Abstract
Autophagy, a catabolic process to remove unnecessary or dysfunctional organelles, is triggered by various signals including nutrient starvation. Depending on the types of the nutrient deficiency, diverse sensing mechanisms and signaling pathways orchestrate for transcriptional and epigenetic regulation of autophagy. However, our knowledge about nutrient type-specific transcriptional regulation during autophagy is limited. To understand nutrient type-dependent transcriptional mechanisms during autophagy, we performed single cell RNA sequencing (scRNAseq) in the mouse embryonic fibroblasts (MEFs) with or without glucose starvation (GS) as well as amino acid starvation (AAS). Trajectory analysis using scRNAseq identified sequential induction of potential transcriptional regulators for each condition. Gene regulatory rules inferred using TENET newly identified CCAAT/enhancer binding protein γ (C/EBPγ) as a regulator of autophagy in AAS, but not GS, condition, and knockdown experiment confirmed the TENET result. Cell biological and biochemical studies validated that activating transcription factor 4 (ATF4) is responsible for conferring specificity to C/EBPγ for the activation of autophagy genes under AAS, but not under GS condition. Together, our data identified C/EBPγ as a previously unidentified key regulator under AAS-induced autophagy.
INTRODUCTION
Maintaining homeostasis is important for cellular function and human health. During the response to nutrient stresses, cells undergo rapid changes to adapt to harmful environments and protect themselves against potential damages. One of the key regulatory mechanisms of maintaining homeostasis is autophagy. Autophagy, which literally means ‘self-eating’, is an intracellular degradation system that removes unnecessary or dysfunctional components (1,2). Under stresses such as nutrient deprivation, autophagy promotes degradation of cellular components to replenish insufficient nutrients to maintain the metabolic homeostasis of cells.
Nutrient scarcity is sensed by distinct mechanisms depending on the nutrient types. Amino acids (AA) deficiency is detected by kinase general control nonderepressible-2 (GCN2) (3), which has high affinity to all uncharged tRNAs that are accumulated during low levels of free AA. The GCN2-dependent phosphorylation of eukaryotic initiation factor 2α (eIF2α) leads to a decrease in the functional complex required for delivering methionine to the ribosome for initiating translation (4). As a result, global protein translation is repressed, trying to overcome the AA deficiency. In the meanwhile, the translation of a subset of mRNAs is still increasing (5). These are associated with AA biosynthesis regulators, AA transporters, and autophagy mediators including ATF4 and CHOP (5). There is also a strategy to overcome AA deficiency through transcriptional regulation. mTORC1 is the key hub coordinating the availability of AA and autophagy (6). Upon amino acid starvation (AAS), mTORC1 becomes inactivated, which subsequently activates the Unc51-like kinase 1 (ULK1) complex, a complex playing central role in initiating as well as terminating autophagy (7,8).
Glucose levels are sensed by proteins including the glucose transporter GLUT2 (SLC2A2) (9,10), and are detected by glucokinase (GCK) (11). When glucose starvation (GS) occurs, autophagy is activated as a mechanism to replenish ATP from cellular components (12). The drop in energy results in an increase in the AMP/ATP ratio, which is sensed by AMPK to induce autophagy (13). GS triggers AMP-activated protein kinase (AMPK) which directly phosphorylates ULK1 at S317 and S777. Once phosphorylated, ULK1 forms a complex with ATG13 and FIP200 leading to an induction of autophagy (14,15).
Under nutrient deficient condition, transcriptional regulation of autophagy in the nucleus as well as cytoplasmic event is important (16,17). A number of transcription factors (TFs) play key roles for autophagy including TFEB, FOXO3a, p53, E2F1 and CREB (18) (Table 1). In response to AAS, TFEB translocates into the nucleus to promote autophagy that recycles unnecessary organelles to increase amino acid availability (19). TFEB subcellular localization is controlled by the AA sensing mTORC1 that phosphorylates TFEB on S211 to enable cytoplasmic sequestration through 14–3–3 protein interaction (19,20). Under glucose starvation, CARM1 is crucial for TFEB-dependent transcriptional activation of autophagy genes with increased histone H3R17 dimethylation (21). Further, CARM1 is responsible for arginine methylation of Pontin, which confers binding ability of Pontin to TUDOR domain of FOXO3a. Thus, the FOXO3a-Pontin-Tip60 complex formed by GS works for enhancer activation of lysosomal and autophagy genes (22). Of note, transcriptional regulation of autophagy has largely been studied without distinguishing nutrient sources. For this reason, controlled studies that consider the source of nutrient deficiency will highly enhance our understanding of transcriptional regulation in autophagy.
Table 1.
