Abstract
The group A Streptococcus (GAS) pilus is a long, flexible, hair‐like structure anchored to the cell surface that facilitates the adherence of GAS to host cells, thus playing a critical role in initiating infections. Because of its important role in GAS virulence, the pilus has become an attractive target for vaccine development. While current research mainly focuses on pilus function and its potential as a vaccine component, there is a lack of knowledge on how the host immune system recognizes and responds to this abundant surface structure. Here we show that both assembled GAS pili and individual pilus proteins induce a potent release of the proinflammatory cytokines tumor necrosis factor and interleukin‐8. We further show that the surface‐exposed backbone pilin and ancillary pilin 1 subunits are Toll‐like receptor 2 (TLR2) agonists. Using reporter cell lines coexpressing human TLR2 in combination with either TLR1 or TLR6, we determined that activation was mediated by the TLR2/TLR6 heterodimer. Finally, we used solid‐phase and flow cytometry binding assays to illustrate a direct interaction between the pilus subunits and TLR2. These results provide further support for the suitability of the pilus as a vaccine component and opens potential avenues for using GAS pili as an adjuvant or immune‐modulation agent.
Keywords: Group A Streptococcus , pilus, Streptococcus pyogenes, Toll‐like receptor
We show that group A Streptococcus (GAS) pilus induces a potent release of the proinflammatory cytokines, and further demonstrate that the surface‐exposed backbone pilin and ancillary pilin 1 subunits are Toll‐like receptor 2 agonists. These results provide further support for the suitability of the pilus as a vaccine component and opens potential avenues for using GAS pili as an adjuvant or immune‐modulation agent.

INTRODUCTION
Streptococcus pyogenes, commonly known as group A Streptococcus (GAS), is a Gram‐positive bacterial pathogen exclusive to humans. GAS is associated with a wide range of diseases from self‐limiting primary infections to life‐threatening invasive diseases, as well as postinfection immune sequelae. 1 GAS causes approximately 700 million cases of pharyngitis annually worldwide. 2 While usually self‐limiting, increasing incidence of these mild symptoms can lead to invasive conditions such necrotizing fasciitis and streptococcal toxic shock syndrome. Furthermore, the greatest burden is attributed to postinfection complications that can cause lifelong injuries and premature deaths, such as acute rheumatic fever and rheumatic heart disease. Estimations of global disease burden show alarming figures of 16–30 million severe and invasive disease cases, and 200 000–300 000 deaths in a given year. 3 , 4 , 5 , 6 Despite being effective in treating infections, antibiotics have failed to reduce disease burden in less privileged areas where invasive GAS infections still cause high mortality rates, and superficial manifestations continue to consume substantial health care resources. 7
The ability of GAS to cause diseases with multiple clinical manifestations is attributed to an array of virulence factors. 2 , 8 , 9 The pilus (plural, pili) is a long, thin, hair‐like surface structure associated with increased virulence in in vivo models. 10 , 11 , 12 , 13 The major part of the pilus shaft is formed by 10–100 repeatedly joined backbone pilins (BPs). Attached to either end of the pilus fiber are one or two ancillary pilins (APs), AP1 and AP2. While AP1 is found at the tip of the pilus structure and has adhesive properties, AP2 is located at the base and serves as anchorage to the bacterial cell wall. 14 As surface‐exposed molecules, BP (also known as the T‐antigen) and AP1 have long been known to stimulate strong antibody responses in humans and animals. 15 , 16 The pilus is therefore considered as a potential vaccine target, 17 , 18 , 19 , 20 , 21 and more recently, as a vaccine carrier. 22 , 23
Immunization of mice and rabbits with multivalent recombinant T‐antigen proteins induces specific and cross‐reactive anti‐T antibodies with bactericidal properties. 18 Similarly, administration of pili expressed on the nonpathogenic bacterium Lactococcus lactis stimulated the production of functional antibodies that possessed the ability to facilitate opsonophagocytosis and interfere with adhesion of GAS to keratinocyte cell lines. 17 Furthermore, when the pilus was utilized as a peptide carrier in a recombinant L. lactis–based mucosal vaccine platform, high levels of antibodies specific to the antigen were generated. 23 , 24 , 25 Current research in the context of vaccine development focuses on the adaptive immune responses elicited by the pilus structure. However, there is a lack of knowledge on the innate immune responses where the immune system initially recognizes the pili and mounts the immediate and broader responses downstream. 26
The innate immune system senses intruding bacteria through recognizing conserved bacterial molecules termed “pathogen‐associated molecular patterns” by the “pattern recognition receptors” mainly found on immune cells. 27 , 28 In response to pathogen‐associated molecular patterns, immune cells initiate critical events for driving both innate and adaptive immunity, including direct crosstalk between different cell types and the production of proinflammatory cytokines. 29 The membrane‐bound Toll‐like receptors (TLRs) are an important subset of pattern recognition receptors that recognize an array of microbe‐derived molecules, including common cell surface components such as lipoproteins (TLR1/2/6), lipopolysaccharide (LPS; TLR4) and flagellin (TLR5), as well as intracellular contents such as microbial nucleic acids (TLR3/7/8/9). 30 Among the 10 different TLRs found in humans, TLR2 has the most diverse selection of ligands, including proteins, lipids and lipid‐modified sugars or peptides. The capacity of TLR2 to recognize such a wide range of bacterial components is attributed to its dimerization with either TLR1 or TLR6. 31 , 32 Advances in genetics and new animal models have helped to confirm the involvement of TLRs in recognizing GAS and activating innate immune responses. For instance, quantitative reverse transcriptase‐PCR (qRT‐PCR) of mouse dendritic cells illustrated a significant increase of TLR2 and TLR4 transcription following treatment with GAS, a trend which was also replicated in vivo. 33 Furthermore, infection models with mice devoid of the TLR adaptor molecule MyD88 illustrated that TLRs facilitate innate immune responses to GAS. Upon administration of GAS to MyD88−/− and wild‐type (WT) mice, the MyD88‐deficient mice demonstrated a severely dampened release of proinflammatory cytokines compared with the WT mice. 33 , 34 Specifically, production of inflammatory cytokines such as interleukin (IL)‐12, interferon‐γ and tumor necrosis factor (TNF) as well as chemoattractants such as monocyte chemoattractant protein 1 (MCP‐1) and keratinocyte‐derived chemokine (KC) was diminished, resulting in reduced recruitment of neutrophils and macrophages to the GAS infection site. 35 Furthermore, MyD88−/− mice had significantly decreased expression of cell surface proteins associated with dendritic cell activation. These results indicated a loss of innate immune responses toward GAS when the TLR pathway was disrupted. 34
Despite the consensus of TLR recognizing and inducing an innate immune response against GAS, the importance and interactions between specific TLRs and bacterial components are yet to be elucidated.
In this study, we examined the ability of GAS pilus to stimulate proinflammatory cytokines and to activate innate immune receptors. We focused on the pilus structure expressed in the globally disseminated M1/T1 strain, which has been shown to induce neutrophil IL‐8 production in murine models. 10 Using both individual recombinant pilus proteins and assembled pili heterogeneously expressed on L. lactis, we show that the surface‐exposed pilus components AP1 and BP induce potent release of the proinflammatory cytokines TNF and IL‐8 in human monocytic cells after engagement with TLR2.
