Abstract
Spinal muscular atrophy (SMA), a pediatric genetic disorder, is characterized by the profound loss of spinal cord motor neurons and subsequent muscle atrophy and death. Although the mechanisms underlying motor neuron loss are not entirely clear, data from our work and others support the idea that glial cells contribute to disease pathology. GATA6, a transcription factor that we have previously shown to be upregulated in SMA astrocytes, is negatively regulated by SMN (survival motor neuron) and can increase the expression of inflammatory regulator NFκB. In this study, we identified upregulated GATA6 as a contributor to increased activation, pro‐inflammatory ligand production, and neurotoxicity in spinal‐cord patterned astrocytes differentiated from SMA patient induced pluripotent stem cells. Reducing GATA6 expression in SMA astrocytes via lentiviral infection ameliorated these effects to healthy control levels. Additionally, we found that SMA astrocytes contribute to SMA microglial phagocytosis, which was again decreased by lentiviral‐mediated knockdown of GATA6. Together these data identify a role of GATA6 in SMA astrocyte pathology and further highlight glia as important targets of therapeutic intervention in SMA.
Keywords: lentivirus, neurodegenerative disease, pro‐inflammatory cytokines
Main Points
GATA6 is upregulated in SMA astrocytes and is associated with increased glial activation, enhanced pro‐inflammatory ligand production, altered microglial morphology and phagocytosis, and motor neuron toxicity.
1. INTRODUCTION
Spinal muscular atrophy (SMA) is a prevalent genetic cause of infant mortality with an incidence of 1 in every 6000–10,000 births (D'Amico et al., 2011; Oskoui & Kaufmann, 2008). SMA is caused by a loss of function mutation in the survival motor neuron‐1 (SMN1) gene, which leads to reduced SMN protein expression, subsequent loss of motor neurons in the spinal cord, skeletal muscle atrophy, and ultimately resulting in respiratory distress and death (Crawford & Pardo, 1996; Lefebvre et al., 1995). A second gene, SMN2, produces some functional SMN, but due to alternative splicing this gene mainly produces a truncated, non‐functional protein (SMNΔ7) (Lorson et al., 1999; Lorson et al., 1998). SMN has been shown to be ubiquitously expressed in all cell and tissue types, though its loss in SMA particularly impacts motor neurons (Chaytow et al., 2018). SMA motor neurons (MNs) show intrinsic deficits in splicing and electrophysiological function; however, restoring SMN to motor neurons alone does not significantly improve disease pathogenesis (Gogliotti et al., 2012; Lee et al., 2012; Martinez et al., 2012; Park et al., 2010). Currently, there are three FDA approved treatments for SMA, two of which focus on correcting or adjusting the splicing of SMN2 (nusinersen, risdiplam), as well as a gene therapy to deliver functional SMN1 (onasemnogene abeparvovec) (Chen, 2020). With recent studies finding potential risk in the AAV9‐SMN gene therapy treatment for less severe SMA patients, the clinical trials to expand this treatment have halted (Hinderer et al., 2018; Pattali et al., 2019; Van Alstyne et al., 2021). The need to identify additional drivers of MN loss remains essential for SMA patient longevity.
Glial cells—including astrocytes and microglia—also lose SMN expression in SMA, which results in aberrant activation. Astrocytes are a highly abundant cell in the central nervous system (CNS), and microglia are the primary immune cells in the CNS. In normal physiological function, both have neuroprotective effects and encourage neuronal development, differentiation, and function. Specifically, astrocytes produce and release growth factors like nerve growth factor (NGF), brain derived neurotrophic factor (BDNF), and glial derived neurotrophic factor (GDNF); maintain extracellular concentrations of ions (H+, K+), free radicals, and neurotransmitters; and provide structural support (Chung et al., 2015; Sidoryk‐Wegrzynowicz et al., 2011). Microglia regulate pro‐ and anti‐inflammatory processes and respond to pathological insults through phagocytosis, but also secrete BDNF, provide metabolic intermediates and neurotransmitters, and respond to neuronal firing through the retraction and extension of their highly motile processes (Fontainhas et al., 2011; Lenz & Nelson, 2018; Rial et al., 2015). The emerging idea of a quad‐partite synapse emphasizes the tightly interdependent and highly responsive nature of communication via secreted factors between astrocytes, microglia, and neurons in the CNS (Schafer et al., 2013).
It is well known that astrocytes and microglia activate in response to stress or injury, and upregulate pro‐inflammatory cytokines, downregulate neurotrophic factor secretion, and can induce neuron death (Siracusa et al., 2019). In disease conditions like SMA, however, the changed interactions between these cells and their contribution to neuronal loss have not yet been fully characterized. Importantly, astrocyte secreted factors that are known to impact motor neuron health and development, such as glial fibrillary acidic protein (GFAP) and pro‐inflammatory cytokines interleukin 1β (IL1β) and interleukin 6 (IL6), are also received by microglial membrane receptors (Domeniconi et al., 2007; Matejuk & Ransohoff, 2020). The activity of these canonical receptors, including IL1R1 and IL6R, can serve to start inflammatory mechanisms and transform ramified microglia with small cell bodies and extensive, ramified branches into large, ameboid cells capable of phagocytosis (Basu et al., 2002; Fu et al., 2014; Klawonn et al., 2021). Complement factors serve as “eat me” tags to activated microglia, and complement proteins C1q and C3 have been found to directly contribute to the loss of viable neurons through primary phagocytosis (phagoptosis) in several neurodegenerative diseases (Brown & Neher, 2012; Heurich et al., 2011; Werneburg et al., 2020; Woodruff et al., 2008). In this way, secreted factors which are essential for astrocytes and microglia to communicate in normal physiological conditions may serve as pathological drivers of activation when dysregulated.
Our lab has previously shown that astrocytes in SMA show activation before overt motor neuron loss and can induce detrimental effects on motor neuron cell health and further astrocyte activation (McGivern et al., 2013; Patitucci & Ebert, 2016). One potential explanation for this effect is the noted change in astrocyte secreted factors, as SMA astrocytes show reduced GDNF release, abnormal microRNA production, and increased NFκB expression (Logan et al., 2019; McGivern et al., 2013; Patitucci & Ebert, 2016; Rindt et al., 2015; Sison et al., 2017). GATA6, a zinc finger transcription factor negatively regulated by SMN and associated with NFkB (Boopathi et al., 2013; Yang et al., 2016), was accordingly found to be increased in SMA mouse and human samples (Yang et al., 2016). Microglial activation has been noted late in the pathology of SMA mouse models (Tarabal et al., 2014), but recent evidence demonstrates microglial phagocytosis of complement‐tagged synapses on proprioceptive neurons in the SMA mouse spinal cord, indicating a role during disease progression (Vukojicic et al., 2019). It is not yet known how upregulated GATA6 changes the astrocyte phenotype in SMA, nor how this impacts astrocyte interactions with motor neurons and microglia. In this study, we utilized spinal‐cord patterned astrocytes differentiated from SMA patient and healthy control induced pluripotent stem cells (iPSCs) to investigate the astrocytic role of GATA6 in aberrant activation and neurotoxicity as well as astrocyte‐driven microglia activation in vitro. We observed an increase in pro‐inflammatory factor production in astrocytes associated with increased GATA6 expression. Lentiviral mediated GATA6 overexpression was found to increase motor neuron cell loss in vulnerable SMA cultures, whereas lentiviral mediated knockdown ameliorated this effect. SMA astrocytes were also found to increase activation and phagocytic ability of SMA microglia, and again GATA6 astrocyte knockdown ameliorated these effects. Together these data indicate a role for GATA6 as a major contributor to the aberrant activation and pathological functions of glial cells in SMA.
2. RESULTS
2.1. Differentiation of spinal cord astrocytes from iPSCs
Though astrocytes in general share a common goal of supporting neuronal function, subtle differences in gene expression, morphology, and function in astrocytes between regions of the brain and spinal cord have been identified (Batiuk et al., 2020; Kerstetter & Miller, 2012). To best model astrocyte subtypes that would interact most directly with degenerating motor neurons in SMA, we differentiated ventral‐caudal patterned neural progenitor cells (NPCs) into spinal cord‐like astrocytes. We utilized several recently updated astrocyte protocols to optimize our own culture conditions (Andersen et al., 2020; de Rus Jacquet, 2019; Roybon et al., 2013; Tcw et al., 2017).
Human iPSCs (iPSCs) from SMA patients and unaffected (healthy control) individuals, hereafter referred to as “SMA” and “HC,” respectively, were differentiated into NPCs through the addition of SMAD inhibitors (SMADi) with retinoic acid (RA) and smoothened agonist (SAG) to mimic spinal cord development (Figure 1a). Plating NPCs at a high density was necessary to achieve robust astrocyte differentiation (Figure 1b; NPC P2 Day 17). These NPCs were found to express known NPC‐associated genes such as NESTIN, SOX2, and PAX6 (Figure 1c). They also expressed spinal cord ventral‐caudal associated genes including ISL1, NKX6.1, OLIG2, HOXB4, and HOXA4, which showed similar gene expression trends to postmortem spinal cord from SMA patients and unaffected individuals (Figure 1c). These spinal cord genes were not upregulated in SMADi only treated NPCs, which instead showed increased expression of forebrain associated genes (TBR2, SIX3, OTX2, Figure 1c).
FIGURE 1.