Functions and target genes of transcription factors by signals
| TF | Signal | Functions | Target genes | References |
|---|---|---|---|---|
| TFEB | GS/AAS | transcriptional activator of genes involved in various steps of the autophagy process. | BECN1, WIPI1, ATG9B, NRBF2, GABARAP, MAP1LC3B, and ATG5 | (19,20) |
| FOXO3a | GS | transcriptional activator of several autophagy genes through enhancer activation | ATG14, LC3, SIRT1, CLN5, and CTNS | (22) |
| p53 | GS | transcriptional activator of Sestrin, which activates AMPK while inhibiting mTORC1 lysosomal recruitment | SESTRIN1/2 and DRAM | (47) |
| E2F1 | Hypoxia | transcriptional activator of BNIP3, which promotes autophagosome biogenesis | BNIP3, ULK1, LC3 and ATG5 | (48) |
| CREB | HBSS | transcriptional activator with CRTC2 of TFEB, which enhances the activity of PPARα | ATG7, ULK1, and TFEB | (49) |
To understand nutrient deficiency dependent gene regulation, we generate single cell RNA sequencing (scRNAseq) data following GS and AAS to mouse embryonic fibroblasts (MEFs). Compared with bulk RNAseq, scRNAseq can provide dynamic changes of gene expression through trajectory analysis (23–25). In particular, gene regulatory rules can be inferred from scRNAseq collected from a number of samples (26). The analysis using scRNAseq for autophagy identified sets of autophagy genes regulated commonly as well as distinctively upon the nutrient deficient condition. Trajectory analysis manifested the sequential induction of TFs such as ATF4 and C/EBPγ followed by autophagy genes. To identify key regulators for each condition, we applied the TENET algorithm (27) which we previously developed to construct gene regulatory networks (GRNs) from scRNAseq data. Interestingly, TENET newly identified C/EBPγ as a key regulator for AAS-, but not GS-induced autophagy. Strikingly, knocking down of C/EBPγ inhibited the induction of the predicted C/EBPγ target genes and attenuated autophagic occurrence in the AAS condition. We further identified that C/EBPγ assisted the recruitment of ATF4 to the binding targets during the AAS-induced autophagic processes. Our results showed C/EBPγ in combination with ATF4 plays an important role in the AAS-induced autophagy process.
MATERIALS AND METHODS
Reagents
The following commercially available antibodies were used: anti-β-actin (A1978) (Sigma-Aldrich); anti-GFP (sc-9996), anti-ATF4 (sc-390063) and anti-C/EBPγ (sc-517003) (Santa Cruz biotechnology); anti-LC3 (#2775) (Cell Signaling technology); anti-C/EBPγ (ab74045) and anti-C/EBPβ (ab264306) (Abcam). Following commercially available fluorescent-labeled secondary antibodies were used: Alexa Fluor 488 donkey anti-rabbit IgG (A21206) and Alexa Fluor 594 donkey anti-mouse IgG (A21203) (Invitrogen). We used antibodies recommended by the manufacturer for the species and application. Bafilomycin A1 (#11038) was from Cayman.
Cell culture
MEFs were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics in a humidified incubator with 5% CO2. MEFs used in the study was regularly tested for mycoplasma contamination. For amino acid starvation (AAS), cells were washed with PBS 2 times, then incubated with all amino acids depleted DMEM supplemented with 10% dialyzed FBS. For glucose starvation (GS), cells were washed with PBS 2 times, then incubated with D-glucose depleted DMEM supplemented with 10% dialyzed FBS. For the control condition, cells were washed with PBS 2 times, then incubated with normal DMEM supplemented with 10% FBS. Transfection was performed with Lipofectamine 3000 (Invitrogen).
Single-cell RNA-seq experiment
5 × 105 of MEFs were plated on culture dish containing high glucose DMEM (Cat. SH30243.01) with FBS and were stabilized for one day. After stabilization, the cells were washed with either glucose starvation media (Cat. LM001-56) or amino acid starvation media (Cat. LM001-90) containing dialyzed 10% FBS. Subsequently, the normal media was changed with appropriate starvation media and cells were incubated in the cell incubator for different duration of time according to starvation signal (amino acid starvation for 3 and 6 hours, glucose starvation for 6 and 12 hours). At the end of starvation, MEFs were collected using trypsin EDTA and resuspended in freezing media (10% DMSO + 90% dialyzed FBS). The control sample without starvation was collected and resuspended in freezing media composed of 90% FBS instead of dialyzed FBS. Vials containing each sample were stored in a deep freezer before proceeding to sequencing. The viability of samples was checked using Countess and 5000 cells from each sample were used for single cell isolation. All scRNAseq libraries were prepared using Single Cell 3′ v2 chemistry 10X chromium Kit and sequenced using Illumina HiSeq 4000.
Immunofluorescence
Cells grown on coverslips at a density of 5 × 104 cells were washed with PBS, then fixed with 2% paraformaldehyde in PBS at room temperature (RT) for 10 min. Fixed cells were permeabilized with 0.5% Triton X-100 in PBS (PBS-T) at RT for 10 min. Blocking was performed with 3% bovine serum in PBS-T for 1 h. For staining, cells were incubated with antibodies at RT for 2 h, followed by incubation with fluorescent-labeled secondary antibodies for 1 h. Cells were mounted and visualized under a confocal microscope (Zeiss, LSM700). For autophagy studies, MEFs were transfected with GFP-LC3 and sub-cultured onto coverslips. The following day, cells were cultured in either complete media or amino acid/glucose-starved media.
