factor for the ADP‐ribosylation factor G protein (ARF‐GEF) (Kitomi et al., 2008). This factor is highly homologous to the Arabidopsis GNOM, which regulates the endosomal recycling of PINFORMED1 (PIN1) auxin efflux protein to the plasma membrane of provascular cells (Richter et al., 2010). PIN1 mediates auxin accumulation at the root primordium founder cells (Benková et al., 2003). The expression of OsPIN genes in gnom1 mutants is altered (S. Liu et al., 2009), while CRL4 expression patterns correlate with OsPIN1 expression (Kitomi et al., 2008; M. Xu et al., 2005). In addition, the formation of an auxin gradient in the crown stem tissues of the crl4 mutant is impaired, and auxin distribution is fainter and less focused than the wild‐type (Kitomi et al., 2008). Taken together, these data indicate that polarized auxin transport is mediated by CRL4/OsGNOM1, and that disruption of the function of the gene results in the inhibition of crown root initiation.
Figure 3.
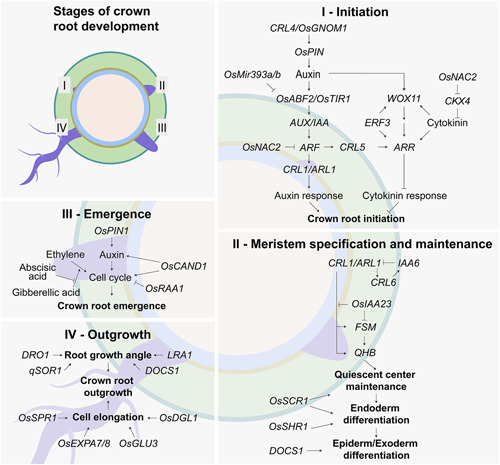
Gene regulatory network controlling crown root initiation, meristem specification and maintenance, emergence, and outgrowth in rice. Arrows indicate positive regulatory action, and the flat‐headed arrows indicate negative regulatory action
Many of the mutants characterized for defects in crown root development are associated with auxin perception and response. In the absence of auxin, members of the AUXIN RESPONSE FACTORS (ARF) transcriptional factors bind tightly to AUXIN/INDOLE‐3‐ACETIC ACID (Aux/IAA) proteins and act as transcriptional repressors (Calderón Villalobos et al., 2012). In the presence of auxin, Aux/IAAs interacts with the TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and AUXIN SIGNALLING F‐BOX (AFB) proteins, which are part of the Skp1‐cullin 1‐F‐box (SCF) E3 ligase complex. TIR1 targets Aux/IAAs for ubiquitylation and degradation. The derepressed ARF is then free to bind to the promoter of auxin‐responsive genes and initiate transcription. OsTIR1 and OsAFB2 are negatively regulated by the microRNAs OsMir393a and OsMir393b. Plants overexpressing OsMIR393a/b showed a reduced number of crown roots and were resistant to exogenous application of 2,4‐D, a synthetic form of auxin (Bian et al., 2012).
Upon Aux/IAA release, OsARF16 binds to the auxin response element in the promoter of CROWN ROOTLESS1 (CRL1) (Inukai et al., 2005). CRL1 encodes an ASYMMETRIC LEAVES2 (AS2)/LATERAL ORGAN BOUNDARIES (LOB) domain (OsLBD3‐2) transcription factor. Characterized at around the same time, the two OsLBD3‐2 mutants crl1 and arl1 have normal primary root development but are devoid of crown roots (Inukai et al., 2005; H. Liu et al., 2005). Crl1 plants can rely on the embryonic root system and grow to maturity given mechanical and nutrient support, and produce a few crown roots at later developmental stages, while arl1 cannot survive more than a few weeks. Closer examination of the crown region revealed that the mutants do not form crown root primordia, which suggests that disruption of CRL1/ARL1 causes a defect in the initiation of crown root development. A gene regulatory network linking CRL1 and several genes involved in crown root initiation, and root apical meristem specification and maintenance, was recently established by analyzing a time series transcriptomic data set and validated in vivo using transient activation assays (Lavarenne et al., 2019).
The crl6 mutant shows a reduction in crown root number by about 50%. These roots, however, have deformed cell structures and lack a normal cell layer organization. CRL6 encodes a chromodomain helicase DNA‐binding (CHD) protein and affects the expression of most OsIAA genes (Y. Wang et al., 2016). Among these is OsIAA23, which plays an important role in quiescent center maintenance (Jun et al., 2011). Osiaa23‐2 has a point mutation in conserved core sequence GWPPV of domain II, which stabilizes the OsIAA23 protein. The wild‐type OsIAA23 protein is expressed uniformly in the early developmental stages of lateral and crown root primordia until it becomes restricted to the quiescent center. In turn, OsIAA23 negatively regulates the expression of FLATTENED SHOOT MERISTEM (FSM). FSM encodes a p150 subunit of chromatin assembly factor‐1 (CAF‐1) and is a putative ortholog of Arabidopsis FASCIATA1 (FAS1) (Abe et al., 2008 ). Fsm mutants show a reduced number of seminal and crown roots; those that develop display an irregular arrangement of cell files along the radial axis. FSM is directly activated by CRL1 and it is required for cell cycle regulation and meristem maintenance and is predicted to regulate QUIESCENT‐CENTER‐SPECIFIC HOMEOBOX (QHB). QHB encodes a homeodomain transcription factor that regulates the specification and maintenance of the quiescent cells in the root apical meristem (Kamiya, Nagasaki, et al., 2003).
SCARECROW (OsSCR1), a transcription factor from the GRAS family, also plays a role in the specification of the quiescent center and, together with SHORTROOT (OsSHR), regulates the radial organization of the root by controlling the division of the endodermis (Cui et al., 2007; Kamiya, Itoh, et al., 2003; Ni et al., 2014). The c68 mutant is defective in the differentiation of the epidermis and exodermis. The gene responsible for this mutation is DEFECTIVE IN OUTERCELL LAYER SPECIFICATION 1 (DOCS1), which encodes a leucine‐rich repeat receptor‐like kinase (LRR RLK). Although a few mutants have been characterized, the molecular mechanisms for the specification of the root outer cell layers (epidermis, exodermis, and sclerenchyma) are still poorly understood.
Crosstalk between auxin and other phytohormones plays an important role in the regulation of crown root development in grasses. For instance, the crown rootless5 (crl5) mutant produces fewer crown roots due to an impairment in crown root initiation (Kitomi et al., 2011). The CRL1 and CRL5 loci show additive phenotypes when combined, which indicates that these two genes might act in different pathways. CRL5 encodes an APETALA/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor that functions downstream of the IAA and ARF signaling pathways to regulate crown root initiation. Plants overexpressing CRL5 displayed a cytokinin‐resistant phenotype for crown root initiation, in addition to upregulating OsRR1 (type‐A RESPONSE REGULATOR1) and OsRR2, two negative regulators of cytokinin signaling. These two genes were downregulated in the crl5 mutant. Overexpression of OsRR1 under the control of the CRL5 promoter in the mutant background rescued the crown root growth phenotype, which indicates that CRL5 promotes crown root growth by repressing cytokinin signaling.
WUSCHEL‐related Homeobox 11 (WOX11) is another gene known to integrate auxin and cytokinin pathways to modulate crown root development in rice (Zhao et al., 2009). WOX11 is a transcription factor primarily expressed in tissues undergoing cell divisions, such as the root apical meristem. Loss‐of‐function mutants produced few to no crown roots two weeks after germination, and plants died before maturity. Downregulation of the gene resulted in few crown roots per plant and a reduction in the growth rate of about 50% compared to wild‐type. Conversely, overexpression of WOX11 resulted in a dramatic increase in the number of crown roots, with roots developing at every node of the stem, including the base of the florets (Zhao et al., 2009). WOX11 expression can be induced by auxin and cytokinin, and the gene affects the expression of auxin‐responsive Aux/IAA and cytokinin type‐A RR genes. More specifically, WOX11 was found to directly repress RR2, which regulates crown root cell proliferation. In vitro and in vivo analyses revealed that WOX11 interacts with the AP2/ERF protein, ERF3 (Zhao et al., 2015), to regulate crown root development. Interestingly, the expression of the two genes only partially overlaps. ERF3 is expressed in crown root primordia and emerging crown roots, while WOX11 is expressed primarily in the latter. ERF3 directly targets RR2 and upregulates its activity, which in turn reduces cytokinin signaling and stimulates crown root development. The interaction between WOX11 and ERF3 seems to occur after crown roots emerge and represses the activity of RR2 to increase cytokinin signaling and stimulate root elongation.
Another gene known to integrate cytokinin and auxin to regulate crown root formation is CYTOKININ OXIDASE/DEHYDROGENASE4 (CKX4) (Gao et al., 2014 ). CKX enzymes are known for catalyzing the irreversible degradation of cytokinins (Frébort et al., 2011). The gain‐of‐function mutation in root enhancer1 (ren1‐D) causes activation of CKX4, which results in a more robust root system with an increase in the number of crown roots. CKX4 has seven cytokinin response elements and two auxin response elements within the 2 kb upstream promoter region. Overexpression of CKX4 reduces auxin biosynthesis while knockdown of CKX4 enhances it, which suggests that this protein plays a role in modulating auxin production downstream of cytokinin. In addition, RR2 expression is downregulated in the ckx4 mutant, which seems to account for the increase in crown root number in the mutant background.