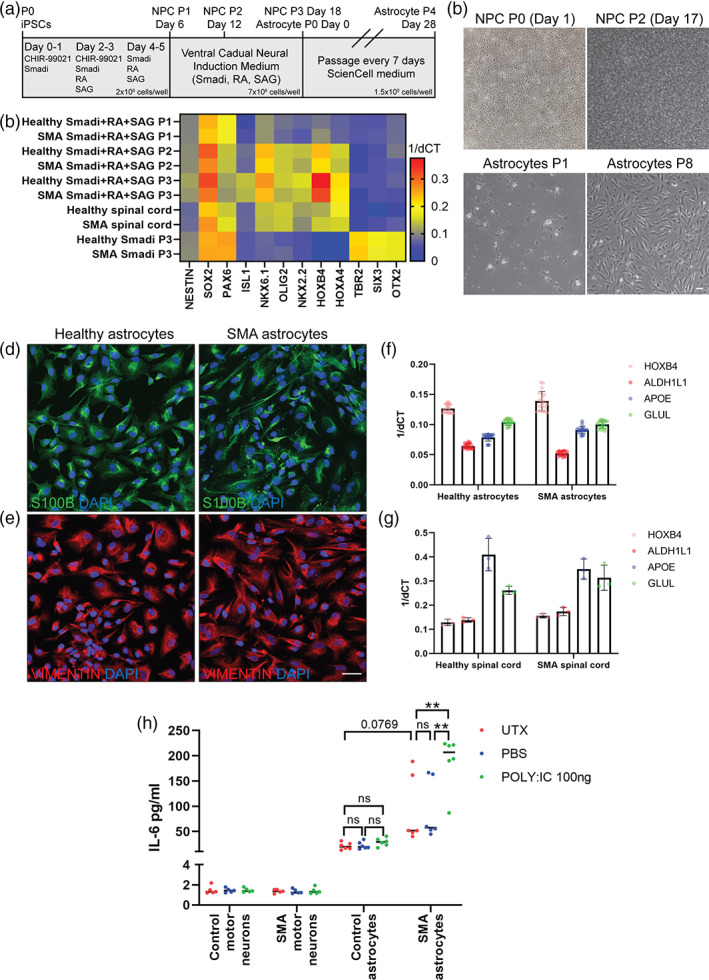
Spinal cord astrocyte differentiation of induced pluripotent stem cells (iPSCs). (a) Schematic of neural progenitor cell (NPC) and astrocyte differentiation protocols from iPSCs. NPCs are patterned and cultured for day 18 before beginning astrocyte differentiation. Astrocytes beyond P4 (day 28) are considered fully differentiated. (b) Representative brightfield images of NPC and astrocyte cultures during early and later stages of culture. Scale bar: 100 μm. (c) Early addition of Wnt agonist (CHIR 99021), SMAD inhibition (SB431542 and LDN‐193189), retinoic acid (RA) and hedgehog smoothened agonist (SAG) during NPC differentiation (P1‐3) increases the expression of general NPC‐associated genes (NESTIN, SOX2, PAX6) and spinal cord ventral caudal‐associated genes (ISL1, NKX6.1, OLIG2, HOXB4 and HOXA4). Postmortem spinal cord and ventral‐caudal patterned NPC (P2 onwards) samples show comparable gene expression profiles, whereas Smadi only treated NPCs show increased forebrain transcription factor gene expression (TBR2, SIX3, OTX2). Markers characteristic of astrocyte identity, including S100β (d) VIMENTIN (e), ALDH1L1, APOE, GLUL (f), and spinal cord ventral caudal transcription factor HOXB4 (D) are readily detected in astrocytes differentiated from NPC cultures and in postmortem spinal cord samples (g). Scale bar: 50 μm. (h) Significantly elevated levels of interleukin 6 (IL6) are detected in conditioned media from stimulated (POLY:IC treated) SMA astrocytes, compared to untreated (UTX) and vehicle treated (PBS) SMA astrocytes, and equivalent healthy astrocyte samples. iPSC‐derived motor neurons served as a negative control for this assay. (2‐way ANOVA, Tukey's multiple comparisons test. **p value<0.005)
SMA and HC astrocytes were next differentiated from the ventral‐caudal NPCs using astrocyte specific medium. A small neuronal population was noted during early stages of astrocyte differentiation (Figure 1b; Astrocyte P1), which diminished at later stages of culture (Figure 1b; Astrocytes P8). Ventral‐caudal patterned astrocytes were found to express characteristic markers of astrocyte identity and function such as S100β (Figure 1d), VIMENTIN (Figure 1e), ALDH1L1, APOE, and GLUL (Figure 1f), which showed similar expression trends to human spinal cord samples (Figure 1g). iPSC‐derived astrocytes and human spinal cord samples also showed equivalent expression of the spinal cord ventral‐caudal transcription factor HOXB4 (Figure 1f, g). The HC and SMA astrocytes were found to secrete interleukin 6 (IL6) into the media, with SMA astrocytes secreting slightly higher levels of IL6 at baseline (p = 0.08, ns, Figure 1h). When stimulated with polyinosinic‐polycytidylic acid (poly:IC), a compound that activates inflammatory responses via toll‐like receptors (De Miranda et al., 2009), HC astrocytes slightly increased IL6 secretion though not significantly, whereas SMA astrocytes significantly upregulated IL6 secretion compared to untreated and PBS‐treated SMA astrocyte samples (p < 0.01, Figure 1h). These data fit with the findings from our previous model (McGivern et al., 2013; Sison et al., 2017), and establish these spinal cord astrocytes as phenotypically and functionally consistent with other in vitro models (de Rus Jacquet, 2019; Perriot et al., 2018; Soubannier et al., 2020; Tcw et al., 2017).
2.2. Differences in SMA astrocyte neurotrophic support and reactivity compared to healthy controls
Our lab previously reported decreased GDNF production and increased production of GATA6 in SMA astrocytes compared to healthy controls (Sison et al., 2017). These findings are consistent with our updated differentiation in which RNA transcripts of GDNF and BDNF were both significantly downregulated in the SMA astrocytes compared to HC (Figure 2a, p = 0.003, 0.022). GATA6 and NFκB transcripts were found to be significantly upregulated in the SMA cultures, also confirming our previous findings (Figure 2b, p < 0.001, p < 0.001). These transcript changes were maintained in protein expression as confirmed by immunocytochemistry (ICC) (p = 0.02, Figure 2c–e).
FIGURE 2.
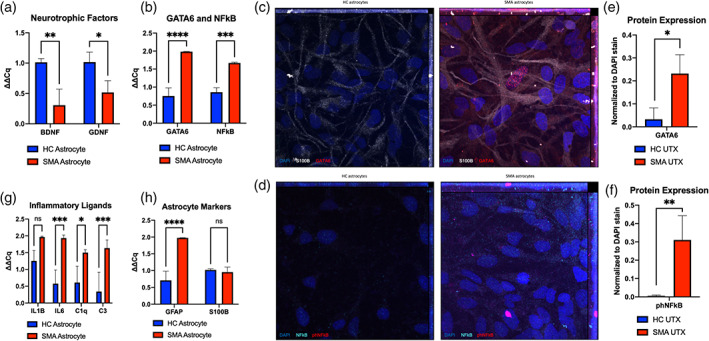
GATA6 and nuclear NFĸB upregulation in SMA astrocytes. (a) Decreased neurotrophic support through BDNF, GDNF and (b) increased GATA6, NFĸB transcript production confirmed in SMA astrocytes compared to healthy controls. (c) Representative images of GATA6 and (d) phNFĸB protein expression in SMA and HC astrocytes by immunocytochemistry ([c] GATA6 = red, S100β = white, DAPI = blue; [d] phNFĸB = pink, NFĸB = teal, DAPI = blue) ([e, f] quantification). (g) Upregulation of transcripts for pro‐inflammatory cytokines and complement factors (IL1β, IL6, C1q, C3) and (h) activation marker GFAP in SMA astrocytes compared to HC, with no changes in general astrocyte marker S100β expression. ICC quantification displays relative fluorescence (total fluorescence for each channel normalized to DAPI) for each image to account for differences in cell density between ROIs. (1‐way ANOVA, Tukey's multiple comparisons test. *p value<0.05, ** < 0.005, *** < 0.0005, **** < 0.0001)
Importantly, phosphorylated NFκB (phNFκB) was also found to be upregulated in the nuclei of SMA astrocytes compared to HC (Figure 2d–f, IHC p < 0.01). phNFκB is known to enter the nucleus and regulate the gene expression of pro‐inflammatory cytokines (Baeuerle & Baltimore, 1988; Christian et al., 2016). Accordingly, we found upregulated transcripts for interleukin 1 beta (IL1β) and interleukin 6 (IL6), as well as upregulated complement cascade members C1q and C3 in the SMA astrocyte cultures (Figure 2g, p = 0.07, 0.001, 0.02, 0.001). Glial fibrillary acidic protein (GFAP), a canonical marker of astrocyte activation, was also found upregulated, though none of these changes were associated with any changes in the general astrocyte marker S100B expression (Figure 2h, p < 0.001, 0.85). Overall, these findings confirm the increased activation of SMA astrocytes compared to HC astrocytes in our updated culture system. They also identify GATA6 and NFκB as potential drivers of increased inflammatory ligand production in SMA.
2.3. Manipulation of GATA6 associated with changes in reactivity and neurotrophic support
To directly examine the influence of GATA6 upregulation on driving astrocyte activation, we infected healthy control astrocytes (HC) with a lentivirus expressing GATA6 (GATA6OE) or a GFP control. We also separately infected SMA astrocytes with a lentivirus expressing an shRNA against GATA6 (GATA6KD) construct or GFP control. We first confirmed GATA6 viral expression or knockdown in RNA transcripts by quantitative real‐time PCR (qRT‐PCR) (Figure 3a) and confirmed its protein expression by ICC (OE fold change of 9.76, KD fold change of −3.22, Figure 3c, e). HC GATA6OE astrocytes showed increased GATA6 levels equivalent to SMA astrocyte levels (HC GATA6OE to SMA GFP qPCR: p = 0.97, ICC p = 0.23), and SMA GATA6KD astrocytes were found to have decreased GATA6 levels similar to HC astrocyte baseline (SMA GATA6KD to HC GFP qPCR: p = 0.06, ICC p = 0.98). Interestingly, no significant changes in NFκB transcript levels were observed in HC GATA6OE samples compared to HC GFP samples (Figure 3b), although nuclear phNFκB protein levels were found significantly increased (ICC p < 0.02, Figure 3f). GATA6KD in the SMA astrocytes did have an associated significant decrease in NFκB transcript levels (p < 0.05) as well as a significant decrease in nuclear phNFκB (ICC p < 0.03, Figure 3b, d, f).
FIGURE 3.
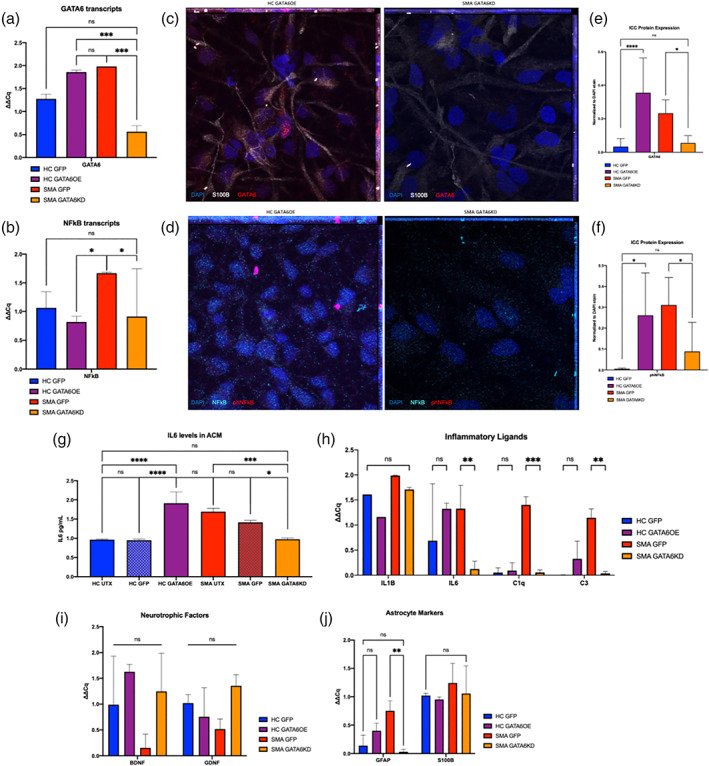
Changes in inflammatory and neurotrophic gene expression after astrocyte GATA6 manipulation. (a) Confirmation of GATA6 transcript reduction (SMA) or overexpression (HC) after exposure to GFP control, GATA6 expressing, or GATA6KD lentivirus and (b) associated change in NFĸB transcript production. (c, d) representative images of GATA6 and phNFĸB protein expression changes after lentiviral manipulation assessed by ICC ([c]: GATA6 = red, S100β = white, DAPI = blue; [d] phNFĸB = pink, NFĸB = teal, DAPI = blue) ([e, f] quantification). (g) Secreted IL6 protein levels confirm transcript changes after GATA6 manipulation. (h) Significant decrease in IL6, C1q, and C3 transcripts after GATA6KD in SMA astrocytes, trends for increases after GATA6OE in HC astrocytes. (i) no significant changes in neurotrophic support after GATA6 manipulation. (j) Significant decrease in reactive GFAP transcripts, but importantly not in characteristic marker S100β. ICC quantification displays relative fluorescence (total fluorescence for each channel normalized to DAPI) for each image to account for differences in cell density between ROIs. (2‐way ANOVA, Tukey's multiple comparisons test for qPCR; 1‐way ANOVA, Tukey's multiple comparisons test for ICC, *p value<0.05, ** < 0.005, *** < 0.0005, **** < 0.0001)
In relation to the NFκB data, we examined canonical pro‐inflammatory cytokines in astrocyte samples, finding a significant increase in secreted IL6 after GATA6OE (HC GFP to HC GATA6OE p < 0.0001, Figure 3g) and a significant decrease after GATA6KD (SMA GFP to SMA GATA6KD p = 0.0002, Figure 3g). No significant changes in IL1β were detected (Figure 3h). Significant decreases in C1q (p < 0.001) and C3 (p < 0.01) transcript levels were also found after GATA6KD, and there was only a slight increase in expression after GATA6OE (fold change = 0.43, 0.99, ns) (Figure 3h). No significant changes in neurotrophic factors or astrocyte marker S100β were found in any condition (Figure 3I, 3j). GFAP transcript was significantly reduced after GATA6KD in SMA astrocytes (p < 0.01) but was not significantly increased in GATA6OE HC astrocytes (fold change = 0.65, ns, Figure 3j). These findings suggest that aberrant GATA6 expression in SMA astrocytes is sufficient to modulate inflammatory ligands, complement components, and GFAP transcripts.