Quantitative RT-PCR
Total RNAs were extracted using Trizol (Invitrogen) and reverse transcription was performed from 2.5 μg of total RNAs using the SuPrimeScript cDNA Synthesis Kit (GeNet Bio). The abundance of mRNA was detected by an ABI prism 7500 system or BioRad CFX384 with SYBR TOPreal qPCR 2X PreMix (Enzynomics). The quantity of mRNA was calculated using ddCt method and Hprt, Gapdh and β-actin were used as controls. All reactions were performed as triplicates. The following mouse primers were used.
β-actin; forward (fwd) 5′-TAGCCATCCAGGCTGTGCTG-3′,
reverse (rev) 5′-CAGGATCTTCATGAGGTAGTC-3′;
Gapdh; fwd 5′-CATGGCCTTCCGTGTTCCTA-3′,
rev 5′-CCTGCTTCACCACCTTCTTGA-3′;
Hprt; fwd 5′-GCTGGTGAAAAGGACCTCTCG-3′,
rev 5′-CCACAGGACTAGAACACCTGC-3′;
Map1lc3b; fwd 5′-CACTGCTCTGTCTTGTGTAGGTTG-3′,
rev 5′-TCGTTGTGCCTTTATTAGTGCATC-3′;
Bnip3; fwd 5′-GCTCCCAGACACCACAAGAT-3′,
rev 5′-TGAGAGTAGCTGTGCGCTTC-3′;
Vps26a; fwd 5′-AGCACCACAACAGAGACAGA-3′,
rev 5′-TCTCAGTTTCTCGGGTGCTT-3′;
Gabarap; fwd 5′-AAGAGGAGCATCCGTTCGAGA-3′,
rev 5′-GCTTTGGGGGCTTTTTCCAC-3′;
Gadd45b; fwd 5′-ACAACGCGGTTCAGAAGATG-3′,
rev 5′-TGTCATTGTCGCAGCAGAAC-3′;
Atg12; fwd 5′-TCCGTGCCATCACATACACA-3′,
rev 5′-TAAGACTGCTGTGGGGCTGA-3′;
Atf4; fwd 5′-TGGCCATCTCCCAGAAAGTT-3′,
rev 5′-TGCTCTGGAGTGGAAGACAG-3′;
Cebpg; fwd 5′-GAGAGCGGAACAATATGGCG-3′,
rev 5′-TGGCTTCCAACCGTTCATTC-3′.
ChIP assay
The ChIP assays were conducted as previously described (28). In brief, cells were crosslinked with 1% formaldehyde for 10 min at room temperature. After glycine quenching, the cells were collected and lysed in a buffer containing 50 mM Tris–HCl (pH 8.1), 10 mM EDTA, 1% SDS, supplemented with complete protease inhibitor cocktail (Roche). After sonication, chromatin extracts containing DNA fragments with an average of 250 bp were then diluted ten times with dilution buffer containing 1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris–HCl (pH 8.1) with complete protease inhibitor cocktail and subjected to immunoprecipitations overnight at 4°C. Immunocomplexes were captured by incubating 40 μl of protein A/G Sepharose for 2 h at 4°C. Beads were washed with TSE I buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.1) and 150 mM NaCl), TSE II buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.1) and 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 10 mM Tris–HCl (pH 8.1) and 1 mM EDTA), three times TE buffer (10 mM Tris–HCl (pH 8.0) and 1 mM EDTA) and eluted in elution buffer (1% SDS and 0.1 M NaHCO3). The supernatant was incubated overnight at 65 °C to reverse-crosslink, and then digested with RNase A for 2 h at 37°C and proteinase K for 2 h at 55°C. ChIP and input DNA were then purified and analyzed for qRT–PCR or used for constructing sequencing libraries. The ChIP assays were conducted with the following primers.
Map1lc3b; fwd 5′-CGGACTGAGACACACACAAG-3′,
rev 5′-ACTCTTTGTTCAAAGCTCCGG-3′;
Bnip3; fwd 5′-GTTCCAGCCTCCGTCTCTAT-3′,
rev 5′-ATCTTGTGGTGTCTGGGAGC-3′;
Atg12; fwd 5′-GCCTCTCAGCAAGCAAAGAT-3′,
rev 5′-GTTTCCGGGGAGAGCTCTG-3′;
Gadd45b; fwd 5′-CCTCTTGGGTTCGTATCTGGA-3′,
rev 5′-CATCTTCTGAACCGCGTTGT-3′;
Vps26a; fwd 5′-CTAGCCTTGCCTGGAGAACT-3′,
rev 5′-ATACCTCAGGCGGACATTGG-3′.
