Abstract
Genome assembly has been benefited from long-read sequencing technologies with higher accuracy and higher continuity. However, most human genome assembly require large amount of DNAs from homogeneous cell lines without keeping cell heterogeneities, since cell heterogeneity could profoundly affect haplotype assembly results. Herein, using single-cell genome long-read sequencing technology (SMOOTH-seq), we have sequenced K562 and HG002 cells on PacBio HiFi and Oxford Nanopore Technologies (ONT) platforms and conducted de novo genome assembly. For the first time, we have completed the human genome assembly with high continuity (with NG50 of ∼2 Mb using 95 individual K562 cells) at single-cell levels, and explored the impact of different assemblers and sequencing strategies on genome assembly. With sequencing data from 30 diploid individual HG002 cells of relatively high genome coverage (average coverage ∼41.7%) on ONT platform, the NG50 can reach over 1.3 Mb. Furthermore, with the assembled genome from K562 single-cell dataset, more complete and accurate set of insertion events and complex structural variations could be identified. This study opened a new chapter on the practice of single-cell genome de novo assembly.
INTRODUCTION
With the increase in single-base accuracy and read length, single molecule long-read sequencing technologies using bulk samples have been widely used in genome assembly (1–6). Because of the advantages (100- to 1000-fold increases) of read length over next generation sequencing (NGS) platforms, long-read sequencing (third generation sequencing, TGS) can better assemble complex genome regions that contain repetitive sequences and chromosomal rearrangements (2), which allows identification of genetic variations and connects them to potential phenotypes. Recently, genomes of many species have been assembled mainly using TGS platform data, such as Vertebrate Genomes Project (VGP), aiming to complete reference genomes for all of the ∼70 000 extant vertebrate species. In 2021, this project assembled 16 species that represent six major vertebrate lineages (7). For the human genome, Telomere-to-Telomere (T2T) consortium has completed and released the first gapless human reference genome using homozygous cell lines (CHM13) (6). Recently, Human Pangenome Reference Consortium (HPRC) generated the first high-quality diploid reference assembly (HG002) (8). In addition, the genomes of many different plants have been assembled with high quality (5,9,10).
Usually, these long-read sequencing assembly require large amounts of DNA (typically several micrograms from millions of cells), and therefore most human genome assembly have been restricted to bulk genome sequencing datasets without keeping the potential genetic heterogeneities among individual cells. However, this is impractical for many situations. In a real assembly application, we need to overcome at least two basic challenges. The first is about cell heterogeneity. Bulk data assembly is based on the premise that all cells in a bulk sample carry the same genome; otherwise it would be difficult to discriminate variations between different genetic clones and variations between different haplotypes within a cell. In fact, somatic copy number alterations (CNAs) could be detected in a number of normal tissue samples (11,12), and the CNAs exhibited strong organ preferences (11). At the same time, in the course of an individual's lifetime, normal human cells accumulate mutations (13–15), and thus normal cell populations can consist of myriad small clones, which contain different mutations (14). In cancer tissue, genetic heterogeneity is even more pronounced (16). In the practical application of assembly, the differences of genomes between cells will greatly affect the accuracy of final assembly. Haplotype assembly results are meaningful only if genetic heterogeneities among different cells in a sample are first addressed.
The second challenge is that quite often we can only get small amount of genomic DNAs for sequencing analysis. Under many situations, to obtain large amounts (several micrograms) of genomic DNA is impractical. For example, in early embryonic development studies (17) and forensic testing, especially in cancer genome research, such as circulating tumor cells (a few CTCs in 1 ml of peripheral blood) (18), tumor biopsy samples(∼ 200 ng DNA for each lung tumor biopsy sample) (19), tumor cells in cerebrospinal fluid (CSF) (about hundreds of breast cancer cells per ml) (20) and tumor cells in ascites (about thousands of ovarian cancer cells per ml) (21). These cells are difficult to culture and amplify in vitro, and even when they can be cultured, there is no guarantee that their genome structure during in vitro culture will remain the same as in vivo.
Single-cell whole-genome sequencing (scWGS) is a powerful tool to reveal cell to cell genetic heterogeneities, especially for cancer research. Identified genomic changes such as CNAs (16), somatic gene mutations (22,23), mitochondrial mutations (24) and other genetic alterations can help to identify cell subclones and their potential contributions to phenotypic changes. Genomic assembly, or haplotype assembly, can be more accurate by using clonal populations that are more genetically similar.
Single-cell NGS genome sequencing technologies are commonly used in microbial genome assembly (25,26). In fact, many bacteria in various environments cannot be cultured in the laboratory, single-cell genome sequencing can reveal genomic and physiological insights into novel organisms that cannot be readily grown in culture, and single-cell genome sequencing can resolve the genetic linkage of sequences within a discrete organism and so can be used in combination with metagenomics approach to complete genome assemblies (27,28). However, NGS platform-based single cell genome sequencing technologies are rarely used in large and complex genome assembly, and even using bulk NGS genome sequencing data the assembly continuity of the contig N50 cannot achieve the megabase level, such as de novo assembly of human genome using bulk Illumina sequencing data alone with SOAPdenovo (29), yielding a contig N50 of 11.1 kb (30); de novo genome assembly for the American pika with contig N50 of ∼42 kb (using the Illumina HiSeq X (PE150bp) platform) (31); de novo genome assembly for the American bison with contig N50 of ∼20 kb (using the Illumina and 454 paired-end libraries) (32). Assembling the human genome using a small amount of genomic DNAs or even a single cell genome sequencing data is much more challenging. It requires not only the support of single-cell TGS platform-based genome sequencing technology, but also a good data analysis strategy and a suitable assembler.
Recently, we developed SMOOTH-seq technology (33), which can sequence the genome of a single-cell on the third-generation sequencing platform. SMOOTH-seq can reliably and effectively detects SVs and ecDNAs in individual human cells, which made it possible to sequence a single cell genome with long reads (around 6kb), providing the prerequisite for human genome assembly from just several individual cells.
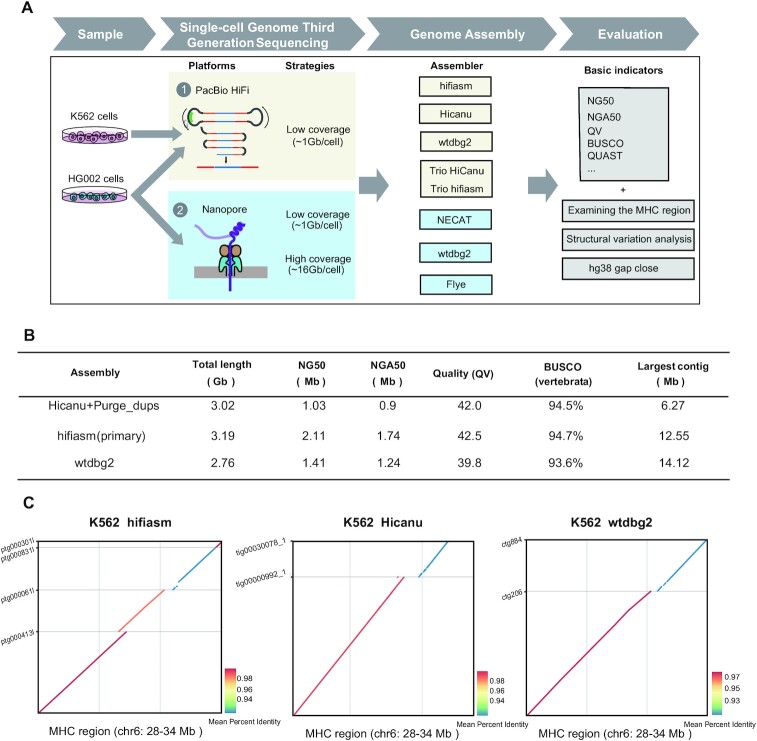
Here we have employed SMOOTH-seq on PacBio HiFi and Oxford Nanopore Technologies (ONT) platforms to sequence K562 (a human chronic myelogenous leukemia (CML) cell line) and HG002 (a normal diploid lymphoblast cell line) and demonstrate the feasibility of genome assembly based on scWGS dataset with different assemblers and rigorous evaluations (Figure 1A). We have systematically explored the factors that affecting the assembly with TGS platform-based single-cell genome sequencing data. Furthermore, to investigate the lower limit of numbers of single cells need to be sequenced for genome assembly, we improved the SMOOTH-seq technology (see methods), and sequenced 30 diploid HG002 cells with relatively high genome coverage on ONT platform, and found that the genome assembly from as low as 30 individual cells (average genome coverage ∼41.7%) can achieve NG50 of ∼1.35 Mb. In addition, through analyzing the structural variations (SVs) of the assembled genome of K562 cells, we found that compared with directly mapping single cell genome sequencing data onto reference genome, many more insertion events could be identified and complex structural variations could be more efficiently and accurately illustrated. Our research gave proof-of-principle evidences to show that it is feasible to assembly human genome with megabase level of NG50 contigs from long-reads single cell genome sequencing data of just a few dozens of individual cells.
