Figure 2.
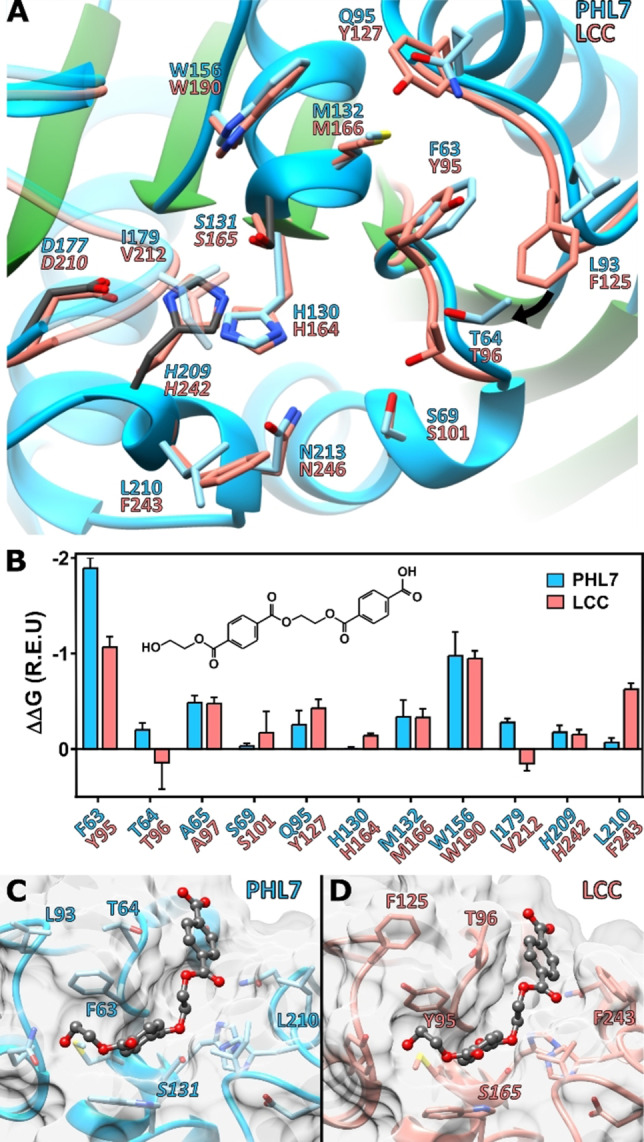
Comparison of the crystal structures of PHL7 and LCC and docking experiments: (A) Active site structures of PHL7 (chain A) and LCC. S131 adopts two different conformations, of which the most occupied is displayed. (B) Predicted per‐residue binding energy contribution based on a docking of 1,2‐ethylene monoterephthalate mono(2‐hydroxyethyl terephthalate) (EMT) in PHL7 and LCC. The best five out of 40000 complexes with the lowest interface binding energy and RMSD lower than 1.5 Å in relation to the p‐nitrophenol and HEMT cocrystal structure of IsPETase are shown. [36] (C,D): Lowest RMSD pose of the 0.25 % best interface binding energy complexes of EMT with PHL7 (C) and LCC (D).