Electron microscopy
Cells were fixed in 0.1 M sodium cacodylate containing 4% glutaraldehyde and 1% paraformaldehyde for 1h at room temperature. After washing three times with 0.1 M sodium cacodylate, cells were dehydrated through a gradient series of ethanol, 20 min each step, starting from 50% ethanol and ending with 100% ethanol. Afterwards, cells were incubated with progressively concentrated propylene oxide dissolved in ethanol, then infiltrated with increasing concentrations of Eponate 8 resin. Samples were baked in a 65°C oven overnight, then sectioned using an Ultra microtom. Sections were viewed with an energy filtering TEM unit (LEO-192AB OMEGA, Carl Zeiss)
Data processing of scRNA-seq data of autophagy
We obtained scRNA-seq data of cells with autophagic reaction using 10× Genomics. Unique molecule identifier (UMI) counts were obtained by aligning raw single-cell reads using Cell Ranger Software. To fill in the missing expressions, we applied one of the imputation algorithms SAVER (29) on the raw UMI counts data. After applied log-transformation on the imputed data, DEGs were identified by comparing each group (control (0 h), AAS 3 hour, AAS 6 h, GS 6 h and GS 12 h) of cells versus every other cells using two-sided Wilcoxon rank sum test followed by Benjamini and Hochberg false discovery rate (FDR) estimate method for multiple hypothesis testing implemented in MATLAB R2018a. Genes which have FDR <0.01 and log2 fold change >0.5 were selected as DEGs. The gene ontology (GO) (30) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (31) were analyzed by using Enrichr 2019 version (32). Subsequently, wishbone, a pseudo-time analysis tool (24) was run on Python 3.5.2. As an input data, we used the union gene set of the DEGs and autophagy-related genes obtained from literature (21). The cell with the highest average expression of the DEGs for the control cell group (0 hour) was selected as the start cell of Wishbone. In order to obtain clear trajectory figures, we ran another pseudo-time analysis tool Monocle 2.6.4 (33) on R 3.4.4 and generated the scatter plot of the cells along with the pseudo-time. We generated all the network figures using Cytoscape 3.6.1 (34).
Public data of ChIP-seq
We downloaded raw read files of ChIP-seq data (GSE75165) of three samples C/EBPγ, ATF4, and GFP control in MEFs under AAS conditions (35). We first removed the adapter sequence using CutAdapt (36) implemented in TrimGalore-0.4.5 and aligned the ChIP-seq reads to the mm10 genome using Bowtie2 (37). Next, we run ChIP-seq peak calling for each TF by comparing each ChIP sample with GFP control using the findPeaks command in the Homer package (38). To identify significant peaks, we used P-value <1.0E–10 provided by the Homer package.
Statistics and reproducibility
All experiments were performed independently at least three times. Statistical analysis was done using GraphPad Prism v5. Student's t-test was used for comparison between two groups. ANOVA analysis with post hoc testing was used for multiple sample comparisons and discrimination of significance relationships. A P-value of <0.05 was considered statistically significant.
RESULTS
scRNAseq generation to understand autophagic process induced by GS and AAS
To identify upstream signal-specific transcriptional regulators and their target genes during the autophagic processes, we generated scRNA-seq data from mouse embryonic fibroblasts (MEFs) in basal condition (0 h), amino acids starvation (AAS) for 3 and 6 h, and glucose starvation (GS) for 6 and 12 h (Figure 1A and Supplementary Figure S1). In total, we obtained around 20 995 cells with an average of 40 000 unique molecule identifier (UMI) counts per cell (Materials and Methods).
Figure 1.
TENET identified master regulators for autophagy process induced by amino acid starvation. (A) A flow chart for gene regulatory network reconstruction based on single cell RNA sequencing (scRNA-seq) data. (B) A heatmap of the scRNA-seq data shows condition-specific differentially expressed genes (DEGs). (C) Pseudo-time analysis shows bifurcating branches from control cells into GS and AAS conditions. We used DDRTree for the embedding, which is implemented in Monocle 2 package. (D) The pseudo-time ordered expression profile of the key regulators C/EBPγ and ATF4 and their target genes in both branches of AAS and GS. We calculated the z-score jointly on all the cells. (E) Key regulatory factors in the GRNS for AAS conditions. (F) The core GRN of the key regulators C/EBPγ and ATF4 in AAS-induced autophagy.
We found that a number of genes were differentially changed by AAS (Figure 1B, clusters 2 and 3) or GS (Figure 1B, clusters 4 and 5 and Supplementary Table S1). Upregulated genes upon AAS include the genes associated with apoptosis (Jun, Gadd45b, Ctsl, Ctsd, Rela, Gadd45g) and macroautophagy (Bnip3l, Rab5a, Lamtor3, Cisd2, Rab39b, Mtdh, Gabarapl2, Gabarapl1, Rab7, Tbc1d15, Gabarap) (Supplementary Figure S2A). Upregulated genes highly observed upon GS include ER-related genes (Rpl30, Rps28, Rpl27a, Rpl36, Rpl22l1) and the genes related to AMPK signaling pathway (Scd2, Eef2, Pck2) (Supplementary Figure S2B and Supplementary Table S1). We repeated the experiment using bulk RNAseq for the matched time points (3, 6, 12 hours for each condition). The expression patterns of the scRNAseq were reproduced in the bulk RNAseq data (Supplementary Figure S3). We also investigated the expression levels of TFs to identify potential transcriptional regulators. The TFs including Atf4, Cebpg and Cebpb were upregulated under the AAS condition. Under the GS condition, we observed Atf4 and Cebpb were mildly upregulated (FDR < 0.01, log ratio > 0.3) (Supplementary Figure S4 and Supplementary Table S2).