More recently, OsNAC2, an upstream integrator of the auxin and cytokinin signaling pathways, has been characterized. OsNAC2 encodes a NAC (NAM, ATAF, and CUC) transcription factor that affects root length and crown root number (Mao et al., 2020). OsNAC2 binds directly to the promoters of OsARF25 (auxin signaling) and OsCKX4 (cytokinin degradation). Furthermore, overexpression of the protein leads to a reduction in auxin biosynthesis and responses and enhanced cytokinin signaling, which results in lower meristematic activity and decreased crown root formation.
The signaling molecules that lead to crown root emergence have been thoroughly studied. In rice, crown root emergence is tightly linked to flooding. When plants are grown in aerated soil, ethylene escapes from roots, but when plants are grown under flooded conditions, water traps ethylene inside the root (Steffens & Rasmussen, 2016). The accumulation of ethylene triggers the production of reactive oxygen species, which in turn triggers programmed cell death in epidermal cells that allows for crown root emergence without damaging the root tip (Steffens et al., 2012). Gibberellic acid enhances the ethylene induction of crown root emergence, while abscisic acid inhibits the effects of gibberellic acid and ethylene (Steffens et al., 2006). Nitric oxide has also been shown to be a critical signaling molecule that induces crown root emergence (Xiong et al., 2009).
Auxin transport also plays an important role in crown root emergence. OsPIN1, an auxin efflux carrier, is required not only for development but for the emergence of crown roots as well (M. Xu et al., 2005). Crown root emergence is significantly reduced in the OsPIN1 knockdown plants, in a similar way to plants treated with the auxin transport inhibitor N‐1‐naphthylphalamic acid. OsCAND1 is one of the few genes characterized in rice that primarily affects the emergence of crown roots rather than their initiation. Mutant plants produce as many crown root primordia as the wild‐type, but these roots fail to emerge, and the seedlings die about two weeks after germination (X. F. Wang et al., 2011). The phenotypes seen in Oscand1 are similar to those of crown rootless mutant2 (crl2) mutant; however, the molecular characterization of the crl2 mutant has not been performed (Yoshiaki et al., 2001). OsCAND1 encodes a CULLIN‐ASSOCIATED AND NEDDYLATION‐DISSOCIATED1 (CAND1) SCF‐TIR1 ubiquitin ligase that is involved in the degradation of AUX/IAA proteins in the presence of auxin. In the Oscand1 mutant, there is significant repression of genes associated with the G2/M transition, as well as an upregulation of a negative regulator of cell division. Interestingly, auxin transport in the mutant seems to be impaired, and the exogenous application of auxin can partially rescue the cell division activity in the meristem of the mutant (X. F. Wang et al., 2011). Oryza sativa ROOT ARCHITECTURE ASSOCIATED 1 (OsRAA1) is another gene involved in cell‐cycle regulation in crown roots. OsRAA1 encodes a homologous protein to the Arabidopsis FLOWERING PROMOTING FACTOR that prevents a transition from metaphase to anaphase (Han et al., 2008). Plants overexpressing OsRAA1 have an increased number of crown roots compared to wild‐type (Ge et al., 2004).
2.4. Brace roots: Overview
Brace roots are adventitious roots that are initiated from nodes aboveground, usually in two to three whorls. This root type is found predominantly in members of the Andropogoneae (maize, sorghum, and sugarcane) and Paniceae (foxtail millet) tribes (Hostetler et al., 2021).
2.4.1. Brace roots: Anatomy and function
Although by definition the difference between crown roots and brace roots is just the position in the plant where they emerge, the morphology, anatomy, and transcriptional signature of brace roots is significantly different from crown roots (Li et al., 2011; Yu et al., 2015). For instance, brace roots have increased root diameter, number of cortical cell layers, and number of metaxylem vessels when compared to crown roots and the primary root (Yu et al., 2015). The mechanism of initiation for brace and crown roots is likely similar; however, no work has been done to support this claim. Research has been conducted to determine the function of brace roots in terms of lodging resistance, water absorption, and mucilage production.
Brace root number, spread width, and the number of brace root whorls were all negatively correlated to root lodging in maize plants grown in two different watering regimes (well‐watered and drought‐stressed) (Sharma & Carena, 2016). More recently, a direct assessment of the role that brace roots play in anchoring the plant has been reported. By using a non‐destructive field‐based mechanical test on CML258 maize inbred plants, it was demonstrated that removal of brace roots significantly increases plant deflection (i.e., plants are more susceptible to lodging) (Reneau et al., 2020). The position of the whorl of brace roots also significantly contributes to anchorage; the whorl closest to the soil contributes the most, and each subsequent whorl shootward contributes less. Brace roots that reach the soil develop lateral roots and can contribute to water and nutrient absorption, as well as to increased lodging resistance. However, knowledge about the role of brace roots growing solely in the air is still scarce.
Maize brace roots produce mucilage, a gelatinous substance rich in polysaccharides exuded by the root tip (Nazari et al., 2020). A recent study has shown the microbiota in the mucilage of a maize landrace from Mexico is enriched in diazotrophic bacteria, which are known to fix atmospheric gas into a usable form for plants, such as ammonium (Van Deynze et al., 2018). Field experiments with plants grown in nitrogen‐depleted soils revealed that atmospheric nitrogen fixation contributed up to 89% of the nitrogen nutrition of the landrace. Not all maize lines produce a significant amount of mucilage; thus, uncovering the genetic network that controls this trait could help increase nitrogen use efficiency and reduce fertilizer application, which would be beneficial both to small and large‐scale agriculture.
2.4.2. Brace roots: Developmental regulation
The mechanisms of how brace roots develop are still poorly understood compared to crown roots. One of the first mutants lacking brace roots identified was rtcs, which shows a complete lack of seminal, crown, and brace roots (Hetz et al., 1996). Its paralog, rtcs‐like (rtcl), is impaired in the elongation of roots, but not initiation and emergence (Taramino et al., 2007; C. Xu et al., 2015). RTCS and RTCL encode LBD transcription factors and are orthologs of CRL1/ARL1, which suggests that the initiation mechanism of adventitious roots in rice and maize is conserved. RTCS and RTCL bind to an LBD motif in the promoter of the auxin response factor (ARF) ZmARF34 to activate downstream genes (Majer et al., 2012).
More recently, a gene involved in brace root development that does not affect crown root growth has been characterized. The corngrass1 mutant, which shows an elevated expression of ZmRAP2.7, also exhibits a six‐fold increase in the number of brace roots, while the number of crown roots is similar to that of wild‐type (Chuck et al., 2007; J. Li et al., 2019). ZmRAP2.7 encodes an AP2 transcription factor, and it is expressed in all root types. A Mu‐transposon insertion (RAP2.7‐Mu) which disrupts the second exon of the gene, results in a lower number of brace roots than wild‐type; mutants have one less whorl of brace roots. The encoded protein acts as a transcriptional activator; however, the downstream pathways targeted by ZmRAP2 are still undetermined.
The MATE transporter BIG EMBRYO1 (BIGE1) regulates lateral organ size and initiation rate in maize (Suzuki et al., 2015). A loss‐of‐function mutation results in enlargement of the embryo scutellum, accelerated leaf formation, and production of brace roots in higher nodes not seen in the wild‐type plant. The ZmCCT10 (CO, CONSTANS, CO‐LIKE, and TIMING OF CAB1) transcription factor family is a key regulator of photoperiod response. Strong overexpression of ZmCCT10 resulted in a prolonged vegetative phase accompanied by dramatic phenotypic changes. Transgenic plants could produce brace roots up to the 37th node, while wild‐type plants usually produced brace roots only up to the 6th node (Stephenson et al., 2019). Similarly, downregulation of the teosinte glume architecture1 (tga1), an SBP box transcription factor that is involved in the regulation of fruitcase and kernel development in maize and teosinte, shows pleiotropic effects in maize plants, with brace roots developing in six more nodes compared to the wild‐type (H. Wang et al., 2015). The molecular mechanisms of how these genes control brace root development are still unclear.
Ethylene plays a major role in the emergence of crown roots in rice; however, the function of this hormone in the emergence of brace roots is largely unknown. To determine the effects of ethylene on brace root development, maize plants were treated daily with different concentrations of 1‐aminocyclopropane1‐carboxylic acid (ACC), an ethylene precursor, which resulted in enhanced emergence of brace roots (Shi et al., 2019). Overexpression of ARGOS8, a gene that reduces ethylene sensitivity, resulted in delayed brace root emergence and decreased response to ACC. This suggests that ethylene is an important hormone for lodging resistance through the regulation of brace root emergence.
Quantitative trait loci mapping has been used to identify loci that contribute to brace root traits in maize (Gu et al., 2017; A. Zhang, Cui, et al., 2018; Z. Zhang, Zhang, et al., 2018). Several candidate genes have been identified for signal transduction and gene expression regulation (A. Zhang, Cui, et al., 2018). Investigation of these candidate genes would open novel directions in the regulation of brace root development.