2.4. Effect of astrocyte GATA6 manipulation on spinal motor neuron survival
We next investigated whether GATA6‐associated reactivity and inflammation in astrocytes contributed to spinal motor neuron loss in vitro. Following an established protocol (Maury et al., 2015) for generating spinal motor neurons (MNs) from human iPSCs, we matured plated MNs for 21–28 days before analysis. To examine the possible contribution of GATA6 to SMA astrocyte neurotoxicity, conditioned media (ACM) from HC, HC‐GFP, HC‐GATA6OE astrocytes, as well as SMA, SMA‐GFP, and SMA‐GATA6KD astrocytes was applied onto HC and SMA MNs. Cell death was then measured using a TUNEL assay and normalized to a live cell counterstain (7AAD, Figure 4a, b, d, e). Quantification of percent change was calculated using normalized values compared to one HC UTX ACM replicate for HC and SMA MNs (Figure 4c, f).
FIGURE 4.
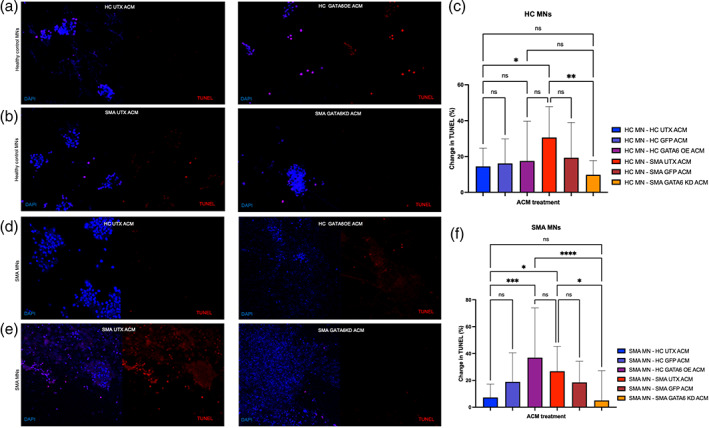
Spinal motor neuron survival after astrocyte GATA6 manipulation. Conditioned media from HC untreated (UTX) or GATA6OE astrocytes applied onto HC (a) and SMA (d) spinal motor neurons (MN). Media from SMA UTX or GATA6KD astrocytes also applied onto HC (b) and SMA (e) MNs. Cell death was assessed with a TUNEL kit. TUNEL stain (red) was normalized to live cell nuclear 7AAD stain (blue) for each image and represented as percent change from the first HC UTX ACM replicate for HC and SMA MNs (c, f). In HC and SMA MNs, SMA UTX was significantly more toxic than HC ACM, an effect which GATA6KD ameliorated. GATA6OE was significantly more toxic than HC UTX only when applied onto SMA MNs. No differences in cell death were found between UTX and GFP conditions. Left panel for each image is merged 7AAD and TUNEL channels, right panel is TUNEL only. (1‐way ANOVA, Tukey's multiple comparisons test. *p value<0.05, ** < 0.005, *** < 0.0005, **** < 0.0001)
Consistent with our previous findings (Sison et al., 2017) in both HC and SMA MNs, ACM from untreated SMA astrocytes induced more cell death than ACM from untreated HC (HC UTX) astrocytes (p < 0.02, 0.04, Figure 4a–c, d–f). No significant differences were found between the untreated and GFP‐control samples in either HC or SMA astrocytes (HC‐HC UTX:GFP p = 0.99, HC‐SMA UTX:GFP p = 0.28, HC‐HC UTX:GFP p = 0.47, SMA‐SMA UTX:GFP p = 0.79). GATA6OE ACM did significantly increase cell death compared to HC UTX ACM in SMA MNs (p < 0.001), though not in HC MNs (p = 0.99). This difference is likely due to the inherent vulnerability of SMA MNs. Strikingly, SMA GATA6KD ACM significantly decreased MN cell death in both HC and SMA MNs compared to SMA UTX ACM (p < 0.01, 0.05). These data support the idea that GATA6 overexpression in SMA astrocytes drives neurotoxicity.
2.5. Effect of astrocyte activation and GATA6 manipulation on microglial activation
Finally, we examined the effect of SMA astrocyte activation on human iPSC‐derived microglia from SMA patients and healthy controls. Many models of neurodegeneration establish a role for activated microglia in driving further neuron loss (Halliday & Stevens, 2011; Hirsch & Hunot, 2009; Hong et al., 2016; Kashon et al., 2004; Stogsdill & Eroglu, 2017; Werneburg et al., 2020; Yanuck, 2019). Because many of the increased pro‐inflammatory cytokines and complement components found within the SMA astrocyte cultures are key signals for microglial activation and phagocytosis, we aimed to examine the possibility of SMA astrocyte‐driven microglial activation contributing to spinal motor neuron loss. We started by generating microglia from SMA and HC iPSC lines using established differentiation protocols and commercial kits (Figure 5a) (Haenseler et al., 2017; McQuade et al., 2018). Generated microglia were confirmed TMEM119+, P2RY12+, and Iba1+ before analyses (Figure 5b).
FIGURE 5.
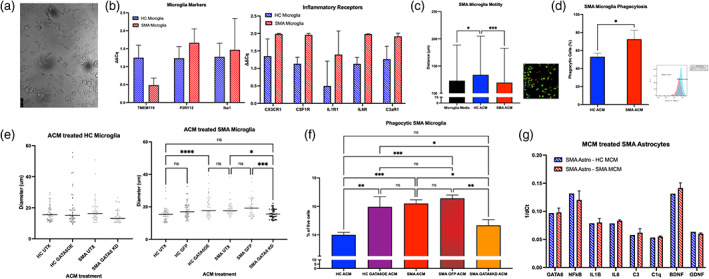
Microglial activation after exposure to astrocyte conditioned media (ACM). iPSC‐derived microglia from HC (blue stripe) and SMA (red stripe) lines express microglia‐specific markers TMEM119, P2RY12, and Iba1 (a representative image, b). (b) SMA microglia show trends for upregulated transcripts for inflammatory receptors including CX3CR1, CSF1R, IL1R1, IL6R, and C3aR1 (ns). (c) SMA microglia exposed to HC ACM (blue) show increased motility compared to SMA ACM (red) in live image tracking (left ‐ quantification, right ‐ representative image). (d) SMA microglia show increased phagocytosis of FluoSpheres after SMA ACM treatment compared to HC ACM. (e, left) no changes in phenotype (soma diameter) after HC microglia treated with GATA6 manipulated ACM. (e, right) a significant increase in soma size of SMA microglia treated with GATA6OE compared to HC UTX was found, as well as a significant decrease after GATA6KD compared to SMA UTX. (f) Significantly increased phagocytosis of FluoSpheres when SMA microglia treated with GATA6OE ACM (purple) compared to HC UTX ACM (blue). Significantly decreased phagocytosis of FluoSpheres when SMA microglia treated with GATA6KD ACM (orange) compared to SMA UTX ACM (red) and SMA GFP ACM (red pattern). (g) no changes found in any transcripts after SMA astrocytes treated with HC (blue stripe) or SMA (red stripe) microglia conditioned media. (1‐way ANOVA, Tukey's multiple comparisons test. *p value<0.05, ** < 0.005, *** < 0.0005, **** < 0.0001)
We first examined differences in inflammatory receptors at baseline between the HC and SMA microglia (Figure 5b). Though no differences were significant, we did find trends for increased pro‐inflammatory receptor transcripts including C3aR1, CXCR1, CSFRA, IL1R1, and IL6R in SMA microglia (Figure 5b). To test if SMA microglia may have heightened sensitivity to astrocyte secreted factors, we first applied HC UTX and SMA UTX ACM onto SMA microglia generated from a yolk‐sac lineage protocol (Figure 5c, d). HC ACM increased microglia mobility—a sign of non‐activated, surveilling microglia (Fontainhas et al., 2011; Franco‐Bocanegra et al., 2019)—compared to control media treated cells as measured by distance traveled through a live‐image tracking assay (p < 0.05, Figure 5c; image). The application of SMA ACM onto SMA microglia significantly reduced this surveilling phenotype (p < 0.001, Figure 5c). To directly examine functional differences, we measured phagocytic ability using fluorescent bead‐based flow cytometry (Figure 5d). We found a significantly increased number of phagocytic SMA microglia after treatment with SMA ACM than HC ACM (p < 0.05, Figure 5d).
We then wanted to examine the role of GATA6 in the SMA astrocyte‐driven microglial activation phenotype and confirm our findings in a hematopoietic lineage model of microglia (Figure 5e, f). First, we found no differences in soma diameter—a sign of activation (Davis et al., 2017)—between HC UTX, HC GATA6OE, or SMA UTX ACM applied onto HC microglia (p = 0.32, 0.69, ns, Figure 5e). A slight but significant decrease in diameter was seen in the SMA GATA6KD treated HC microglia compared to HC GATA6OE (p < 0.02, Figure 5e). In the SMA microglia treatments, a significant increase in size was seen in the HC GATA6OE condition, comparable to SMA UTX treated microglia (HC UTX:GATA6OE p < 0.0001, GATA6OE:SMA UTX p = 0.71, Figure 5e). SMA microglia treated with SMA GATA6KD ACM showed a significant decrease in diameter, reducing sizes to HC UTX levels (SMA UTX:GATA6KD p < 0.05, HC UTX: GATA6KD p = 0.99). No differences were found between HC UTX and HC GFP, or SMA UTX and SMA GFP conditions (p = 0.13, 0.84). Functionally, a significant decrease in phagocytic cells was found in SMA microglia treated with the SMA GATA6KD ACM compared to SMA UTX or SMA GFP ACM (p < 0.02, 0.002, Figure 5f). A significant increase in phagocytic cells was also found in SMA microglia after treatment with the HC GATA6OE ACM compared to HC UTX ACM (p < 0.002, Figure 5f).
Finally, we aimed to test if the activating or toxic effect was unique to SMA astrocytes or if SMA microglia had similar activating effects. Therefore, we treated SMA and control astrocytes with SMA or control microglia conditioned medium (MCM). Interestingly, no differences in activation were found when astrocytes were treated with media conditioned by HC or SMA microglia (ns, Figure 5g). GATA6 transcripts were not differentially expressed between SMA and HC microglia or between SMA and HC motor neurons (data not shown) suggesting aberrant GATA6 expression is more astrocyte restricted.
2.6. Proposed role of GATA6 in SMA astrocyte‐mediated neuron loss
Overall, these data suggest a role for SMA astrocytes in SMA microglial activation and support a role of aberrant GATA6 expression as a driving factor in astrocyte mediated neurotoxicity and microglial phagocytosis (Figure 6). Other recent data from the lab (Welby et al., 2021) give rise to the theory that the loss of neurotrophic support and/or upregulation of pro‐inflammatory signals in SMA astrocytes may be downstream consequences of upregulated microRNAs, including miR‐146a. Future work will focus on validating these proposed mechanisms and identifying mediators such as microRNAs between GATA6 and neurotrophic factors or pro‐inflammatory molecules. These mediators may help identify other disease‐specific changes in SMA astrocytes and explain why GATA6 OE alone did not perfectly replicate all aspects of disease phenotype in the HC astrocytes.
FIGURE 6.
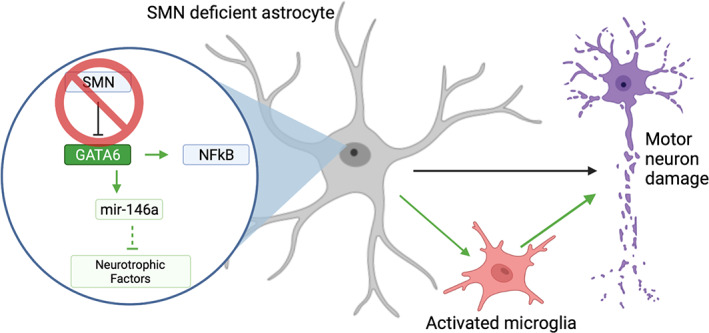
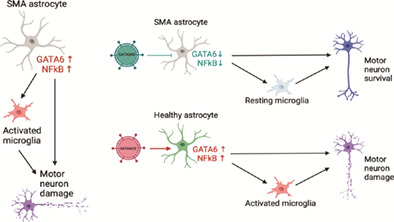
Schematic of proposed role of GATA6 in relation to NFĸB‐mediated astrocyte neurotoxicity. (a) in SMA astrocytes, the loss of SMN allows for GATA6 overexpression through the loss of a negative regulator. GATA6 overexpression may contribute to NFĸB overexpression leading to an increase in pro‐inflammatory cytokine production. These secreted factors lead to motor neuron damage directly, as well as serve to activate microglia to drive additional neurotoxicity. Black lines represent accepted mechanisms in published literature; green lines represent relationships proposed by this manuscript. Created using biorender.com
3. DISCUSSION
The loss of SMN in SMA causes selective lower motor neuron loss in patients, and this is replicated in vivo with SMA mouse models (Sleigh et al., 2011). However, in purified motor neuron cultures in vitro, SMA motor neurons do not show increased cell death compared to healthy controls (Rossoll et al., 2003; Sison et al., 2017). This is in contrast to other neurodegenerative diseases such as ALS, where iPSC‐models of ALS motor neurons do show increased TUNEL staining over healthy controls by day 20 of differentiation (Chen et al., 2014). This recent distinction, along with a body of evidence showing that targeting SMN restoration to only motor neurons has limited effect, suggest a role for non‐cell autonomous contributions to SMA motor neuron vulnerability (Gogliotti et al., 2012; Hua et al., 2011; Lee et al., 2012; Martinez et al., 2012). In parallel, pre‐synaptic deficits and aberrant activation of glial cells in SMA have been observed before overt motor neuron loss (McGivern et al., 2013). Specifically, SMA astrocytes have been shown to decrease neurotrophic and growth factor production and increase the pro‐inflammatory production of GFAP, IL6, and IL1β (McGivern et al., 2013; Rindt et al., 2015; Sison et al., 2017). The application of SMA astrocyte conditioned media has been shown to have detrimental effects on both healthy control MNs and those with reduced SMN (Martin et al., 2017; Sison et al., 2017). Activated (Iba1+, M1) microglia have also been found to be increased at early symptomatic stages and localized to motor neurons in SMA rodents (Cerveró et al., 2018; Ling et al., 2010; Tarabal et al., 2014). Synapse loss has been associated with microglial phagocytosis, with complement factors C1q and C3 implicated in targeting microglia removal of healthy synapses (Ling et al., 2010; Vukojicic et al., 2019). Together, these data robustly indicate a role for glia in contributing to motor neuron loss in SMA.
Here, we utilized human iPSC‐derived astrocytes, microglia, and spinal motor neurons from healthy control and SMA patient lines to examine disease properties in human cells. Though all astrocytes are thought to share common features (e.g., upregulation of GFAP after activation) and functions (e.g., formation and maintenance of the blood–brain barrier), recent studies have identified differences in gene expression, morphology, and function between astrocytes in distinct regions of the CNS (Batiuk et al., 2020; Morel et al., 2019). Even within the spinal cord, three distinct populations of astrocytes are hypothesized to preferentially support neurons from the same progenitor subtypes (Hochstim et al., 2008; Kerstetter & Miller, 2012; Tsai et al., 2012). To address these emerging ideas, we used a more disease‐relevant astrocyte culture by patterning NPCs with small molecules to mimic spinal cord development, then differentiated these into functional spinal cord astrocytes (Batiuk et al., 2020; Kerstetter & Miller, 2012; Sofroniew, 2000). We found that SMA astrocytes upregulated classical markers of activation, decreased neurotrophic support, and increased pro‐inflammatory ligands and complement components compared to controls. These data establish our spinal cord astrocytes as a disease‐relevant and functional model concurrent with others being used in the field (de Rus Jacquet, 2019; Perriot et al., 2018; Soubannier et al., 2020; Tcw et al., 2017). They also allow us to examine astrocyte activation and interactions with spinal motor neurons and microglia in an SMA‐specific context throughout the rest of this study.
In order to more specifically investigate the novel relationship between SMN, GATA6, and NFκB in the context of astrocyte activation, we infected astrocytes with a lentivirus to drive GATA6 expression in HC astrocytes or knock it down in SMA astrocytes. Interestingly, we found that though the transcript and protein levels of GATA6 were successfully manipulated, only the knockdown of GATA6 in SMA astrocytes, not the overexpression in HC astrocytes, was associated with a significant change in NFκB transcript levels. Both, however, were associated with increased nuclear phNFκB protein levels. Knocking down GATA6 in the SMA astrocytes significantly decreased inflammatory and activation marker transcripts, though it did not restore neurotrophic support through BDNF or GDNF. Within these SMN‐deficient SMA astrocytes, our data indicate that GATA6 is a main contributor of aberrant activation, confirming what transcriptome analyses, in vivo studies, and our own previous findings predicted (Sison et al., 2017; Yang et al., 2016). In contrast, overexpressing GATA6 in the HC astrocytes showed trending—but not significant—increases in these transcripts. This suggests that these inflammatory ligands are more tightly regulated by NFκB than GATA6, with GATA6 potentially competing with functional alternative regulators of NFκB in healthy SMN‐expressing astrocytes (Neumann & Naumann, 2007). The relationship between GDNF, BDNF, and GFAP may be similarly complicated. Histone deacetylase (HDAC), an enzyme which can increase DNA susceptibility to transcription factor (TF) regulation, has shown inverse associations with GFAP compared to BDNF and GDNF. In different contexts, low HDAC (DNA more accessible for TF regulation) is associated with decreased GFAP, whereas high HDAC (DNA less accessible for regulation) is associated with increased BDNF and GDNF (Kumar et al., 2018; Wu et al., 2008). These complicated associations with the disease‐specific genetic changes may begin to illuminate why the impact of GATA6KD was more striking in the SMA astrocytes than its overexpression in controls.
We have previously found that SMA iPSC‐derived motor neurons die through a Fas‐dependent and caspase‐8 mediated apoptotic mechanism when in mixed cultures with SMA astrocytes (Sareen et al., 2012). Interestingly, pro‐inflammatory cytokines, including TNFα and IL1β, can stimulate Fas‐mediated apoptosis (Kondo et al., 2009). Expectedly, SMA ACM was found to increase motor neuron cell death in SMA and HC cultures compared to HC ACM, and the knockdown of GATA6 in SMA astrocytes ameliorated this effect in both SMA and HC MNs. Strikingly, the overexpression of GATA6 in HC astrocytes increased cell death only in the SMA MNs, not in the healthy controls. This again could be due to regulatory mechanisms—like those involving SMN—which are intact in the HC MNs but are absent in SMA MNs leaving them more vulnerable to even slight changes in astrocyte secreted factors. One example of this is the SMN‐deficiency‐linked activation of p53 found in vulnerable motor neurons at pre‐symptomatic stages in SMA mice (Murray et al., 2015). This distinction between GATA6OE toxicity to HC and SMA MNs emphasizes the importance of therapeutically targeting the early‐activating SMA astrocytes, especially in patients not receiving the AAV therapy to restore SMN. It also provides an interesting topic of future study regarding the resistance of AAV9‐SMN treated motor neurons to astrocyte toxicity, specifically regarding GATA6‐driven toxicity.