TENET predicted C/EBPγ as a key regulator in AAS-induced autophagy
We investigated potential transcriptional regulators for autophagy. Trajectory analysis using Monocle (23) against scRNAseq data identified bifurcating branches from control cells into GS and AAS conditions (Figure 1C and Supplementary Figure S5). The expression profiles across the pseudo-time for AAS confirmed that transcriptional regulators such as ATF4 (5,39) were induced prior to the induction of autophagy genes (Figure 1D), suggesting a potential role as a transcriptional regulator for autophagy genes. These genes were not actively regulated in the GS condition (Figure 1D).
To identify potential upstream signal-specific transcriptional regulators for autophagy, we applied TENET which reconstructs gene regulatory networks (GRNs) from scRNAseq data (27). Especially, TENET uniquely showed an ability to suggest key regulators from scRNAseq data (27). Applying TENET along the two branches of AAS and GS focusing on the autophagy-related genes reported previously (21), we reconstructed two GRNs of autophagic processes caused by AAS and GS (Supplementary Figure S6 and Supplementary Tables 3 and 4), respectively. TENET identified key regulators by investigating hub nodes by calculating the number of potential target genes (40,41). Importantly, C/EBPγ was identified as the top hub TF followed by Mafg and ATF4 (Figure 1E). These TFs were not shown in the GS-specific networks (Supplementary Figure S7). The autophagy genes identified as the target of C/EBPγ showed increased expression following the induction of Cebpg (Figure 1D). C/EBPγ was predicted to regulate various autophagy genes including Map1lc3b, Bnip3l and Atg12 together with ATF4 (Figure 1F). For GS-induced autophagy, we identified Ddit3 and Nupr1 as the top ranked regulators among the up-regulated genes by GS (Supplementary Figure S8). Ddit3 and Nupr1 have already been identified to play a role in transcriptionally activating target genes to protect cells by overcoming ER stress (42,43).
C/EBPγ is a key regulator in AAS-induced autophagy
The results of TENET provided the potential regulators specific to GS- or AAS-induced autophagy. Since the role of C/EBPγ was not reported in autophagy, we examined whether C/EBPγ is responsible for the starvation-induced autophagy. We first performed immunoblot analysis to check the change of the level of lipidated LC3-II formation as a marker for autophagic occurrence after C/EBPγ knockdown. Immunoblot analysis showed that the conversion of non-lipidated LC3-I to lipidated LC3-II upon AAS as well as in GS (Figure 2A). However, knockdown of C/EBPγ in MEFs attenuated the lipidated LC3-II levels under the AAS- but not GS- condition (Figure 2A). In parallel, autophagic flux analysis using Bafilomycin A1, an autophagy inhibitor, revealed that the accumulation of LC3-II upon AAS, but not GS, was significantly attenuated in C/EBPγ knockdown MEFs compared to control MEFs (Figure 2B). Further, transmission electron microscopy (TEM) analysis revealed an increase in the number of autophagic vesicles including autophagosomes, autolysosomes, and multilamellar bodies in WT MEFs, but not in the C/EBPγ knockdown MEFs upon AAS (Supplementary Figure S9). However, knockdown of C/EBPγ showed little or no effect on MEFs upon GS (Supplementary Figure S9). These data indicate that C/EBPγ is responsible for the AAS-induced autophagic occurrence.
Figure 2.
Loss of C/EBPγ impair amino acid starvation-induced autophagy. (A) Immunoblot analysis of LC3 levels in MEFs with or without C/EBPγ knockdown upon amino acid starvation for 6 hours and glucose starvation for 12 hours. The LC3-II/β-actin ratio is indicated. (B) Autophagic flux was analyzed in MEFs after knocking down with siRNA for C/EBPγ (siC/EBPγ) or non-specific siNS in the presence or absence of Bafilomycin A1 (200 nM; 2 h) under amino acid starvation for 6 hours and glucose starvation for 12 h. The LC3-II/β-actin ratio is indicated. (C) Representative confocal images of GFP-LC3 puncta formation under amino acid starvation for 6 h and glucose starvation for 12 hours. Scale bar, 20 μm. The graph shows the quantification of LC3-positive punctate cells. Bars, mean ± s.e.m.; n = 3 with over 150 cells; one-way ANOVA with post hoc Tukey's test. ***P < 0.001. (D) Representative confocal images of mCherry-GFP–LC3 assays in MEFs by knocking down with either siNS or siC/EBPγ in the absence or presence of amino acid starvation for 6 hours and glucose starvation for 12 hours. Colocalization between mCherry and GFP signal (yellow puncta) denotes autophagosomal vesicles that have not fused with a lysosomal compartment (phagophores or autophagosomes). mCherry signal without GFP signal (red puncta) denotes acidic autophagosomal vesicles (acidic amphisomes or autolysosomes). Scale bar, 20 μm. The graph indicates the number of yellow and red puncta per cell. Bars, mean ± s.e.m.; n = 10 biologically independent cells; ***P < 0.001.