RESULTS
The pilus from serotype M1/T1 group A Streptococcus induces inflammatory responses
Our investigations began with evaluating whether the pilus has the ability to induce cellular inflammatory responses. To focus on the immune responses against the complete pilus structure, we employed the nonpathogenic surrogate species L. lactis to express the fully assembled pilus from an M1/T1 serotype GAS strain (PilM1). 23 The L. lactis gain‐of‐function strain with cell surface expression of the whole pilus structure (PilM1 L. lactis) was incubated with human monocytic THP‐1 cells, and secretion of the proinflammatory cytokines IL‐8 and TNF quantified by ELISA (Figure 1a). Compared with the untreated THP‐1 cells, incubation with PilM1 L. lactis significantly increased the level of TNF release; whereas the cells incubated with WT L. lactis did not show increased TNF production [157 pg mL–1 versus 12 pg mL–1, P < 0.001 at multiplicity of infection (MOI) 10; 69 pg mL–1 versus 4 pg mL–1, P = 0.003 at MOI 1]. A similar trend was observed with IL‐8 expression, where a threefold increase in cytokine production was seen in cells incubated with PilM1 L. lactis compared with the untreated negative control, or with WT L. lactis at the same MOI (Figure 1b). A recombinant form of the TLR‐5 agonist flagellin was used as a positive control 36 and triggered comparable TNF and IL‐8 responses at a much higher concentration (25 nM).
Figure 1.
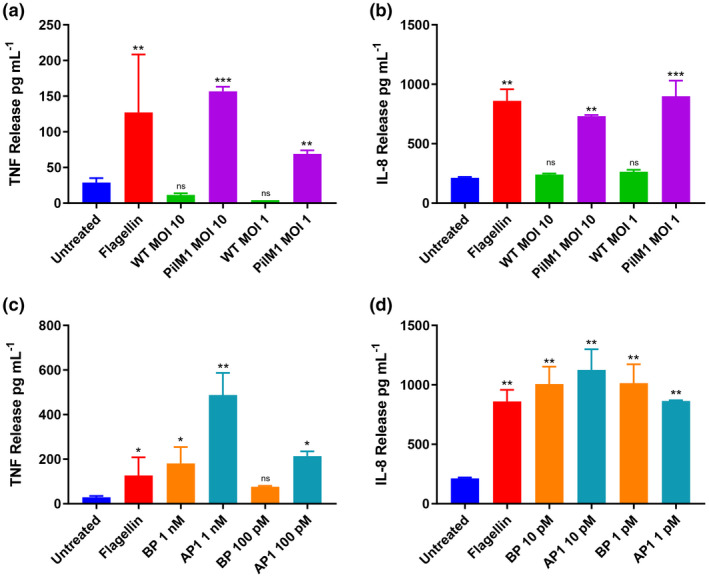
The M1/T1 group A Streptococcus pilus induces an inflammatory response in THP‐1 cells. THP‐1 cells were incubated overnight (a, b) with wild‐type (WT) or PilM1 Lactococcus lactis strains at a multiplicity of infection (MOI) of 10 or 1, or (c, d) with rBP or rAP1at varying concentrations. The Toll‐like receptor 5 agonist flagellin (1 µg mL−1) was used as the positive control and untreated cells as the negative control. The concentration of tumor necrosis factor (TNF) or interleukin 8 (IL‐8) in cell supernatants was measured by ELISA. Experiments were performed in duplicates and data are shown as mean ± s.d. One representative of three independently performed experiments is shown. Statistical significance was determined by one‐way ANOVA, and the P‐values were calculated by Holm–Šídák’s multiple comparisons test. *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 compared with the negative control. AP, ancillary pilin; BP, backbone pilin; ns, not significant; s.d., standard deviation.
Recombinant AP1 and rBP stimulate cytokine release
Our next focus was to determine the specific component(s) of the pilus structure responsible for the stimulation of proinflammatory cytokines. In the pilus structure, AP1 and BP are more likely to be recognized by the host immune response because of their accessibility on the cell surface, whereas AP2 is embedded in the cell wall and thus lacks direct contact with the host. Therefore we focused this part of the study on these two surface‐exposed pilus proteins only. THP‐1 cells were incubated with rBP or rAP1, and the production of TNF and IL‐8 was quantified by ELISA (Figure 1c). At either high (1 nM) or low (100 pM) concentrations, both rBP and rAP1 induced the release of TNF. The concentration of TNF detected from cells incubated with rBP was sixfold higher than that from untreated cells (180 pg mL–1 versus 29 pg mL–1, P < 0.05). Cells incubated with rAP1 released 17‐fold more TNF than untreated cells (488 pg mL–1 versus 29 pg mL–1, P < 0.001). Furthermore, a difference was observed in the TNF‐stimulating ability between BP and AP1. TNF production was 2.7‐fold higher in cells incubated with 1 nM rAP1, compared with cells treated with an equimolar concentration of rBP (P ≤ 0.001). In line with these results, IL‐8 secretion was also highly upregulated in response to both pilus subunits (Figure 1d). Notably, IL‐8 expression was more sensitive to pilin stimulation compared with TNF, where incubation with just 1 pM of rBP or rAP1 resulted in significantly higher levels of IL‐8 induction than the negative control (P < 0.01 for rAP1 and P < 0.003 for rBP). A range of concentrations were tested; however, because of the hypersensitivity in IL‐8 expression, a difference in response between the two pilins could not be discerned (data not shown).
Serotype M1/T1 GAS pilus proteins activate TLR2
TLRs have been shown to mediate a range of inflammatory responses to GAS; however, the specific TLRs involved in these interactions are not yet identified. 35 To examine whether the proinflammatory properties of M1/T1 pilus stems from its engagement with any TLRs, a collection of human embryonic kidney cells that are stably co‐transfected to overexpress individual human TLR genes and a reporter gene encoding an nuclear factor‐kappa B‐inducible secreted embryonic alkaline phosphatase 33 , 37 was employed. The HEK‐Blue hTLR cell lines expressing either human TLR2, TLR4 or TLR5 were selected for the purpose of this study owing to their reported ability to recognize bacterial cell surface components. 38 Incubation of both WT and PilM1 strains of L. lactis caused a colorimetric shift in the detection media of the HEK‐Blue hTLR2 cells, indicative of TLR2 activation (Figure 2a). As expected, the absorbance of HEK‐Blue hTLR2 cells incubated with WT L. lactis was higher compared with untreated cells, as the cell wall of Gram‐positive bacteria contains large amounts of peptidoglycan, a known ligand for TLR2. However, the extent of TLR2 stimulation in cells exposed to PilM1 L. lactis was substantially greater than that in cells incubated with WT L. lactis (1.3‐fold higher at MOI 100, P = 0.02; 1.3‐fold higher at MOI 10, P = 0.02). When these assays were repeated with either TLR4‐ or TLR5‐expressing HEK‐Blue hTLR cells, neither WT L. lactis nor pili expressing L. lactis were able to elicit TLR activation (Figure 2b, c). These results indicate that despite the background stimulation from L. lactis, the activation of TLR2 was enhanced in the presence of pili, suggesting that the pilus structure contains a TLR2 agonist.
Figure 2.
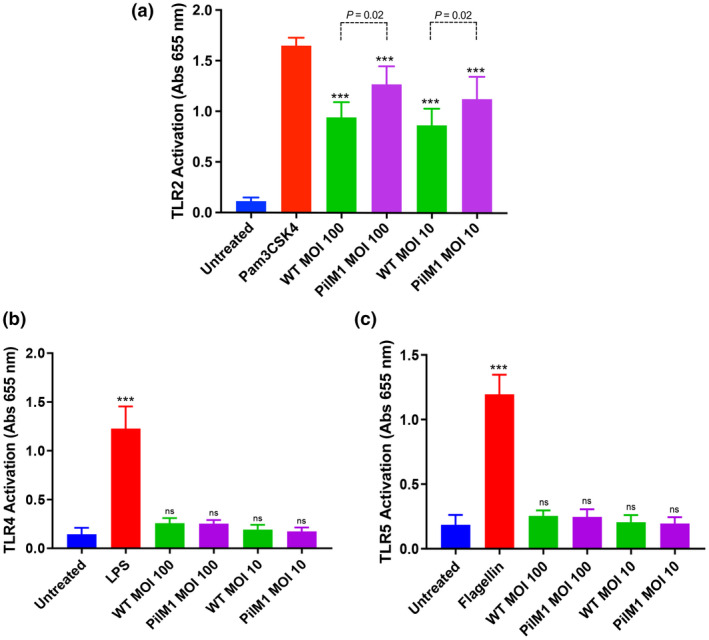
TLR2 is activated in response to the M1/T1 group A Streptococcus pilus. (a) HEK‐Blue hTLR2, (b) HEK‐Blue hTLR4 and (c) HEK‐Blue hTLR5 cells were incubated with wild‐type (WT) or PilM1 Lactococcus lactis strains, positive control (1 μg mL−1 Pam3CSK4 for TLR2, 100 ng mL−1 lipopolysaccharide for TLR4 or 1 μg mL−1 flagellin for TLR5) or left untreated. Levels of TLR activation were determined by absorbance at 655 nm. Comparisons with negative control denoted on experimental groups. Average of three independently performed experiments is shown. Experiments were performed in duplicates and data are shown as mean ± s.d. Statistical significance was determined by one‐way ANOVA, and P‐values were calculated by Holm–Šídák’s multiple comparisons test. ***P ≤ 0.001 compared with the negative control. MOI, multiplicity of infection; ns, not significant; s.d., standard deviation; TLR, Toll‐like receptor.
AP1 and BP proteins from M1/T1 GAS activate TLR2
To evaluate the TLR2 stimulatory abilities of individual pilus proteins, and to identify the main contributor of TLR2 activation in the pilus structure, purified rAP1 and rBP were employed in the HEK‐Blue hTLR assays. Incubation with rBP or rAP1 triggered significantly higher responses in HEK‐Blue hTLR2 cells compared with untreated cells (P < 0.001; Figure 3a), indicating that both rBP and rAP1 play a role in pilus‐mediated activation of TLR2. At 1 pM, both proteins equally caused a 1.6‐fold increase in the activation level relative to untreated cells. However, at 10 pM rAP1 showed a more potent stimulation, causing a threefold higher TLR2 activation compared with untreated cells, whereas rBP triggered a 2.5‐fold higher activation (P < 0.001; Figure 3a).
Figure 3.
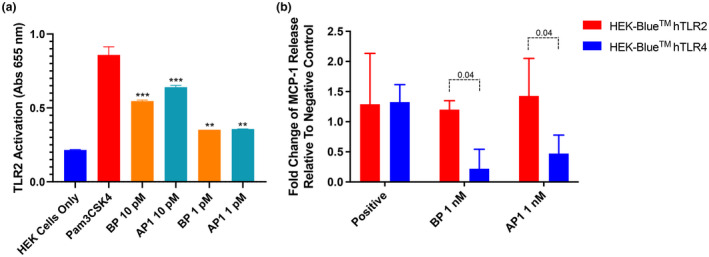
The M1/T1 group A Streptococcus pilus and pilus subunits induce TLR2 activation. (a) HEK‐Blue hTLR 2 cells were incubated with rBP or rAP1, 1 μg mL−1 Pam3CSK4 (positive control) or left untreated (negative control). Levels of TLR2 activation were determined by measuring the absorbance at 655 nm. (b) Alternatively, HEK‐Blue hTLR2 cells and HEK‐Blue hTLR4 cells were incubated with rBP or rAP1, positive control [1 μg mL−1 Pam3CSK4 for TLR2 or 100 ng mL−1 lipopolysaccharide for TLR4] or left untreated. Levels of MCP‐1 secretion in the culture supernatant were measured as the absorbance at 450–570 nm. Absorbance values of the treated cells were expressed as fold change from the absorbance value of untreated cells. Experiments were performed in duplicates and data are shown as mean ± s.d. The average of two independently performed experiments is shown. Statistical significance was determined by two‐way ANOVA, and P‐values were calculated by Holm–Šídák’s multiple comparisons test. **P ≤0.01, ***P ≤ 0.001 compared with the negative control. AP, ancillary pilin; BP, backbone pilin; MCP‐1, monocyte chemoattractant protein 1; s.d., standard deviation; TLR, Toll‐like receptor.
To further verify the involvement of TLR2 in the innate immune response to individual GAS pilus proteins, cytokine secretion from HEK‐Blue hTLR2 incubated with rBP or rAP1 was investigated. HEK‐Blue hTLR4 cells, which did not undergo activation following exposure to pili expressing L. lactis, were included as a negative control for comparison. MCP‐1 was selected as the read‐out for this experiment as it is upregulated downstream of both TLR2 and TLR4 activation. 39 As the sensitivity of pilus subunit–induced MCP‐1 production was lower than TNF and IL‐8 production, HEK‐Blue hTLR2 cells were treated with 1 nM of either rBP1 or rAP1 before cytokine levels were measured. HEK‐Blue hTLR2 treated with rBP and rAP1 released significantly higher levels of MCP‐1 than the untreated cells (Figure 3b). Conversely, incubation with rBP or rAP1 did not result in any marked difference in MCP‐1 production in the treated HEK‐Blue hTLR4 cells compared with their untreated counterparts. Specifically, there was a sixfold difference between BP‐induced hTLR2 and hTLR4 activation (P = 0.04), and a 3.5‐fold difference between rAP1‐induced hTLR2 and hTLR4 activation (P = 0.04). This confirmed that the pilus subunits specifically stimulated MCP‐1 secretion via TLR2, further supporting the implication that the pilus from M1/T1 serotype GAS is a TLR2 agonist.
Pilus proteins from M1/T1 GAS physically interact with TLR2
The results presented in the preceding sections suggest that the M1/T1 pilus interacts with TLR2‐expressing cells. To further establish the specific interaction between pilus proteins and TLR2, and to assess whether direct binding between pilus proteins and TLR2 occurs, we first examined the physical interaction using a solid‐phase binding assay. Binding of varying concentrations of rBP or rAP1 to immobilized recombinant TLR2 was detected using purified antibodies against rBP or rAP1 (Figure 4a). rAP1 demonstrated notable dose‐dependent binding to rTLR2, with the measured absorbance increasing 10‐fold when the concentration of rAP1 was elevated from 5 to 100 pM. Conversely, rBP of the same concentration range only caused a negligible change in absorbance, indicating little to no binding between rBP and rTLR2. The interaction between the pilus proteins and cell‐associated TLR2 was further evaluated by flow cytometry. HEK‐Blue hTLR2 was incubated with fluorescently labeled rBP or rAP1. After performing washing steps to remove unbound proteins, the fluorescence signals were measured. These values were used to calculate the mean fluorescence intensity, which correlated with the amount of cell‐bound pilus protein. When the HEK‐Blue hTLR2 cells were incubated with labeled rAP1, a significant increase in fluorescence intensity was observed compared with the untreated negative control (mean fluorescence intensities of 1,493 and 583, respectively, P < 0.001; Figure 4b), whereas incubation with labeled rBP did not result in a significant shift in mean fluorescence intensity when compared with control cells (Figure 4c). As expected, incubation with either labeled rBP or rAP1 did not alter the fluorescence signals in HEK‐Blue hTLR4 cells, indicating the lack of interaction between pilus proteins and TLR4 (Figure 4b, c).
Figure 4.
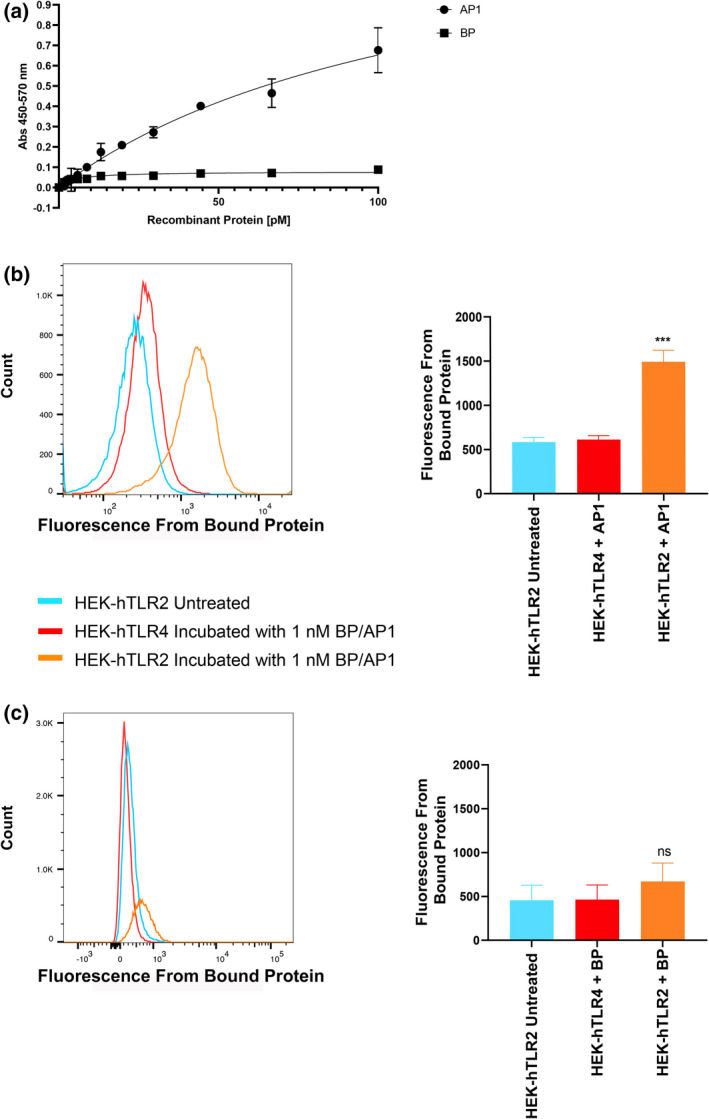
M1/T1 group A Streptococcus pilus proteins physically interact with TLR2. (a) Solid‐phase binding assay of rBP and rAP1 to recombinant TLR2. rBP and rAP1 at various concentrations were incubated with 1 µg mL−1 rTLR2 immobilized on a 96‐well plate. Protein binding was detected by ELISA using purified rabbit antibodies against rAP1 or rBP. A representative of three independent experiments is shown. Experiment was performed in duplicates and data are shown as mean ± s.d. Binding to TLRs expressed on HEK‐293 cells was also analyzed on flow cytometry, using fluorescence‐labeled (b) rAP1 and (c) rBP. Untreated HEK‐Blue hTLR2 was used as a negative control. Fluorescence intensities were measured from 30,000 events for each group. Mean fluorescence intensity data are shown as mean ± s.d. from three independent experiments. Statistical significance was determined by one‐way ANOVA followed by Holm–Šídák’s multiple comparisons test. ***P < 0.001 compared with the untreated negative control. AP, ancillary pilin; BP, backbone pilin; ns, not significant; s.d., standard deviation; TLR, Toll‐like receptor.
Pilus proteins from M1/T1 GAS activate the TLR2/TLR6 heterodimer
Endogenous TLR2 requires heterodimerization with either TLR1 or TLR6 for activation. 32 Inspection of the crystal structure of the heterodimer suggests that upon binding, the ligand stabilizes the two receptors and facilitates interactions between their intracellular moieties to trigger downstream signaling. 40 , 41 HEK cells endogenously express TLR1 and TLR6, and therefore the HEK‐Blue hTLR2 cells are able to dimerize with TLR1 or TLR6 prior to receptor activation. 42 To differentiate specific pathways in pili‐mediated TLR2 activation, we utilized the HEK‐Blue hTLR cell lines that have been modified by a double knockout of endogenous TLR1 and TLR2 genes, followed by coexpression of TLR2 with either TLR1 or TLR6 and the nuclear factor‐kappa B–inducible secreted embryonic alkaline phosphatase reporter. Recombinant BP or rAP1 were incubated with these cell lines and absorbance was read at 655 nm to identify the level of secreted embryonic alkaline phosphatase released downstream of heterodimer activation. In HEK‐Blue hTLR2/hTLR6 cells exposed to either rBP or rAP1, there was a significant increase in absorbance compared with untreated cells (Figure 5a), indicating that both pilus subunits activated TLR2 dimerized to TLR6. In addition, the activation level of cells exposed to rAP1 was significantly higher than that of cells exposed to rBP (twofold difference at 100 pM, P < 0.001; 1.6‐fold difference at 10 pM, P = 0.02). This is consistent with the previous observation that rAP1 was a more potent activator of TLR2. By contrast, no activation of the HEK‐Blue hTLR2/hTLR1 cells was observed in the presence of either rBP or rAP1 (Figure 5b).
Figure 5.
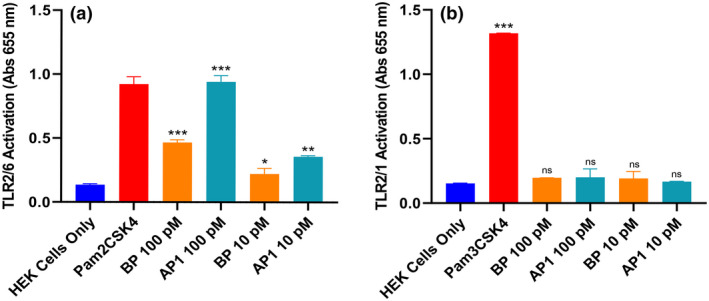
The M1/T1 group A Streptococcus pilus subunits activate TLR2/TLR6 heterodimers. (a) HEK‐Blue hTLR2/hTLR6 cells or (b) HEK‐Blue hTLR2/hTLR1 cells were incubated overnight with rBP or rAP1, positive control (1 μg mL−1 Pam2CSK4 for TLR2/6, or 1 μg mL−1 Pam3CSK4 for TLR2/1) or left untreated. Levels of TLR activation were determined by absorbance at 655 nm. Experiments were performed in duplicates and data are shown as mean ± s.d. One representative of three independently performed experiments is shown. Statistical significance was determined by one‐way ANOVA, and P‐values were calculated by Holm–Šídák’s multiple comparisons test. *P ≤ 0.05, **P ≤ 0.01, ***P < 0.001 compared with the negative control. AP, ancillary pilin; BP, backbone pilin; ns, not significant; s.d., standard deviation; TLR, Toll‐like receptor.
DISCUSSION
TLR‐mediated recognition of bacterial components is an effective mechanism for mounting a rapid defense against pathogens. Moreover, the interaction between pathogen components and the TLRs have implications beyond immediate nonspecific protection, as the activation also serves the role of bridging innate and adaptive immune responses. 26 , 43 Here we show that the GAS pili and individual pilus proteins are potent TLR2 agonists with the ability to induce a strong proinflammatory cytokine response.
GAS is known to induce a range of inflammatory responses from cells. To isolate the effect of pilus from other GAS components, we expressed the pilus structure of an M1/T1 serotype GAS on the surface of L. lactis (PilM1). The PilM1 L. lactis strain elicited significant release of the proinflammatory cytokines IL‐8 and TNF by THP‐1 monocytes when compared with the WT bacterium. Furthermore, individual pilus proteins BP and AP1 induced high levels of cytokine production, suggesting that the assembled pili and individual pilin subunits are potent stimulators of proinflammatory cytokine responses. These results are in line with the findings from previous studies of other Gram‐positive bacteria including Streptococcus agalactiae and Streptococcus pneumoniae, which showed that the piliated strains induced significantly higher levels of chemokine release from immune cells compared with their respective pili deletion mutants. 44 , 45 Interestingly, we noted that the AP1 subunit stimulated a higher level of cytokine release compared with the BP subunit. This trend was echoed in studies investigating Gram‐positive Lacticaseibacillus rhamnosus (previously Lactobacillus rhamnosus), which indicated that deletion of the tip pilus subunit resulted in a substantial reduction in cytokine release from dendritic cells exposed to the bacterium. 46
Numerous studies have implied the involvement of TLRs in proinflammatory cytokine production in response to GAS. For example, mice devoid of TLR signaling protein MyD88 showed severely dampened proinflammatory cytokine release upon GAS infection. 35 Using the HEK‐Blue hTLR cell assays, we confirmed that the M1/T1 pilus, either as a fully assembled structure or as individual subunits, stimulated pronounced TLR2 activation. Notably, AP1 appeared to be a stronger TLR2 agonist than BP, which corresponds to its superior ability to induce cytokine production in THP‐1 cells. These results are congruent with the investigations of Basset et al. 47 on the type 1 pilus of S. pneumoniae, which demonstrated that the tip subunit RrgA, which shares over 70% amino acid sequence similarity with AP1 from M1/T1 GAS, 46 is the main determinant of TLR2‐dependent IL‐8 stimulation in both HEK cells and murine macrophages. 34
Using a plate‐based binding assay and a flow cytometry binding assay, we confirmed the physical interaction between AP1 and TLR2. The lack of association between the pilus proteins and HEK‐Blue hTLR4 further demonstrated that the interaction with, and the activation of, HEK‐Blue hTLR2 was specific to TLR2 rather than nonspecific binding to other cell surface components of the HEK‐Blue hTLR cell lines. TLR2 is one of the most promiscuous of the TLRs, largely owing to its characteristics of heterodimerizing with either TLR1 or TLR6 upon interacting with a ligand. 32 While most of the previously identified TLR2 ligands are either lipids or lipid‐modified polysaccharides or proteins, nonlipidated bacterial proteins including the meningococcal porin PorB 48 and the staphylococcal SSL3 49 have been reported to directly bind to TLR2. However, SSL3 is a TLR2 antagonist that inhibits cytokine production in murine macrophages, 49 and PorB interacts with TLR2 in its TLR2/1 heterodimeric form. 50 To our knowledge, the AP1 from M1/T1 GAS is the first bacterial protein recognized by the TLR2/6 heterodimer without a lipid moiety, emphasizing the unique properties of the GAS pilus.
In GAS, the pilus subunits are encoded by the pilus operon within the FCT (fibronectin‐ and collagen‐binding T‐antigen region) pathogenicity island, of which nine different types have been identified. 51 , 52 The M1/T1 GAS pilus investigated in this study is of the FCT‐2 type, which shares higher sequence similarity to the FCT‐3 and FCT‐4 pili, but are less similar to pili of other FCT types. 51 Interestingly, GAS strains associated with distinct clinical presentations often possess different pilus variants. 9 Therefore, it is reasonable to hypothesize that different GAS pilus variants may exhibit different properties in TLR signaling and cytokine stimulation, and the varying immune‐potentiating effects may link to the different levels of disease manifestations caused by different GAS strains. Further studies are warranted to verify whether there is such connection between the magnitude of cytokine stimulation and disease severity between strains harboring distinct pilus types.
From the results we have amassed, it can be concluded that the M1/T1 GAS pilus is a TLR2 ligand with the ability to induce an innate immune response. The ability of pili to intensify immune responses provides an advantage over other GAS antigens with less intrinsic immunogenicity, supporting the investigation of GAS pili as a vaccine candidate. 53 As nonlipidated molecules with potent TLR2 activity, the AP1 from M1/T1 GAS exhibits activities that suggest utility as a vaccine adjuvant. The capacity to stimulate innate immune cells through TLR2 may also explain the intrinsic immunogenicity of an emerging mucosal vaccine delivery platform based on an L. lactis vector that expresses the serotype M1/T1 GAS pilus. 24 , 25
Further studies are ongoing to explore the molecular basis for the interaction between AP1 and TLR2, and the potential implications in the context of the role of pili in GAS virulence and host immune responses.
METHODS
Bacterial strains and reagents
Bacterial strains are listed in Table 1. L. lactis MG1363 was grown under static conditions at 28°C, in M17 broth supplemented with 5% glucose and 200 µg mL–1 kanamycin where appropriate. The plasmid containing the complete pilus operon from the genome of S. pyogenes serotype M1/T1 strain SF370 (ATCC 700294) under the control of the lactococcal promoter P23 23 was introduced into L. lactis by electroporation as described previously. 23 Escherichia coli BL21 was grown at 37°C in LB containing 50 µg mL–1 ampicillin and 30 µg mL–1 chloramphenicol. The TLR2 agonist Pam3CKS4, the TLR2/6 agonist Pam2CKS4 and the TLR4 agonist LPS from E. coli K12 strain (LPS‐EK) were obtained from InvivoGen (San Diego, CA, USA). ToxOut Endotoxin‐Free Water from BioVision (San Francisco, CA, USA) was utilized as a negative control and diluent in the cell‐based assays.
Table 1.
Bacterial strains used in this study.
| Species | Strain | Relevant characteristics |
|---|---|---|
| Lactococcus lactis | MG1363 | Wild‐type strain |
| MG1363 plZ12‐Km2:P23R:PilM1 | Expresses fully assembled pilus from GAS SF370 (serotype M1/T1 strain) | |
| Escherichia coli | BL21 pET32a3c:bp | Expresses rBP from GAS SF370, fused to a thioredoxin and N‐terminal His6 tag and a protease 3C site 54 |
| BL21 pPROEX‐Htb:ap1 | Expresses rAP1 from GAS SF370 with N‐terminal His6 tag | |
| BL21 pPROEX‐Htb:Fla | Expresses flagellin from Salmonella enterica subsp. enterica serovar Typhimurium strain |
AP, ancillary pilin; GAS, group A Streptococcus.
Protein cloning, expression and purification
Open reading frames encoding BP and AP1 without the N‐terminal signal peptide were amplified from genomic S. pyogenes SF370 (ATCC 700294) DNA using primers M1_bp.fw + M1_bp.rev, M1_ap1.fw + M1_ap1.rev listed in Table 2, and subsequently cloned into the pET32a vector and the pROEx‐Htb vector, respectively, to be expressed in E. coli BL21(DE3) pLysS (Novagen, San Diego, CA, USA). DNA encoding full‐length flagellin was amplified from Salmonella enterica subsp. enterica serovar Typhimurium strain 13‐931 using the primers Fla_BamHI.fw and Fla_XhoI.rev listed in Table 2 and cloned into the pROEx‐Htb vector. Cultures were induced mid‐log phase (OD600nm = 0.6) using 0.1 mM IPTG for 4 h at 28°C. The E. coli cells were harvested and, following resuspension in lysis buffer (20 mM Tris, 0.5 M NaCl, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 0.1% Triton X‐100), lysed by sonication. The His6‐tagged proteins were purified from the lysates on Ni2+‐nitrilotriacetic acid Sepharose 6 Fast Flow nickel resin (GE Healthcare, Chicago, IL, USA) according to the manufacturer’s instructions. All purified proteins were pretreated with 100 µg mL–1 Polymyxin B (Sigma‐Aldrich, St. Louis, MO, USA) for 15 min at room temperature (RT) prior to use in assays to inactivate any contaminating endotoxins.
Table 2.
PCR primers used in this study. Restriction enzyme sequences are underlined.
| Primer name | Sequence (5’‐3’) |
|---|---|
| M1_bp.fw | CGGGATCCGCTACAACAGTTCACGG |
| M1_bp.rev | CGGAATTCTTATTCAAAGACTTTTTTATTTG |
| M1_ap1.fw | GGATCCAAGACTGTTTTTGGTTTAG |
| M1_ap1.rev | CTCGAGCTAACCAGTTTCTGGCAAAGGCTCTTTATTATTTTC |
| Fla_BamHI.fw | GCGGATCCATGGCACAAGTCATTAATAC |
| Fla_XhoI.rev | GAGAGAGACTCGAGACGCAGTAAAGAGAGGACG |
Fluorescently tagged recombinant pilus proteins were prepared by incubating recombinant AP1 (rAP1) and rBP in bicarbonate buffer (50 mM NaHCO3) with equimolar amounts of Red Mega 485 NHS fluorescent label (Sigma‐Aldrich, St. Louis, MO, USA) for 1 h at RT. Labeled proteins were separated from unbound fluorescent tags by passing through a G‐25 Sephadex desalting column (Cytiva, Marlborough, MA, USA).
Cell lines
Human myelomonocytic THP‐1 cells (ATCC TIB‐202) were maintained in Roswell Park Memorial Institute‐1640 (RPMI‐1640) medium supplemented with 10% fetal bovine serum, 0.05 mM 2‐mercaptoethanol, 100 U mL–1 penicillin and 100 µg mL–1 streptomycin. The commercially available HEK‐Blue hTLR cell lines (InvivoGen, San Diego, CA, USA) were maintained in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum and the antibiotics specified in the manufacturer’s instructions, grown at 37°C with 5% CO2 and kept below 15 passages.
Cytokine assay
THP‐1 cells were seeded into a 96‐well plate at a density of 1 × 105 cells per well in antibiotic‐free RPMI‐1640 medium. L. lactis cells were grown overnight and resuspended in RPMI‐1640, using the conversion rate of OD600nm of 1.0 = ~1 × 108 CFU mL–1 to determine the cell density. THP‐1 cells were treated with 1 × 105 or 1 × 106 L. lactis cells to achieve an MOI of 1 or 10, respectively. Alternatively, rBP or rAP1 were added at a final concentration of 1 nM or 100 pM. RMPI‐1640 medium was used as a negative control and 2.5 µM recombinant flagellin protein or 1 µg mL–1 LPS‐EK was used as a positive control where appropriate. The plate was incubated for 20 h at 37°C in the presence of 5% CO2 before culture supernatants were collected and assessed for presence of cytokines with the Crux Biolab human cytokine ELISA kits (Bayswater, VIC, Australia) as per the manufacturer’s instructions.
HEK‐Blue hTLR cells were grown until 80% confluency was achieved, then detached and resuspended in HEK‐Blue Detection medium (InvivoGen, San Diego, CA, USA). L. lactis cells were prepared as above and added to the HEK‐Blue hTLR cells at an MOI of 10. Alternatively, rBP or rAP1 was added to cells at a concentration of 2 nM. Pam3CSK4 or LPS at 1 µg mL–1 was added where appropriate as a positive control, and RPMI‐1640 was used as a negative control. The plate was then incubated for 20 h at 37°C and 5% CO2, and culture supernatants were assayed for cytokines with the Crux Biolab human cytokine ELISA kits as per the manufacturer’s instructions.
Toll‐like receptor reporter assay
The HEK‐Blue hTLR cells grown to a confluency of 80% were detached and suspended in HEK‐Blue Detection medium at a density of 2.8 × 105 cells mL–1. Overnight L. lactis cultures were washed in phosphate‐buffered saline (PBS), and the cell density was determined as earlier. Aliquots of L. lactis were stored in 10% glycerol at −80°C and used across assays for consistency. Immediately prior to setting up the assay, aliquots were thawed completely on ice, washed in PBS and resuspended in endotoxin‐free water at 2.5 × 108 CFU mL–1 and 2.5 × 107 CFU mL–1. Recombinant BP or rAP1 was diluted in endotoxin‐free water at concentrations of 1 nM or 100 pM. Where appropriate, 1 µg mL–1 Pam3CSK4, 10 ng mL–1 Pam2CSK4, 100 ng mL–1 LPS‐EK or 25 nM recombinant flagellin was prepared in endotoxin‐free water for use as positive controls, and endotoxin‐free water was used as a negative control. Test or control samples were added to each well of a 96‐well plate at 20 µL followed by 180 µL per well of the HEK‐Blue cell suspension. The plate was then incubated at 37°C, 5% CO2 for 6–9 h before the A655nm was read.
Solid‐phase binding assays
Recombinant TLR2 (Sino Biological, Beijing, China) was diluted to 1 µg mL–1 in carbonate–bicarbonate buffer and added to a 96‐well plate at 100 µL per well. The plate was incubated overnight at 4°C, then washed with PBS‐T buffer (1× PBS/0.05% Tween‐20) and blocked with 3% bovine serum albumin in PBS‐T. After a washing step, the plate was incubated for 3 h at RT with 100 µL per well of rBP or rAP1 in PBS at concentrations between 100 pM and 1.73 pM. This was followed by plate washing and the addition of 100 µL per well of purified primary rabbit antibodies against rBP or rAP1 23 for 2 h at RT. The plate was subsequently washed and incubated with HRP‐conjugated goat anti‐rabbit IgG antibody (1:1000; Abcam, Cambridge, UK) for 1 h at RT. After a final wash step, the plate was incubated for 10 min with 100 μL per well of 3,3,5,5‐tetramethylbenzidine, before 100 μL per well of 1 M HCl was added to stop the reaction prior to taking A450/570nm readings.
Flow cytometry binding assay
HEK‐Blue hTLR2 cells and HEK‐Blue hTLR4 cells were detached using 10 mM ethylenediaminetetraacetic acid, washed with PBS and suspended in cold fluorescence‐activated cell sorting buffer (1× PBS/1% fetal bovine serum/5 mM ethylenediaminetetraacetic acid) at a density of 1 × 107 cells mL–1. Recombinant BP or rAP1 labeled with Fluorescent Red Mega 485 NHS‐ester (Sigma‐Aldrich, St. Louis, MO, USA) was diluted in fluorescence‐activated cell sorting buffer to a concentration of 2 nM, and incubated with the HEK‐Blue hTLR2 or HEK‐Blue hTLR4 cell suspension at a 1:1 ratio for 30 min. HEK‐Blue hTLR2 with fluorescence‐activated cell sorting buffer only was also prepared as a negative control. Cells were washed two times, resuspended in 0.5 mL fluorescence‐activated cell sorting buffer and strained before fluorescence data were acquired on an LSR II Flow Cytometer set to record 30 000 events and analyzed with FlowJo version 10.7 (BD Biosciences, San Francisco, CA, USA).
Statistical analysis
Statistical significance was determined by one‐way ANOVA (two‐way ANOVA for the MCP‐1 assay) followed by Holm–Šídák’s multiple comparisons test using PRISM (version 8.0; GraphPad Software Inc.).
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTION
Risa Takahashi: Formal analysis; Investigation; Writing—original draft; Writing—review and editing. Fiona J Radcliff: Methodology; Writing—review and editing. Thomas Proft: Methodology; Resources; Supervision; Writing—review and editing. Catherine J‐Y Tsai: Conceptualization; Funding acquisition; Investigation; Supervision; Validation; Writing—original draft; Writing—review and editing.
ACKNOWLEDGMENTS
Ms Takahashi is a recipient of the University of Auckland Health Research Doctoral Scholarship. Dr Tsai is an Auckland Medical Research Foundation Postdoctoral Fellow. The rabbit anti‐BP and anti‐AP1sera were kindly provided by Dr Jacelyn Loh at the University of Auckland. The protein fluorescent labeling was conducted with the help of Dr Ries Langley at the University of Auckland. Open access publishing facilitated by The University of Auckland, as part of the Wiley ‐ The University of Auckland agreement via the Council of Australian University Librarians.
References
- 1. Manetti AGO, Zingaretti C, Falugi F, et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol 2007; 64: 968–983. [DOI] [PubMed] [Google Scholar]
- 2. Castro SA, Dorfmueller HC. A brief review on Group A Streptococcus pathogenesis and vaccine development. R Soc Open Sci 2021; 8: 201991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sims Sanyahumbi A, Colquhoun S, Wyber R, Carapetis JR. Global Disease Burden of Group A Streptococcus. In: Ferretti JJ, Stevens DL, Fischetti, eds. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City, OK: University of Oklahoma Health Sciences Center: 2016: 1–44. [PubMed] [Google Scholar]
- 4. Cannon JW, Zhung J, Bennett J, et al. The economic and health burdens of diseases caused by group A Streptococcus in New Zealand. Int J Infect Dis 2021; 103: 176–181. [DOI] [PubMed] [Google Scholar]
- 5. Efstratiou A, Lamagni T. Epidemiology of Streptococcus pyogenes . In: Ferretti JJ, Stevens DL, Fischetti, eds. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City, OK: University of Oklahoma Health Sciences Center: 2016: 1–27. [PubMed] [Google Scholar]
- 6. Carapetis JR. Rheumatic heart disease in developing countries. N Engl J Med 2007; 357: 439–441. [DOI] [PubMed] [Google Scholar]
- 7. Ralph AP, Carapetis JR. Group A streptococcal diseases and their global burden. Curr Top Microbiol Immunol 2013; 368: 1–27. [DOI] [PubMed] [Google Scholar]
- 8. Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis 2003; 3: 191–200. [DOI] [PubMed] [Google Scholar]
- 9. Walker MJ, Barnett TC, McArthur JD, et al. Disease manifestations and pathogenic mechanisms of group a Streptococcus . Clin Microbiol Rev 2014; 27: 264–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crotty Alexander LE, Maisey HC, Timmer AM, et al. M1T1 group A streptococcal pili promote epithelial colonization but diminish systemic virulence through neutrophil extracellular entrapment. J Mol Med (Berl) 2010; 88: 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lizano S, Luo F, Bessen DE. Role of streptococcal T antigens in superficial skin infection. J Bacteriol 2007; 189: 1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ton‐That H, Schneewind O. Assembly of pili in gram‐positive bacteria. Trends Microbiol 2004; 12: 228–234. [DOI] [PubMed] [Google Scholar]
- 13. Tsai JYC, Loh JMS, Clow F, Lorenz N, Proft T. The Group A Streptococcus serotype M2 pilus plays a role in host cell adhesion and immune evasion. Mol Microbiol 2017; 103: 282–298. [DOI] [PubMed] [Google Scholar]
- 14. Proft T, Baker EN. Pili in Gram‐negative and Gram‐positive bacteria ‐ structure, assembly and their role in disease. Cell Mol Life Sci 2009; 66: 613–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mora M, Bensi G, Capo S, et al. Group A Streptococcus produce pilus‐like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA 2005; 102: 15641–15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young PG, Moreland NJ, Loh JM, et al. Structural conservation, variability, and immunogenicity of the T6 backbone Pilin of serotype M6 Streptococcus pyogenes . Infect Immun 2014; 82: 2949–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loh JMS, Lorenz N, Tsai CJ‐Y, Khemlani AHJ, Proft T. Mucosal vaccination with pili from Group A Streptococcus expressed on Lactococcus lactis generates protective immune responses. Sci Rep 2017; 7: 7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loh JMS, Rivera‐Hernandez T, McGregor R, et al. A multivalent T‐antigen‐based vaccine for Group A Streptococcus. Sci Rep 2021; 11: 4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soriani M, Telford JL. Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol 2010; 5: 735–747. [DOI] [PubMed] [Google Scholar]
- 20. Moreland NJ, Waddington CS, Williamson DA, et al. Working towards a group A streptococcal vaccine: report of a collaborative Trans‐Tasman workshop. Vaccine 2014; 32: 3713–3720. [DOI] [PubMed] [Google Scholar]
- 21. Steemson JD, Moreland NJ, Williamson D, Morgan J, Carter PE, Proft T. Survey of the bp/tee genes from clinical group A streptococcus isolates in New Zealand – implications for vaccine development. J Med Microbiol 2014; 63: 1670–1678. [DOI] [PubMed] [Google Scholar]
- 22. Chamcha V, Jones A, Quigley BR, Scott JR, Amara RR. Oral immunization with a recombinant Lactococcus lactis expressing HIV‐1 antigen on GAS pilus induces strong mucosal immunity in the gut. J Immunol 2015; 195: 5025–5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wagachchi D, Tsai JYC, Chalmers C, Blanchett S, Loh JMS, Proft T. PilVax ‐ A novel peptide delivery platform for the development of mucosal vaccines. Sci Rep 2018; 8: 2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blanchett S, Tsai CJ, Sandford S, et al. Intranasal immunization with Ag85B peptide 25 displayed on Lactococcus lactis using the PilVax platform induces antigen‐specific B‐ and T‐cell responses. Immunol Cell Biol 2021; 99: 767–781. [DOI] [PubMed] [Google Scholar]
- 25. Clow F, Peterken K, Pearson V, Proft T, Radcliff FJ. PilVax, a novel Lactococcus lactis‐based mucosal vaccine platform, stimulates systemic and mucosal immune responses to Staphylococcus aureus . Immunol Cell Biol 2020; 98: 369–381. [DOI] [PubMed] [Google Scholar]
- 26. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140: 805–820. [DOI] [PubMed] [Google Scholar]
- 27. Janeway CA. A primitive immune system. Nature 1989; 341: 108. [DOI] [PubMed] [Google Scholar]
- 28. Kawai T, Akira S. The role of pattern‐recognition receptors in innate immunity: Update on Toll‐like receptors. Nat Immunol 2010; 11: 373–384. [DOI] [PubMed] [Google Scholar]
- 29. Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol 2015; 16: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 31. Oliveira‐Nascimento L, Massari P, Wetzler LM. The role of TLR2 in infection and immunity. Front Immunol 2012; 3: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll‐like receptors. Proc Natl Acad Sci USA 2000; 97: 13766–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X, Fan X, Bi S, Li N, Wang B. Toll‐like receptors 2 and 4‐mediated reciprocal Th17 and antibody responses to group A streptococcus infection. J Infect Dis 2017; 215: 644–652. [DOI] [PubMed] [Google Scholar]
- 34. Loof TG, Goldmann O, Medina E. Immune recognition of Streptococcus pyogenes by dendritic cells. Infect Immun 2008; 76: 2785–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loof TG, Goldmann O, Gessner A, Herwald H, Medina E. Aberrant inflammatory response to Streptococcus pyogenes in mice lacking myeloid differentiation factor 88. Am J Pathol 2010; 176: 754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ciacci‐Woolwine F, Blomfield IC, Richardson SH, Mizel SB. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect Immun 1998; 66: 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu J, Ma C, Wang H, et al. A MyD88‐JAK1‐STAT1 complex directly induces SOCS‐1 expression in macrophages infected with Group A Streptococcus . Cell Mol Immunol 2015; 12: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van der Aar AMG, Sylva‐Steenland RMR, Bos JD, Kapsenberg ML, de Jong EC, Teunissen MBM. Cutting edge: Loss of TLR2, TLR4, and TLR5 on langerhans cells abolishes bacterial recognition. J Immunol 2007; 178: 1986–1990. [DOI] [PubMed] [Google Scholar]
- 39. Schaefer TM, Desouza K, Fahey JV, Beagley KW, Wira CR. Toll‐like receptor (TLR) expression and TLR‐mediated cytokine/chemokine production by human uterine epithelial cells. Immunology 2004; 112: 428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin MS, Kim SE, Heo JY, et al. Crystal structure of the TLR1‐TLR2 heterodimer induced by binding of a tri‐acylated lipopeptide. Cell 2007; 130: 1071–1082. [DOI] [PubMed] [Google Scholar]
- 41. Kang JY, Nan X, Jin MS, et al. Recognition of lipopeptide patterns by toll‐like receptor 2‐toll‐like receptor 6 heterodimer. Immunity 2009; 31: 873–884. [DOI] [PubMed] [Google Scholar]
- 42. Kirschning CJ, Wesche H, Merrill Ayres T, Rothe M. Human toll‐like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med 1998; 188: 2091–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 2009; 22: 240–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Banerjee A, Kim BJ, Carmona EM, et al. Bacterial Pili exploit integrin machinery to promote immune activation and efficient blood‐brain barrier penetration. Nat Commun 2011; 2: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barocchi MA, Ries J, Zogaj X, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA 2006; 103: 2857–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Izoré T, Contreras‐Martel C, El Mortaji L, et al. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae . Structure 2010; 13: 106–115. [DOI] [PubMed] [Google Scholar]
- 47. Basset A, Turner K, Boush E, Sayeed S, Dove S, Malley R. Expression of the Type 1 Pneumococcal Pilus Is Bistable and Negatively Regulated by the Structural Component RrgA. Infect Immun 2011; 79: 2974–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Massari P, Visintin A, Gunawardana J, et al. Meningococcal porin PorB binds to TLR2 and requires TLR1 for signaling. J Immunol 2006; 176: 2373–2380. [DOI] [PubMed] [Google Scholar]
- 49. Yokoyama R, Itoh S, Kamoshida G, et al. Staphylococcal superantigen‐like protein 3 binds to the toll‐like receptor 2 extracellular domain and inhibits cytokine production induced by Staphylococcus aureus, cell wall component, or Lipopeptides in murine macrophages. Infect Immun 2012; 80: 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wetzler LM. Innate immune function of the neisserial porins and the relationship to vaccine adjuvant activity. Future Microbiol 2010; 5: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Falugi F, Zingaretti C, Pinto V, et al. Sequence variation in group A Streptococcus pili and association of pilus backbone types with lancefield T serotypes. J Infect Dis 2008; 198: 1834–1841. [DOI] [PubMed] [Google Scholar]
- 52. Nakata M, Köller T, Moritz K, et al. Mode of Expression and Functional Characterization of FCT‐3 Pilus Region‐Encoded Proteins in Streptococcus pyogenes Serotype M49. Infect Immun 2009; 77: 32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Azuar J, Mukaida H, Toth S. Recent Advances in the Development of Peptide Vaccines and Their Delivery Systems against Group A Streptococcus. Vaccines 2019; 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Proft T, Webb PD, Handley V, Fraser JD. Two Novel Superantigens Found in Both Group A and Group C Streptococcus . Infect Immun 2003; 71: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]