Next, we began to investigate the emerging idea of pathologically activated microglia in SMA by differentiating microglia from SMA and healthy control iPSCs. At baseline, we observed differences between the HC and SMA microglia in pro‐inflammatory receptors known to activate microglia from a neurotrophic, surveilling phenotype—characterized by small soma and highly motile processes—towards an ameboid, phagocytic phenotype (Franco‐Bocanegra et al., 2019; Fu et al., 2014; Klawonn et al., 2021). We also identified upregulated complement receptor C3aR1, a receptor which has been specifically tied to microglial activation and phagocytosis of neurons or synapses in several models of neurodegeneration (Harder et al., 2020; Litvinchuk et al., 2018; Werneburg et al., 2020; Zhang et al., 2020). The upregulation of these receptors in SMA microglia may prime them for the upregulated ligands produced by SMA astrocytes to encourage activation. Encouragingly, we found that SMA ACM applied onto SMA microglia increased activated phenotypes compared to HC ACM or no treatment in both yolk‐sac lineage and hematopoietic progenitor lineage microglia. These data first confirmed the ability of iPSC‐derived astrocytes and microglia to communicate via secreted factors. Additionally, they suggest that microglia‐mediated phagocytosis could contribute to viable neuron loss in SMA, as has been suggested in neurodegenerative models such as Alzheimer's and Parkinson's disease (Fricker et al., 2012; Neher et al., 2011; Neniskyte et al., 2011). Importantly, the use of individually differentiated cultures and ACM treatments in this study suggest that microglia activation in SMA may be fueled by activated astrocyte secreted factors.
Finally, we examined the potential impact of GATA6 knockdown on this astrocyte‐driven microglial activation. Morphologically and functionally, we found that treatment with SMA UTX and HC GATA6OE ACM increased activated phenotypes of SMA microglia compared to HC UTX or SMA GATA6KD ACM. The role of the complement proteins—especially C3—within this process is one promising area for future studies. C3 is upstream of complement cascade products which can be secreted to act as tags for phagocytosis (C3b, C5a), activators for immune cells (C3a), or initiators of the membrane attack complex (C5b) (Ricklin et al., 2016). In SMA, already vulnerable MNs receiving neurotoxic conditioning and C5b signals from activated astrocytes may be upregulating their own C3 in addition to initiating the terminal pathway towards cell lysis (Carpanini et al., 2019; Peterson et al., 2017). Excess complement products then may be additionally tagging nearby healthy neurons for phagocytosis‐mediated cell death (i.e., phagoptosis) as well as functional synapses for removal by astrocyte‐activated microglia. More experiments to examine the production of C3 by SMA MNs before and after ACM treatment, as well as in MN‐microglia co‐cultures, are needed to investigate this hypothesis. Nevertheless, these data demonstrate the ability of iPSC‐derived astrocytes and microglia to communicate via secreted factors and provide compelling evidence for astrocyte‐driven activation of microglia in SMA. Of particular interest, this activation appears to be an astrocyte‐first mechanism, as the application of microglia conditioned media onto SMA astrocytes showed no effect. This is in contrast to microglia‐first activation theories which have been proposed in the context of Alzheimer's, Huntington's, and Parkinson's disease (Park et al., 2021; Savage et al., 2020; Yun et al., 2018). This is an area of future research, as C3‐expressing activated astrocytes were observed in postmortem tissue from patients with Alzheimer's, Huntington's, and Parkinson's disease, as well as multiple sclerosis (MS) and amyotrophic lateral sclerosis (ALS) (Liddelow et al., 2017). It would be especially of interest to compare the activation timelines of SMA and ALS, as motor neurons are particularly affected in these disorders—despite quite dissimilar genetic causes—and microglia have been directly linked to motor neuron loss in ALS (Frakes et al., 2014; Mejzini et al., 2019). Using our isolated iPSC‐derived culture system and treatments of ACM or MCM, we may be able to parse out if this astrocyte‐first activation is specific to SMA, or if in vivo and postmortem studies are missing early mechanisms of neuron loss (Franklin et al., 2021).
Together, these data identify GATA6 as a novel inducer of NFκB and inflammatory signaling in SMA astrocytes and further suggest a role for astrocyte‐driven microglia activation, which may contribute to MN death in SMA. More consideration for astrocytes and microglia when designing therapeutics for SMA may positively impact MN health to improve lifespan and function for patients.
4. MATERIALS AND METHODS
4.1. Pluripotent stem cells
Two healthy control iPSC lines (21.8, 4.2) and three SMA patient iPSC (7.12, 3.6, 8.3) lines were utilized in these experiments (Sison et al., 2017). All pluripotent stem cells were maintained on Matrigel (Corning) in Essential 8 (Gibco) and passaged every 4–6 days. The iPSCs and differentiated cells were confirmed mycoplasma negative.
4.2. Astrocyte differentiation
Spinal cord patterned astrocytes were generated from iPSC‐derived NPCs. Catalog numbers for each reagent are listed in supplemental materials (Table S1) and a protocol schematic is shown in Figure 1a. Briefly, iPSCs were grown to confluency, dissociated with Accutase, and plated at 2 million cells/well into Matrigel (Corning) coated 6 well‐plates in NPC base medium (50% DMEM/F12, 50% Neurobasal, 2% B27, 1% N2, 1% Antibiotic/Antimycotic, 0.1% B‐mercaptoethanol, 50 ng/ml Laminin, and 0.5uM Ascorbic Acid) supplemented with 10uM Y‐27632, 3uM Chir‐99021, 40uM SB431542, and 0.2uM LDN193189. On day 1, NPC media was changed and supplemented with 3uM Chir‐99021, 40uM SB431542, and 0.2uM LDN193189. Cells were washed with PBS if a lot of cell death was observed. Days 2 and 3 involved NPC media changes supplemented with 3uM Chir‐99021, 40uM SB431542, 0.2uM LDN193189, 100 nM retinoic acid (RA), and 500 nM hedgehog smoothened agonist (SAG). From day 4 onwards, NPC media was supplemented with 40uM SB431542, 0.2uM LDN193189, 100 nM RA, and 500 nM SAG and changed daily. Cells were passaged via Accutase treatment on Day 6 (P1), Day 12 (P2), and Day 18 (P3). Plating density for NPC P1 and P2 is 7 million cells per well (of a six well plate) and plating density for NPC P3 to begin astrocyte differentiation (P0) is 150,000 cells/well (of a six well plate).
Astrocyte differentiations were cultured with ScienCell Astrocyte Medium containing 1% Astrocyte Growth Supplement, 1% Penicillin/Streptomycin, and 2% B27. Cells were fed every 48 h and passaged with Accutase every 6–9 days upon confluency (minimum of three passages). Astrocyte P1, P2, and P3 were all plated at 150,000 cells/well, at P4 cells were considered differentiated and split onto coverslips for immunocytochemistry, into six well plates for RNA or protein, or into T75 tissue culture treated flasks for ACM generation and collection. Astrocyte conditioned media was collected upon each media change after P4, spun to remove any cells or debris, and stored in sterile Falcon tubes at –20°C. Frozen medias were slowly thawed on ice before use. Poly:IC treatments were diluted to 100 ng/ml in supplemented Astrocyte media and left for 24 h. Cells were then rinsed with PBS and fed with supplemented Astrocyte media. Media was collected from stimulated astrocytes after 48 h, and pellets were collected for analyses. IL‐6 ELISA assay was performed on collected media (Affymetrix eBioscience, #88–7066‐22).
4.3. Lentivirus production and usage
GFP control virus (titer–1.0e8 TU/ml) was acquired from Cellomics Technology (cat: PLV‐10077). Plasmids for GATA6OE and KD viruses were generously donated by Dr. Stephen Duncan at the Medical University of South Carolina. GATA6OE (titer–8.91e8 TU/ml) and GATA6KD (titer–4.48e8 TU/ml) viruses were produced by the Viral Vector Core at the Blood Research Institute/MCW. Astrocytes were infected with appropriate virus (MOI = 10) for 48 h at Astrocyte p.1. After 48 h, virus was removed, and cells were fed as normal for 48 h. Astrocytes were passaged three times after infection (to Astrocyte p.4) before degree of knockdown (71.59% KD) or overexpression (246.91% OE) was assessed.
4.4. Motor neuron differentiation and treatments
Spinal motor neurons were differentiated based on the Maury et al. (2015) protocol (Maury et al., 2015). Briefly, embryoid bodies were generated from iPSCs and patterned in the presence of Chir‐99021 with dual SMAD inhibition (SB 431542 and LDN 1931899) followed by treatment with retinoic acid (RA), smoothened agonist (SAG), and DAPT. Spinal motor neuron progenitor cells were then dissociated and plated on Matrigel‐coated glass coverslips for terminal differentiation and maturation in growth factor supplemented medium for 28–42 days in vitro. Treatments with ACM were performed with 20% ACM in MN Maturation media after day 28 and left for 48 h at 37°C before fixing, collection, or analyses.
4.5. Microglia differentiations
Microglia were differentiated using a previously published protocol (Haenseler et al., 2017) or the commercially available differentiation kit by STEMCELL Technologies (Haenseler et al., 2017; McQuade et al., 2018) (#05310, #100–0019, #100–0020). Briefly, iPSCs were patterned into macrophages (pMacs) using BMP4, VEGF, SCF, IL3, and M‐CSF. pMacs were matured into microglia (pMGL) using N2, IL34, and GM‐CSF (experiments shown in Figure 5c, d). Alternatively, iPSCs were differentiated into hematopoietic progenitor cells (HPCs) using the STEMdiff Hematopoietic Kit (StemCell Technologies). Floating HPCs were then collected and plated at 50,000 cells/ml in STEMdiff Microglia Differentiation media (StemCell Technologies) for 24 days, followed by rapid maturation in STEMdiff Microglia Maturation media (StemCell Technologies) for at least 4 days before usage (experiments shown in 5B, 5E, 5F, 5G). Microglia were confirmed TMEM119+, CD45+, and CD11b + (Figure 5b).
4.6. Quantitative real‐time PCR
RNA was isolated from control and SMA cell pellets using the RNeasy Mini Kit (Qiagen) following manufacturer's instructions, quantified using a Nanodrop Spectrophotometer, treated with RQ1 Rnase‐free Dnase (Promega), and converted to cDNA using the Promega Reverse Transcription system (Promega). SYBR green qRT‐PCR was performed in triplicate using cDNA and run on Bio‐Rad CFX384 real time thermocycler. Primers for each target are available in Supplemental materials (Table S2). Cq values for each target gene were normalized to housekeeping gene (GAPDH) values and Ct values for each sample were normalized to a human cortex sample to calculate relative fold change (ddCT) in gene expression.
4.7. Immunocytochemistry
Plated cells were fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature and rinsed with PBS. Nonspecific labeling was blocked and the cells permeabilized with 0.25% Triton X‐100 in PBS with 1% BSA and 0.1% Tween 20 for 15 min at room temperature. Cells were incubated with primary antibodies overnight at 4°C, then labeled with appropriate fluorescently‐tagged secondary antibodies. Hoechst nuclear dye was used to label nuclei. Primary antibodies used were chicken anti‐Vimentin (abcam, ab24525), mouse anti‐S100β (Sigma, S2532), rabbit anti‐GATA6 (Cell Signaling #5851), mouse anti‐NFκB p65 (Cell Signaling #6956), and rabbit anti‐phospho‐NFκB p65 (Cell Signaling #3033). Secondary antibodies used were donkey anti‐mouse AF488 (Invitrogen), goat anti‐chicken AF568 (Invitrogen), donkey anti‐rabbit AF546 (Invitrogen), and donkey anti‐mouse AF647 (Invitrogen). All primaries were used at a 1:500 dilution, and all secondaries were used at 1:1000 dilution. Analyses were performed on three randomly selected fields per coverslip using standard fluorescent microscopy using equivalent exposure conditions. Total fluorescence in each channel was measured using FIJI (ImageJ) software. Bar graphs in Figures 2 and 3 show relative expression (total stain of interest divided by total DAPI for that field) to account for variable number of cells in each ROI. Representative images were taken on Zeiss confocal microscope at 63× as a z‐stack and displayed as a maximum intensity projection (MIP).
4.8. TUNEL assay
Plated cells were fixed in 4% PFA for 20 min at room temperature, rinsed with PBS, and then stained using a TUNEL Assay Kit ‐ BrdU‐Red (abcam, ab66110). Manufacturer's instructions were adapted for coverslips instead of Coplin jars. Briefly, cells were permeabilized as described for ICC, washed, and incubated with DNA labeling solution for 1 h at 37°C. Cells were washed and optional 7AAD nuclear counterstain was then applied for 30 min at room temperature. Coverslips were imaged within 3 h of staining with standard fluorescent microscopy. Three images were acquired from randomly selected fields for each coverslip. Images were analyzed for total fluorescence in either channel using FIJI (ImageJ) software. Relative expression for each condition (total TUNEL stain divided by total 7AAD stain) was analyzed to account for variable number of cells in each ROI. This high‐throughput analysis was validated through analysis of individual TUNEL+ cells divided by individual 7AAD+ cells (Supplemental Figure S3). Representative images were acquired on a Zeiss confocal microscope within 3 h of staining.
4.9. Microglia assays
Microglia were treated with 1:2 astrocyte conditioned media with microglia maturation media for 24 h at 37°C. Live image tracking was performed with 1:6000 Invitrogen Cell Tracker Green CMFDA dye with 5000 cells per condition. Images were acquired over 16 h using an Olympus IX83 Inverted Light Microscope and analyzed on ImageJ using OlympusViewer, TrackMate, and Chemotaxis Tool plugins. Soma size measures were generated by fixing cells onto glass coverslips and staining with Iba1 (abcam, ab5076), then acquiring three random images per coverslip. Images were blindly analyzed using Nikon Elements Advanced Research package to measure soma diameter of total n = 50 cells per condition. Phagocytosis assay was performed by treating cells with astrocyte conditioned media or microglia maturation media (control) for 24 h at 37°C. Cells were then incubated with FluoSpheres Sulfate Microspheres (ThermoFisher #F8851) for 15 min at 37°C. Unbound beads were removed from cell suspension, then cells and bound beads were suspended in FACS flow buffer on ice. Flow cytometry was performed immediately and gated on live cells to remove any unbound beads from downstream analyses. Flow cytometry was performed on an LSRFortessa X‐20 Special Order Research Product or BD Accuri C6 (BD Biosciences, Franklin Lakes, NJ).
4.10. Statistical analyses
Experimental conditions within each experiment were performed in technical triplicates for a minimum of three independent experiments. Data were analyzed using GraphPad Prism software and the appropriate statistical tests including the Student's t test, 1‐way ANOVA, and 2‐way ANOVA followed by Tukey's post hoc analysis of significance. Changes were considered statistically significant when p < 0.05.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Reilly L. Allison, Emily Welby, and Guzal Khayrullina performed and analyzed experiments. Reilly L. Allison, Emily Welby, Guzal Khayrullina, Barrington G. Burnett, and Allison D. Ebert designed experiments and interpreted data. Allison D. Ebert and Barrington G. Burnett supervised the study and provided funding. Reilly L. Allison wrote the manuscript, Emily Welby and Reilly L. Allison created figures and legends, and Emily Welby and Allison D. Ebert edited the manuscript. All authors approved the manuscript.
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGMENTS
We would like to acknowledge the Children's Research Institute FLOW Core at the Medical College of Wisconsin (MCW) and thank Melissa Whyte for her help with the microglia flow cytometry assays. We thank Dr. Michele Battle for helpful GATA6 discussions and assistance with figure generation. GATA6 knock down and overexpression plasmids were generously provided by Dr. Stephen Duncan (Medical University of South Carolina). Lentiviruses were produced by the Viral Vector Core at the Blood Research Institute/MCW. This project was supported by grants from the National Institutes of Health R21NS102911 (ADE), 1R01NS119594 (BGB), and CureSMA (ADE).
Allison, R. L. , Welby, E. , Khayrullina, G. , Burnett, B. G. , & Ebert, A. D. (2022). Viral mediated knockdown of GATA6 in SMA iPSC‐derived astrocytes prevents motor neuron loss and microglial activation. Glia, 70(5), 989–1004. 10.1002/glia.24153
Funding information CureSMA; National Institute of Neurological Disorders and Stroke, Grant/Award Numbers: R01NS119594, R21NS102911
DATA AVAILABILITY STATEMENT
Data Accessibility The data supporting the findings of this study are not applicable for depositing into a public database; however, raw data are available from the corresponding author upon reasonable request.
REFERENCES
- Andersen, J. , Revah, O. , Miura, Y. , Thom, N. , Amin, N. D. , Kelley, K. W. , Singh, M. , Chen, X. , Thete, M. V. , Walczak, E. M. , Vogel, H. , Fan, H. C. , & Paşca, S. P. (2020). Generation of functional human 3D Cortico‐motor Assembloids. Cell, 183(7), 1913–1929.e1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle, P. A. , & Baltimore, D. (1988). I kappa B: A specific inhibitor of the NF‐kappa B transcription factor. Science, 242(4878), 540–546. [DOI] [PubMed] [Google Scholar]
- Basu, A. , Krady, J. K. , O'Malley, M. , Styren, S. D. , DeKosky, S. T. , & Levison, S. W. (2002). The type 1 interleukin‐1 receptor is essential for the efficient activation of microglia and the induction of multiple proinflammatory mediators in response to brain injury. The Journal of Neuroscience, 22(14), 6071–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk, M. Y. , Martirosyan, A. , Wahis, J. , de Vin, F. , Marneffe, C. , Kusserow, C. , Koeppen, J. , Viana, J. F. , Oliveira, J. F. , Voet, T. , Ponting, C. P. , Belgard, T. G. , & Holt, M. G. (2020). Identification of region‐specific astrocyte subtypes at single cell resolution. Nature Communications, 11(1), 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, E. , Hypolite, J. A. , Zderic, S. A. , Gomes, C. M. , Malkowicz, B. , Liou, H. C. , Wein, A. J. , & Chacko, S. (2013). GATA‐6 and NF‐κB activate CPI‐17 gene transcription and regulate Ca2+ sensitization of smooth muscle contraction. Molecular and Cellular Biology, 33(5), 1085–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G. C. , & Neher, J. J. (2012). Eaten alive! Cell death by primary phagocytosis: 'phagoptosis'. Trends in Biochemical Sciences, 37(8), 325–332. [DOI] [PubMed] [Google Scholar]
- Carpanini, S. M. , Torvell, M. , & Morgan, B. P. (2019). Therapeutic inhibition of the complement system in diseases of the central nervous system. Frontiers in Immunology, 10, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveró, C. , Blasco, A. , Tarabal, O. , Casanovas, A. , Piedrafita, L. , Navarro, X. , Esquerda, J. E. , & Calderó, J. (2018). Glial activation and central synapse loss, but not Motoneuron degeneration, are prevented by the Sigma‐1 receptor agonist PRE‐084 in the Smn2B/− mouse model of spinal muscular atrophy. Journal of Neuropathology and Experimental Neurology, 77(7), 577–597. [DOI] [PubMed] [Google Scholar]
- Chaytow, H. , Huang, Y. T. , Gillingwater, T. H. , & Faller, K. M. E. (2018). The role of survival motor neuron protein (SMN) in protein homeostasis. Cellular and Molecular Life Sciences, 75(21), 3877–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Qian, K. , Du, Z. , Cao, J. , Petersen, A. , Liu, H. , Blackbourn, L. W. , Huang, C. L. , Errigo, A. , Yin, Y. , Lu, J. , Ayala, M. , & Zhang, S. C. (2014). Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell, 14(6), 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. H. (2020). New and developing therapies in spinal muscular atrophy: From genotype to phenotype to treatment and where do we stand? International Journal of Molecular Sciences, 21(9), 3297. 10.3390/ijms21093297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, F. , Smith, E. L. , & Carmody, R. J. (2016). The regulation of NF‐κB subunits by phosphorylation. Cell, 5(1), 12. 10.3390/cells5010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, W. S. , Allen, N. J. , & Eroglu, C. (2015). Astrocytes control synapse formation, function, and elimination. Cold Spring Harbor Perspectives in Biology, 7(9), a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, T. O. , & Pardo, C. A. (1996). The neurobiology of childhood spinal muscular atrophy. Neurobiology of Disease, 3(2), 97–110. [DOI] [PubMed] [Google Scholar]
- D'Amico, A. , Mercuri, E. , Tiziano, F. D. , & Bertini, E. (2011). Spinal muscular atrophy. Orphanet Journal of Rare Diseases, 6, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, B. M. , Salinas‐Navarro, M. , Cordeiro, M. F. , Moons, L. , & De Groef, L. (2017). Characterizing microglia activation: A spatial statistics approach to maximize information extraction. Scientific Reports, 7(1), 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda, J. , Yaddanapudi, K. , Hornig, M. , & Lipkin, W. I. (2009). Astrocytes recognize intracellular polyinosinic‐polycytidylic acid via MDA‐5. The FASEB Journal, 23(4), 1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rus Jacquet, A. (2019). Preparation and co‐culture of iPSC‐derived dopaminergic neurons and astrocytes. Current Protocols in Cell Biology, 85(1), e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeniconi, M. , Hempstead, B. L. , & Chao, M. V. (2007). Pro‐NGF secreted by astrocytes promotes motor neuron cell death. Molecular and Cellular Neurosciences, 34(2), 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontainhas, A. M. , Wang, M. , Liang, K. J. , Chen, S. , Mettu, P. , Damani, M. , Fariss, R. N. , Li, W. , & Wong, W. T. (2011). Microglial morphology and dynamic behavior is regulated by ionotropic glutamatergic and GABAergic neurotransmission. PLoS One, 6(1), e15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes, A. E. , Ferraiuolo, L. , Haidet‐Phillips, A. M. , Schmelzer, L. , Braun, L. , Miranda, C. J. , Ladner, K. J. , Bevan, A. K. , Foust, K. D. , Godbout, J. P. , Popovich, P. G. , Guttridge, D. C. , & Kaspar, B. K. (2014). Microglia induce motor neuron death via the classical NF‐κB pathway in amyotrophic lateral sclerosis. Neuron, 81(5), 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco‐Bocanegra, D. K. , McAuley, C. , Nicoll, J. A. R. , & Boche, D. (2019). Molecular mechanisms of microglial motility: Changes in ageing and Alzheimer's disease. Cell, 8(6), 15955. 