Figure 1.
The assembly workflow and K562 assembly metrics. (A) The workflow illustrates the samples, sequencing platforms, assemblers and evaluation indicators we used to demonstrate the feasibility of genome assembly based on scWGS dataset. (B) K562 assembly (95 cells) benchmarking results for Pacbio HiFi data (primary contigs). N50 is the sequence length of the shortest contig at half of the total assembly size; NG50 is the sequence length of the shortest contig at half of the reference genome size; NGA50 is the sequence length of the shortest aligned block at half of the reference genome size; BUSCO is a tool that assess the completeness of benchmarking universal single-copy orthologs present in an assembly; Per-base consensus quality values (QV) represents a log-scaled probability of errors for assembly, higher QVs indicate more accurate consensus. (C) K562 cells MHC assemblies were compared with the reference human genome (hg38). Only contigs longer than 500kb are displayed.
MATERIALS AND METHODS
Culture of HG002
NA24385 cells (HG002) were purchased from the Coriell Institute (https://www.coriell.org/). Cells were cultured in RPMI1640 (Gibco, cat#11875093) with 15% fetal bovine serum (FBS, Gibco, cat# 26140079) at 37°C with 5% CO2.
Single-cell preparation
For FACS sorting, 7-AAD viability staining solution (BioLegend, cat#420404) were used in single cell suspension and then single cells were sorted into 96-well plates by FACS, each well contained 2.5 μl cell lysis buffer (0.25 μl 100 mM Tris–EDTA, 0.125 μl 20 mg/μl Qiagen protease, 0.075 μl 10% Triton X-100, 0.05 μl 1 M KCl and 2 μl H2O). And then the reaction was carried out at 50°C for 3 h to digest the proteins binding on the gDNAs and then 70°C for 30 min to inactivate the protease.
SMOOTH-seq
The description of SMOOTH-seq
As described in our previous work (33), SMOOTH-seq (single-molecule real-time sequencing of long fragments amplified through transposon insertion) was the single-cell genome sequencing method based on TGS platform, which could reliably detect structural variations (SVs), extrachromosomal circular DNAs (ecDNAs), etc.
In SMOOTH-seq, the long tagmentation fragments could be recovered through adjusting the concentration of Tn5 transposase. Tn5 transposition has been widely applied to construct shotgun fragment libraries for NGS and in these methods Tn5 usually contains two different adaptor sequences which makes it losing 50% of the genomic fragments. Instead, SMOOTH-seq embedded commercialized Tn5 transposase with just one adaptor sequence which could recover all of the original DNA fragments through transposition-PCR. Additionally, SMOOTH-seq was able to amplify long DNA fragments in an individual human cell through optimizing the reaction conditions, including concentration of the adaptor conjunct transposase, transposition reaction buffer and DNA polymerase. In our previous paper we only showed that SMOOTH-seq worked for PacBio sequel II platform.
The principle of SMOOTH-seq
First, genomic DNA from a single cell was randomly fragmented by low-density Tn5 transposon insertion after cell lysis and proteinase digestion. Then, the produced fragments underwent strand displacement and amplification using Tks DNA polymerase which was able to amplify long DNA fragments with PCR primers which contained 16 bp-PacBio-barcodes. Next, the amplified single cell genomic DNAs of different barcodes were pooled together. After library construction, the purified amplicons were around 6 kb long. Finally, the libraries were sequenced on PacBio Sequel II System using CCS mode.
SMOOTH-seq library preparation of single cells for PacBio platform
In this study, we used previous SMOOTH-seq protocol on PacBio sequel II platform to sequence HG002 cells, the detailed experimental steps of SMOOTH-seq protocol was described in method part of our previous published paper (33). In order to sequence HG002 cells on Nanopore platform, we improved the initial SMOOTH-seq protocol through replacing 16 bp-PacBio-barcode with 24bp-Nanopore-barcode for fitting Nanopore platform. The detailed protocol of SMOOTH-seq for Nanopore platform was as below.
SMOOTH-seq amplification of single cells for Nanopore platform
After cell lysis, a 7.5 μl tagmentation mixture including 2 μl 5 × TAPS_PEG8K (50 mM TAPS–NaOH (or KOH), pH 8.3 (RT), 25 mM MgCl2, 40% PEG8K), and 1μl 0.2 ng/μl adaptor conjuncted Tn5 enzyme (Vazyme, Cat#S601-01) were added into each cell lysate. The tagmentation reaction was carried out at 55°C for 10min, followed by adding 2.5 μl 0.2% SDS and standing at room temperature for 5 min to stop tagmentation, releasing the fragmented gDNAs. After tagmentation, strand displacement of the Tn5 adaptors and amplification of the fragmented gDNA was carried out using 1 μl 1.25U/μl Tks Gflex DNA Polymerase (TAKARA, Cat#R060B), 25 μl 2× Gflex PCR Buffer,6.5 μl H2O and 560 nM I5-nano PCR primer which containing 24 bp cell barcode (5′ AATGATACGGCGACCACCGAGATCTNNNNNNNNNNNNNNNNNNNNNNNNTCGTCGGCAGCGTC 3′) (the I5 PCR primers shown in Supplemental Table S10). The PCR program was carried out at 72°C for 3 min, 98°C for 1 min, and then 20-22 cycles of 98°C for 15 s, 60°C for 30 s, and 68°C for 5 min. After that, gDNA amplicons using different barcode primers were pooled together and purified with 0.4 volume of Ampure XP beads (Beckman, Cat#A63882) for twice. These purified amplicons were quantified using Qubit, and about total of 1–2 ug amplicon products were used for further library construction.
To improve the coverage of HG002 cells, we optimized our protocol. First, we designed 24 types of conjuncted Tn5 enzyme with 24 bp barcodes (the Tn5 adapter primers was shown in Supplemental Table S10). After tagmentation, we pooled cells with different barcodes together and then purified them with 0.8 volume of Ampure XP beads and finally eluted with 50 μl H2O. These purified genomic DNAs were then used for amplification and library construction (The detailed of tagmentation and amplification protocol the same as above).
SMOOTH-seq library preparation for Nanopore platform and sequencing
A total amount of 1 μg DNA per sample was used as input material for the DNA library preparations. SQK-LSK109 Kit (Nanopore, UK) was used to construct 1D library. The DNA library was constructed by standard ligation method without DNA fragmentation, after end repaired, added the sequencing adaptor, motor protein and tether protein were connected to prepare the DNA library. After that, each amplicon fragment library was loaded into 1 R9 flow cell and sequenced on PromethION HAC (high accuracy) model.
Culture of K562 cell line and K562 bulk DNA extraction
K562 cells were maintained in RPMI1640 with 10% fetal bovine serum, 1× l-glutamine and 1× Pen/Strep(Gibco, cat#10378016) and were cultured at 37°C with 5% CO2. K562 genomic DNA (gDNA) was extracted using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN,cat# 69504) following the manufacturer's Quick-StartProtocol. Then the extracted K562 gDNA was quantified using the Qubit dsDNA HS Assay Kit (Invitrogen, cat#Q32851).
Validation of structure variations in K562
Insertion and deletion events in K562 that can be detected from de novo genome assembly approach but not by single cell direct mapping approach were selected for PCR validation. PCR primers were designed by Primer designing tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and the PCR amplicons need span the breakpoints and produce PCR products. Tks Gflex DNA Polymerase were used in PCR amplification, and the details of SV validation primers and PCR parameter were shown as Supplemental Table S7. All PCR amplicons were analyzed on 1% agarose gels.