To further evaluate the role of C/EBPγ in the autophagic process, the formation of green fluorescent protein (GFP)-tagged LC3-positive autophagosomes was examined. The increase in GFP-LC3 punctate cells was significantly attenuated in the C/EBPγ knockdown MEFs compared to control MEFs upon AAS-, but not GS-, induced autophagy (Figure 2C). The mCherry-GFP-LC3 reporter was applied to assess the overall number of LC3 puncta formation as well as autophagic flux. Because the GFP signal is attenuated in the acidic lysosomal condition, whereas the mCherry is not, this reporter allows discrimination between autophagosomal vesicles that have not fused with a lysosomal compartment (yellow puncta) and acidic autophagosomal vesicles (red puncta). Induction of the total numbers of both red and yellow puncta upon AAS, but not GS, was notably attenuated in C/EBPγ knockdown MEFs in contrast to control MEFs (Figure 2D). These data strongly indicate that C/EBPγ is required for AAS-induced autophagy.
Induction of ATF4 is required for the function of C/EBPγ during AAS
The trajectory analysis shows the sequential induction of Atf4, Cebpg and autophagy genes (Figure 1D), suggesting potential roles for these two TFs under AAS-induced autophagy. The network analysis revealed that the majority of the ATF4 targets (91 out of 106, P-value < 1.0e–130) overlapped with a larger number of target genes for C/EBPγ (Figure 3A). To confirm this, we investigated their occupancy across the genome using public chromatin immunoprecipitation assay followed by sequencing (ChIPseq) data against ATF4 and C/EBPγ upon AAS in MEFs (35). The binding occupancy identified by ChIPseq analysis also showed that the majority of ATF4 peaks (215 out of 233) overlapped with the C/EBPγ peaks (Figure 3B). The genes co-regulated by ATF4 and C/EBPγ include the autophagy-associated genes including Map1lc3b, Mbnl2 and Cdkn1a (Figure 3C, Supplementary Figure S10 and Supplementary Table S3).
Figure 3.
TENET identified C/EBPγ and ATF4 share common GRNs in AAS-induced autophagy. (A) C/EBPγ and ATF4 share significantly overlapped target genes, predicted by TENET. (B) C/EBPγ and ATF4 share a significant number of peaks in a public ChIP-seq data in MEFs upon amino acid starvation. (C) A gene locus shows the common peaks.
To further investigate the role for C/EBPγ and ATF4, we performed quantitative RT-PCR (qRT-PCR) analysis of potential target genes and examined whether they are coregulated by C/EBPγ and ATF4 in autophagy upon AAS and GS. Both AAS and GS induced the mRNA levels of autophagy genes including Map1lc3b, Bnip3, Vps26a, Gabarap, Gadd45b and Atg12, whereas knockdown of either C/EBPγ or ATF4 significantly attenuated the mRNA levels of these genes in AAS, but not GS condition (Figure 4A, B). These data indicate that both C/EBPγ and ATF4 are required for AAS-induced autophagy. We further questioned the relationships between ATF4 and C/EBPγ. Upon AAS in MEFs, knockdown of C/EBPγ did not affect the mRNA expression of ATF4 (Figure 4A), while knockdown of ATF4 clearly attenuated the induction of the C/EBPγ mRNA expression, indicating that ATF4 regulates C/EBPγ at transcriptional level (Figure 4B). We also checked whether exogenous C/EBPγ prior to GS can mimic the effect of AAS on induction of autophagy genes. When C/EBPγ was overexpressed under AAS and GS conditions, the expression of autophagy genes was not affected (Supplementary Figure S11). We also performed ChIP assay to determine whether Cebpg gene expression is directly dependent on ATF4. ChIP assay revealed that ATF4 was recruited to the Cebpg promoter after 3 hours of AAS condition (Supplementary Figure S12). However, C/EBPγ failed to be recruited to the Cebpg promoter under AAS. These data indicate that ATF4 directly regulates Cebpg gene expression irrespective of binding to C/EBPγ.
Figure 4.
C/EBPγ is amino acid starvation-specific transcription factor in autophagy. (A) Quantitative RT-PCR analysis of C/EBPγ-dependent genes in MEFs by knocking down with either siNS or siC/EBPγ in the absence or presence of amino acid starvation and glucose starvation for the indicated times. Bars, mean ± s.e.m.; n = 3; *P < 0.05, **P < 0.01, ***P < 0.001. Statistics by two-tailed t-test. (B) Quantitative RT-PCR analysis of C/EBPγ-dependent genes in MEFs by knocking down with either siNS or siATF4 in the absence or presence of amino acid starvation and glucose starvation for the indicated times. Bars, mean ± s.e.m.; n = 3; *P < 0.05, **P < 0.01, ***P < 0.001. Statistics by two-tailed t-test.
C/EBPγ forms a heterodimer with ATF4 for transcriptional activation of target genes in AAS-induced autophagy
ATF forms heterodimer with C/EBPγ to bind the C/EBP-ATF response element (CARE); ATF4 binds C/EBPγ upon stress response (35) but C/EBPβ during the adipogenesis (44). The expression levels of both C/EBPγ and C/EBPβ were induced under AAS in MEFs (Figure 1B). Intriguingly, TENET did not find C/EBPβ as a core node in the AAS networks. Immunoblot analysis revealed that ATF4 bound C/EBPγ but not C/EBPβ upon AAS, whereas ATF4 failed to bind C/EBPγ upon GS (Figure 5A).
Figure 5.