3. ENVIRONMENTAL REGULATION OF ROOT DEVELOPMENT AND GROWTH: DROUGHT
Drought is one of the major yield‐limiting factors affecting crop plants. In the United States alone, the annual losses in 2018 were estimated to be between 10 and 14 billion dollars (IPCC, 2014). These numbers have likely increased, as droughts have become more frequent and severe due to climate change (Reidmiller et al., 2017). Grasses have evolved mechanisms that allow them to respond quickly and adapt to changes in their environment to survive periods of drought. Through a series of experiments conducted in the greenhouse, field, and using the luminescence‐based imaging system (GLO‐Roots), it was shown in Setaria viridis, a model for panicoid grasses, that the crown locally senses water availability (Rellán‐Álvarez et al., 2015; Sebastian et al., 2016). Under ideal growing conditions, crown roots emerge periodically and make up the bulk of the root system of a grass plant; however, under drought‐stressed conditions, crown root postemergence growth is suppressed, and the emerged roots ultimately senesce (Figure 4a,b). Plants that are grown under drought‐stressed conditions can rapidly produce new crown roots upon rewatering. New crown roots emerge de novo, not by the resumption of roots that had their growth suppressed, but by newly emerged crown roots, and growth only occurred if the water was applied directly to the crown. Plants in which water was applied to the bottom of the pot recovered their water status but did not develop new crown roots (Sebastian et al., 2016). This dynamic response prevents overdrawing of the limited water resources in the soil and confirms the crown's importance in sensing water and triggering crown root growth.
Figure 4.
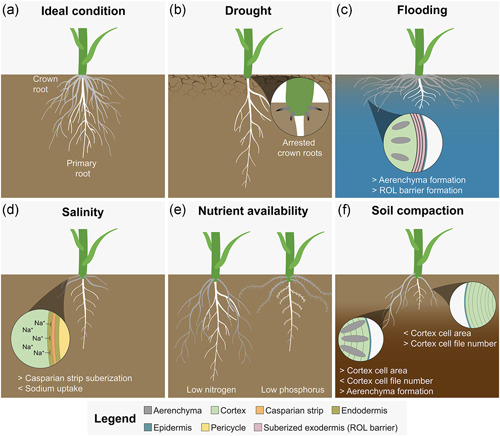
Root system architecture responses to different environmental conditions. (a) Ideal condition: crown roots make up the bulk of the mature grass root system. (b) Drought: Proliferation of the primary root system branches, with steeper root angles. Crown root growth is arrested to conserve water. (c) Flooding: Shallow root system to avoid growing in deeper areas where oxygen is less available. Ethylene accumulation stimulates additional aerenchyma formation, which allows oxygen transport in roots. In addition, the formation of radial oxygen loss barriers helps to reduce the loss of oxygen transported via the aerenchyma. (d) Salinity: Reduction of root elongation. Formation of suberized barriers for salt exclusion from transport into the shoot. (e) Nutrient availability: Foraging for mobile (e.g., nitrogen) and immobile (e.g., phosphorus) nutrients either through searching far or through exploring local areas by increased branching. (f) Soil compaction: Thickening of primary root and older crown roots facilitates soil penetration but is not sufficient to allow for growth in more compacted areas. Crown roots from newer nodes are thicker from the time of emergence due to increased cortex cell area and are less sensitive to soil compaction. Increased aerenchyma counteracts the effects of hypoxia in roots growing in compacted soil
Steep, cheap, and deep is a hypothetical ideotype in which water use and nitrogen acquisition are most efficient, particularly in soils where the resources are limited (Lynch, 2013). An ideal scenario was proposed for each type of root, using maize as a model. For conditions where water is the limiting factor, the primary root and seminal roots should have a larger diameter for penetration of more compacted soils and a lower number of lateral roots, which allows for better exploration of water resources without depletion. Crown root number should be intermediate, with a steep growth angle and very few lateral roots. The low number of laterals is beneficial because it allows for a better allocation of internal resources. The plant can invest its carbon towards the elongation of the crown root, which can explore even deeper soils. Plants should invest in one node of brace roots with high occupancy. Brace roots that emerge from higher nodes are less likely to reach the soil and might be a waste of resources. The brace roots that reach the soil should have a less steep growth angle, to avoid competition with the already established crown roots. It should also have less laterals for the reasons already mentioned.
This hypothetical ideotype was supported with empirical evidence. Maize recombinant inbred lines with significant diversity in lateral root number and length were grown in three different environments (greenhouse mesocosms, field rainout shelters, field with natural drought) (Zhan et al., 2015). In general, genotypes that had fewer and longer lateral roots grown under drought‐stressed conditions showed deeper rooting, increased shoot biomass, increased yield, and less lateral root respiration. These results supported the hypothesis that making fewer lateral roots improves rooting depth and hence improves drought tolerance. In rice grown under progressive drought, having a deeper root system with long lateral root branching and a reduced number of crown roots enhances drought tolerance (Hazman & Brown, 2018).
Root system architecture is a complex trait governed by root number, length, growth angle, volume, among other traits. A few quantitative trait loci have been isolated for some of these: root length (Kitomi et al., 2018; Obara et al., 2010; H. Wang et al., 2013), root growth angle (Uga et al., 2012; Uga et al., 2011) and root volume (Uga, Yamamoto, et al., 2013). However, to this date, only a few of these QTLs have been cloned. DEEP ROOTING 1 (DRO1) controls root system architecture by causing asymmetric root growth and bending of the root tip downwards in response to gravity, resulting in deeper root systems (Uga, Sugimoto, et al., 2013). Introduction of DRO1 in a drought susceptible rice cultivar resulted in increased drought tolerance due to deeper soil exploration. More recently, quantitative trait locus for SOIL SURFACE ROOTING 1 (qSOR1), a homolog of DRO1, has been cloned (Kitomi et al., 2020). Similar to DRO1, qSOR1 regulates root system architecture by changing the gravitropic responses of roots. A loss‐of‐function allele results in a shallow root system, which allows plants to be more resistant to salinity stress. DOCS1 and LARGE ROOT ANGLE1 also mediate gravitropic responses in roots to control root system architecture (Bettembourg et al., 2017; L. Wang et al., 2018).
Anatomical changes in the root can also improve drought tolerance in grasses. Conversion of root cortical tissue into cortical aerenchyma reduces metabolic costs of soil exploration and improves drought tolerance in maize (Zhu et al., 2010). Recombinant inbred lines with high or low amounts of root cortical aerenchyma were grown in greenhouse mesocosms and in the field. Genotypes that had more root cortical aerenchyma grown under drought‐stress conditions had more shoot biomass, greater root length, and were deeper rooting. In line with these results, reduced living cortical area, reduced cell file number, and large cortical cell size are also anatomical changes that influence drought tolerance in maize (Chimungu et al., 2014a, 2014b; Jaramillo et al., 2013). In rice, the stele of crown roots becomes more lignified during drought stress, which is suggested to improve drought tolerance by maintaining root function as a water conductant while reducing loss of ions and water to the soil (Hazman & Brown, 2018).
3.1. Environmental regulation of root development and growth: Flooding
Except for rice, which is tolerant to flooding, other grass crops have varying levels of susceptibility to flooding, which causes moderate to severe yield losses in agriculture. When facing saturated soil conditions, plants usually respond by producing new adventitious roots that are better suited for the environment (Colmer, 2003; Colmer & Voesenek, 2009). An anatomical adaptation seen in roots of crop species is the formation of aerenchyma (Figure 4c), a modified tissue derived from parenchyma cells that create air gaps within the roots and allows better gas exchange (Yamauchi et al., 2013). Rice roots constitutively develop aerenchyma, but depletion of oxygen in soil due to waterlogging further induces the development of the tissue. Maize, however, only develops aerenchyma when grown in oxygen‐deficient soils or when certain nutrients are limiting.
The phytohormone ethylene plays a major role in flooding responses in plants. Under waterlogged conditions, the gaseous hormone cannot diffuse out of the plant cells and is then trapped within the root. Accumulation of ethylene in the roots triggers reactive oxygen species production, which subsequently triggers programmed cell death in the epidermis. These changes in tissue integrity allow for new adventitious roots to emerge and for the formation of lysigenous aerenchyma in the cortex (Steffens & Sauter, 2009). Besides ethylene, two other environmental factors play a role in the acclimation of rice to flooding. Gravity and light (or the lack of) determine the angle for crown root growth in rice. When crown roots emerge in the light, they grow downwards to avoid desiccation, but when emerged in the dark they grow upwards. A possible explanation for this behavior is that it allows roots to avoid growth in deeper parts of the soil where oxygen is less available (Lin & Sauter, 2018).
Another anatomical response to waterlogging is the synthesis of radial oxygen loss (ROL) barriers, which reduce the loss of oxygen transported via the aerenchyma (Colmer, 2003). In rice, ROL starts decreasing at about 20 mm from the root tip and 50 mm from the root tip, the barrier efficiency significantly increases, likely due to the deposition of suberin in the root hypodermis/exodermis (Colmer et al., 2019). More recently, ROL barriers were also found to be present not only in crown roots but also in lateral roots. ROL and barrier formation were compared between a teosinte species that thrives in flooded conditions (Zea nicaraguensis), a maize introgression line that carries the locus for ROL barrier formation from the aforementioned teosinte, and the maize inbred that was used for the introgression. It was shown that the lateral roots of the teosinte and the introgression line formed ROL barriers, while the maize inbred did not (Pedersen et al., 2021). While ROL barriers prevent the loss of oxygen, only lateral root tissues in teosinte showed a higher oxygen status. This is likely due to the significantly higher amount of aerenchyma formation in teosinte. This shows that both ROL barrier and aerenchyma formation work in synergy to improve tissue oxygen status under flooded conditions. Furthermore, modeling showed that lateral roots that have ROL barriers could grow up to 74 mm, more than double of what lateral roots without ROL barriers can grow (33 mm). Uncovering the genetic basis for this trait could help to improve the flooding tolerance of susceptible crops, such as maize.
3.2. Environmental regulation of root development and growth: Salinity
Salinity poses a significant challenge to agriculture. Low rainfall and irrigation can cause the accumulation of salt in the soil. Additionally, saltwater intrusion into freshwater aquifers due to excessive groundwater extraction for irrigation can cause increased salt levels in irrigation and drinking water (Greene et al., 2016). Salinity challenges plants in two ways: (1) Accumulation of salt causes a decrease in osmotic potential, causing osmotic stress to the plant and making uptake of water and nutrients from the soil more challenging (2) Ion toxicity causing an imbalance in other essential ions and interfering with growth.
While high salinity has a significant effect on root growth and development, shoot growth is more affected by high salinity. Of the different grasses that serve as cereal crops, barley is considered the most salt‐tolerant (Munns et al., 2006). It can sustain growth up to a concentration of 250 mM NaCl, which is equivalent to 50% seawater levels. Ranked by their salt tolerance, barley is followed by wheat, maize and rice, which has a low tolerance to salinity (Tanji & Kielen, 2002).
While barley is more salt‐tolerant, a significant reduction of seminal root emergence and growth was observed in different barley genotypes when exposed to 100 and 150 nM NaCl at the seedling stage (Shelden et al., 2013). Interestingly, this reduction did not correlate with root and shoot ion content. Similar to barley, reduction of root growth due to high salinity was observed in other grasses such as maize and rice. In maize, a stronger reduction of crown and seminal root elongation was observed compared to the primary root when seedlings were transferred to a liquid culture containing 100 mM NaCl (M. Zhang, Kong, et al., 2015). This stress response may be modulated through specific transcription factors, for example, OsMADS25 in rice (N. Xu et al., 2018). It was observed that overexpression of this transcription factor improves primary root elongation under saline conditions by modulating abscisic acid and reactive oxygen species‐related stress response pathways.
A way to cope with high salinity is avoidance of soil regions that contain high concentrations of salt. For example, in rice, the loss‐of‐function of a OsDRO1 homolog in the quantitative trait locus for SOIL SURFACE ROOTING 1 (qSOR1) led to shallower root growth angles with more roots near the soil surface (Kitomi et al., 2020). It was discovered that these plants had higher yields in saline paddy soil compared to their wild‐type by avoiding the more saline subsoil. Other responses such as halotropism allow root tips of sorghum to sense the direction of a salinity gradient and grow in the other direction (Galvan‐Ampudia et al., 2013).
Another way to cope with high salinity is the exclusion of sodium ions from the shoot. For example, it was found in maize that the high‐affinity K+ transporter (HKT) ZmHKT1, which is expressed in the root stele, is involved in the exclusion of sodium ions from the shoot (M. Zhang, Cao, et al., 2018). In addition to pumping sodium ions, the formation of root apoplastic barriers block sodium movement to the shoot as shown in rice (Krishnamurthy et al., 2011) (Figure 4d).
This shows that salinity causes stunting of root growth. While molecular mechanisms of salt tolerance and exclusion play an important role, changes in root system architecture and root anatomy can also contribute to achieving more salt tolerance. As we move to agriculture on more marginal land, improved salinity tolerance will be an important factor in breeding.
3.3. Environmental regulation of root development and growth: Nutrient availability
Roots show a strong response to differences in nutrient distribution in the soil. In a classic experiment, high concentrations of phosphate, nitrate, ammonium, or potassium were locally applied to root systems of barley seedlings (Drew, 1975). Except for potassium, lateral root emergence and elongation were promoted by this treatment. Since the application of fertilizer is both expensive and potentially damaging to the environment, much research has gone into understanding root responses to nutrients to improve overall plant nutrient‐use efficiency. The three main nutrients that are often yield limiting in agricultural systems are considered to be nitrogen, phosphorus, and potassium. Additionally, calcium, magnesium, and sulfur are consumed in large quantities; hence all these are called macronutrients. However, all other elements, called micronutrients since they are consumed in smaller quantities, play important roles as well, and plants often suffer from micronutrient deficiencies.
While nitrogen is a mobile nutrient, many other nutrients are immobile, including phosphorus and potassium. Plants need to grow towards immobile nutrients to extract them from the soil. For this reason, different optimal root growth strategies were observed for nitrogen and phosphorus. In maize, extensive work was done on understanding these differences. As previously discussed, one main concept that has been developed is the “steep, cheap, and deep” root ideotype, which optimizes for water and nitrogen uptake (Lynch, 2013) (Figure 4e). This idea is being supported by research showing that maize plants with a steeper root angle, due to a mutation in the CBL‐interacting serine/threonine‐protein kinase 15 (ZmCIPK15), showed improved nitrogen uptake under low‐nitrogen conditions. Additionally, it was found that a higher quantity of root cortical aerenchyma may improve rooting depth and subsequently nitrogen uptake due to a reduced cost for root growth (Saengwilai et al., 2014). Similarly, larger cortical cells and reduced cortical cell layer numbers may also lower the metabolic burden due to a reduced living cortical area and lead to better soil exploration and improved phosphorus capture (Galindo‐Castañeda et al., 2018). In rice it was found that the auxin transporter OsPIN1b might be involved in the response of seminal roots to low nitrogen and phosphorus leading to increased root elongation and, hence, nutrient acquisition (Sun et al., 2018).
While lower lateral root densities would increase rooting depth and nitrogen acquisition, higher lateral root densities were found to be beneficial for uptake of phosphorus under low phosphorus conditions (Jia et al., 2018; Zhu & Lynch, 2004). Increased lateral root production leads to more soil exploration and helps in improving phosphorus capture. This also aligns with findings that increased numbers of seminal roots in domesticated maize improved nutrient capture (Perkins & Lynch, 2021).
While there appears to be a trade‐off for lateral root densities when comparing nitrogen and phosphorus uptake efficiency, a screening of a very large panel of maize lines yielded an average lateral root branching density of six branches per centimeter (Postma et al., 2014). The authors concluded that it might indicate that this number is optimal to capture both nutrients without wasting energy.
Much is now understood about nutrient capture by grass roots. However, most studies have been done on root systems without particular attention to the dynamic aspects of root system architecture. Less is known on how dynamic responses of root systems influence nutrient capture and how these more dynamic responses could be harnessed for breeding.
3.4. Environmental regulation of root development and growth: Soil compaction
Soil compaction occurs when soil particles are pressed together and results in increased soil density with reduced pore space. This phenomenon has become more prevalent in modern agriculture due to the increased size and weight of the machinery used to cultivate the land. As a consequence, yield losses due to poor root system development are increasing as well.
The most common root response to heavily compacted soils is the stunting of root growth (Figure 4f). Increasing soil bulk density decreased root length in maize, with the highest density completely suppressing root elongation (Lucas et al., 1954, 2019). Ethylene, not just mechanical impedance itself, is responsible for this response. Reduction in air pores lowers gas diffusion in the soil layers and traps ethylene, which triggers responses that suppress root growth (Pandey et al., 2021). Disruption of ethylene response in rice allows roots to grow in compacted soils. One mechanism by which plants can grow past compacted soils is trematotropism, the ability of roots to locate pores in the soil (Dexter, 1986). When grown in compacted soil, near‐isogenic lines of wheat had a dramatic increase in macropore root colonization compared to plants grown in loose soil (Atkinson et al., 2020).
4. CONCLUSIONS AND FUTURE DIRECTIONS
Despite the absolute dependency of global society on the cultivation of cereal crops and the importance of plant root systems for providing the essential nutrients and water that supports crop yields, our understanding of the developmental mechanisms that pattern the diverse collection of root types in grasses is still in its infancy. Beyond the practical value of such knowledge, the origin of the grass and monocot root system is also of importance from an evolutionary perspective, as members of this plant clade are distinguished by a number of anatomical features such as the loss of secondary thickening growth (Roodt et al., 2019; Spicer & Groover, 2010). This important developmental constraint may have required innovations in how plants establish new conduits for water and nutrient uptake, as the expansion of the vascular system would be impossible otherwise. From this perspective, the development of the nodal root system, characterized by the seminal, crown, and brace roots of grasses, represents a diversification in the function of this multi‐axial vascular system. Are these root types, which have been defined largely by their position of origin, specified in a developmental sense as distinct organ types, as floral organ identity is determined (Coen & Meyerowitz, 1991)? Or is their anatomical and physiological diversity a consequence of the unique environmental contexts in which they initiate and emerge? Other outstanding questions pertain to the earliest stages of lateral root formation. While extensive mechanistic details are being elucidated for Arabidopsis, their relevance for any other species is a mystery. It may be that the initiation of lateral roots opposite the phloem pole, instead of the xylem pole, occurs due to the inherent differences in vascular patterning that occurs in monocots, or it may simply be that this trait is plastic and varies extensively between species. In any case, digging deeper into the secret lives of grass roots will provide the answers to many of these long‐standing questions and the necessary means to do something useful with this knowledge in hand.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGEMENTS
We would like to thank members of the Dinneny lab for their comments and suggestions during the drafting of this manuscript. This study was supported by the Advanced Research Projects Agency‐Energy Grant DE‐AR0000825 and the U.S. Department of Energy's Biological and Environmental Research program (Grant DE‐SC0008769 to J.R.D.). José Dinneny is supported in part by a Faculty Scholar grant from Howard Hughes Medical Institute and the Simons Foundation (55108515).
Viana, W.G. , Scharwies, J.D. & Dinneny, J.R. (2022) Deconstructing the root system of grasses through an exploration of development, anatomy and function. Plant, Cell & Environment, 45, 602–619. 10.1111/pce.14270
Willian G. Viana and Johannes D. Scharwies are contributed equally to this study.
REFERENCES
- Abbe, E.C. & Stein, O.L. (1954) The growth of the shoot apex in maize: embryogeny. American Journal of Botany, 41, 285–293. [Google Scholar]
- Abe, M. , Kuroshita, H. , Umeda, M. , Itoh, J.‐I. & Nagato, Y. (2008) The rice flattened shoot meristem, encoding CAF‐1 p150 subunit, is required for meristem maintenance by regulating the cell‐cycle period. Developmental Biology, 319, 384–393. [DOI] [PubMed] [Google Scholar]
- Ahmed, M.A. , Zarebanadkouki, M. , Kaestner, A. & Carminati, A. (2016) Measurements of water uptake of maize roots: the key function of lateral roots. Plant and Soil, 398, 59–77. [Google Scholar]
- Ahmed, M.A. , Zarebanadkouki, M. , Meunier, F. , Javaux, M. , Kaestner, A. & Carminati, A. (2018) Root type matters: measurement of water uptake by seminal, crown, and lateral roots in maize. Journal of Experimental Botany, 69, 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, J.A. , Hawkesford, M.J. , Whalley, W.R. , Zhou, H. & Mooney, S.J. (2020) Soil strength influences wheat root interactions with soil macropores. Plant, Cell & Environment, 43, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, J.A. , Rasmussen, A. , Traini, R. , Voß, U. , Sturrock, C. , Mooney, S.J. et al. (2014) Branching out in roots: uncovering form, function, and regulation. Plant Physiology, 166, 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babé, A. , Lavigne, T. , Séverin, J.‐P. , Nagel, K.A. , Walter, A. , Chaumont, F. et al. (2012) Repression of early lateral root initiation events by transient water deficit in barley and maize. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 367, 1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Y. , Aggarwal, P. , Robbins, N.E. , Sturrock, C.J. , Thompson, M.C. , Tan, H.Q. et al. (2014) Plant roots use a patterning mechanism to position lateral root branches toward available water. Proceedings of the National Academy of Sciences, 111, 9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková, E. , Michniewicz, M. , Sauer, M. , Teichmann, T. , Seifertová, D. , Jürgens, G. et al. (2003) Local, efflux‐dependent auxin gradients as a common module for plant organ formation. Cell, 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bettembourg, M. , Dal‐Soglio, M. , Bureau, C. , Vernet, A. , Dardoux, A. , Portefaix, M. et al. (2017) Root cone angle is enlarged in docs1 LRR‐RLK mutants in rice. Rice, 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, H. , Xie, Y. , Guo, F. , Han, N. , Ma, S. , Zeng, Z. et al. (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). The New Phytologist, 196, 149–161. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos, L.I. , Lee, S. , De Oliveira, C. , Ivetac, A. , Brandt, W. , Armitage, L. et al. (2012) A combinatorial TIR1/AFB‐Aux/IAA co‐receptor system for differential sensing of auxin. Nature Chemical Biology, 8, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero, P.J. , Casimiro, I. & Lloret, P.G. (1995) Lateral root initiation by asymmetrical transverse divisions of pericycle cells in four plant species: raphanus sativus, Helianthus annuus, Zea mays, and Daucus carota. Protoplasma, 188, 49–58. [Google Scholar]
- Chen, R. , Xu, N. , Yu, B. , Wu, Q. , Li, X. , Wang, G. et al. (2020) The WUSCHEL‐related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice. Plant Science, 298, 110575. [DOI] [PubMed] [Google Scholar]
- Chimungu, J.G. , Brown, K.M. & Lynch, J.P. (2014a) Reduced root cortical cell file number improves drought tolerance in maize. Plant Physiology, 166, 1943–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimungu, J.G. , Brown, K.M. & Lynch, J.P. (2014b) Large root cortical cell size improves drought tolerance in maize. Plant Physiology, 166, 2166–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Liang, W. , Li, J. , Hong, F. , Wu, Y. , Wang, L. et al. (2013) A CLE‐WOX signalling module regulates root meristem maintenance and vascular tissue development in rice. Journal of Experimental Botany, 64, 5359–5369. [DOI] [PubMed] [Google Scholar]
- Chuck, G. , Mark Cigan, A. , Saeteurn, K. & Hake, S. (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nature Genetics, 39, 544–549. [DOI] [PubMed] [Google Scholar]
- Clark, L.H. & Harris, W.H. (1981) Observations on the root anatomy of rice (Oryza sativa L.). American Journal of Botany, 68, 154–161. [Google Scholar]
- Coen, E.S. & Meyerowitz, E.M. (1991) The war of the whorls: genetic interactions controlling flower development. Nature, 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Colmer, T.D. (2003) Aerenchyma and an inducible barrier to radial oxygen loss facilitate root aeration in upland, paddy and deep‐water rice (Oryza sativa L.). Annals of Botany, 91 Spec No, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer, T.D. , Kotula, L. , Malik, A.I. , Takahashi, H. , Konnerup, D. , Nakazono, M. et al. (2019) Rice acclimation to soil flooding: low concentrations of organic acids can trigger a barrier to radial oxygen loss in roots. Plant, Cell & Environment, 42, 2183–2197. [DOI] [PubMed] [Google Scholar]
- Colmer, T.D. & Voesenek, L.A.C.J. (2009) Flooding tolerance: suites of plant traits in variable environments. Functional Plant Biology, 36, 665–681. [DOI] [PubMed] [Google Scholar]
- Colombi, T. , Herrmann, A.M. , Vallenback, P. & Keller, T. (2019) Cortical cell diameter is key to energy costs of root growth in wheat. Plant Physiology, 180, 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Levesque, M.P. , Vernoux, T. , Jung, J.W. , Paquette, A.J. , Gallagher, K.L. et al. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science, 316, 421–425. [DOI] [PubMed] [Google Scholar]
- Dexter, A.R. (1986) Model experiments on the behaviour of roots at the interface between a tilled seed‐bed and a compacted sub‐soil. Plant and Soil, 95, 149–161. [Google Scholar]
- Dickinson, A.J., Zhang, J., Luciano, M., Wachsman, G., Sandoval, E., Schnermann, M., Dinneny, J.R. et al. (2021) A plant lipocalin promotes retinal‐mediated oscillatory lateral root initiation. Science, 373, 1532–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer, H.J. (1937) A quantitative study of the roots and root hairs of a winter rye plant (Secale cereale). American Journal of Botany, 24, 417–420. [Google Scholar]
- Drew, M.C. (1975) Comparison of the effects of a localised supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. The New Phytologist, 75, 479–490. [Google Scholar]
- Dubrovsky, J.G. , Doerner, P.W. , Colón‐Carmona, A. & Rost, T.L. (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiology, 124, 1648–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan, C. , Meda, S. & Varotto, S. (2010) ZmPIN1‐mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiology, 152, 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frébort, I. , Kowalska, M. , Hluska, T. , Frébortová, J. & Galuszka, P. (2011) Evolution of cytokinin biosynthesis and degradation. Journal of Experimental Botany, 62, 2431–2452. [DOI] [PubMed] [Google Scholar]
- Fusseder, A. (1987) The longevity and activity of the primary root of maize. Plant and Soil, 101, 257–265. [Google Scholar]
- Galindo‐Castañeda, T. , Brown, K.M. & Lynch, J.P. (2018) Reduced root cortical burden improves growth and grain yield under low phosphorus availability in maize. Plant, Cell & Environment, 41, 1579–1592. [DOI] [PubMed] [Google Scholar]
- Galvan‐Ampudia, C.S. , Julkowska, M.M. , Darwish, E. , Gandullo, J. , Korver, R.A. , Brunoud, G. et al. (2013) Halotropism is a response of plant roots to avoid a saline environment. Current Biology, 23, 2044–2050. [DOI] [PubMed] [Google Scholar]
- Gao, S. , Fang, J. , Xu, F. , Wang, W. , Sun, X. , Chu, J. et al. (2014) Cytokinin oxidase/Dehydrogenase4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiology, 165, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, L. , Chen, H. , Jiang, J.‐F. , Zhao, Y. , Xu, M.‐L. , Xu, Y.‐Y. et al. (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiology, 135, 1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, R. , Timms, W. , Rengasamy, P. , Arshad, M. & Cresswell, R. (2016) Soil and aquifer salinization: toward an integrated approach for salinity management of groundwater, In: Jakeman, A.J., Barreteau, O., Hunt, R.J., Rinaudo, J.‐D. & Ross, A. Integrated groundwater management. Cham: Springer, pp. 377–412. [Google Scholar]
- Gu, D. , Mei, X. , Yu, T. , Sun, N. , Xu, D. , Liu, C. et al. (2017) QTL identification for brace‐root traits of maize in different generations and environments. Crop Science, 57, 13–21. [Google Scholar]
- Guyomarc'h, S. , Leran, S. , Auzon‐Cape, M. , Perrine‐Walker, F. , Lucas, M. & Laplaze, L. (2012) Early development and gravitropic response of lateral roots in Arabidopsis thaliana. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 367, 1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T. , Mayer, U. & Jürgens, G. (1999) The auxin‐insensitive bodenlos mutation affects primary root formation and apical‐basal patterning in the Arabidopsis embryo. Development, 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Han, Y. , Cao, H. , Jiang, J. , Xu, Y. , Du, J. , Wang, X. et al. (2008) Rice root architecture associated1 binds the proteasome subunit RPT4 and is degraded in a D‐box and proteasome‐dependent manner. Plant Physiology, 148, 843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazman, M. & Brown, K.M. (2018) Progressive drought alters architectural and anatomical traits of rice roots. Rice, 11, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herud, O. , Weijers, D. , Lau, S. & Jürgens, G. (2016) Auxin responsiveness of the MONOPTEROS‐BODENLOS module in primary root initiation critically depends on the nuclear import kinetics of the Aux/IAA inhibitor BODENLOS. The Plant Journal, 85, 269–277. [DOI] [PubMed] [Google Scholar]
- Hetherington, A.J. , Berry, C.M. & Dolan, L. (2020) Multiple origins of dichotomous and lateral branching during root evolution. Nature Plants, 6, 454–459. [DOI] [PubMed] [Google Scholar]
- Hetherington, A.J. & Dolan, L. (2018) Stepwise and independent origins of roots among land plants. Nature, 561, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington, A.J. & Dolan, L. (2019) Evolution: diversification of angiosperm rooting systems in the early cretaceous. Current Biology, 29, R1081–R1083. [DOI] [PubMed] [Google Scholar]
- Hetz, W., Hochholdinger, F., Schwall, M. & Feix G. (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. The Plant Journal, 10(5), 845–857. 10.1046/j.1365-313x.1996.10050845.x [DOI] [Google Scholar]
- Hochholdinger, F. & Feix, G. (1998) Early post‐embryonic root formation is specifically affected in the maize mutant lrt1. The Plant Journal, 16, 247–255. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F. , Woll, K. , Sauer, M. & Dembinsky, D. (2004) Genetic dissection of root formation in maize (Zea mays) reveals root‐type specific developmental programmes. Annals of Botany, 93, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger, F. & Zimmermann, R. (2008) Conserved and diverse mechanisms in root development. Current Opinion in Plant Biology, 11, 70–74. [DOI] [PubMed] [Google Scholar]
- Hoppe, D.C. , McCully, M.E. & Wenzel, C.L. (1986) The nodal roots of Zea: their development in relation to structural features of the stem. Canadian Journal of Botany/Journal canadien de botanique, 64, 2524–2537. [Google Scholar]
- Hostetler, A.N. , Khangura, R.S. , Dilkes, B.P. & Sparks, E.E. (2021) Bracing for sustainable agriculture: the development and function of brace roots in members of Poaceae. Current Opinion in Plant Biology, 59, 101985. [DOI] [PubMed] [Google Scholar]
- Husakova, E. , Hochholdinger, F. & Soukup, A. (2013) Lateral root development in the maize (Zea mays) lateral rootless1 mutant. Annals of Botany, 112, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inahashi, H. , Shelley, I.J. , Yamauchi, T. , Nishiuchi, S. , Takahashi‐Nosaka, M. , Matsunami, M. et al. (2018) OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiologia Plantarum, 164, 216–225. [DOI] [PubMed] [Google Scholar]
- Inukai, Y. , Sakamoto, T. , Ueguchi‐Tanaka, M. , Shibata, Y. , Gomi, K. , Umemura, I. et al. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell, 17, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . (2014) Climate change 2014: summary for policymakers.
- Itoh, J.‐I. , Nonomura, K.‐I. , Ikeda, K. , Yamaki, S. , Inukai, Y. , Yamagishi, H. et al. (2005) Rice plant development: from zygote to spikelet. Plant & Cell Physiology, 46, 23–47. [DOI] [PubMed] [Google Scholar]
- Jackson, R.B. & Caldwell, M.M. (1993) The scale of nutrient heterogeneity around individual plants and its quantification with geostatistics. Ecology, 74, 612–614. [Google Scholar]
- Jansen, L. , Roberts, I. , De Rycke, R. & Beeckman, T. (2012) Phloem‐associated auxin response maxima determine radial positioning of lateral roots in maize. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 367, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo, R.E. , Nord, E.A. , Chimungu, J.G. , Brown, K.M. & Lynch, J.P. (2013) Root cortical burden influences drought tolerance in maize. Annals of Botany, 112, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, X. , Liu, P. & Lynch, J.P. (2018) Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. Journal of Experimental Botany, 69, 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun, N. , Gaohang, W. , Zhenxing, Z. , Huanhuan, Z. , Yunrong, W. & Ping, W. (2011) OsIAA23‐mediated auxin signaling defines postembryonic maintenance of QC in rice. The Plant Journal, 68, 433–442. [DOI] [PubMed] [Google Scholar]
- Kamiya, N. , Itoh, J.‐I. , Morikami, A. , Nagato, Y. & Matsuoka, M. (2003) The SCARECROW gene's role in asymmetric cell divisions in rice plants. The Plant Journal, 36, 45–54. [DOI] [PubMed] [Google Scholar]
- Kamiya, N. , Nagasaki, H. , Morikami, A. , Sato, Y. & Matsuoka, M. (2003) Isolation and characterization of a rice WUSCHEL‐type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. The Plant Journal, 35, 429–441. [DOI] [PubMed] [Google Scholar]
- Kawata, S.‐I. & Shibayama, H. (1965) On the lateral root Primordia formation in the crown roots of rice plants. Japanese Journal of Crop Science, 33, 423–431. [Google Scholar]
- Kiesselbach, T.A. (1949) The structure and reproduction of corn. Bulletin of the Agricultural Experiment Station of Nebraska. 161. [Google Scholar]
- Kitomi, Y. , Hanzawa, E. , Kuya, N. , Inoue, H. , Hara, N. , Kawai, S. et al. (2020) Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proceedings of the National Academy of Sciences of the United States of America, 117, 21242–21250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi, Y. , Inahashi, H. , Takehisa, H. , Sato, Y. & Inukai, Y. (2012) OsIAA13‐mediated auxin signaling is involved in lateral root initiation in rice. Plant Science, 190, 116–122. [DOI] [PubMed] [Google Scholar]
- Kitomi, Y. , Ito, H. , Hobo, T. , Aya, K. , Kitano, H. & Inukai, Y. (2011) The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type‐A response regulator of cytokinin signaling. The Plant Journal, 67, 472–484. [DOI] [PubMed] [Google Scholar]
- Kitomi, Y. , Nakao, E. , Kawai, S. , Kanno, N. , Ando, T. & Fukuoka, S. et al. (2018) Fine mapping of QUICK ROOTING 1 and 2, quantitative trait loci increasing root length in rice. G3, 8, 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi, Y. , Ogawa, A. , Kitano, H. & Inukai, Y. (2008) CRL4 regulates crown root formation through auxin transport in rice. Plant Root, 2, 19–28. [Google Scholar]
- Krishnamurthy, P. , Ranathunge, K. , Nayak, S. , Schreiber, L. & Mathew, M.K. (2011) Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.). Journal of Experimental Botany, 62, 4215–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavarenne, J. , Gonin, M. , Guyomarc'h, S. , Rouster, J. , Champion, A. & Sallaud, C. et al. (2019) Inference of the gene regulatory network acting downstream of CROWN ROOTLESS 1 in rice reveals a regulatory cascade linking genes involved in auxin signaling, crown root initiation, and root meristem specification and maintenance. The Plant Journal, 100, 954–968. [DOI] [PubMed] [Google Scholar]
- Li, J. , Chen, F. , Li, Y. , Li, P. , Wang, Y. & Mi, G. (2019) ZmRAP2.7, an AP2 transcription factor, is involved in maize brace roots development. Frontiers in Plant Science, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.‐J. , Fu, Y.‐R. , Huang, J.‐G. , Wu, C.‐A. & Zheng, C.‐C. (2011) Transcript profiling during the early development of the maize brace root via Solexa sequencing. The FEBS Journal, 278, 156–166. [DOI] [PubMed] [Google Scholar]
- Lin, C. & Sauter, M. (2018) Control of adventitious root architecture in rice by darkness, light, and gravity. Plant Physiology, 176, 1352–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Wang, S. , Yu, X. , Yu, J. , He, X. , Zhang, S. et al. (2005) ARL1, a LOB‐domain protein required for adventitious root formation in rice. The Plant Journal, 43, 47–56. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Wang, J. , Wang, L. , Wang, X. , Xue, Y. , Wu, P. et al. (2009) Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Research, 19, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Lucas, M. , Schlüter, S. , Vogel, H.‐J. & Vetterlein, D. (2019) Roots compact the surrounding soil depending on the structures they encounter. Scientific Reports, 9, 16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J.P. (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Annals of Botany, 112, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer, C., Xu, C., Berendzen, K.W. & Hochholdinger, F. (2012) Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot‐borne root initiation in maize (Zea maysL.). Philosophical Transactions of the Royal Society B: Biological Sciences, 367(1595), 1542–1551. 10.1098/rstb.2011.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, C. , He, J. , Liu, L. , Deng, Q. , Yao, X. , Liu, C. et al. (2020) OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnology Journal, 18, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns, R. , James, R.A. & Läuchli, A. (2006) Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany, 57, 1025–1043. [DOI] [PubMed] [Google Scholar]
- Nakajima, K. , Sena, G. , Nawy, T. & Benfey, P.N. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature, 413, 307–311. [DOI] [PubMed] [Google Scholar]
- Nazari, M. , Riebeling, S. , Banfield, C.C. , Akale, A. , Crosta, M. , Mason‐Jones, K. et al. (2020) Mucilage polysaccharide composition and exudation in maize from contrasting climatic regions. Frontiers in Plant Science, 11, 587610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J. , Shen, Y. , Zhang, Y. & Wu, P. (2014) Definition and stabilisation of the quiescent centre in rice roots. Plant Biology, 16, 1014–1019. [DOI] [PubMed] [Google Scholar]
- Obara, M. , Tamura, W. , Ebitani, T. , Yano, M. , Sato, T. & Yamaya, T. (2010) Fine‐mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4(+) concentrations in hydroponic conditions. Theoretical and Applied Genetics. Theoretische und angewandte Genetik, 121, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Ramírez, C. , Guillotin, B. , Xu, X. , Rahni, R. , Zhang, S. , Yan, Z. et al. (2021) Ground tissue circuitry regulates organ complexity in maize and Setaria . Science (New York, N.Y.), 374, 1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, B.K. , Huang, G. , Bhosale, R. , Hartman, S. , Sturrock, C.J. , Jose, L. et al. (2021) Plant roots sense soil compaction through restricted ethylene diffusion. Science, 371, 276–280. [DOI] [PubMed] [Google Scholar]
- Pedersen, O. , Nakayama, Y. , Yasue, H. , Kurokawa, Y. , Takahashi, H. , Heidi Floytrup, A. et al. (2021) Lateral roots, in addition to adventitious roots, form a barrier to radial oxygen loss in Zea nicaraguensis and a chromosome segment introgression line in maize. The New Phytologist, 229, 94–105. [DOI] [PubMed] [Google Scholar]
- Péret, B. , De Rybel, B. , Casimiro, I. , Benková, E. , Swarup, R. , Laplaze, L. et al. (2009) Arabidopsis lateral root development: an emerging story. Trends in Plant Science, 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Perianez‐Rodriguez, J. , Rodriguez, M. , Marconi, M. , Bustillo‐Avendaño, E. , Wachsman, G. , Sanchez‐Corrionero, A. et al. (2021) An auxin‐regulable oscillatory circuit drives the root clock in Arabidopsis. Science Advances, 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, A.C. & Lynch, J.P. (2021) Increased seminal root number associated with domestication improves nitrogen and phosphorus acquisition in maize seedlings. Annals of Botany, 128, 453–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka, J.J. , Winter, C.M. & Benfey, P.N. (2012) Control of Arabidopsis root development. Annual Review of Plant Biology, 63, 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placido, D.F. , Sandhu, J. , Sato, S.J. , Nersesian, N. , Quach, T. , Clemente, T.E. et al. (2020) The LATERAL ROOT DENSITY gene regulates root growth during water stress in wheat. Plant Biotechnology Journal, 18, 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, J.A. , Dathe, A. & Lynch, J.P. (2014) The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiology, 166, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidmiller, D. , Avery, C.W. , Easterling, D.R. , Kunkel, K.E. , Lewis, K.L.M. & Maycock, T.K. et al. (2017) Fourth national climate assessment. Volume II: Impacts, Risks, and Adaptation in the United States, Report‐in‐Brief.
- Rellán‐Álvarez, R. , Lobet, G. & Dinneny, J.R. (2016) Environmental control of root system biology. Annual Review of Plant Biology, 67, 619–642. [DOI] [PubMed] [Google Scholar]
- Rellán‐Álvarez, R. , Lobet, G. , Lindner, H. , Pradier, P.‐L. , Sebastian, J. & Yee, M.‐C. et al. (2015) GLO‐Roots: an imaging platform enabling multidimensional characterization of soil‐grown root systems. eLife, 4, e07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneau, J.W. , Khangura, R.S. , Stager, A. , Erndwein, L. , Weldekidan, T. , Cook, D.D. et al. (2020) Maize brace roots provide stalk anchorage. Plant Direct, 4, e00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, S. , Anders, N. , Wolters, H. , Beckmann, H. , Thomann, A. , Heinrich, R. et al. (2010) Role of the GNOM gene in Arabidopsis apical‐basal patterning—from mutant phenotype to cellular mechanism of protein action. European Journal of Cell Biology, 89, 138–144. [DOI] [PubMed] [Google Scholar]
- Robbins, N.E. & Dinneny, J.R. (2018) Growth is required for perception of water availability to pattern root branches in plants. Proceedings of the National Academy of Sciences, 115, E822–E831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, G.P. , Klingensmith, K.M. , Klug, M.J. , Paul, E.A. , Crum, J.R. & Ellis, B.G. (1997) Soil resources, microbial activity, and primary production across an agricultural ecosystem. Ecological Applications, 7, 158–170. [Google Scholar]
- Roodt, D. , Li, Z. , Van de Peer, Y. & Mizrachi, E. (2019) Loss of wood formation genes in monocot genomes. Genome Biology and Evolution, 11, 1986–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhry, S. , Del Bianco, M. , Kieffer, M. & Kepinski, S. (2013) Auxin controls gravitropic setpoint angle in higher plant lateral branches. Current Biology, 23, 1497–1504. [DOI] [PubMed] [Google Scholar]
- Saengwilai, P. , Nord, E.A. , Chimungu, J.G. , Brown, K.M. & Lynch, J.P. (2014) Root cortical aerenchyma enhances nitrogen acquisition from low‐nitrogen soils in maize. Plant Physiology, 166, 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, H.M. , Postma, J.A. , Kochs, J. , Pflugfelder, D. , Lynch, J.P. & van Dusschoten, D. (2020) Spatio‐temporal variation in water uptake in seminal and nodal root systems of barley plants grown in soil. Frontiers in Plant Science, 11, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian, J. , Yee, M.‐C. , Goudinho Viana, W. , Rellán‐Álvarez, R. , Feldman, M. , Priest, H.D. et al. (2016) Grasses suppress shoot‐borne roots to conserve water during drought. Proceedings of the National Academy of Sciences of the United States of America, 113, 8861–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. & Carena, M.J. (2016) BRACE: a method for high throughput maize phenotyping of root traits for short‐season drought tolerance. Crop Science, 56, 2996–3004. [Google Scholar]
- Shelden, M.C. , Roessner, U. , Sharp, R.E. , Tester, M. & Bacic, A. (2013) Genetic variation in the root growth response of barley genotypes to salinity stress. Functional Plant Biology, 40, 516–530. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Drummond, B.J. , Habben, J.E. , Brugire, N. , Weers, B.P. , Hakimi, S.M. et al. (2019) Ectopic expression of ARGOS8 reveals a role for ethylene in root‐lodging resistance in maize. The Plant Journal, 97, 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer, R. & Groover, A. (2010) Evolution of development of vascular cambia and secondary growth. The New Phytologist, 186, 577–592. [DOI] [PubMed] [Google Scholar]
- Steffens, B. , Kovalev, A. , Gorb, S.N. & Sauter, M. (2012) Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. The Plant Cell, 24, 3296–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens, B. & Rasmussen, A. (2016) The physiology of adventitious roots. Plant Physiology, 170, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens, B. & Sauter, M. (2009) Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. The Plant Cell, 21, 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens, B. , Wang, J. & Sauter, M. (2006) Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta, 223, 604–612. [DOI] [PubMed] [Google Scholar]
- Stephenson, E. , Estrada, S. , Meng, X. , Ourada, J. , Muszynski, M.G. , Habben, J.E. et al. (2019) Over‐expression of the photoperiod response regulator ZmCCT10 modifies plant architecture, flowering time and inflorescence morphology in maize. PloS one, 14, e0203728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Tao, J. , Bi, Y. , Hou, M. , Lou, J. , Chen, X. et al. (2018) OsPIN1b is involved in rice seminal root elongation by regulating root apical meristem activity in response to low nitrogen and phosphate. Scientific Reports, 8, 13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M. , Sato, Y. , Wu, S. , Kang, B.‐H. & McCarty, D.R. (2015) Conserved functions of the MATE transporter BIG EMBRYO1 in regulation of lateral organ size and initiation rate. The Plant Cell, 27, 2288–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, H. , Lu, X. , Opitz, N. , Marcon, C. , Paschold, A. , Lithio, A. et al. (2016) Transcriptomic and anatomical complexity of primary, seminal, and crown roots highlight root type‐specific functional diversity in maize (Zea mays L.). Journal of Experimental Botany, 67, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji, K.K. & Kielen, N.C. (2002) Agricultural drainage water management in arid and semi‐arid areas. Roma (Italia): FAO. [Google Scholar]
- Taramino, G. , Sauer, M. , Stauffer, J.L., Jr. , Multani, D. , Niu, X. et al. (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post‐embryonic shoot‐borne root initiation: map‐based cloning of the maize RTCS gene. The Plant Journal, 50, 649–659. [DOI] [PubMed] [Google Scholar]
- Torres‐Martínez, H.H. , Hernández‐Herrera, P. , Corkidi, G. & Dubrovsky, J.G. (2020) From one cell to many: morphogenetic field of lateral root founder cells in Arabidopsis thaliana is built by gradual recruitment. Proceedings of the National Academy of Sciences of the United States of America, 117, 20943–20949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga, Y. , Hanzawa, E. , Nagai, S. , Sasaki, K. , Yano, M. & Sato, T. (2012) Identification of qSOR1, a major rice QTL involved in soil‐surface rooting in paddy fields. Theoretical and Applied Genetics. Theoretische und angewandte Genetik, 124, 75–86. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Okuno, K. & Yano, M. (2011) Dro1, a major QTL involved in deep rooting of rice under upland field conditions. Journal of Experimental Botany, 62, 2485–2494. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics, 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Yamamoto, E. , Kanno, N. , Kawai, S. , Mizubayashi, T. & Fukuoka, S. (2013) A major QTL controlling deep rooting on rice chromosome 4. Scientific Reports, 3, 3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze, A. , Zamora, P. , Delaux, P.‐M. , Heitmann, C. , Jayaraman, D. , Rajasekar, S. et al. (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage‐associated diazotrophic microbiota. PLoS Biology, 16, e2006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman, J.M. , Xuan, W. , Beeckman, T. & Benfey, P.N. (2013) To branch or not to branch: the role of pre‐patterning in lateral root formation. Development, 140, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens, I. , Komatsu, M. , Zhang, Y. , Berendzen, K.W. , Niu, X. , Sakai, H. et al. (2011) Rootless with undetectable meristem 1 encodes a monocot‐specific AUX/IAA protein that controls embryonic seminal and post‐embryonic lateral root initiation in maize. The Plant Journal, 66, 341–353. [DOI] [PubMed] [Google Scholar]
- Waidmann, S. , Ruiz Rosquete, M. , Schöller, M. , Sarkel, E. , Lindner, H. , LaRue, T. et al. (2019) Cytokinin functions as an asymmetric and anti‐gravitropic signal in lateral roots. Nature Communications, 10, 3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Studer, A.J. , Zhao, Q. , Meeley, R. & Doebley, J.F. (2015) Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics, 200, 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Xu, X. , Zhan, X. , Zhai, R. , Wu, W. , Shen, X. et al. (2013) Identification of qRL7, a major quantitative trait locus associated with rice root length in hydroponic conditions. Breeding Science, 63, 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Guo, M. , Li, Y. , Ruan, W. , Mo, X. , Wu, Z. et al. (2018) LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. Journal of Experimental Botany, 69, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.F. , He, F.‐F. , Ma, X.‐X. , Mao, C.‐Z. , Hodgman, C. , Lu, C.‐G. et al. (2011) OsCAND1 is required for crown root emergence in rice. Molecular Plant, 4, 289–299. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, D. , Gan, T. , Liu, L. , Long, W. , Wang, Y. et al. (2016) CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiology and Biochemistry: PPB/Societe francaise de physiologie vegetale, 105, 185–194. [DOI] [PubMed] [Google Scholar]
- Woll, K. , Borsuk, L.A. , Stransky, H. , Nettleton, D. , Schnable, P.S. & Hochholdinger, F. (2005) Isolation, characterization, and pericycle‐specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiology, 139, 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , Lee, C.‐M. , Hayashi, T. , Price, S. , Divol, F. , Henry, S. et al. (2014) A plausible mechanism, based upon Short‐Root movement, for regulating the number of cortex cell layers in roots. Proceedings of the National Academy of Sciences of the United States of America, 111, 16184–16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J. , Lu, H. , Lu, K. , Duan, Y. , An, L. & Zhu, C. (2009) Cadmium decreases crown root number by decreasing endogenous nitric oxide, which is indispensable for crown root primordia initiation in rice seedlings. Planta, 230, 599–610. [DOI] [PubMed] [Google Scholar]
- Xu, C. , Tai, H. , Saleem, M. , Ludwig, Y. , Majer, C. , Berendzen, K.W. et al. (2015) Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot‐borne root formation. The New Phytologist, 207, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Zhu, L. , Shou, H. & Wu, P. (2005) A PIN1 family gene, OsPIN1, involved in Auxin‐dependent adventitious root emergence and tillering in rice. Plant & Cell Physiology, 46, 1674–1681. [DOI] [PubMed] [Google Scholar]
- Xu, N. , Chu, Y. , Chen, H. , Li, X. , Wu, Q. , Jin, L. et al. (2018) Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA‐mediated regulatory pathway and ROS scavenging. PLoS genetics, 14, e1007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, T. , Shimamura, S. , Nakazono, M. & Mochizuki, T. (2013) Aerenchyma formation in crop species: a review. Field Crops Research, 152, 8–16. [Google Scholar]
- Yamauchi, T. , Tanaka, A. , Inahashi, H. , Nishizawa, N.K. , Tsutsumi, N. , Inukai, Y. et al. (2019) Fine control of aerenchyma and lateral root development through AUX/IAA‐ and ARF‐dependent auxin signaling. Proceedings of the National Academy of Sciences of the United States of America, 116, 20770–20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiaki, I. , Masami, M. , Yasuo, N. , Hidemi, K. & Akira, Y. (2001) Characterization of rice mutants deficient in the formation of crown roots. Breeding Science, 51, 123–129. [Google Scholar]
- Yu, P. , Hochholdinger, F. & Li, C. (2015) Root‐type‐specific plasticity in response to localized high nitrate supply in maize (Zea mays). Annals of Botany, 116, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, A. , Schneider, H. & Lynch, J.P. (2015) Reduced lateral root branching density improves drought tolerance in maize. Plant Physiology, 168, 1603–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A. , Cui, Z. , Li, C. , Luo, J. , Guan, Y. , Liu, L. et al. (2018) Identification of maize brace‐root quantitative trait loci in a recombinant inbred line population. Euphytica/Netherlands Journal of Plant Breeding, 214, 168. [Google Scholar]
- Zhang, M. , Cao, Y. , Wang, Z. , Wang, Z. , Shi, J. , Liang, X. et al. (2018) A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. The New Phytologist, 217, 1161–1176. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Kong, X. , Xu, X. , Li, C. , Tian, H. & Ding, Z. (2015a) Comparative transcriptome profiling of the maize primary, crown and seminal root in response to salinity stress. PloS One, 10, e0121222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , von Behrens, I. , Zimmermann, R. , Ludwig, Y. , Hey, S. & Hochholdinger, F. (2015) LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the Aux/IAA gene rootless with undetectable meristem 1. Journal of Experimental Botany, 66, 3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Marcon, C. , Tai, H. , von Behrens, I. , Ludwig, Y. , Hey, S. et al. (2016) Conserved and unique features of the homeologous maize Aux/IAA proteins ROOTLESS WITH UNDETECTABLE MERISTEM 1 and RUM1‐like 1. Journal of Experimental Botany, 67, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Paschold, A. , Marcon, C. , Liu, S. , Tai, H. , Nestler, J. et al. (2014) The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. Journal of Experimental Botany, 65, 4919–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Zhang, X. , Lin, Z. , Wang, J. , Xu, M. , Lai, J. et al. (2018c) The genetic architecture of nodal root number in maize. The Plant Journal, 93, 1032–1044. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Cheng, S. , Song, Y. , Huang, Y. , Zhou, S. , Liu, X. et al. (2015) The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. The Plant Cell, 27, 2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Hu, Y. , Dai, M. , Huang, L. & Zhou, D.‐X. (2009) The WUSCHEL‐related homeobox gene WOX11 is required to activate shoot‐borne crown root development in rice. The Plant Cell, 21, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Brown, K.M. & Lynch, J.P. (2010) Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant, Cell & Environment, 33, 740–749. [DOI] [PubMed] [Google Scholar]
- Zhu, J. & Lynch, J.P. (2004) The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays) seedlings. Functional Plant Biology, 31, 949–958. [DOI] [PubMed] [Google Scholar]