Data pre-processing
For HiFi reads, we used ccs v4.0.0 (https://github.com/PacificBiosciences/ccs) to generate CCS reads with –minPasses 1, and then used lima v1.10.0 (https://github.com/PacificBiosciences/barcoding) to demultiplex and trim barcoded CCS reads for each cell. For ONT reads, we used nanoplexer v0.1 (https://github.com/hanyue36/nanoplexer) and cutadapt v3.4 (https://github.com/marcelm/cutadapt) to demultiplex and trim barcoded ONT reads for each cell. Cleaned HiFi and ONT reads were then used for assembly. To estimate genome coverage, clean ONT reads were then mapped to hg38 human reference genome using minimap2 v2.24 (https://github.com/lh3/minimap2), while HiFi reads used pbmm2 v1.1.0 (https://github.com/PacificBiosciences/pbmm2) for genome alignment.
Genome assembly
For the genomic assembly of single-cell HiFi data, we used hifiasm, Hicanu, and wtdbg2. We then used Purge_dups (34) to identify primary contigs from Hicanu result, and to improve the base-level quality and continuity of assembly, we used wtpoa-cns (35) to polish the initial assemblies of wtdbg2 as it was recommended by wtdbg2. For the genomic assembly of single-cell ONT data, we used NECAT, Flye and wtdbg2. Codes for the above processes can be found at https://github.com/hlxie/sc-assembly.
The assessment of genome assembly quality
To evaluate assembly continuity, QUAST v5.0.2 (https://github.com/ablab/quast) was used to calculate N50, NG50, NGA50 largest contig length and total contig length. N50 is the sequence length of the shortest contig at half of the total assembly size; NG50 is the sequence length of the shortest contig at half of the reference genome size; NGA50 is the sequence length of the shortest aligned block at half of the reference genome size. To assess the completeness of benchmarking universal single-copy orthologs present in an assembly, we used BUSCO v5.3 (https://busco.ezlab.org) with the vertebrata odb10 dataset. Per-base consensus quality values (QV) was estimated by Merqury with Illumina data. It represents a log-scaled probability of error for assembly (based on the shared k-mers between the assembly and the Illumina read set). Higher QVs indicate a more accurate consensus. QUAST diffs reported the number of large mis-assembly (>5 kb) normalized by the assembly size (in Mb) with QUAST parameters ‘-x 5000 -m 10000 -i 10000’. The collapsed and expandable sequences were evaluated by SDA (https://github.com/mrvollger/SDA). In brief, we aligned single-cell sequencing raw reads (for samples with raw data greater than 300 Gb, we conducted downsampling and extracted 10 million reads) to each assembly of HG002. The regions in the assembly without common repeats collapses (identified by RepeatMasker (v4.1.0) and TRF (v4.09)), and with higher coverage (mean + at least three standard deviations) and the length was >15 kb was defined as collapsed. Expandable regions were estimated by multiplying the length of each collapse against the read depth divided by the average genome coverage.
Closing gaps in the human reference genome hg38
The telomere-to-telomere (T2T) Consortium has released the first gapless human genome (https://github.com/marbl/CHM13), and we used it to evaluate how many gaps our assembly closed for hg38. We first aligned de novo assembled contigs to T2T-CHM13 reference, and transformed mapping contigs (mapping quality > 30) to hg38 coordinates using Liftover. Finally, the hg38 gaps (download in UCSC https://genome-euro.ucsc.edu/cgi-bin/hgTables?db=hg38&hgta_track=gap&hgta_table=gap&hgta_doSchema=describe+table+schema) that spanned by contigs were identified as the assembly-closed gaps. We next used RIdeogram (https://github.com/cran/RIdeogram) to mark the gaps closed region in the assembled genome and visualize the assemblies of the HG002 genome. Only contigs labeled as ‘correct’ from QUAST will be displayed, and the QUAST parameters were ‘-x 10000 -m 10000 -i 10000’.
Structural variation analysis of K562 cell assembly
Since some SVs are heterozygous in K562, we used primary and alternate contig sets to find more heterozygous SVs. For hifiasm, we used the phased haplotype1 and haplotype2 contigs (*.hap?.p_ctg.gfa). For Hicanu, we used both primary and alternate contig sets. Since the results of wtdbg2 contain only primary contig, we did not use it to identify SVs. The primary and alternate contig sets were mapped to human reference genome (hg38) using minimap2 with parameters ‘asm5 -r 2k –cs’, and then paftools.js was used to identify insertions and deletions (≥100 bp). We used precision = TP/(TP + FP), recall = TP/(TP + FN), and F1 = 2 × precision × recall/(precision + recall) to quantify the performance of insertion and deletion detection with bulk SVs treated as ground truth (download in https://github.com/cyang235/Smooth-seq). Bedtools v2.30.0 was used to calculate the intersection number of SVs.
Examining the MHC locus
The PAF format results from assembled contigs that aligned to hg38 reference with minimap2 were used to accomplish the MHC dotPlot, using pafCoordsDotPlotly.R (https://github.com/tpoorten/dotPlotly) with parameters ‘-m 2000 -q 500000 -k 10 -s -t -l -p 8’ for K562 cells, and ‘-m 1000 -q 80000 -k 10 -s -t -l -p 8’ for HG002 haplotype. HLA*LA was used to finish the MHC gene typing analysis with HLA-ASM.pl (https://github.com/DiltheyLab/HLA-LA/blob/master/HLA-ASM.md).
RESULTS
Single-cell genome assembly of K562 cells
To demonstrate the feasibility of genome assembly at single-cell levels, we first analyzed the SMOOTH-seq PacBio HiFi data of K562 cells, where 95 single cells were sequenced, with median genome coverage of ∼17.2%. The average length of HiFi reads were around 6.6kb, and the total sequencing depth for these 95 cells was ∼37×.
To find a reliable assembly approach for single-cell genome sequencing HiFi data, we have used hifiasm (36), Hicanu (37) and wtdbg2 (35) to assemble the sequences from 95 individual cells. Hifiasm and Hicanu were designed for HiFi data with high base-accuracy, and these assemblers can help to work toward haplotype-resolved assembly related to the k-mer distributions. However, K562 cell line has a partial triploid genome and the reads have been exponentially amplified before sequencing, the k-mer distribution had skewed, causing the ‘pseudo-haplotype’ assembly results generated by hifiasm and Hicanu potentially biased. Therefore, we focused on un-phased primary assembly contigs. For hifiasm, the assembly graph of primary contig sets were used to evaluate the assembly quality. For Hicanu, the total assembly size (including primary and alternate contig sets) is larger than human reference genome, where we used Purge_dups (34) to identify primary contigs. As for wtdbg2, we used wtpoa-cns (35) to polish the initial assemblies, and the polished contigs was used to evaluate the quality of the assembly (see Methods).
The summary statistics of primary contigs for these three assemblers are presented in Figure 1B. Per-base consensus quality values (QV) was estimated by Merqury (38) using K562 bulk Illumina data. Hifiasm assembly has the longest NG50 with 2.11Mb, the highest QV value of 42.5, the longest total primary contigs (3.19 Gb, exceeding human genome size), and the highest BUSCO (39) completeness (94.7%). Wtdbg2 assembly has the lowest QV, but it has the longest contig with 14.12Mb. Although K562 is a human CML cell line with many heterozygous structural variations (SVs) and point mutations, it takes no more than 100 individual cells with hifiasm to get NG50 of over 2 Mb and BUSCO completeness close to 95%.
To further evaluate the accuracy of assembly, we examined whether the contigs yielded by the above assemblers spanned the major histocompatibility complex (MHC) locus, which is difficult to resolve using NGS-based short reads sequencing data due to its repetitive and highly polymorphic nature (40). For single cell genome sequencing HiFi data, spanning the whole MHC region is still challenging since the ∼6 kb read length is still not long enough and K562 has many heterozygous SVs, which may affect the assembly continuity in complex genomic regions. For hifiasm assembly, primary contig spans most of the MHC region with four contigs (contig length > 500 kb). Hicanu assembly spans MHC region with two contigs while some remaining regions were not assembled. Wtdbg2 assembly spans most of the MHC region with two contigs (Figure 1C). Although the length of single-cell genome sequencing HiFi reads is only about 6 kb and K562 cell line have many heterozygous SVs, with the relative uniform coverage of genome and the appropriate assemblers we can still assemble K562 genome with high continuity, which will be helpful for cancer research.
Single-cell haplotype assembly of HG002 cells
The NG50 of K562 cell assembly is over 2 Mb. However, K562 cell line has a partial triploid genome, and different to normal diploid cells, sequencing triploid cells can result in higher genome coverage under the same sequencing depth. To demonstrate the feasibility of genome assembly based on normal diploid cells, we used SMOOTH-seq to sequence HG002 cell line on HiFi platform, and obtained 157 sequenced cells with in total ∼156 Gb data. The read length ranges from 3329 bp to 11 344 bp, and the median genome coverage of each individual cell is ∼12.4%, with the average sequencing depth of ∼1 Gb per cell (Supplemental Table S1).
We also used hifiasm, Hicanu and wtdbg2 to assemble HG002 genome. QV was estimated with Illumina data sets (download in https://github.com/human-pangenomics/HG002_Data_Freeze_v1.0). Figure 2A displayed primary contig statistics from these three assemblers. Hifiasm has the highest QV value and the least expandable repeats, but there are a few more mis-assemblies and more collapsed repeats. Hicanu has the lowest continuity with NG50 of 0.27Mb, the less collapsed repeats and fewer mis-assemblies. Wtdbg2 has the highest continuity with NG50 of 0.65 Mb, the largest contig of 6.82 Mb and the highest BUSCO completeness (91.1%).
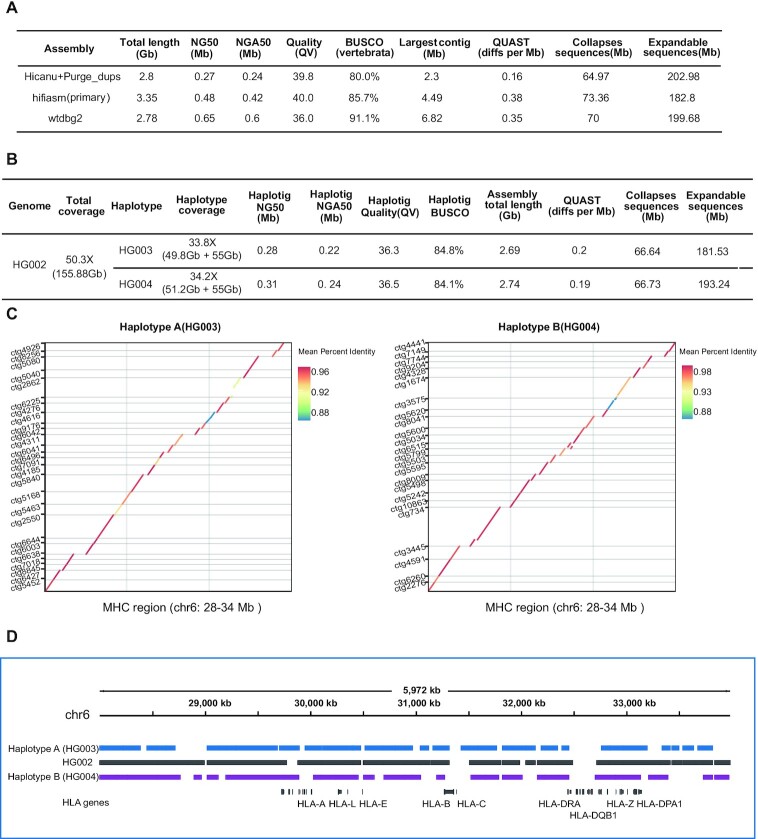
Figure 2.
Single-cell haplotype assembly metrics of HG002 cells. (A) HG002 assembly (157 cells) benchmarking results for Pacbio HiFi mode (primary contigs). QUAST diffs reports the number of large structural discrepancies (>5 kb) observed between the assemblies and phased HG002 reference genome normalized by the assembly size (in Mb). The total base of Collapsed sequences and Expandable sequences report the amount of bp that are collapsed and potentially expandable in each assembly (smaller is better). (B) Trio Hicanu HG002 trio heterozygosity assembly statistics. Using two parental genome specific k-mers, trio HiCanu separated parental haplotype reads, and then we used wtdbg2 to assemble parental haplotypes. QUAST diffs reports the number of large structural discrepancies (>5 kb) observed between the HG003/HG004 haplotype assemblies and HG003/HG004 haplotype reference genome normalized by the assembly size (in Mb). (C) Trio Hicanu HG002 MHC haplotype assemblies were compared with the reference human genome (hg38), and only contigs longer than 80 kb are displayed. (D) Trio Hicanu HG002 primary contigs and associated haplotigs (from wtdbg2) spanning the MHC region were displayed and annotated along with various HLA gene.
Compared with K562 cells, the assembly continuity of HG002 was lower, which may be due to the fact that 95 K562 cells we sequenced have better coverage uniformity than 157 HG002 cells we sequenced (Supplemental Figure S1). K562 has a near-triploid genome (41), therefore, the initial genomic DNA content of an individual K562 cell is higher than individual HG002 cell. For single-cell genome amplification method, higher genomic DNA input will reduce the amplification bias and yield more uniform amplification, resulting in more even genome coverage (42), which was not a random result according to our observations.
Next, we tried to explore whether single cell genome sequencing data from just a small number of diploid cells can be used for haplotype assembly. Since our reads were exponentially amplified before sequencing, and the distributions of k-mer were skewed, the ‘pseudo-haplotype’ assembly was not reliable. Therefore, we used trio HiCanu (43) and trio hifiasm to do the trio-based haplotype assembly. For trio HiCanu, we separated parental haplotype reads with two parental genome specific k-mers (from ∼30 × Illumina data and download in https://github.com/human-pangenomics/HG002_Data_Freeze_v1.0), and then used wtdbg2 to assemble parental haplotypes. As shown in Figure 2B, the parental genomes’ specific k-mers were used to partition long reads from the offspring into paternal and maternal sets (HG003:49.8Gb; HG004:51.2Gb), and un-assignable reads (55 Gb) that are homozygous and can be assigned to both sets. Through the assembly of wtdbg2, the NG50 size of the trio HiCanu F1 haplotigs was 0.28 Mb (HG003 haplotype) and 0.31 Mb (HG004 haplotype), and both parental haplotypes’ BUSCO completeness was greater than 84%, and QV value >36. For trio hifiasm, it does not partition reads upfront, and can get haplotype assembly result directly. Trio hifiasm have almost the same haplotype assembly continuity with trio Hicanu (HG003 NG50: 0.23 Mb; HG004 NG50: 0.24 Mb), and trio hifiasm had higher QV value and less collapsed and expandable repeats than trio HiCanu. For BUSCO completeness trio HiCanu showed better performance than trio hifiasm (Supplemental Table S3).
To further evaluate the accuracy of haplotype assembly, we examined the MHC/HLA locus (Figure 2C). We compared assembly typing results for the six classical human leukocyte antigen (HLA) genes, which have been well characterized by previous studies (44). Trio HiCanu failed to capture an HLA-B gene for HG003 haplotype but the rest of the HLA typing results are correct and only a single base error happened in HLA-DQB1. HG004 haplotype has one error in HLA gene typing and two base errors in HLA-C, with the rests correctly identified (Figure 2D; Supplemental Table S2). Trio hifiasm failed to capture HLA-A and incorrectly captured HLA-B genes for HG003 haplotype, and failed to capture HLA-A, incorrectly capturing HLA-B and HLA-DQB1 genes for HG004 haplotype (Supplemental Table S4).
Single-cell genome assembly of HG002 cells using ONT dataset
Although Pacbio HiFi reads show high accuracy (>99.9%) and can produce around 400 Gb data per run (Pacbio Sequel II platform), after the Circular Consensus Sequencing workflow only about 15 Gb CCS data was retained (33). In other words, if we sequence 1 Gb per cell, the cost for individual cell was ∼260$; if we attempt to obtain deeper sequencing data per cell (higher than 5×), sequencing one run per cell was needed, and the cost for only one individual cell would be ∼3900$, which is a very expensive and unaffordable strategy. Compared to Pacbio HiFi platform, ONT platform was much more cost-effective and affordable. ONT platform can produce around 110 Gb data per run, and if we sequence 1 Gb per cell, the cost for an individual cell was ∼14$, and if sequencing 5 Gb per cell, the cost for one individual cell is ∼70$. To find a better strategy for assembly using a small number of cells and demonstrate the feasibility of our method on different third-generation sequencing platforms, we also assembled HG002 cell line based on ONT platform data.
First, we used the strategy of multi-cells with low sequencing depth. We sequenced 192 single cells and obtained ∼221.7 Gb data for assembly, with ∼1.1 Gb for each individual cell. The average read length was ∼6.4 kb and the median genome coverage of a cell was ∼7.9% (Supplemental Table S5). Unlike PacBio HiFi reads, the error rates of ONT reads are much higher. Previous studies showed that the average error rates of ONT reads for eight species ranged from 12.0% (for S. cerevisiae) to 20.1% (for A. thaliana) and the error rates in ONT reads are more broadly distributed (45). Many of the current de novo assemblers for ONT reads require error correction steps, and some of them are corrected before assembly (e.g. Flye (46), wtdbg2 (35)), while others first corrected raw reads and then assembled them using corrected reads (e.g. NECAT (45)). Herein, we used Flye, wtdbg2 and NECAT to complete the de novo genome assembly of HG002 with ONT reads (∼71×) from 192 single cells.
Primary contig statistics for these three assemblers are presented in Figure 3A. Both corrected-before-assembly assemblers (Flye and wtdbg2) performed similarly well and are more consistent and accurate than NECAT. Flye has the highest continuity with NG50 of 1.38 Mb, and its BUSCO completeness was 93.1% and the largest contig is 11.42 Mb. According to our results, for the assembly of single-cell ONT dataset, the choice of assembler can drastically affect the quality of the final results, and an appropriate tool can increase assembly continuity up to 4-fold. However, due to the high error rates of ONT reads, even the best assembly results have lower QV values than those from HiFi data.
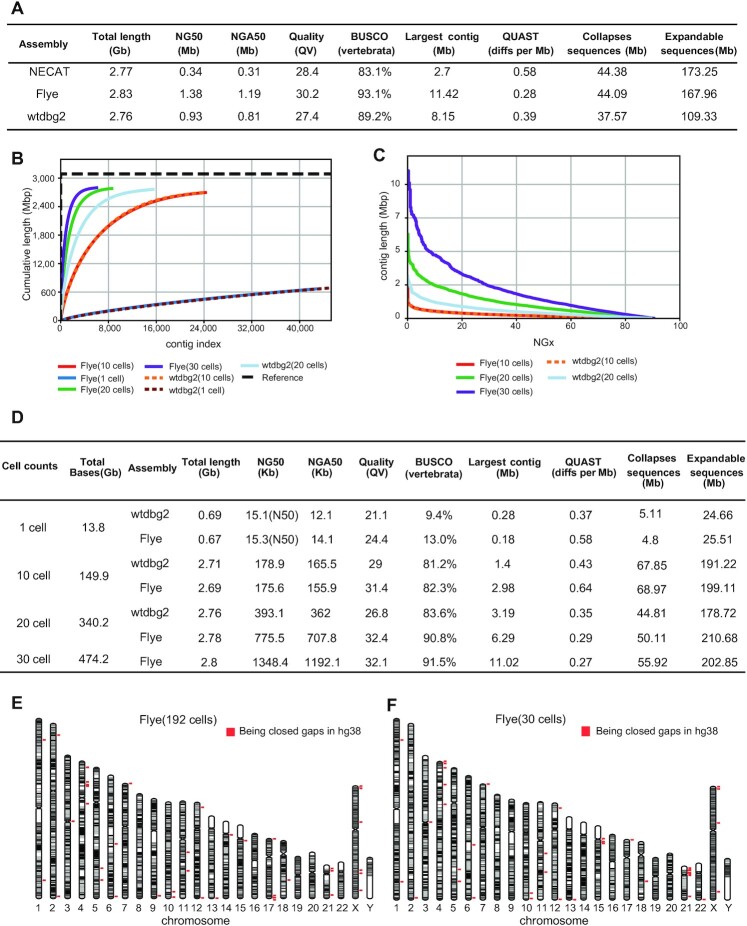
Figure 3.
Assembly metrics for HG002 192 cells at a lower sequencing depth per cell and 30 cells at a higher genome coverage. (A) HG002 assembly (192 cells) benchmarking results for ONT mode (primary contigs). Per-base consensus quality values (QV) was estimated by Merqury. QUAST diffs reports the number of large structural discrepancies (>5 kb) observed between the assemblies and phased HG002 reference genome normalized by the assembly size (in Mb). The total base of Collapsed sequences and Expandable sequences report the amount of bp that are collapsed and potentially expandable in each assembly (smaller is better). (B) Cumulative plot illustrates the growth rate of assemblies’ length. (C) NGx plot showing contig length distribution (NG50: contigs equal or larger than this represent 50% of the estimated genome size). Since the total length of contigs assembled from one cell did not exceed 1.5 Gb, NGx was not shown. (D) HG002 assembly (1, 10, 20, 30 cells) benchmarking results for ONT mode (primary contigs). Only contigs of length greater than 10 kb will be taken into accounting basic indicators. Per-base consensus quality values (QV) was estimated by Merqury. QUAST diffs reports the number of large structural discrepancies (>5 kb) observed between the assemblies and phased HG002 reference genome normalized by the assembly size (in Mb). The total base of Collapsed sequences and Expandable sequences report the amount of bp that are collapsed and potentially expandable in each assembly (smaller is better) (E) Visual representation of the most contigs from ONT assembly results with 192 cells at a lower sequencing depth per cell. Each gray and black block indicates a continuous contig alignment, which was calculated by QUAST, and only contigs labed ‘correct’ from QUAST will be displayed (the QUAST parameter of ‘lower threshold for the relocation’ was 10 kb). The red dots mark the gap-closed regions in the assembled genome. (F) Visual representation of the most contigs from ONT assembly results with 30 cells at a higher genome coverage. Each gray and black block indicates a continuous contig alignment, which was calculated from QUAST, and only contigs labeled as ‘correct’ from QUAST will be displayed (the QUAST parameter of ‘lower threshold for the relocation’ was 10 kb). The red dots mark the gap-closed regions in the assembled genome.
Single-cell genome assembly of HG002 cells using high coverage ONT datasets
In the practical application of assembly, we might encounter the situation of very few cells available. To investigate the lower limit of numbers of single cells need to be sequenced for genome assembly, we deeply sequenced 30 HG002 single cells (average sequencing depth of a single cell was 5×) with high genome coverage (ranging from 27.9% to 53.4%) on ONT platform (Supplemental Table S6). We then explored the results of assembly of 30 cells, 20 cells, 10 cells and even just a single cell. Since NECAT performed not well in single cell datasets, we used wtdbg2 and Flye for the subsequent assembly.
First, we assembled a cell with the highest genome coverage of 53.4%, and the sequencing depth was about 4.5× (∼13.8 Gb), with average read length of 7.1 kb. Contig statistics are presented in Figure 3B, D. Flye has the highest continuity with N50 of 15.3 kb. Wtdbg2 has the longest contig of ∼280 kb. Genome assembly from just one individual cell is an extreme case, and in practice such continuity of genome assembly is far from sufficient.
Second, we used the top 10 sequenced cells with the highest coverage for assembly (the total sequencing depth was about 48×, and average genome coverage was 49.1%). Wtdbg2 and Flye showed similar continuity with NG50 of about 175 kb, similar BUSCO completeness of ∼81%, and the longest contig in Flye is 2.98 Mb (Figure 3B–D).
Third, we used the top 20 sequenced cells with the highest coverage for assembly (sequencing depth ∼109×; average genome coverage ∼45.2%). At this sequencing depth and number of cells sequenced, Flye began to show its fitness in single cell datasets with NG50 of ∼776 kb (Figure 3C, D), the longest contig was ∼6.3 Mb, and the BUSCO completeness was 90.8% (Figure 3D). Such continuity exceeded HiFi assembly results with 157 cells at a lower sequencing depth per cell (NG50: 0.65 Mb).
Finally, we used Flye to assemble all 30 cells (sequencing depth ∼153×; average genome coverage ∼41.7%) with NG50 ∼1.35 Mb, the longest contig was ∼11 Mb and BUSCO completeness was ∼92% (Figure 3B–D). Such continuity was comparable to ONT assembly results with 192 cells at a lower sequencing depth per cell (NG50 ∼1.38 Mb) (Figure 3E, F). These results suggest that the genome assembly from as low as 30 individual cells was in line with that from 192 individual cells at a lower sequencing depth per cell.
Identification of SVs using K562 cell genome assembly
As for the assembly results of K562, it is important to know whether the original SV information can still be retained in the assembled K562 genome, and whether new SVs can be identified after assembly, which were well concerned questions in the genome assembly of cancer cells. Since some SVs are heterozygous in K562, it is not appropriate to detect SVs using only the primary contig sets, so we also used the alternate contig sets, allowing us to find more heterozygous SVs.