C/EBPγ-ATF4 heterodimers functions as key transcriptional activators for amino acid starvation-induced autophagy. (A) Co-immunoprecipitation assay was performed to detect the interaction between ATF4 and C/EBPγ in MEFs upon amino acid starvation and glucose starvation. (B) ChIP assays were performed using anti-C/EBPγ, anti-ATF4 and anti-C/EBPβ antibodies on CARE of Map1lc3b, Bnip3, Atg12, Gadd45b, and Vps26a genes in MEFs by knocking down with either siNS or siC/EBPγ in the absence or presence of amino acid starvation. Bars, mean ± s.e.m.; n = 3; *P < 0.05, **P < 0.01, ***P < 0.001, NS non-significant. Statistics by two-tailed t-test. (C) ChIP assays were performed using anti-C/EBPγ, anti-ATF4 and anti-C/EBPβ antibodies on CARE of Map1lc3b, Bnip3, Atg12, Gadd45b and Vps26a genes in MEFs by knocking down with either siNS or siC/EBPγ in the absence or presence of glucose starvation.
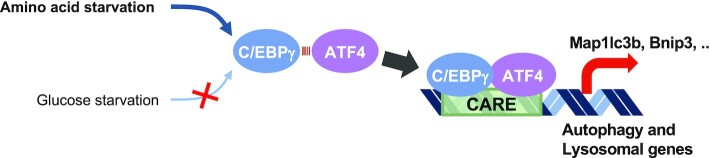
To further examine whether the binding of C/EBPγ to CARE is required for the additional binding to ATF4, we performed ChIP assays on CARE of C/EBPγ target genes using anti-C/EBPγ, ATF4 and C/EBPβ antibodies to determine whether C/EBPγ-ATF4 complex is recruited to the target promoters along with ATF4 for the activation of AAS-induced autophagy genes. ChIP assays revealed that C/EBPγ-ATF4 complex was co-recruited on CARE of target genes including Map1lc3b, Bnip3, Atg12, Gadd45b and Vps26a in AAS, but not GS conditions (Figure 5B, C). Remarkably, knockdown of C/EBPγ almost completely abolished recruitment of ATF4 upon AAS-, but not GS-induced autophagy (Figure 5B, C). Together, these data indicate that C/EBPγ-ATF4 heterodimers are recruited on CARE of autophagy genes and that C/EBPγ functions as an AAS-specific key regulator of autophagy along with ATF4 (Figure 6).
Figure 6.
Schematic model depicting that C/EBPγ-ATF4 heterodimers function as AAS-specific transcriptional regulators of autophagy.
DISCUSSION
Autophagy is a highly conserved intracellular process that allows the degradation of cytoplasmic proteins and organelles by lysosome in response to nutrient and energy starvation. Therefore, the ability of cells to respond to nutrient withdrawal including AAS and GS is essential for the maintenance of metabolic homeostasis and viability. The detected nutrient deficiency triggers diverse signaling pathways depending on the source of nutrients in the cytoplasm (45). It is reported that there are nutrient deficiency-induced transcriptional regulatory mechanisms of autophagy genes in the nucleus (18).
We hypothesized that specific upstream nutrient signals to induce autophagy are involved in the determination of specific downstream target gene regulation of autophagy. To explore the importance of nuclear events in autophagy in response to different upstream nutrient signals, we induced autophagy in MEFs by GS and AAS and sought to identify specific TFs for each condition. We selected 3 time points (including WT) for GS- and AAS-induced autophagy, respectively, to obtain scRNAseq data. Without measuring transcription in a number of time points (which has been used for bulk RNAseq), we observed dynamic changes in gene expression that differed during autophagy induced by GS and AAS.
We did not apply any batch correction for the scRNAseq dataset. Previous benchmarking study showed that batch effect can be corrected for even for temporal ordering of scRNAseq data (46). However, batch correction can also lose relevant biologically meaningful trajectory (46). The repeated experiment using bulk RNAseq data indicates that our analysis on scRNAseq data is robust against batch effect.
Sequential induction of gene expression has been used to identify potential regulators (24,27). Trajectory analysis for the scRNAseq showed sequential induction of ATF4, C/EBPγ and autophagy genes, suggesting their key roles during autophagy upon AAS. Accordingly, TENET identified ATF4 and C/EBPγ as key regulators for AAS-induced autophagy. It is intriguing that ATF4 requires the assistance of C/EBPγ to activate autophagy genes under AAS. ATF4 is upregulated in both GS- and AAS-induced autophagy, but ATF4 is responsible for the induction of C/EBPγ upon AAS, but not GS. Although previous studies have shown that C/EBPγ binds ATF4 for antioxidant functions (35), C/EBPγ specifically affects cell viability under AAS (Supplementary Figure S13). Our results reveal that heterodimers of ATF4 and C/EBPγ induce the expression of genes involved in the autophagy process in AAS-, but not GS-induced autophagy.
We obtained both AAS- and GS-specific networks, and focused on the function of C/EBPγ, an AAS-specific TF. It is remarkable that C/EBPγ was identified from the AAS-specific network by TENET and functionally validated in cell-based and molecular mechanistic studies. Compared with other GRN reconstructors, TENET has a property to predict key regulators for the underlying biological processes (27). We could not find the networks involved with other factors such as TFEB (19,20) and FOXO3a (22). It is because these factors were expressed less than 10% of cell populations potentially due to dropouts. Even with this limitation, TENET successfully identified key regulators for AAS-induced autophagy in our experiments.