10.1038/s41598-021-95535-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, H. , Clarke, B. E. , & Patani, R. (2021). Astrocytes and microglia in neurodegenerative diseases: Lessons from human in vitro models. Progress in Neurobiology, 200, 101973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker, M. , Neher, J. J. , Zhao, J. W. , Théry, C. , Tolkovsky, A. M. , & Brown, G. C. (2012). MFG‐E8 mediates primary phagocytosis of viable neurons during neuroinflammation. The Journal of Neuroscience, 32(8), 2657–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, R. , Shen, Q. , Xu, P. , Luo, J. J. , & Tang, Y. (2014). Phagocytosis of microglia in the central nervous system diseases. Molecular Neurobiology, 49(3), 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti, R. G. , Quinlan, K. A. , Barlow, C. B. , Heier, C. R. , Heckman, C. J. , & Didonato, C. J. (2012). Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory‐motor defects are a consequence, not a cause, of motor neuron dysfunction. The Journal of Neuroscience, 32(11), 3818–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler, W. , Sansom, S. N. , Buchrieser, J. , Newey, S. E. , Moore, C. S. , Nicholls, F. J. , Chintawar, S. , Schnell, C. , Antel, J. P. , Allen, N. D. , Cader, M. Z. , Wade‐Martins, R. , James, W. S. , & Cowley, S. A. (2017). A highly efficient human pluripotent stem cell microglia model displays a neuronal‐co‐culture‐specific expression profile and inflammatory response. Stem Cell Reports, 8(6), 1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, G. M. , & Stevens, C. H. (2011). Glia: Initiators and progressors of pathology in Parkinson's disease. Movement Disorders, 26(1), 6–17. [DOI] [PubMed] [Google Scholar]
- Harder, J. M. , Williams, P. A. , Braine, C. E. , Yang, H. S. , Thomas, J. M. , Foxworth, N. E. , John, S. W. M. , & Howell, G. R. (2020). Complement peptide C3a receptor 1 promotes optic nerve degeneration in DBA/2J mice. Journal of Neuroinflammation, 17(1), 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich, B. , El Idrissi, N. B. , Donev, R. M. , Petri, S. , Claus, P. , Neal, J. , Morgan, B. P. , & Ramaglia, V. (2011). Complement upregulation and activation on motor neurons and neuromuscular junction in the SOD1 G93A mouse model of familial amyotrophic lateral sclerosis. Journal of Neuroimmunology, 235(1–2), 104–109. [DOI] [PubMed] [Google Scholar]
- Hinderer, C. , Katz, N. , Buza, E. L. , Dyer, C. , Goode, T. , Bell, P. , Richman, L. K. , & Wilson, J. M. (2018). Severe toxicity in nonhuman primates and piglets following high‐dose intravenous Administration of an Adeno‐Associated Virus Vector Expressing Human SMN. Human Gene Therapy, 29(3), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, E. C. , & Hunot, S. (2009). Neuroinflammation in Parkinson's disease: A target for neuroprotection? Lancet Neurology, 8(4), 382–397. [DOI] [PubMed] [Google Scholar]
- Hochstim, C. , Deneen, B. , Lukaszewicz, A. , Zhou, Q. , & Anderson, D. J. (2008). Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell, 133(3), 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S. , Beja‐Glasser, V. F. , Nfonoyim, B. M. , Frouin, A. , Li, S. , Ramakrishnan, S. , Merry, K. M. , Shi, Q. , Rosenthal, A. , Barres, B. A. , Lemere, C. A. , Selkoe, D. J. , & Stevens, B. (2016). Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science, 352(6286), 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Y. , Sahashi, K. , Rigo, F. , Hung, G. , Horev, G. , Bennett, C. F. , & Krainer, A. R. (2011). Peripheral SMN restoration is essential for long‐term rescue of a severe spinal muscular atrophy mouse model. Nature, 478(7367), 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashon, M. L. , Ross, G. W. , O'Callaghan, J. P. , Miller, D. B. , Petrovitch, H. , Burchfiel, C. M. , Sharp, D. S. , Markesbery, W. R. , Davis, D. G. , Hardman, J. , Nelson, J. , & White, L. R. (2004). Associations of cortical astrogliosis with cognitive performance and dementia status. Journal of Alzheimer's Disease, 6(6), 595–604. [DOI] [PubMed] [Google Scholar]
- Kerstetter, A. E. , & Miller, R. H. (2012). Isolation and culture of spinal cord astrocytes. Methods in Molecular Biology, 814, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klawonn, A. M. , Fritz, M. , Castany, S. , Pignatelli, M. , Canal, C. , Similä, F. , Tejeda, H. A. , Levinsson, J. , Jaarola, M. , Jakobsson, J. , Hidalgo, J. , Heilig, M. , Bonci, A. , & Engblom, D. (2021). Microglial activation elicits a negative affective state through prostaglandin‐mediated modulation of striatal neurons. Immunity, 54(2), 225–234.e226. [DOI] [PubMed] [Google Scholar]
- Kondo, M. , Murakawa, Y. , Harashima, N. , Kobayashi, S. , Yamaguchi, S. , & Harada, M. (2009). Roles of proinflammatory cytokines and the Fas/Fas ligand interaction in the pathogenesis of inflammatory myopathies. Immunology, 128(1 Suppl), e589–e599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. , Godbole, N. M. , Chaturvedi, C. P. , Singh, R. S. , George, N. , Upadhyay, A. , Anjum, B. , Godbole, M. M. , & Sinha, R. A. (2018). Mechanisms involved in epigenetic down‐regulation of Gfap under maternal hypothyroidism. Biochemical and Biophysical Research Communications, 502(3), 375–381. [DOI] [PubMed] [Google Scholar]
- Lee, A. J. , Awano, T. , Park, G. H. , & Monani, U. R. (2012). Limited phenotypic effects of selectively augmenting the SMN protein in the neurons of a mouse model of severe spinal muscular atrophy. PLoS One, 7(9), e46353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, S. , Burglen, L. , Reboullet, S. , Clermont, O. , Burlet, P. , Viollet, L. , Benichou, B. , Cruaud, C. , Millasseau, P. , & Zeviani, M. (1995). Identification and characterization of a spinal muscular atrophy‐determining gene. Cell, 80(1), 155–165. [DOI] [PubMed] [Google Scholar]
- Lenz, K. M. , & Nelson, L. H. (2018). Microglia and beyond: Innate immune cells as regulators of brain development and behavioral function. Frontiers in Immunology, 9, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow, S. A. , Guttenplan, K. A. , Clarke, L. E. , Bennett, F. C. , Bohlen, C. J. , Schirmer, L. , Bennett, M. L. , Munch, A. E. , Chung, W. S. , Peterson, T. C. , Wilton, D. K. , Frouin, A. , Napier, B. A. , Panicker, N. , Kumar, M. , Buckwalter, M. S. , Rowitch, D. H. , Dawson, V. L. , Dawson, T. M. , … Barres, B. A. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541(7638), 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, K. K. , Lin, M. Y. , Zingg, B. , Feng, Z. , & Ko, C. P. (2010). Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One, 5(11), e15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinchuk, A. , Wan, Y. W. , Swartzlander, D. B. , Chen, F. , Cole, A. , Propson, N. E. , Wang, Q. , Zhang, B. , Liu, Z. , & Zheng, H. (2018). Complement C3aR inactivation attenuates tau pathology and reverses an immune network deregulated in Tauopathy models and Alzheimer's disease. Neuron, 100(6), 1337–1353.e1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, S. , Arzua, T. , Canfield, S. G. , Seminary, E. R. , Sison, S. L. , Ebert, A. D. , & Bai, X. (2019). Studying human neurological disorders using induced pluripotent stem cells: From 2D monolayer to 3D Organoid and blood brain barrier models. Comprehensive Physiology, 9(2), 565–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson, C. L. , Hahnen, E. , Androphy, E. J. , & Wirth, B. (1999). A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proceedings of the National Academy of Sciences of the United States of America, 96(11), 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson, C. L. , Strasswimmer, J. , Yao, J. M. , Baleja, J. D. , Hahnen, E. , Wirth, B. , Le, T. , Burghes, A. H. , & Androphy, E. J. (1998). SMN oligomerization defect correlates with spinal muscular atrophy severity. Nature Genetics, 19(1), 63–66. [DOI] [PubMed] [Google Scholar]
- Martin, J. E. , Nguyen, T. T. , Grunseich, C. , Nofziger, J. H. , Lee, P. R. , Fields, D. , Fischbeck, K. H. , & Foran, E. (2017). Decreased motor neuron support by SMA astrocytes due to diminished MCP1 secretion. The Journal of Neuroscience, 37(21), 5309–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, T. L. , Kong, L. , Wang, X. , Osborne, M. A. , Crowder, M. E. , Van Meerbeke, J. P. , Xu, X. , Davis, C. , Wooley, J. , Goldhamer, D. J. , Lutz, C. M. , Rich, M. M. , & Sumner, C. J. (2012). Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. The Journal of Neuroscience, 32(25), 8703–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matejuk, A. , & Ransohoff, R. M. (2020). Crosstalk between astrocytes and microglia: An overview. Frontiers in Immunology, 11, 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury, Y. , Come, J. , Piskorowski, R. A. , Salah‐Mohellibi, N. , Chevaleyre, V. , Peschanski, M. , Martinat, C. , & Nedelec, S. (2015). Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nature Biotechnology, 33(1), 89–96. [DOI] [PubMed] [Google Scholar]
- McGivern, J. V. , Patitucci, T. N. , Nord, J. A. , Barabas, M. A. , Stucky, C. L. , & Ebert, A. D. (2013). Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia, 61(9), 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade, A. , Coburn, M. , Tu, C. H. , Hasselmann, J. , Davtyan, H. , & Blurton‐Jones, M. (2018). Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Molecular Neurodegeneration, 13(1), 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejzini, R. , Flynn, L. L. , Pitout, I. L. , Fletcher, S. , Wilton, S. D. , & Akkari, P. A. (2019). ALS genetics, mechanisms, and therapeutics: Where are we now? Frontiers in Neuroscience, 13, 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, L. , Men, Y. , Chiang, M. S. R. , Tian, Y. , Jin, S. , Yelick, J. , Higashimori, H. , & Yang, Y. (2019). Intracortical astrocyte subpopulations defined by astrocyte reporter mice in the adult brain. Glia, 67(1), 171–181. [DOI] [PubMed] [Google Scholar]
- Murray, L. M. , Beauvais, A. , Gibeault, S. , Courtney, N. L. , & Kothary, R. (2015). Transcriptional profiling of differentially vulnerable motor neurons at pre‐symptomatic stage in the Smn (2b/−) mouse model of spinal muscular atrophy. Acta Neuropathologica Communications, 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher, J. J. , Neniskyte, U. , Zhao, J. W. , Bal‐Price, A. , Tolkovsky, A. M. , & Brown, G. C. (2011). Inhibition of microglial phagocytosis is sufficient to prevent inflammatory neuronal death. Journal of Immunology, 186(8), 4973–4983. [DOI] [PubMed] [Google Scholar]
- Neniskyte, U. , Neher, J. J. , & Brown, G. C. (2011). Neuronal death induced by nanomolar amyloid β is mediated by primary phagocytosis of neurons by microglia. The Journal of Biological Chemistry, 286(46), 39904–39913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, M. , & Naumann, M. (2007). Beyond IkappaBs: Alternative regulation of NF‐kappaB activity. The FASEB Journal, 21(11), 2642–2654. [DOI] [PubMed] [Google Scholar]
- Oskoui, M. , & Kaufmann, P. (2008). Spinal muscular atrophy. Neurotherapeutics, 5(4), 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, G. H. , Maeno‐Hikichi, Y. , Awano, T. , Landmesser, L. T. , & Monani, U. R. (2010). Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. The Journal of Neuroscience, 30(36), 12005–12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. S. , Kam, T. I. , Lee, S. , Park, H. , Oh, Y. , Kwon, S. H. , Song, J. J. , Kim, D. , Kim, H. , Jhaldiyal, A. , Na, D. H. , Lee, K. C. , Park, E. J. , Pomper, M. G. , Pletnikova, O. , Troncoso, J. C. , Ko, H. S. , Dawson, V. L. , & Dawson, T. M. (2021). Blocking microglial activation of reactive astrocytes is neuroprotective in models of Alzheimer's disease. Acta Neuropathologica Communications, 9(1), 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patitucci, T. N. , & Ebert, A. D. (2016). SMN deficiency does not induce oxidative stress in SMA iPSC‐derived astrocytes or motor neurons. Human Molecular Genetics, 25(3), 514–523. [DOI] [PubMed] [Google Scholar]
- Pattali, R. , Mou, Y. , & Li, X. J. (2019). AAV9 vector: A novel modality in gene therapy for spinal muscular atrophy. Gene Therapy, 26(7–8), 287–295. [DOI] [PubMed] [Google Scholar]
- Perriot, S. , Mathias, A. , Perriard, G. , Canales, M. , Jonkmans, N. , Merienne, N. , Meunier, C. , El Kassar, L. , Perrier, A. L. , Laplaud, D. A. , Schluep, M. , Déglon, N. , & Du Pasquier, R. (2018). Human induced pluripotent stem cell‐derived astrocytes are differentially activated by multiple sclerosis‐associated cytokines. Stem Cell Reports, 11(5), 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, S. L. , Nguyen, H. X. , Mendez, O. A. , & Anderson, A. J. (2017). Complement protein C3 suppresses axon growth and promotes neuron loss. Scientific Reports, 7(1), 12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial, D. , Lemos, C. , Pinheiro, H. , Duarte, J. M. , Gonçalves, F. Q. , Real, J. I. , Prediger, R. D. , Gonçalves, N. , Gomes, C. A. , Canas, P. M. , Agostinho, P. , & Cunha, R. A. (2015). Depression as a glial‐based synaptic dysfunction. Frontiers in Cellular Neuroscience, 9, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin, D. , Reis, E. S. , Mastellos, D. C. , Gros, P. , & Lambris, J. D. (2016). Complement component C3 ‐ the "Swiss Army knife" of innate immunity and host defense. Immunological Reviews, 274(1), 33–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindt, H. , Feng, Z. , Mazzasette, C. , Glascock, J. J. , Valdivia, D. , Pyles, N. , Crawford, T. O. , Swoboda, K. J. , Patitucci, T. N. , Ebert, A. D. , Sumner, C. J. , Ko, C. P. , & Lorson, C. L. (2015). Astrocytes influence the severity of spinal muscular atrophy. Human Molecular Genetics, 24(14), 4094–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll, W. , Jablonka, S. , Andreassi, C. , Kroning, A. K. , Karle, K. , Monani, U. R. , & Sendtner, M. (2003). Smn, the spinal muscular atrophy‐determining gene product, modulates axon growth and localization of beta‐Actin mRNA in growth cones of motoneurons. The Journal of Cell Biology, 163(4), 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon, L. , Lamas, N. J. , Garcia, A. D. , Yang, E. J. , Sattler, R. , Lewis, V. J. , Kim, Y. A. , Kachel, C. A. , Rothstein, J. D. , Przedborski, S. , Wichterle, H. , & Henderson, C. E. (2013). Human stem cell‐derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Reports, 4(5), 1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen, D. , Ebert, A. D. , Heins, B. M. , McGivern, J. V. , Ornelas, L. , & Svendsen, C. N. (2012). Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS One, 7(6), e39113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, J. C. , St‐Pierre, M. K. , Carrier, M. , El Hajj, H. , Novak, S. W. , Sanchez, M. G. , Cicchetti, F. , & Tremblay, M. (2020). Microglial physiological properties and interactions with synapses are altered at presymptomatic stages in a mouse model of Huntington's disease pathology. Journal of Neuroinflammation, 17(1), 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, D. P. , Lehrman, E. K. , & Stevens, B. (2013). The "quad‐partite" synapse: Microglia‐synapse interactions in the developing and mature CNS. Glia, 61(1), 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoryk‐Wegrzynowicz, M. , Wegrzynowicz, M. , Lee, E. , Bowman, A. B. , & Aschner, M. (2011). Role of astrocytes in brain function and disease. Toxicologic Pathology, 39(1), 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa, R. , Fusco, R. , & Cuzzocrea, S. (2019). Astrocytes: Role and functions in brain pathologies. Frontiers in Pharmacology, 10, 1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison, S. L. , Patitucci, T. N. , Seminary, E. R. , Villalon, E. , Lorson, C. L. , & Ebert, A. D. (2017). Astrocyte‐produced miR‐146a as a mediator of motor neuron loss in spinal muscular atrophy. Human Molecular Genetics, 26(17), 3409–3420. [DOI] [PubMed] [Google Scholar]
- Sleigh, J. N. , Gillingwater, T. H. , & Talbot, K. (2011). The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Disease Models & Mechanisms, 4(4), 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew, M. V. (2000). Astrocyte failure as a cause of CNS dysfunction. Molecular Psychiatry, 5(3), 230–232. [DOI] [PubMed] [Google Scholar]
- Soubannier, V. , Maussion, G. , Chaineau, M. , Sigutova, V. , Rouleau, G. , Durcan, T. M. , & Stifani, S. (2020). Characterization of human iPSC‐derived astrocytes with potential for disease modeling and drug discovery. Neuroscience Letters, 731, 135028. [DOI] [PubMed] [Google Scholar]
- Stogsdill, J. A. , & Eroglu, C. (2017). The interplay between neurons and glia in synapse development and plasticity. Current Opinion in Neurobiology, 42, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabal, O. , Caraballo‐Miralles, V. , Cardona‐Rossinyol, A. , Correa, F. J. , Olmos, G. , Llado, J. , Esquerda, J. E. , & Caldero, J. (2014). Mechanisms involved in spinal cord central synapse loss in a mouse model of spinal muscular atrophy. Journal of Neuropathology and Experimental Neurology, 73(6), 519–535. [DOI] [PubMed] [Google Scholar]
- Tcw, J. , Wang, M. , Pimenova, A. A. , Bowles, K. R. , Hartley, B. J. , Lacin, E. , Machlovi, S. I. , Abdelaal, R. , Karch, C. M. , Phatnani, H. , Slesinger, P. A. , Zhang, B. , Goate, A. M. , & Brennand, K. J. (2017). An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports, 9(2), 600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H. H. , Li, H. , Fuentealba, L. C. , Molofsky, A. V. , Taveira‐Marques, R. , Zhuang, H. , Tenney, A. , Murnen, A. T. , Fancy, S. P. , Merkle, F. , Kessaris, N. , Alvarez‐Buylla, A. , Richardson, W. D. , & Rowitch, D. H. (2012). Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science, 337(6092), 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alstyne, M. , Tattoli, I. , Delestree, N. , Recinos, Y. , Workman, E. , Shihabuddin, L. S. , Zhang, C. , Mentis, G. Z. , & Pellizzoni, L. (2021). Gain of toxic function by long‐term AAV9‐mediated SMN overexpression in the sensorimotor circuit. Nature Neuroscience, 24(7), 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojicic, A. , Delestrée, N. , Fletcher, E. V. , Pagiazitis, J. G. , Sankaranarayanan, S. , Yednock, T. A. , Barres, B. A. , & Mentis, G. Z. (2019). The classical complement pathway mediates microglia‐dependent remodeling of spinal motor circuits during development and in SMA. Cell Reports, 29(10), 3087–3100.e3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welby, E. , Rehborg, R. J. , Harmelink, M. , & Ebert, A. D. (2021). Assessment of cerebral spinal fluid biomarkers and microRNA‐mediated disease mechanisms in spinal muscular atrophy patient samples. Human Molecular Genetics. (in press). 10.1093/hmg/ddab365 [DOI] [PubMed] [Google Scholar]
- Werneburg, S. , Jung, J. , Kunjamma, R. B. , Ha, S. K. , Luciano, N. J. , Willis, C. M. , Gao, G. , Biscola, N. P. , Havton, L. A. , Crocker, S. J. , Popko, B. , Reich, D. S. , & Schafer, D. P. (2020). Targeted complement inhibition at synapses prevents microglial synaptic engulfment and synapse loss in demyelinating disease. Immunity, 52(1), 167–182 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, T. M. , Costantini, K. J. , Taylor, S. M. , & Noakes, P. G. (2008). Role of complement in motor neuron disease: Animal models and therapeutic potential of complement inhibitors. Advances in Experimental Medicine and Biology, 632, 143–158. [PubMed] [Google Scholar]
- Wu, X. , Chen, P. S. , Dallas, S. , Wilson, B. , Block, M. L. , Wang, C. C. , Kinyamu, H. , Lu, N. , Gao, X. , Leng, Y. , Chuang, D. M. , Zhang, W. , Lu, R. B. , & Hong, J. S. (2008). Histone deacetylase inhibitors up‐regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum, 11(8), 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. W. , Chen, C. L. , Chou, W. C. , Lin, H. C. , Jong, Y. J. , Tsai, L. K. , & Chuang, C. Y. (2016). An integrative Transcriptomic analysis for identifying novel target genes corresponding to severity Spectrum in spinal muscular atrophy. PLoS One, 11(6), e0157426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanuck, S. F. (2019). Microglial phagocytosis of neurons: Diminishing neuronal loss in traumatic, infectious, inflammatory, and autoimmune CNS disorders. Frontiers in Psychiatry, 10, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, S. P. , Kam, T. I. , Panicker, N. , Kim, S. , Oh, Y. , Park, J. S. , Kwon, S. H. , Park, Y. J. , Karuppagounder, S. S. , Park, H. , Oh, N. , Kim, N. A. , Lee, S. , Brahmachari, S. , Mao, X. , Lee, J. H. , Kumar, M. , An, D. , Kang, S. U. , … Ko, H. S. (2018). Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson's disease. Nature Medicine, 24(7), 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. Y. , Pan, J. , Mamtilahun, M. , Zhu, Y. , Wang, L. , Venkatesh, A. , Shi, R. , Tu, X. , Jin, K. , Wang, Y. , Zhang, Z. , & Yang, G. Y. (2020). Microglia exacerbate white matter injury via complement C3/C3aR pathway after hypoperfusion. Theranostics, 10(1), 74–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
Data Accessibility The data supporting the findings of this study are not applicable for depositing into a public database; however, raw data are available from the corresponding author upon reasonable request.