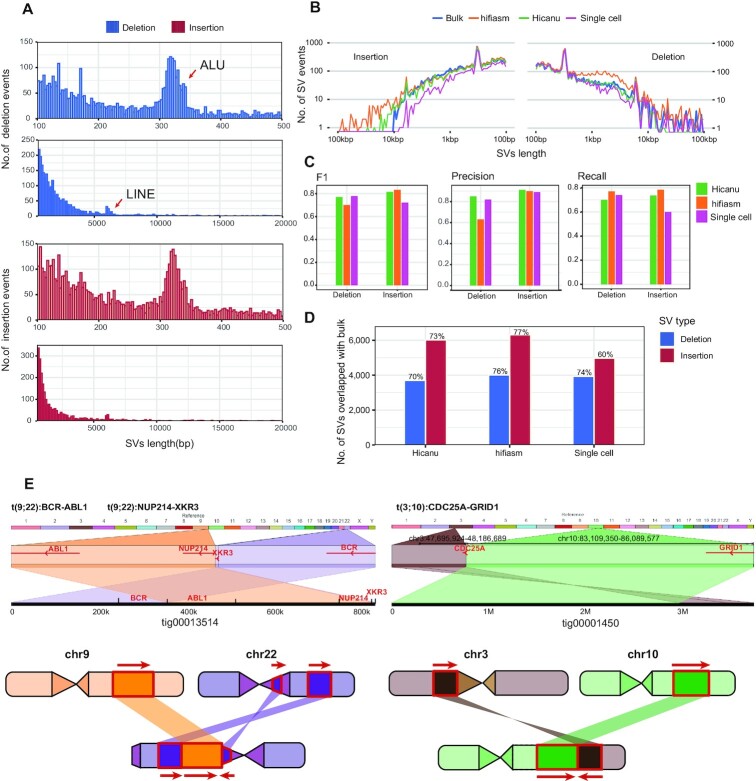
For hifiasm, we used the phased haplotype1 and haplotype2 contigs (*.hap?.p_ctg.gfa), and for Hicanu, we used both primary and alternate contig sets to detect SVs. After mimimap2 alignment and variants calling by paftools.js (47), we finally identified 7132 insertions and 6365 deletions from hifiasm, 6604 insertions and 4355 deletions from Hicanu. The size distribution of SVs detected from hifiasm contigs show 300 bp peak for ALUs and 6 kb peak for LINEs (Figure 4A, B). We then compared the distributions of insertion lengths detected by hifiasm assembly, Hicanu assembly, bulk Pacbio CLR reads and single-cell HiFi reads (2-cell supported) (33), where bulk CLR SVs were treated as ground truth. Insertions detected by assemblies were longer (some insertions exceed 10 kb) and have almost the same length distribution with bulk CLR reads (Figure 4B). To evaluate the accuracy of insertion detection, we used three metrics: precision, recall and F1-score. The precision of insertion detection for these two assemblers was >0.9 (Figure 4C). Hifiasm assembly can identify more insertions with the highest recall rate, and its F1-score was higher than that of Hicanu. The true positive insertions identified by hifiasm accounted for ∼77% of insertions identified from bulk samples, exceeding those of single cell data direct mapping and Hicanu assembly (Figure 4D).
Figure 4.
SVs discovery and distribution from K562 cells assembly. (A) Size distribution of SVs identified from K562 cells with hifiasm assembly, 300 bp peak for ALU and 6kb peak for LINE. (B) Length distribution of SVs identified from Hicanu assembly, hifiasm assembly, raw single cell HiFi reads and bulk CLR reads. (C) The precision, recall and F1-score of SVs identified from Hicanu assembly, hifiasm assembly, raw single cell HiFi reads, where bulk CLR SVs were treated as ground truth. (D) The percentage of true positive SVs identified from Hicanu assembly, hifiasm assembly, raw single cell HiFi reads, where bulk CLR SVs were treated as ground truth. (E) Ribbon (56) visualization the translocations in K562 cells. The diagram on the left indicates the detailed positions and directions of the translocation of BCR-ABL1 locus and NUP214-XKR3 locus. The right diagram indicates the translocation of chr3 with chr10 generates CDC25A-GRID1 fusion gene, which is not detected in single cell direct mapping result.
Since for majority of the single cells the length of HiFi reads were around 6 kb, most insertions found in single cells are less than 6 kb. By assembling, we can identify longer and more complete insertion events. For example, we observed a homozygous insertion event of ∼4100 bp (chr13:89 668 900–89 668 990), which was found in both bulk data and assembled contigs but not in the single-cell reads direct mapping result (Supplemental Figure S2). We also detected a heterozygous insertion in the ITGA11 gene from assembled contig, without supported reads in the single-cell reads direct mapping result. This is because when reads are not long enough to span the whole insertion events, unaligned sub-sequence will appear at the end of the reads, and the unaligned portion will be masked by a process termed ‘soft-clipping’ (48). The software we used to find SVs will not recognize it as an SV event, but when we assemble sequencing reads, the ‘soft-clipping’ of reads overlaps into a contig, the unaligned portion will appear in the middle of the contig and then will be recognized as insertion events. Therefore, if targeting detecting insertions, using assembly's data is a better choice.
For detection of deletion events, the length distribution from single cell HiFi data was more similar with bulk Hifi data (Figure 4B). Hifiasm assembly can identify more deletions with the highest recall rate, and the true positive detections identified by hifiasm is also the highest, but the precision and F1-score is a little lower. There was no significant advantage in deletion detection using the assembled contigs. But we still got some deletion events not detected in the single cell direct mapping result but could be detected in both bulk data and assembled contigs (Supplemental Figure S3).
To further verify our results, we selected 20 SVs (6 deletion events and 14 insertion events) detected from both HiCanu and hifiasm as candidates for SV validation. These SVs are not detected at the single cell direct mapping data but can be detected by de novo genome assembly. In addition, we used K562 cell line gDNAs as PCR templates for SV validation. 100% (6 out of 6) selected deletion events were successfully amplified with expected size. Of these deletion events, three were homozygous and the remaining three were heterozygous (Supplemental Figure S4a; Table S7). For insertion events, 71% (10 out of 14) selected events were validated with the exact insertion sizes and the remaining four insertion events failed to amplify any bands with two different primer pairs. Of these insertion events, two were heterozygous and the remaining eight were homozygous (Supplemental Figure S4b; Table S7). These results indicated that structure variations identified using the de novo genome assembly approach are convincing.
For some complex structural variations, especially for detecting complex translocation events, it is more dependent on reads length. In line with our expectations, translocation events could be more reliably identified after genome assembly. For example, translocation of chr3 with chr10 generates CDC25A-GRID1 fusion gene, which is not detected in single cell direct mapping result, but after assembly, there have a contig with 3.7 Mb length permitting us to find this event robustly (Figure 4E). Other known translocation events in the K562 cell line like BCR-ABL1 locus and NUP214-XKR3 locus (49) were also reliably identified using assembly data, and both of which can be revealed by just one contig (Figure 4E). These results indicated that, after assembly, most of the SVs found in bulk sequencing dataset were recovered robustly, especially for many longer insertion events and complex structural variations, which could be identified more accurately and more completely.
Assembling single cell ONT data permit closing gaps in human reference genome
The current hg38 reference genome contains ∼151 Mb of unknown sequences distributed throughout the human genome (6), and these breaks in the assembly span many complex repeats. Recently, T2T has finished the first truly complete human reference genome (3.055 billion base pairs) using CHM13 cell line, providing gapless assemblies for all 22 autosomes plus ChrX (6). Aligned to T2T-CHM13 reference, our de novo assembled contigs using the ONT dataset with 192 single cells of diploid HG002 line closed 39 gaps, where the length of 14 gaps is greater than or equal to 50 kb according to the annotation of hg38 (Supplemental Table S8, Figure 3E). For example, an unresolved 50 kb gap on ChrX is spanned by a single contig with the length of 352 868 bp in our assembly (contig_6500), however it turned out that this gap is only 3653 bp long (according to T2T-CHM13 reference. Supplemental Figure S5a). For the assembly contigs using 30 single HG002 cells’ high coverage (average coverage ∼41.7%) ONT dataset, 38 gaps have been spanned, of which 15 gaps’ lengths are greater than or equal to 50 kb (Supplemental Table S9, Figure 3F). For instance, an unresolved 50 kb gap on Chr13 (according to hg38) is spanned by a single contig with the length of 205 180 bp in our assembly (contig_16890), which spans GRK1 gene whose mutation is known causing Oguchi disease (50) (Supplemental Figure S5a).
Single cell genome de novo assembly can be used to cancer research (high cellular heterogeneity) and embryonic development research (only a small amount of DNAs available). The HOX genes are major regulators of embryonic development. One of their most conserved functions is to coordinate the formation of specific body structures along the anterior-posterior (AP) axis in Bilateria (51), and mutations in HOX genes can lead to increased cancer predisposition, and HOX genes might mediate the effect of many other cancer susceptibility factors by recognizing or executing altered genetic information (52). Therefore, we examined the HOX gene region with assembled contigs from ONT dataset with 192 single cells of HG002 line. For the HOX gene clusters (HOXA, HOXB, HOXC and HOXD), two or three assembled contigs can cover each of them and most of the genes are completely assembled (Supplemental Figure S5b).
DISCUSSION
In this study, we have presented a systematic analysis of de novo human genome assembly with a small number of single cells sequenced on the HiFi or ONT platforms, and subsequently conducted de novo human genome assembly with 1, 10, 20 and 30 normal diploid cells respectively. First, we assembled the genome of human CML cell line K562 using 95 individual cells, where the NG50 can reach 2.11 Mb with hifiasm. Second, we used the strategy of multi-cell low-depth sequencing to sequence 157 individual cells (Pacbio HiFi platform) and 192 individual cells (ONT platform) to assemble the genome of HG002 cell line separately. The results have shown that the HiFi dataset with 157 single cells can assemble the HG002 genome with the NG50 of 0.65 Mb, and owing to the high accuracy of HiFi data, haplotype assembly can still be completed as demonstrated by satisfying HLA typing results. Using the ONT dataset with 192 single cells we can assemble the HG002 genome with NG50 of 1.38 Mb, closing 39 gaps in hg38 reference genome. Third, we sequenced 30 HG002 single cells with high genome coverage on the ONT platform, and assembled the genome using 1, 10, 20, 30 individual cells respectively. The results showed that the assembly continuity with sequencing data from as few as 20 individual cells (average genome coverage ∼ 45.2%) can achieve the NG50 of 0.7 Mb, exceeding that of HiFi dataset with 157 single cells (median genome coverage ∼ 12.4%). The assembly continuity with sequencing data from 30 individual cells (average genome coverage ∼ 41.7%) can achieve the NG50 of 1.35 Mb, comparable to that of ONT dataset with 192 single cells (median genome coverage ∼ 7.9%). Our results have demonstrated the practicability of single-cell human genome assembly.
Through analyzing the structural variations of the assembled genome of K562 cells, we found that compared to single cell direct mapping strategy, insertion events and complex structural variations could be identified more accurately and more completely. In addition, our results demonstrated that the choice of different assemblers can affect the assembly results profoundly, especially in the cases with very few individual cells sequenced and analyzed, where Flye is more suitable for single-cell genome de novo assembly with the high coverage datasets from ONT platform.
The small amount of cells available and the large genetic heterogeneity within a population of cells are two sets of difficulties that hinder the application of genome assembly in biomedical research. Single cell whole genome long-read sequencing technology can help us solve these problems. Here, we have used different sequencing platforms and strategies to explore the feasibility of single-cell de novo genome assembly and identified the factors affecting the assembly results, and finally improved the resolution of genome assembly to single-cell levels.
In fact, assemblers are also important for the genome assembly of single-cell dataset. Due to highly non-uniform read coverage generated by single-cell whole genome amplification (53), the assembler needs to adjust its strategy to achieve better assembly results, such as Velvet-SC (53), SPAdes (28), IDBA-UD (54) and SOAPdenovo2 (55), which are designed for single-cell genome NGS data. However, since they are designed for the assembly of a single microbial genome, which may not be suitable for our single-cell human genome assembly. The algorithm design of these tools is instructive, and we hope new assemblers can be developed to assemble human genome with the single-cell TGS data.
Although limited by the cost and available sequencing techniques, assembling human genome from just one single cell with great continuity can still not be achieved currently. We believe that with further development of single-cell whole genome long-read sequencing technologies, restoring genome structure from just a single cell will finally be achieved, and will promote related biomedical researches.
DATA AVAILABILITY
The analysis code is deposited in github (https://github.com/hlxie/sc-assembly). The HG002 Data have been deposited in the Sequence Read Archive (SRA) under BioSample accession: SAMN25232871; BioProject: PRJNA800164 (https://www.ncbi.nlm.nih.gov/bioproject/800164).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Beijing Advanced Innovation Centre for Genomics for support, and part of the analysis was performed on the High Performance Computing Platform of the Center for Life Sciences(Peking University).
Authors contributions: F.T. conceived the project and supervised the overall experiments. H.X. was in charge of the bioinformatics analysis with the help of C.Y., and S.J. assisted in figure plotting, and Y.G assisted in data sorting. W.L. was in charge of the experimental part and developed the SMOOTH-seq protocol for Nanopore platform. Y.H. helped in a part of HG002 library preparation for PacBio platform. H.X., W.L., C.Y. and F.T. wrote the manuscript with help from all authors.
Contributor Information
Haoling Xie, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Peking University-Tsinghua University-National Institute of Biological Sciences Joint Graduate Program (PTN), School of Life Sciences, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Wen Li, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Yuqiong Hu, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Cheng Yang, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Jiansen Lu, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Yuqing Guo, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Lu Wen, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
Fuchou Tang, School of Life Sciences, Biomedical Pioneering Innovation Center, Peking University, Beijing 100871, China; Peking University-Tsinghua University-National Institute of Biological Sciences Joint Graduate Program (PTN), School of Life Sciences, Peking University, Beijing 100871, China; Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China; Beijing Advanced Innovation Center for Genomics (ICG), Ministry of Education Key Laboratory of Cell Proliferation and Differentiation, Beijing 100871, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Beijing AdvancedInnovation Center for Genomics at Peking University, and National Key Research and Development Program of China [2018YFA0107601].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T.et al.. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tyson J.R., O’Neil N.J., Jain M., Olsen H.E., Hieter P., Snutch T.P.. MinION-based long-read sequencing and assembly extends the Caenorhabditis elegans reference genome. Genome Res. 2018; 28:266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A.et al.. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020; 585:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Logsdon G.A., Vollger M.R., Hsieh P.H., Mao Y., Liskovykh M.A., Koren S., Nurk S., Mercuri L., Dishuck P.C., Rhie A.et al.. The structure, function and evolution of a complete human chromosome 8. Nature. 2021; 593:101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belser C., Baurens F.C., Noel B., Martin G., Cruaud C., Istace B., Yahiaoui N., Labadie K., Hřibová E., Doležel J.et al.. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun. Biol. 2021; 4:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V, Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A.et al.. The complete sequence of a human genome. 2021; bioRxiv doi:27 May 2021, preprint: not peer reviewed 10.1101/2021.05.26.445798. [DOI] [PMC free article] [PubMed]
- 7. Rhie A., McCarthy S.A., Fedrigo O., Damas J., Formenti G., Koren S., Uliano-Silva M., Chow W., Fungtammasan A., Kim J.et al.. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021; 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jarvis E.D., Formenti G., Rhie A., Guarracino A., Yang C., Tracey A., Thibaud-nissen F., Vollger M.R., Porubsky D., Cheng H.. Automated assembly of high-quality diploid human reference genomes. 2022; bioRxiv doi:06 March 2022, preprint: not peer reviewed 10.1101/2022.03.06.483034. [DOI] [PMC free article] [PubMed]
- 9. Lin G., He C., Zheng J., Koo D.H., Le H., Zheng H., Tamang T.M., Lin J., Liu Y., Zhao M.et al.. Chromosome-level genome assembly of a regenerable maize inbred line A188. Genome Biol. 2021; 22:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiao W.B., Schneeberger K.. The impact of third generation genomic technologies on plant genome assembly. Curr. Opin. Plant Biol. 2017; 36:64–70. [DOI] [PubMed] [Google Scholar]
- 11. Li R., Di L., Li J., Fan W., Liu Y., Guo W., Liu W., Liu L., Li Q., Chen L.et al.. A body map of somatic mutagenesis in morphologically normal human tissues. Nature. 2021; 597:398–403. [DOI] [PubMed] [Google Scholar]
- 12. Zhou Y., Bian S., Zhou X., Cui Y., Wang W., Wen L., Guo L., Fu W., Tang F.. Single-Cell multiomics sequencing reveals prevalent genomic alterations in tumor stromal cells of human colorectal cancer. Cancer Cell. 2020; 38:818–828. [DOI] [PubMed] [Google Scholar]
- 13. Martincorena I., Campbell P.J.. Somatic mutation in cancer and normal cells. Science. 2015; 349:1483–1489. [DOI] [PubMed] [Google Scholar]
- 14. Moore L., Cagan A., Coorens T.H.H., Neville M.D.C., Sanghvi R., Sanders M.A., Oliver T.R.W., Leongamornlert D., Ellis P., Noorani A.et al.. The mutational landscape of human somatic and germline cells. Nature. 2021; 597:381–386. [DOI] [PubMed] [Google Scholar]
- 15. Abascal F., Harvey L.M.R., Mitchell E., Lawson A.R.J., Lensing S.V., Ellis P., Russell A.J.C., Alcantara R.E., Baez-Ortega A., Wang Y.et al.. Somatic mutation landscapes at single-molecule resolution. Nature. 2021; 593:405–410. [DOI] [PubMed] [Google Scholar]
- 16. Bian S., Hou Y., Zhou X., Li X., Yong J., Wang Y., Wang W., Yan J., Hu B., Guo H.et al.. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018; 362:1060–1063. [DOI] [PubMed] [Google Scholar]
- 17. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A.et al.. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009; 6:377–382. [DOI] [PubMed] [Google Scholar]
- 18. Maheswaran S., Haber D.A.. Circulating tumor cells: a window into cancer biology and metastasis. Curr. Opin. Genet. Dev. 2010; 20:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kage H., Kohsaka S., Shinozaki-Ushiku A., Hiraishi Y., Sato J., Nagayama K., Ushiku T., Takai D., Nakajima J., Miyagawa K.et al.. Small lung tumor biopsy samples are feasible for high quality targeted next generation sequencing. Cancer Sci. 2019; 110:2652–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J.S., Melisko M.E., Magbanua M.J.M., Kablanian A.T., Scott J.H., Rugo H.S., Park J.W.. Detection of cerebrospinal fluid tumor cells and its clinical relevance in leptomeningeal metastasis of breast cancer. Breast Cancer Res. Treat. 2015; 154:339–349. [DOI] [PubMed] [Google Scholar]
- 21. Peterson V.M., Castro C.M., Chung J., Miller N.C., Ullal A.V., Castano M.D., Penson R.T., Lee H., Birrer M.J., Weissleder R.. Ascites analysis by a microfluidic chip allows tumor-cell profiling. Proc. Natl. Acad. Sci. U.SA. 2013; 110:E4978–E4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lodato M.A., Woodworth M.B., Lee S., Evrony G.D., Mehta B.K., Karger A., Lee S., Chittenden T.W., D’Gama A.M., Cai X.et al.. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015; 350:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zafar H., Tzen A., Navin N., Chen K., Nakhleh L.. SiFit: inferring tumor trees from single-cell sequencing data under finite-sites models. Genome Biol. 2017; 18:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ludwig L.S., Lareau C.A., Ulirsch J.C., Christian E., Muus C., Li L.H., Pelka K., Ge W., Oren Y., Brack A.et al.. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell. 2019; 176:1325–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciobanu D., Clum A., Ahrendt S., Andreopoulos W.B., Salamov A., Chan S., Quandt C.A., Foster B., Meier-Kolthoff J.P., Tang Y.T.et al.. A single-cell genomics pipeline for environmental microbial eukaryotes. Iscience. 2021; 24:102290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bowers R.M., Doud D.F.R., Woyke T.. Analysis of single-cell genome sequences of bacteria and archaea. Emerg. Top. Life Sci. 2017; 1:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lasken R.S. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 2012; 10:631–640. [DOI] [PubMed] [Google Scholar]
- 28. Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D.et al.. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012; 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li R., Zhu H., Ruan J., Qian W., Fang X., Shi Z., Li Y., Li S., Shan G., Kristiansen K.et al.. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010; 20:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mostovoy Y., Levy-Sakin M., Lam J., Lam E.T., Hastie A.R., Marks P., Lee J., Chu C., Lin C., Dzakula Z.et al.. A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods. 2016; 13:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sjodin B.M.F., Galbreath K.E., Lanier H.C., Russello M.A.. Chromosome-level reference genome assembly for the American Pika (Ochotona princeps). J. Hered. 2021; 112:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dobson L.K., Zimin A., Bayles D., Fritz-Waters E., Alt D., Olsen S., Blanchong J., Reecy J., Smith T.P.L., Derr J.N.. De novo assembly and annotation of the North American bison (Bison bison) reference genome and subsequent variant identification. Anim. Genet. 2021; 52:263–274. [DOI] [PubMed] [Google Scholar]
- 33. Fan X., Yang C., Li W., Bai X., Zhou X., Xie H., Wen L., Tang F.. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021; 22:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guan D., Guan D., McCarthy S.A., Wood J., Howe K., Wang Y., Durbin R., Durbin R.. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020; 36:2896–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruan J., Li H.. Fast and accurate long-read assembly with wtdbg2. Nat. Methods. 2020; 17:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng H., Concepcion G.T., Feng X., Zhang H., Li H.. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021; 18:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nurk S., Walenz B.P., Rhie A., Vollger M.R., Logsdon G.A., Grothe R., Miga K.H., Eichler E.E., Phillippy A.M., Koren S.. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 2020; 30:1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rhie A., Walenz B.P., Koren S., Phillippy A.M.. Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020; 21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manni M., Berkeley M.R., Seppey M., Simão F.A., Zdobnov E.M.. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021; 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brandt D.Y.C., Aguiar V.R.C., Bitarello B.D., Nunes K., Goudet J., Meyer D.. Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 genomes project phase I data. G3 Genes Genomes Genet. 2015; 5:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naumann S., Reutzel D., Speicher M., Decker H.J.. Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk. Res. 2001; 25:313–322. [DOI] [PubMed] [Google Scholar]
- 42. Fitz-gibbon S., Tomida S., Chiu B., Nguyen L., Du C., Miller J.F., Sodergren E., Craft N., Weinstock G.M.. Highly multiplexed targeted DNA sequencing from single nuclei. Nat. Protoc. 2014; 133:2152–2160. [Google Scholar]
- 43. Koren S., Rhie A., Walenz B.P., Dilthey A.T., Bickhart D.M., Kingan S.B., Hiendleder S., Williams J.L., Smith T.P.L., Phillippy A.M.. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol. 2018; 36:1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chin C.S., Wagner J., Zeng Q., Garrison E., Garg S., Fungtammasan A., Rautiainen M., Aganezov S., Kirsche M., Zarate S.et al.. A diploid assembly-based benchmark for variants in the major histocompatibility complex. Nat. Commun. 2020; 11:4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen Y., Nie F., Xie S.Q., Zheng Y.F., Dai Q., Bray T., Wang Y.X., Xing J.F., Huang Z.J., Wang D.P.et al.. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 2021; 12:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kolmogorov M., Yuan J., Lin Y., Pevzner P.A.. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019; 37:540–546. [DOI] [PubMed] [Google Scholar]
- 47. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang J., Mullighan C.G., Easton J., Roberts S., Heatley S.L., Ma J., Rusch M.C., Chen K., Harris C.C., Ding L.et al.. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods. 2011; 8:652–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Engreitz J.M., Agarwala V., Mirny L.A.. Three-Dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS One. 2012; 7:e44196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mucciolo D.P., Sodi A., Murro V., Passerini I., Palchetti S., Pelo E., Virgili G., Rizzo S.. A novel GRK1 mutation in an Italian patient with Oguchi disease. Ophthalmic Genet. 2018; 39:137–138. [DOI] [PubMed] [Google Scholar]
- 51. Merabet S., Galliot B.. The TALE face of Hox proteins in animal evolution. Front. Genet. 2015; 6:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li B., Huang Q., Wei G.H.. The role of hox transcription factors in cancer predisposition and progression. Cancers (Basel). 2019; 11:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chitsaz H., Yee-Greenbaum J.L., Tesler G., Lombardo M.J., Dupont C.L., Badger J.H., Novotny M., Rusch D.B., Fraser L.J., Gormley N.A.et al.. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat. Biotechnol. 2011; 29:915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peng Y., Leung H.C.M., Yiu S.M., Chin F.Y.L.. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012; 28:1420–1428. [DOI] [PubMed] [Google Scholar]
- 55. Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y.et al.. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012; 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nattestad M., Aboukhalil R., Chin C.S., Schatz M.C.. Ribbon: intuitive visualization for complex genomic variation. Bioinformatics. 2021; 37:413–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analysis code is deposited in github (https://github.com/hlxie/sc-assembly). The HG002 Data have been deposited in the Sequence Read Archive (SRA) under BioSample accession: SAMN25232871; BioProject: PRJNA800164 (https://www.ncbi.nlm.nih.gov/bioproject/800164).