Differential upstream signal-specific gene regulation for autophagy has been poorly understood due to lack of controlled experiment. Our study identified C/EBPγ as an AAS-specific TF, by combining scRNAseq with a systemic approach. This finding provides an example of nutrient type deficiency-specific gene regulation, further expecting the existence of upstream signal-specific transcriptional regulators for autophagy.
DATA AVAILABILITY
The scRNAseq (GSE189710) and bulk RNAseq (GSE199923) data can be downloaded from Gene Expression Omnibus database.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: K.J.W. and S.H.B. conceived and designed the research. D.K., J.K., Y.S.U. and Y.R.K. performed the experiments and analyzed the data. D.K., J.K., S.H.B. and K.J.W. wrote the paper.
Contributor Information
Dongha Kim, Creative Research Initiatives Center for Epigenetic Code and Diseases, School of Biological Sciences, Seoul National University, Seoul 08826, Republic of Korea; Department of Anatomy, College of Medicine, The Catholic University of Korea, Seoul 06591, Republic of Korea.
Junil Kim, Biotech Research and Innovation Centre (BRIC), University of Copenhagen, 2200 Copenhagen, Denmark; School of Systems Biomedical Science, Soongsil University, 369 Sangdo-Ro, Dongjak-Gu, Seoul 06978, Republic of Korea.
Young Suk Yu, Creative Research Initiatives Center for Epigenetic Code and Diseases, School of Biological Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Yong Ryoul Kim, Creative Research Initiatives Center for Epigenetic Code and Diseases, School of Biological Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Sung Hee Baek, Creative Research Initiatives Center for Epigenetic Code and Diseases, School of Biological Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Kyoung-Jae Won, Biotech Research and Innovation Centre (BRIC), University of Copenhagen, 2200 Copenhagen, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Creative Research Initiatives Programs (Research Center for Epigenetic Code and Diseases) [2017R1A3B1023387 to S.H.B.]; Science Research Center program [NRF-2016R1A5A1010764 to S.H.B., 2021R1A4A3031875, 2022R1A2C4002022 to D.K.]; Bio & Medical Technology Development Program [2021R1F1A1063914, 2021M3H9A2096988 to J.K.] from the National Research Foundation (NRF) by the Korea government; Novo Nordisk Foundation [NNF17CC0027852 to K.J.W.]; Lundbeck Foundation [R313-2019-421 to K.J.W.]; Independent Research Fund Denmark [0135-00243B to K.J.W.]. Funding for open access charge: Det Frie Forskningsråd [0135-00243B].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chun Y., Kim J.. Autophagy: an essential degradation program for cellular homeostasis and life. Cells. 2018; 7:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glick D., Barth S., Macleod K.F.. Autophagy: cellular and molecular mechanisms. J. Pathol. 2010; 221:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A.G.. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000; 6:269–279. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y., Wang Y., Kanyuka K., Parry M.A.J., Powers S.J., Halford N.G.. GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in arabidopsis. J. Exp. Bot. 2008; 59:3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimball S.R., Jefferson L.S.. Amino acids as regulators of gene expression. Nutr. Metab. 2004; 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rabanal-Ruiz Y., Otten E.G., Korolchuk V.I.. 2017) mTORC1 as the main gateway to autophagy. Essays Biochem. 61:565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng L., Zhang W., Zhou Y., Li F., Wei H., Peng J.. Recent advances in understanding amino acid sensing mechanisms that regulate mTORC1. Int. J. Mol. Sci. 2016; 17:1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng X., Su H., Wang L., Yao R., Ma Y., Bai L., Wang Y., Guo X., Wang Z.. Phosphoproteomics analysis reveals a pivotal mechanism related to amino acid signals in goat fetal fibroblast. Front. Vet. Sci. 2021; 8:685548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thorens B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia. 2015; 58:221–232. [DOI] [PubMed] [Google Scholar]
- 10. Marín-Juez R., Rovira M., Crespo D., Van Der Vaart M., Spaink H.P., Planas J.V. GLUT2-mediated glucose uptake and availability are required for embryonic brain development in zebrafish. J. Cereb. Blood Flow Metab. 2015; 35:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matschinsky F.M., Wilson D.F.. The central role of glucokinase in glucose homeostasis: a perspective 50 years after demonstrating the presence of the enzyme in islets of langerhans. Front. Physiol. 2019; 10:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moruno F., Pérez-Jiménez E., Knecht E.. Regulation of autophagy by glucose in mammalian cells. Cells. 2012; 1:372–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mihaylova M.M., Shaw R.J.. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011; 13:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Löffler A.S., Alers S., Dieterle A.M., Keppeler H., Franz-Wachtel M., Kundu M., Campbell D.G., Wesselborg S., Alessi D.R., Stork B.. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011; 7:696–706. [DOI] [PubMed] [Google Scholar]
- 15. Alers S., Löffler A.S., Wesselborg S., Stork B.. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012; 32:2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He C., Klionsky D.J.. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009; 43:67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hale A.N., Ledbetter D.J., Gawriluk T.R., Rucker E.B.. Autophagy: regulation and role in development. