Abstract
Ribosome serves as a universal molecular machine capable of synthesis of all the proteins in a cell. Small-molecule inhibitors, such as ribosome-targeting antibiotics, can compromise the catalytic versatility of the ribosome in a context-dependent fashion, preventing transpeptidation only between particular combinations of substrates. Classic peptidyl transferase center inhibitor chloramphenicol (CHL) fails to inhibit transpeptidation reaction when the incoming A site acceptor substrate is glycine, and the molecular basis for this phenomenon is unknown. Here, we present a set of high-resolution X-ray crystal structures that explain why CHL is unable to inhibit peptide bond formation between the incoming glycyl-tRNA and a nascent peptide that otherwise is conducive to the drug action. Our structures reveal that fully accommodated glycine residue can co-exist in the A site with the ribosome-bound CHL. Moreover, binding of CHL to a ribosome complex carrying glycyl-tRNA does not affect the positions of the reacting substrates, leaving the peptide bond formation reaction unperturbed. These data exemplify how small-molecule inhibitors can reshape the A-site amino acid binding pocket rendering it permissive only for specific amino acid residues and rejective for the other substrates extending our detailed understanding of the modes of action of ribosomal antibiotics.
INTRODUCTION
The emerging bacterial antimicrobial resistance represents a global challenge for public health (1). The incredible adaptability of bacterial cells causes the drugs used to treat infections to become inactive and/or ineffective, thereby making the design of new drugs, active against resistant bacteria, increasingly demanding. Detailed mechanistic understanding of currently used drugs may greatly facilitate the rational design of the new compounds active against drug-resistant bacteria. The bacterial ribosome is an essential drug target as many classes of antibiotics bind to its functional centers and interfere with different aspects of protein synthesis. In part due to the extreme conservation of the translation apparatus as well as the utmost importance of protein synthesis for cell survival, ribosome inhibitors are among the most potent antimicrobial drugs and constitute more than half of all medicines used to treat bacterial infections in the clinic. Ribosome-targeting antibiotics are indispensable not only as therapeutic agents but also as tools for basic research. A lot of what we know about the mechanisms of action of ribosome inhibitors has been elucidated with the help of structural methods: X-ray crystallography and cryo-electron microscopy. Most of the early structures of translation inhibitors were obtained in complex with either a vacant ribosome or individual ribosomal subunits (2–8). While laying the initial foundation for understanding the mechanisms of action of many ribosome-targeting inhibitors (9), most of these early structures fail to explain selective inhibition of translating ribosomes by several classes of drugs (10–12), suggesting that the actual mechanisms of action of many ribosome inhibitors could be much more sophisticated than has been thought for decades. The high-resolution structures of various inhibitors bound to ribosomes in functionally meaningful states in combination with results of modern biochemical techniques can supply information essential for uncovering their true mechanisms of action.
Perhaps, one of the best examples of such antibiotics with poorly understood mechanisms of action, despite being studied for decades, is chloramphenicol (CHL). This drug targets the catalytic peptidyl transferase center (PTC) located in the large ribosomal subunit. Early biochemical studies suggested that CHL acts as a classic competitive inhibitor that prevents accommodation of any incoming aminoacyl-tRNA (aa-tRNA) into the ribosomal A site, resulting in inhibition of peptide bond formation (13). Consistent with this hypothesis, the initial structures of CHL bound to the vacant large ribosomal subunit (3,5) or vacant ribosomes (14,15) located its binding site in the A site of the PTC. However, this initially widely accepted model of CHL action as an indiscriminate inhibitor of the formation of every peptide bond failed to explain some experimental data: (i) inhibition of translation of polylysine but not polyphenylalanine templates (16–18); (ii) strong dependency of the CHL-mediated inhibition of puromycin reaction on the nature of the donor substrate (18,19); (iii) CHL-promoted arrest of translation at specific codons of the regulatory leader open reading frames of the CHL-resistance genes (20,21). A more recent in vivo ribosome profiling data, together with the results of in vitro primer extension inhibition assay (toe-printing) (11), as well as single-molecule FRET data (22), clearly demonstrated that CHL does not block the formation of every peptide bond with the same efficiency but instead interferes with translation in a context-specific manner. The nature of specific C-terminal residues of the nascent peptide, as well as of the A-site acceptor, was found to strongly influence the ability of CHL to inhibit peptidyl transfer: the presence of alanine, and to a lesser extent of serine and threonine, in the penultimate position of the peptide is conducive to CHL-induced ribosome stalling (11,22). In contrast, glycine in the P or A sites of the PTC strongly counteracts the inhibitory effect of CHL (11,22).
Our recent structural study rationalized why the presence of Ala, Ser, or Thr in the penultimate position of the growing peptide is required for efficient CHL-induced ribosome stalling (23). The structures of CHL in complex with the ribosome carrying short peptidyl-tRNA analogs conducive to the drug's action revealed that the high-affinity CHL binding site is formed not only by the ribosome alone but also by the C-terminal residues of the growing polypeptide. According to our structural data, the CHL-induced ribosome stalling occurs because the aminoacyl moiety of an incoming A-site tRNA is unable to displace the tightly bound CHL molecule from its canonical binding site when drug binding is stabilized by interactions with the proper penultimate residue of the growing protein, and, therefore, cannot get accommodated into the ribosomal A site making peptide bond formation unattainable (23). However, these structural data did not explain why CHL is unable to inhibit peptide bond formation between the incoming glycyl-tRNA and a nascent peptide that otherwise is conducive to the drug action (i.e. containing Ala, Ser, or Thr in the penultimate position) in combination with any other incoming aa-tRNA.
In this work, we set to close this gap and solved two high-resolution structures of Thermus thermophilus (Tth) 70S ribosomes with or without CHL and carrying glycyl- and formyl-methionyl-tRNAs in the A and P sites, respectively. Moreover, using our recently developed approach for the semisynthesis of non-hydrolyzable peptidyl-tRNAs (24), we have also solved a similar structure with CHL, glycyl-tRNA in the A site and tripeptidyl-tRNA in the P site. The new structures demonstrate that, due to the lack of a side chain, fully accommodated incoming glycine residue does not clash sterically with the ribosome-bound CHL molecule in the A site, explaining previous biochemical findings. Moreover, we show why, in the case of A-site glycyl-tRNA, peptide bond formation remains insensitive to CHL even if the drug is additionally anchored on the ribosome through direct interactions with the peptidyl-tRNA in the P site. Altogether, our results provide the critical missing piece of evidence for our comprehensive understanding of the context-specificity of action of classical PTC inhibitor CHL.
MATERIALS AND METHODS
Preparation of full-length tRNAiMet and tRNAGly
All reagents and chemicals were obtained from MilliporeSigma (USA). Wild-type deacylated initiator tRNAiMet was overexpressed and purified from Escherichia coli as described previously (25). Glycine-specific tRNAGly was purified according to (26) with modifications. In brief, tRNAGly (CCC anticodon) encoding sequence from E. coli was cloned into the pBSTNAV plasmid vector (AddGene, USA) using EcoRI and PstI restriction sites. Transformants carrying the resulting pBSTNAV-Gly vector and expressing tRNAGly were selected by aminoacylation of the total tRNA preparations (50 ng/μl) with 10 μM [14C]-glycine (222 dpm/pmol) and glycine-specific aminoacyl-tRNA-synthetase (GlyRS) from Thermus themophilus (60 ng/μl) in a buffer containing 60 mM HEPES–KOH pH 7.5, 10 mM MgCl2, 2.5 mM ATP, 5 mM DTT, 0.5 μg/μl BSA. For large-scale preparation of tRNAGly, tRNA-expressing E. coli cells were grown overnight in LB media with 100 μg/μl ampicillin. The cells were harvested and resuspended in buffer containing 1 mM Tris–HCl pH 7.4, 10 mM Mg (CH3COO)2, and treated with 0.5 volumes of acidic phenol pH 4.5. The aqueous phase was then precipitated by the addition of two volumes of ethanol and NaCl to 500 mM final concentration. The resulting pellet was resuspended in 1 M NaCl, and the soluble fraction was precipitated with ethanol again. The pellet was resuspended in 200 mM Tris–HCl pH 9.0, and the resulting solution of total tRNA was incubated for 2 h at 37°C to promote deacylation of any remaining aa-tRNAs. Deacylated total tRNA was precipitated with ethanol, resuspended in buffer A (40 mM sodium phosphate buffer pH 7.0), and subjected to anion exchange chromatography on an 8-ml MonoQ column (10/100 GL, GE Healthcare) using 300-ml 0–100% linear gradient of buffer B (40 mM sodium phosphate buffer pH 7.0, 1M NaCl) for elution. Fractions containing tRNAGly were identified by aminoacylation with [14C]-glycine, pooled together, precipitated with ethanol, resuspended in buffer C (20 mM NH4CH3COO pH 5.5, 400 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% methanol), and subjected to reversed-phase chromatography on 20-ml C5 column (C5-5, 250 × 10 mM, Discovery BIO Wide Pore, Supelco) using 300-ml 0–60% linear gradient of buffer C supplemented with 40% methanol. Fractions containing tRNAGly were identified, pooled, and precipitated with ethanol as before. The final tRNAGly preparation was resuspended in 10 mM NH4CH3COO pH 5.5 and assessed for purity and ability to accept glycine using denaturing PAGE and aminoacylation assay, respectively. The efficiency of aminoacylation of the final tRNAGly preparation was estimated to be >95%.
Tailing of tRNAiMet and tRNAGly
Tailing of tRNAiMet or tRNAGly (replacement of the 3′-terminal A76 nucleotide carrying 3′-OH group with the one carrying 3′-NH2) was performed as described previously with minor modifications (27). Briefly, deacylated tRNAiMet or tRNAGly(40 μM) was incubated at 37°C for 1 h in a buffer containing 100 mM glycine–NaOH pH 9.0, 1 mM DTT, 1 mM pyrophosphate, 1 mM 3′-NH2-ATP (Axxora, USA), and 10 μM of the CCA-adding enzyme from E. coli. The reaction was terminated by the addition of EDTA to 20 mM, treated with 1:1 phenol-chloroform mixture (pH 8.0), and precipitated with ethanol. The resulting tRNA pellet was dissolved in 20–40 μl of 10 mM NH4CH3COO pH 5.5 and desalted via Sephadex G-25 (MilliporeSigma, USA) spin columns (20–40 μl of tRNA solution per 500 μl of G-25 media).
Aminoacylation of tailed tRNAiMet and tRNAGly
Aminoacylation and formylation of the 3’-aminotailed-tRNAiMet with methionine was accomplished as described previously (28,29). For aminoacylation of the tailed 3′-NH2-tRNAiMet with cysteine, 40 μM tRNA was incubated at 25°C for 80 min in a buffer containing 100 mM HEPES–KOH pH 8.2, 20 mM MgCl2, 7.5 mM KCl, 1 mM DTT, 10 mM ATP and 1 mM cysteine together with 1 mg/ml methionine-specific aminoacyl-tRNA-synthetase (MetRS) from E.coli (24). Aminoacylation reaction was terminated by the addition of EDTA to 30 mM concentration and then treated with phenol and precipitated with ethanol. The pellet was dissolved in a buffer containing 20 mM HEPES–KOH pH 8.2 and 4 mM TCEP to a final tRNA concentration of 20 μg/μl. Based on the analytical aminoacylation assay with [14C]-glycine as well as denaturing PAGE with 7M urea, the total yield of non-hydrolyzable aminoacylated Cys-NH-tRNAiMet was >90%.
For aminoacylation of the tailed 3′-NH2-tRNAGly with glycine, 2 μM tRNA was incubated at 50°C for 4 h in a buffer containing 60 mM HEPES–KOH pH 7.2, 15 mM MgCl2, 20 mM KCl, 5 mM DTT, 5 mM ATP and 100 μM glycine together with 400 ng/μl glycine-specific aminoacyl-tRNA-synthetase (GlyRS) from T. thermophilus. The reaction mixture was then treated as described above for the aminoacylation of tRNAiMet with cysteine, but the pellets were dissolved in 10 mM NH4CH3COO pH 5.5 to 200 μM final concentration. The aliquots were flash-frozen in liquid nitrogen and stored at –80°C until further use in crystallization experiments. The total yield of non-hydrolyzable aminoacylated Gly-NH-tRNAGly was > 95%.
Preparation of fMAC-peptidyl-tRNAiMet
Preparation of hydrolysis-resistant peptidyl-tRNA carrying fMet-Ala-Cys tripeptide was accomplished according to our recently developed methodology based on native chemical ligation (NCL) reaction (24). For NCL, dry thioester-activated peptides fMAC-TBZ (98% purity) carrying thio-benzyl group as the C-terminal modification (NovoPro Biosciences, China) was dissolved in a buffer containing 1M HEPES–KOH pH 7.4 and 6M guanidine–HCl to obtain 50 mM final concentration. Next, 5 μl of the peptide solution were mixed with 15 μl of 400 mM 4-mercaptophenylacetic acid (MPAA) titrated to pH 7.0 with NaOH, and 20 μl of Cys-NH-tRNAiMet. Also, TCEP pH 6.8 was added to the reaction mixture to 100 mM final concentration. The NCL reaction mixture was incubated for 16 h at room temperature. The NCL products were purified by HPLC on a 1.7-ml C4 reversed-phase column (Proteo 300, 100 × 4.6 mm, Higgins Analytical) using 20-ml 0–60% linear gradient of 40% methanol in the buffer C (see above) supplemented with 10 mM β-mercaptoethanol. Before applying onto the C4 column, the NCL mixture underwent a buffer-exchange procedure using Amicon Ultra 10K centrifugal filter units (MilliporeSigma, USA). The fractions from the C4 column that contained the desired peptidyl-tRNAs were pooled, ethanol-precipitated, and dissolved in a buffer containing 10 mM NH4CH3COO pH 5.5, 5 mM DTT to 200 μM final concentration. The aliquots were flash-frozen in liquid nitrogen and stored at –80°C until further use in crystallization experiments.
X-ray crystallographic structure determination
Wild-type 70S ribosomes from T. thermophilus (strain HB8) were prepared as described previously (30–32,28). Synthetic mRNA with the sequence 5′-GGC-AAG-GAG-GUA-AAA-AUG-GGG-UAA-3′ containing Shine-Dalgarno sequence followed by the P-site methionine and the A-site glycine codons was obtained from Integrated DNA Technologies (USA). Complexes of the wild-type T. thermophilus 70S ribosomes with mRNA and hydrolysis-resistant A-site aminoacyl (Gly-tRNAGly) and P-site aminoacyl (fMet-tRNAiMet) or peptidyl (fMAC-tRNAiMet) tRNAs were formed as described previously for deacylated (32) or aminoacylated tRNAs (28,29). For Tth 70S ribosome complexes with CHL, the antibiotic was added to the stabilization buffers to 500 μM each final concentration.
Collection and processing of the X-ray diffraction data, model building and structure refinement were performed as described in our previous reports (23,24,28,29,32,33). Diffraction data were collected at beamlines 24ID-C and 24ID-E at the Advanced Photon Source (Argonne National Laboratory). A complete dataset for each complex was collected using 0.979 Å irradiation at 100 K from multiple regions of the same crystal, using 0.3-degree oscillations. Raw data were integrated and scaled using XDS software (5 February 2021) (34). Molecular replacement was performed using PHASER from the CCP4 program suite (version 7.0) (35). The search model was generated from the previously published structures of T. thermophilus 70S ribosome with bound mRNA and aminoacylated tRNAs (PDB entry 6XHW (29)). Initial molecular replacement solutions were refined by rigid-body refinement with the ribosome split into multiple domains, followed by positional and individual B-factor refinement using PHENIX software (version 1.17) (36). Non-crystallographic symmetry restraints were applied to four parts of the 30S ribosomal subunit (head, body, spur, and helix 44) and four parts of the 50S subunit (body, L1-stalk, L10-stalk and C-terminus of the L9 protein). Structural models were built in Coot (version 0.8.2) (37). The statistics of data collection and refinement are compiled in Supplementary Table S1. All figures showing atomic models were rendered using PyMol software (www.pymol.org).
RESULTS
Glycine residue adopts the same conformation in the A site as other amino acids
The main goal of this study was to uncover structural bases for the inability of PTC-targeting antibiotic CHL to inhibit peptide bond formation with the incoming glycine acceptor. Ribosome profiling data demonstrated that glycine codon is prevalent at the sites where CHL is expected to arrest the ribosome but, for an unknown reason, fails to halt translation (11,22), suggesting that out of all possible A-site substrates, only glycine can form peptide bond in the presence of CHL. This could be due to (i) an alternative placement of the accommodated glycine residue in the ribosomal A site compared to other amino acids; (ii) an alternative position of the ribosome-bound CHL in the presence of glycine in the A site; (iii) displacement of ribosome-bound CHL by glycyl-tRNA despite the stabilization of the drug provided by its favorable interactions with the penultimate residue of the growing peptide or (iv) ability of the glycine residue to fit into the A site together with CHL. Any of these scenarios could make peptide bond formation reaction immune to the presence of CHL. To discriminate between these possibilities, we first set to check whether the glycine residue accommodated in the A site adopts an unusual conformation different from the one typically observed for other amino acids by determining the ribosome structure in the pre-attack state of the peptide bond formation.
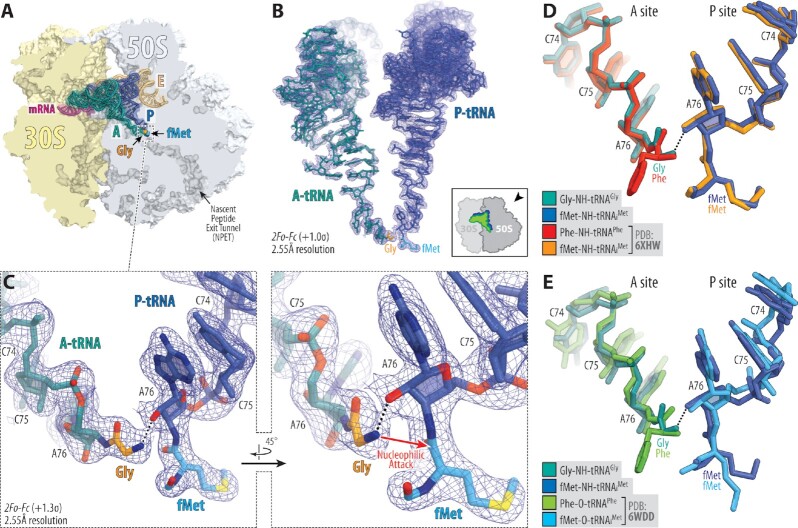
We have assembled Thermus thermophilus (Tth) 70S ribosome complex harboring Gly-tRNAGly and fMet-tRNAiMet in the A and P sites, respectively. The amino acid moieties for both tRNAs were attached to the ribose of the A76 nucleotide via non-hydrolyzable amide bonds (see Online Methods), allowing us to capture the pre-attack state in the structures (Figure 1C). We then determined its structure at 2.55Å resolution (Figure 1A; Supplementary Table S1). The observed high-quality electron density map allowed unambiguous modeling of both amino acid residues attached to the A- and P-site tRNAs (Figure 1B, C). We aligned our new structure containing A-site Gly-tRNAGly with the previously published structures of the 70S ribosome in the pre-attack state containing either the hydrolysis-resistant amide-linked (Figure 1D) (28,29) or native ester-linked (Figure 1E) (38) full-length Phe-tRNAPhe and fMet-tRNAiMet in the A and P sites, respectively. We found no major structural differences in the positions of the main-chain atoms between Gly and Phe residues in the A site as well as between the CCA-ends of the tRNAs, indicating that, despite being the smallest amino acid residue with the highest degree of flexibility, Gly accommodates in the ribosomal A site the same way as other amino acids. Moreover, the orientation of the attacking α-amino group of the aa-tRNA relative to the carbonyl carbon of the P-site substrate is nearly identical not only between the structures harboring A-site Gly versus Phe residues but also between the structures with non-hydrolyzable amide-linked versus native ester-linked tRNAs (Figure 1D, E), suggesting that stable amide-linked aa-tRNAs are adequate mimics of their native ester-linked counterparts. However, it is important to note that, despite having similar positions in the A site, glycyl-tRNA is a poor A-site substrate for peptide bond formation in comparison to other aa-tRNAs (39).
Figure 1.
Structure of the 70S ribosome in complex with glycyl-tRNA in the A site. (A) Overview of the T. thermophilus 70S ribosome structure containing full-length Gly-tRNAGly and fMet-tRNAiMet in the A and P sites, respectively, viewed as a cross-cut section through the ribosome. The 30S subunit is shown in light yellow, the 50S subunit is in light blue, the mRNA is in magenta, and the A-, P-, and E-site tRNAs are colored teal, blue, and light orange, respectively. The glycyl (Gly) and formylmethionyl (fMet) moieties of the A- and P-site tRNAs are colored orange and light blue, respectively. (B) High-resolution (2.55Å) 2Fo–Fc electron difference Fourier map (blue mesh) of the ribosome-bound A- and P-site tRNAs. The refined models of tRNAs are displayed in their respective electron density maps contoured at 1.0σ. The entire bodies of the A- and P-site tRNAs are viewed from the back of the 50S subunit, as indicated by the inset. Ribosome subunits are omitted for clarity. (C) Close-up views of the CCA-ends of the A- and P-site tRNAs carrying Gly (orange) and fMet (light blue) moieties, respectively. Nitrogens are colored blue; oxygens are red. H-bonds are shown by black dotted lines. Note that the H-bond between the α-amino group and the 2′-OH of the A76 of the P-site tRNA is pivotal for optimal orientation of α-amine for an in-line nucleophilic attack onto the carbonyl carbon of the P-site substrate. (D, E) Superpositioning of our 70S ribosome structure carrying Gly-NH-tRNAGly (teal) and fMet-NH-tRNAiMet (blue) in the A and P sites, respectively, with the previously reported structures of ribosome-bound full-length aminoacyl-tRNAs featuring either non-hydrolyzable amide linkages (D, PDB entry 6XHW (29)) or native ester bonds (E, PDB entry 6WDD (38)) between the amino acid moieties and the ribose of nucleotide A76 of A- and P-site tRNAs. All structures were aligned based on domain V of the 23S rRNA.
Superpositioning of this structure with the previous high-resolution structures of ribosome-bound CHL in the presence of either full-length deacylated tRNAs in both A and P sites (Supplementary Figure S1A) (40) or short deacylated A-site tRNA and P-site peptidyl-tRNA analogs (Supplementary Figure S1B) (23) reveals a relatively small clash between the main-chain Cα atom of the Gly residue and the nitrobenzyl moiety of CHL. This observation suggests that CHL might be preventing the accommodation of Gly residue into the A site, or the drug would adopt a previously unseen conformation within its canonical site or could even be displaced.
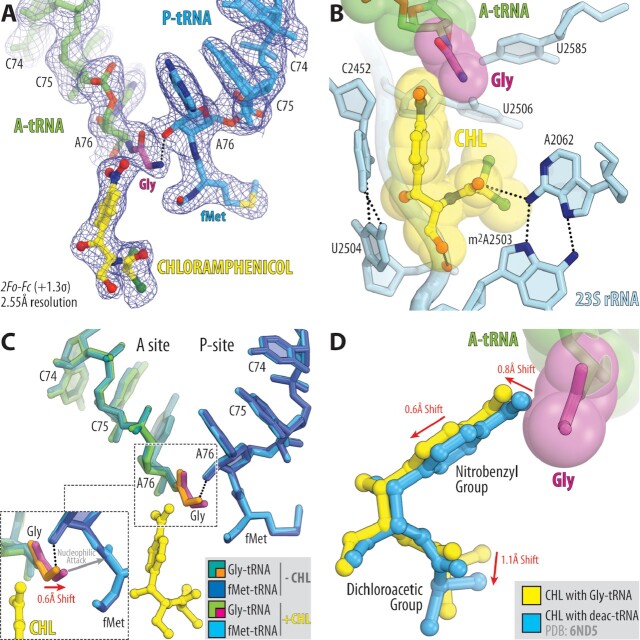
Fully accommodated glycyl-tRNA can co-exist with the ribosome-bound CHL
Next, in order to directly address the possibility of CHL displacement or binding in the pocket adjacent to or other than its canonical site within the PTC in the presence of Gly, we have determined the structure of the same complex of the Tth 70S ribosome carrying Gly-tRNAGly and fMet-tRNAiMet in the A and P sites but now also bound with CHL (Figure 2A; Supplementary Table S1). Contrary to the in silico predictions (Supplementary Figure S1), the structure solved to 2.55 Å revealed fully accommodated Gly-tRNAGly and CHL both bound to the ribosome side-by-side (Figure 2B). Unexpectedly, neither of the two molecules moved significantly away from their canonical binding pockets in the PTC in order to allow enough space for the other one to bind. Instead, to avoid the clash shown in Supplementary Figure S1, each of the molecules shifts just a fraction of one Å away from each other – Cα atom of the Gly residue moves by ∼0.6Å, while the attacking α-amino group moves by ∼0.4 Å (Figure 2C), and the entire CHL molecule moves by ∼0.8Å in the opposite direction (Figure 2D) – providing enough space for both molecules to fit snuggly side-by-side in the PTC (Figure 2B). Given the relatively small range of observed CHL relocation and the fact that all of its interacting partners remain the same, we do not consider the observed slightly shifted position of the ribosome-bound CHL in the presence of glycyl-tRNA as an alternative binding site (Figure 2D). This result also indicates that CHL exhibits some degree of conformational flexibility while being bound to the ribosome. Importantly, the observed minimal shift of the attacking α-amino group of the glycyl-tRNA towards the donor carbonyl carbon of the P-site substrate as a result of CHL binding is consistent with the poor sensitivity of translation elongation to inhibition by CHL in the presence of A-site glycine residue (Figure 2C) (11,22). Our structure clearly shows that due to the absence of the side chain, glycine residue can be accommodated in the PTC A site in the presence of CHL, providing the first insight into how the incoming glycine allows the ribosome to overcome the inhibitory action of CHL and rationalizing previous in vitro and in vivo findings (11,22).
Figure 2.
Structure of the 70S ribosome in complex with CHL and glycyl-tRNA in the A site. (A) 2Fo–Fc electron density map (blue mesh) of the A-site Gly-NH-tRNAGly (green with glycine residue in magenta) and the P-site fMet-NH-tRNAiMet (blue) bound to the T. thermophilus 70S ribosome in the presence of CHL (yellow). (B) Close-up view of CHL bound in the PTC, highlighting H-bond interactions (dashed lines) of the drug with the nucleotides of the 23S rRNA (light blue). P-site tRNA substrate is omitted for clarity. Note that the tip of the nitrobenzyl moiety of CHL forms Van-der-Waals interactions with the glycine residue accommodated in the A site. (C) Superposition of the two new 70S ribosome structures carrying A-site Gly-tRNAGly and P-site fMet-tRNAiMet in the presence and absence of CHL. The structures were aligned based on domain V of the 23S rRNA. Note that binding of CHL causes a 0.6Å shift of the accommodated glycine residue towards the carbonyl carbon of the P-site tRNA substrate (red arrow). (D) Superposition of the previous structure of CHL bound to the 70S ribosome in the presence of deacylated A- and P-site tRNAs (blue, PDB entry 6ND5 (40)) with the new structure of CHL together with the A-site Gly-tRNAGly. Note that, due to the presence of glycine residue, CHL is slightly shifted in its canonical binding pocket to avoid steric hindrance with glycine residue.
Although our data reveal a previously unknown ability of CHL to remain bound at the PTC in the presence of glycyl-tRNA, nevertheless, this structure utilizes fMet-tRNAiMet in the P site, which is too short to be conducive to the inhibitory action of CHL regardless of the nature of aa-tRNA in the A site. Indeed, in vitro toe-printing experiments show that a growing peptide needs to be at least three amino acids in length before CHL can cause translation arrest (11,23,41). Most likely, this happens because formyl-methionyl moiety does not directly interact with the ribosome-bound CHL and, hence, is unable to provide added stability to the drug, unlike longer peptides carrying Ala, Ser, or Thr in the penultimate position (23). Therefore, there is a possibility that the structure carrying fMet-tRNAiMet in the P site might not reflect the true physiological state of the ribosome, in which the inhibitory action of CHL is trumped by the incoming glycyl-tRNA.
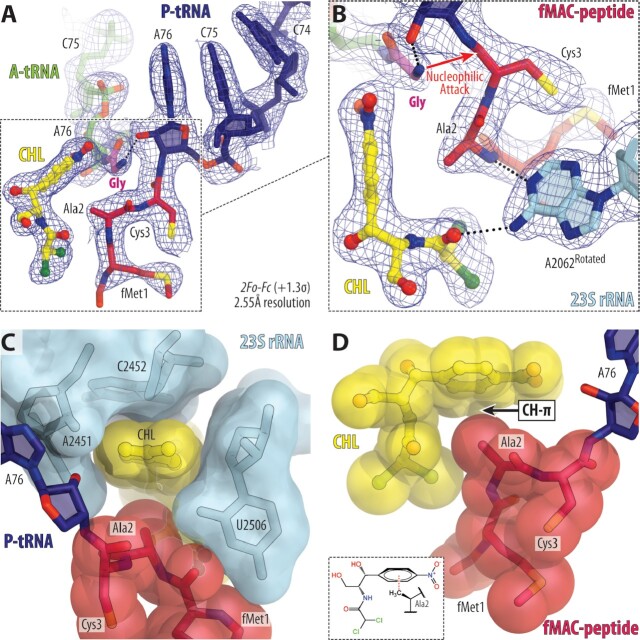
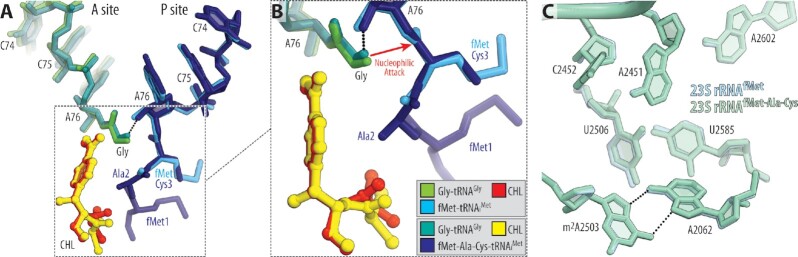
To check whether the presence of a peptidyl-tRNA instead of aminoacyl-tRNA in the P site can affect the placement of A-site Gly-tRNAGly in the CHL-bound ribosome, we set to determine a similar structure but now featuring peptidyl-tRNA in the P site. To this end, we used our recently developed chemoenzymatic approach based on native chemical ligation (NCL) reaction (24) to produce a non-hydrolyzable peptidyl-tRNA carrying formyl-Met-Ala-Cys (fMAC) peptide sequence that is conducive to CHL action due to the presence of Ala residue in the penultimate position. Using this peptidyl-tRNA as the P-site substrate, we assembled a complex of Tth 70S ribosome containing CHL and Gly-tRNAGly in the A site and solved its structure at 2.55Å resolution (Figure 3A; Supplementary Table S1). The observed electron density map revealed the presence of CHL in its canonical position as well as glycine residue in the A site and allowed unambiguous modeling of all three amino acid residues of the fMAC peptide moiety attached to the CCA-end of the P-site tRNA (Figure 3A, B). In both of our structures containing CHL, the drug molecule is clearly visible in its canonical site in the PTC (Figure 2A; Figure 3A) so that the nitrobenzyl group of the drug intercalates into the A-site cleft (Figure 3C), a hydrophobic pocket formed by the 23S rRNA residues A2451, C2452 and U2506, whereas its dichloroacetic moiety interacts with the nucleobase of A2062 (Figure 2B; Figure 3B). Similar to our recent structures showing interactions of CHL with short peptidyl-tRNA analogs carrying Met-Thr-Ile or Met-Ala-Ile tripeptide sequences (23), the penultimate alanine of the fMAC-tRNA stabilizes ribosome-bound CHL by establishing an energetically favorable CH–π interaction (42) between the side chain of alanine and the nitrobenzyl moiety of CHL (Figure 3D). Moreover, this structure shows that nucleotides G2061, A2062 and U2506 tightly coordinate the main chain groups of the fMAC peptide in the ribosomal exit tunnel (Supplementary Figure S2), fully consistent with our recent findings (24). Most importantly, alignment of the two structures of the ribosome-bound CHL, the one with fMet-tRNAiMet versus fMAC-tripeptidyl-tRNAiMet, revealed no substantial differences in the positions of the P-site donor carbonyl group or the attacking α-amino group of the acceptor A-site Gly-tRNAGly (Figure 4A, B), or the positions of the key 23S rRNA nucleotides around the PTC (Figure 4C). Since fMet moiety is too far and does not interact with CHL directly, while fMAC-peptidyl moiety does (Figure 4A, B), efficient binding of CHL to the ribosome in the presence of Gly-tRNAGly occurs independently of the stabilizing interaction(s) with the P-site tRNA. This means that the observed small shifts in the positions of the glycyl moiety as well as the CHL itself do not result in significant changes in the CHL binding affinity, underlining robust co-binding of CHL and Gly-tRNAGly in the PTC of the 70S ribosome. Thus, both of our CHL-containing structures show how the fully accommodated Gly-tRNAGly can co-exist in the ribosomal A site together with the CHL molecule.
Figure 3.
Structure of the 70S ribosome in complex with CHL, A-site glycyl-tRNA, and P-site fMAC-peptidyl-tRNA. (A, B) 2Fo–Fc electron density map (blue mesh) of the A-site Gly-tRNAGly (green with glycine residue in magenta) and the P-site peptidyl-tRNAiMet (navy with peptide in crimson) bound to the T. thermophilus 70S ribosome in the presence of CHL (yellow). (C, D) Close-up views of the ribosome-bound CHL, highlighting intercalation of its nitrobenzyl group into the A-site cleft formed by nucleotides A2451 and C2452 of the 23S rRNA (C), and the CH-π interactions with the side chain of Ala2 in the peptidyl-tRNA (D). Note that the side chain of the Ala2 residue of the fMAC tripeptide directly interacts with the ribosome-bound CHL.
Figure 4.
Comparison of the two new structures of ribosome-bound CHL in the presence of P-site aminoacyl versus peptidyl-tRNA substrates. (A, B) Superpositioning of the T. thermophilus 70S ribosome structures carrying Gly-tRNAGly (green or teal) and either fMet-tRNAiMet (blue) or fMet-Ala-Cys-tRNAiMet (navy) in the A and P sites, respectively, both in the presence of CHL (red or yellow). The structures were aligned based on domain V of the 23S rRNA. (C) Comparison of the positions of the key 23S rRNA nucleotides around the PTC in the same structures. Note that there are no significant differences in the positions of A- or P-site substrates or the PTC nucleotides.
DISCUSSION
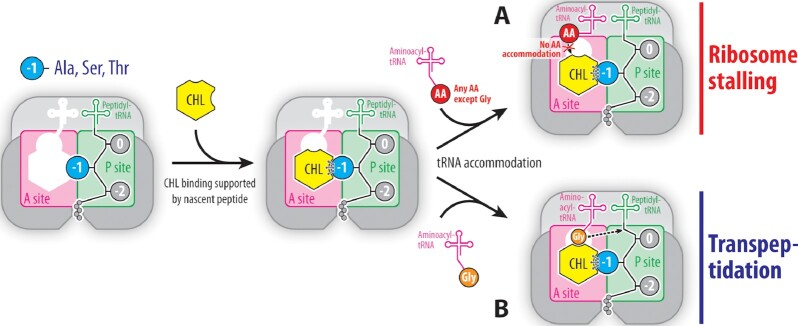
The nature of the penultimate residue in the growing polypeptide chain was known to pre-determine the ability of CHL to inhibit peptide bond formation (11,22). The presence of Ala, Ser or Thr in the penultimate position of the nascent peptide is required for efficient CHL-induced ribosome stalling because these residues directly interact with the ribosome-bound CHL, thereby stabilizing it and increasing its affinity for the ribosome (23). As a result, the tightly bound CHL molecule can no longer be displaced from its canonical binding site by the competing aminoacyl moiety of an incoming A-site tRNA, making the next peptide bond formation reaction impossible and leading to the CHL-induced translation arrest (Figure 5A) (23). However, if CHL is firmly anchored at the PTC through interactions with the nascent peptide and the incoming aa-tRNA carries glycine residue, CHL-dependent ribosome stalling becomes much less pronounced, and peptide bond formation can occur because there is no steric hindrance between the drug and the fully accommodated A-site glycine residue as discovered in this study (Figure 5B). The new structural data complement and expand our understanding of the site-specificity of CHL action (23), being now able to explain not only the critical role of the penultimate residue of the nascent peptide in CHL-mediated ribosome stalling (Figure 5A) but also rationalize the role of the incoming aa-tRNA in evading the inhibitory effect of the drug (Figure 5B).
Figure 5.
Schematic diagram illustrating the expanded increased affinity model. (A) When the growing polypeptide chain carries alanine, serine, or threonine (blue circle) in the penultimate position, the affinity of CHL (yellow hexagon) for the ribosome increases due to direct interactions with the side chains of these amino acids preventing accommodation of the aminoacyl moiety (red circle) of the aa-tRNA into the A site and, thereby, making peptidyl transferase reaction impossible, which results in ribosome stalling. (B) Due to the absence of the side chain, glycine residue (orange circle) can be accommodated into the PTC even in the presence of bound CHL and normally react with the P-site substrate (dashed arrow), sustaining transpeptidation reaction. Small and large ribosomal subunits are highlighted in shades of grey. A and P sites are highlighted in magenta and green, respectively. Amino acids of the growing polypeptide chain are shown by grey circles with the penultimate amino acid residue highlighted in blue.
Even though CHL does not stall ribosomes when the incoming aa-tRNA carries glycine residue, the drug, apparently, still affects the elongation step. Namely, smFRET data shows that CHL binding increases ribosome residence time in the non-rotated state after the accommodation of glycyl-tRNA (22). Given that CHL and glycyl-tRNA co-habit the ribosomal A site, this can be interpreted as a slow-down effect of CHL on transpeptidation reaction because ribosome ratcheting physically cannot happen until after transpeptidation has occurred and P-site tRNA has become deacylated. Therefore, in the case of only the glycyl-tRNA, CHL can actually inhibit peptide bond formation simply by stabilizing the pre-attack state of both aa-tRNA and peptidyl-tRNA in the PTC. Apparently, the inevitable change of the nascent chain conformation in the PTC immediately after the transpeptidation reaction must destroy the drug-stabilizing interactions of the penultimate alanine residue in the peptidyl-tRNA and the ribosome-bound CHL molecule, resulting in fast vacant-ribosome-like kinetics of CHL dissociation from the ribosome (or maybe even faster, in the case, when the subsequent ribosome subunit ratcheting places bulky amino acid in the penultimate position of the growing peptide chain). The loss of CHL stabilization provided by the nascent peptide as a consequence of transpeptidation and concomitant re-orientation of the entire nascent chain should be sufficient to promote drug dissociation (43). However, it is still unclear at which exact step of the elongation cycle the drug dissociates from the PTC and to which extent transpeptidation by itself facilitates drug removal. Anyhow, even if CHL causes slow-down in transpeptidation rate with incoming glycyl-tRNAs, this effect is rather secondary, while the primary effect is the CHL-dependent blockage of aa-tRNA accommodation for all other (non-glycine) amino acid residues into the A site (23).
While CHL was introduced in clinical practice nearly seven decades ago, its medical use has been diminished with the development of newer antibiotics due to its side effects (44,45). Other drugs of this class, such as florfenicol, are still used in the United States to treat infections in farm animals (46). Interestingly, members of the chemically unrelated and newest class of clinically important PTC-targeting protein synthesis inhibitors—the oxazolidinones—bind to the PTC of a bacterial ribosome at a location largely overlapping with that of the CHL binding site (Supplementary Figure S3A) (47,48). Similar to CHL, oxazolidinones, such as linezolid (LZD), exhibit a pronounced context-specificity of action inhibiting peptide bond formation one step after Ala residues appear in the nascent chain (11,22). However, unlike CHL, LZD normally inhibits peptide bond formation, even if the incoming aa-tRNA carries glycine as the acceptor (11,22). Superpositioning of our structure containing A-site Gly-tRNAGly and P-site fMet-tRNAiMet with the available structures of LZD bound to Haloarcula marismortui 50S subunit complexed with a P-site tRNA analog carrying N-AcPhe residue (Supplementary Figure S3B) (47) or Escherichia coli ribosome carrying P-site peptidyl-tRNA conducive to the inhibitory action of LZD (Supplementary Figure S3C) (48) reveals a significant overlap between the main-chain atoms of the Gly residue and the morpholino moiety of LZD. The observed overlap is unlikely to be resolved by small shifts of the drug and/or Gly residue in the A site, suggesting that LZD is unable to co-exist in the A site with any incoming amino acid residues (even the smallest Gly), thereby rationalizing biochemical in vivo and in vitro data (11,22).
The inability of CHL to inhibit transpeptidation between the A-site glycine residue and peptidyl-tRNA substrate in the P site represents an extreme manifestation of the so-called drug-mediated restriction of the A-site, a concept that was previously proposed for the chemically unrelated macrolide antibiotics (49). Ribosome exhibits amazing catalytic versatility toward the incoming aa-tRNAs being able to utilize the entire repertoire of proteinogenic amino acids as substrates. However, ribosome-bound drug molecules can sterically restrict access of the aa-tRNAs to the ribosomal A site, thereby limiting the allowable set of acceptor aa-tRNAs to only a few. For example, during macrolide-induced ribosome stalling on +X + motifs (positively charged residue followed by any residue in the –1 and 0 positions of the P-site substrate and positively charged residue in the A site), the ribosomal A site becomes restrictive to only the positively charged amino acid residues – lysine or arginine, while other residues can still be accommodated and preserve reactivity (49). Based on the recent cryo-EM structures of the ribosome nascent chain complexes stalled on the + X + motifs, it has been suggested that selectivity of the A site stems from the macrolide-induced conformational rearrangements of PTC nucleotides and/or nascent peptide chain, resulting in problematic accommodation of lysyl or arginyl aa-tRNAs (50). In contrast to macrolides, which bind to the ribosomal exit tunnel away from the PTC, CHL does not allosterically induce structural rearrangements of the key PTC nucleotides or the growing peptide chain and, instead, acts directly as a steric restrictor for all incoming amino acid residues except only for the glycine.
Our new data attest to the critical role of biochemically driven empiricism in structural biology, as the ability of CHL and Gly-tRNAGly to co-exist at the PTC could not have been predicted on the basis of existing ribosome-antibiotic co-crystal structures. In fact, in silico comparisons of the previous structures resulted in the opposite, obviously incorrect, predictions (Supplementary Figure S1). Moreover, the set of new structures reported here yet again illustrates that molecular mechanisms underlying context-specific inhibitory action of ribosome-targeting antibiotics, such as CHL, could be understood only from the structures of functionally meaningful complexes of the ribosomes that include all the key players: the antibiotic, aa- and peptidyl-tRNA substrates conducive or non-conducive to the drug action. Our structure featuring peptidyl-tRNA shows that CHL employs chemical moieties of the nascent chain for stable interaction with the target, and hence, its binding site is formed not only by the ribosome alone but also by the growing polypeptide. This finding raises the possibility of exploiting a wide range of chemical functionalities provided by the growing peptide, especially those of the side chains of the uniformly oriented C-terminal residues, for the rational design of novel antibacterials having better affinity to the ribosome. Altogether, the results from this study are complementary to our recent related work (23) and show that the interplay between the ribosome, the nascent peptide chain, and the ribosome-bound drug defines whether or not translation arrest would occur in the presence of the PTC-targeting antibiotics such as CHL.
DATA AVAILABILITY
Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession codes:
7U2H for the T. thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-tRNAGly, aminoacylated P-site fMet-tRNAiMet, and deacylated E-site tRNAGly;
7U2I for the T. thermophilus 70S ribosome in complex with CHL, mRNA, aminoacylated A-site Gly-tRNAGly, aminoacylated P-site fMet-tRNAiMet, and deacylated E-site tRNAGly;
7U2J for the T. thermophilus 70S ribosome in complex with CHL, mRNA, aminoacylated A-site Gly-tRNAGly, peptidylated P-site fMet-Ala-Cys-tRNAiMet, and deacylated E-site tRNAGly;
All previously published structures that were used in this work for structural comparisons were retrieved from the RCSB Protein Data Bank: PDB entries 6XHW, 6WDD, 6ND5, 3CPW, 7S1G.
No sequence data were generated in this study.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Alexander Mankin and Dr Nora Vazquez-Laslop for their critical reading of the manuscript, as well as Dr Maxim Svetlov for valuable suggestions. We thank the staff at NE-CAT beamlines 24ID-C and 24ID-E for help with data collection, especially Drs Malcolm Capel, Frank Murphy, Igor Kourinov, Anthony Lynch, Surajit Banerjee, David Neau, Jonathan Schuermann, Narayanasami Sukumar, James Withrow, Kay Perry, Ali Kaya and Cyndi Salbego.
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health [P30-GM124165 to NE-CAT]. The Eiger 16M detector on 24ID-E beamline is funded by an NIH-ORIP HEI grant [S10-OD021527 to NE-CAT]. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Contributor Information
Egor A Syroegin, Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.
Elena V Aleksandrova, Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.
Yury S Polikanov, Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA; Department of Pharmaceutical Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA; Center for Biomolecular Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-GM132302 to Y.S.P.]; National Science Foundation [MCB-1907273 to Y.S.P.]; Illinois State startup funds [to Y.S.P.]. Funding for open access charge: National Institutes of Health [R01-GM132302 to Y.S.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hansen J.L., Ippolito J.A., Ban N., Nissen P., Moore P.B., Steitz T.A.. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell. 2002; 10:117–128. [DOI] [PubMed] [Google Scholar]
- 3. Hansen J.L., Moore P.B., Steitz T.A.. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003; 330:1061–1075. [DOI] [PubMed] [Google Scholar]
- 4. Brodersen D.E., Clemons W.M. Jr, Carter A.P., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin b on the 30S ribosomal subunit. Cell. 2000; 103:1143–1154. [DOI] [PubMed] [Google Scholar]
- 5. Schlunzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F.. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001; 413:814–821. [DOI] [PubMed] [Google Scholar]
- 6. Schlunzen F., Harms J.M., Franceschi F., Hansen H.A., Bartels H., Zarivach R., Yonath A.. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003; 11:329–338. [DOI] [PubMed] [Google Scholar]
- 7. Tu D., Blaha G., Moore P.B., Steitz T.A.. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005; 121:257–270. [DOI] [PubMed] [Google Scholar]
- 8. Schroeder S.J., Blaha G., Moore P.B.. Negamycin binds to the wall of the nascent chain exit tunnel of the 50S ribosomal subunit. Antimicrob. Agents Chemother. 2007; 51:4462–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin J., Zhou D., Steitz T.A., Polikanov Y.S., Gagnon M.G.. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 2018; 87:451–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kannan K., Vazquez-Laslop N., Mankin A.S.. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012; 151:508–520. [DOI] [PubMed] [Google Scholar]
- 11. Marks J., Kannan K., Roncase E.J., Klepacki D., Kefi A., Orelle C., Vazquez-Laslop N., Mankin A.S.. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12150–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sothiselvam S., Liu B., Han W., Ramu H., Klepacki D., Atkinson G.C., Brauer A., Remm M., Tenson T., Schulten K.et al.. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9804–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drainas D., Kalpaxis D.L., Coutsogeorgopoulos C.. Inhibition of ribosomal peptidyltransferase by chloramphenicol. Kinetic studies. Eur. J. Biochem. 1987; 164:53–58. [DOI] [PubMed] [Google Scholar]
- 14. Dunkle J.A., Xiong L., Mankin A.S., Cate J.H.. Structures of the escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bulkley D., Innis C.A., Blaha G., Steitz T.A.. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. USA. 2010; 107:17158–17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kucan Z., Lipmann F.. Differences in chloramphenicol sensitivity of cell-free amino acid polymerization systems. J. Biol. Chem. 1964; 239:516–520. [PubMed] [Google Scholar]
- 17. Vazquez D. Antibiotics affecting chloramphenicol uptake by bacteria. Their effect on amino acid incorporation in a cell-free system. Biochim. Biophys. Acta. 1966; 114:289–295. [DOI] [PubMed] [Google Scholar]
- 18. Cannon M. The puromycin reaction and its inhibition by chloramphenicol. Eur. J. Biochem. 1968; 7:137–145. [DOI] [PubMed] [Google Scholar]
- 19. Rheinberger H.J., Nierhaus K.H.. Partial release of AcPhe-Phe-tRNA from ribosomes during poly(U)-dependent poly(Phe) synthesis and the effects of chloramphenicol. Eur. J. Biochem. 1990; 193:643–650. [DOI] [PubMed] [Google Scholar]
- 20. Lovett P.S. Translation attenuation regulation of chloramphenicol resistance in bacteria -a review. Gene. 1996; 179:157–162. [DOI] [PubMed] [Google Scholar]
- 21. Lovett P.S. Translational attenuation as the regulator of inducible cat genes. J. Bacteriol. 1990; 172:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi J., Marks J., Zhang J., Chen D.H., Wang J., Vazquez-Laslop N., Mankin A.S., Puglisi J.D.. Dynamics of the context-specific translation arrest by chloramphenicol and linezolid. Nat. Chem. Biol. 2020; 16:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Syroegin E.A., Flemmich L., Klepacki D., Vazquez-Laslop N., Micura R., Polikanov Y.S.. Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol. Nat. Struct. Mol. Biol. 2022; 29:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Syroegin E.A., Aleksandrova E.V., Polikanov Y.S.. Insights into the ribosome function from the structures of non-arrested ribosome nascent chain complexes. 2022; bioRxiv doi:21 February 2022, preprint: not peer reviewed 10.1101/2022.02.21.480960. [DOI] [PMC free article] [PubMed]
- 25. Schmitt E., Blanquet S., Mechulam Y.. Crystallization and preliminary X-ray analysis of escherichiacoli methionyl-tRNAMet(f) formyltransferase complexed with formyl-methionyl-tRNAMet(f). Acta Crystallogr. D Biol. Crystallogr. 1999; 55:332–334. [DOI] [PubMed] [Google Scholar]
- 26. Degut C., Monod A., Brachet F., Crepin T., Tisne C.. In vitro/in vivo production of tRNA for X-ray studies. Methods Mol. Biol. 2016; 1320:37–57. [DOI] [PubMed] [Google Scholar]
- 27. Gamper H., Hou Y.M.. tRNA 3′-amino-tailing for stable amino acid attachment. RNA. 2018; 24:1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Polikanov Y.S., Steitz T.A., Innis C.A.. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014; 21:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Svetlov M.S., Syroegin E.A., Aleksandrova E.V., Atkinson G.C., Gregory S.T., Mankin A.S., Polikanov Y.S.. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. 2021; 17:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 31. Polikanov Y.S., Blaha G.M., Steitz T.A.. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012; 336:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A.. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015; 22:342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitcheltree M.J., Pisipati A., Syroegin E.A., Silvestre K.J., Klepacki D., Mason J.D., Terwilliger D.W., Testolin G., Pote A.R., Wu K.J.Y.et al.. A synthetic antibiotic class overcoming bacterial multidrug resistance. Nature. 2021; 599:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kabsch W xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Loveland A.B., Demo G., Korostelev A.A.. Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading. Nature. 2020; 584:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johansson M., Ieong K.W., Trobro S., Strazewski P., Aqvist J., Pavlov M.Y., Ehrenberg M.. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Svetlov M.S., Plessa E., Chen C.W., Bougas A., Krokidis M.G., Dinos G.P., Polikanov Y.S.. High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition. RNA. 2019; 25:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orelle C., Carlson S., Kaushal B., Almutairi M.M., Liu H., Ochabowicz A., Quan S., Pham V.C., Squires C.L., Murphy B.T.et al.. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 2013; 57:5994–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishio M. The CH/pi hydrogen bond in chemistry. Conformation, supramolecules, optical resolution and interactions involving carbohydrates. Phys. Chem. Chem. Phys. 2011; 13:13873–13900. [DOI] [PubMed] [Google Scholar]
- 43. Crowe-McAuliffe C., Murina V., Turnbull K.J., Huch S., M. Kasari., Takada H., Nersisyan L., Sundsfjord A., Hegstad K., Atkinson G.C.et al.. Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics. Nat. Commun. 2022; 13:1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li C.H., Cheng Y.W., Liao P.L., Yang Y.T., Kang J.J.. Chloramphenicol causes mitochondrial stress, decreases ATP biosynthesis, induces matrix metalloproteinase-13 expression, and solid-tumor cell invasion. Toxicol. Sci. 2010; 116:140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cohen B.H., Saneto R.P.. Mitochondrial translational inhibitors in the pharmacopeia. Biochim. Biophys. Acta. 2012; 1819:1067–1074. [DOI] [PubMed] [Google Scholar]
- 46. Syriopoulou V.P., Harding A.L., Goldmann D.A., Smith A.L.. In vitro antibacterial activity of fluorinated analogs of chloramphenicol and thiamphenicol. Antimicrob. Agents Chemother. 1981; 19:294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ippolito J.A., Kanyo Z.F., Wang D., Franceschi F.J., Moore P.B., Steitz T.A., Duffy E.M.. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 2008; 51:3353–3356. [DOI] [PubMed] [Google Scholar]
- 48. Tsai K., Stojković V., Lee D.J., Young I.D., Szal T., Vazquez-Laslop N., Mankin A.S., Fraser J.S., Fujimori D.G.. Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics. Nat. Struct. Mol. Biol. 2022; 29:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sothiselvam S., Neuner S., Rigger L., Klepacki D., Micura R., Vazquez-Laslop N., Mankin A.S.. Binding of macrolide antibiotics leads to ribosomal selection against specific substrates based on their charge and size. Cell Rep. 2016; 16:1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beckert B., Leroy E.C., Sothiselvam S., Bock L.V., Svetlov M.S., Graf M., Arenz S., Abdelshahid M., Seip B., Grubmuller H.et al.. Structural and mechanistic basis for translation inhibition by macrolide and ketolide antibiotics. Nat. Commun. 2021; 12:4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession codes:
7U2H for the T. thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-tRNAGly, aminoacylated P-site fMet-tRNAiMet, and deacylated E-site tRNAGly;
7U2I for the T. thermophilus 70S ribosome in complex with CHL, mRNA, aminoacylated A-site Gly-tRNAGly, aminoacylated P-site fMet-tRNAiMet, and deacylated E-site tRNAGly;
7U2J for the T. thermophilus 70S ribosome in complex with CHL, mRNA, aminoacylated A-site Gly-tRNAGly, peptidylated P-site fMet-Ala-Cys-tRNAiMet, and deacylated E-site tRNAGly;
All previously published structures that were used in this work for structural comparisons were retrieved from the RCSB Protein Data Bank: PDB entries 6XHW, 6WDD, 6ND5, 3CPW, 7S1G.
No sequence data were generated in this study.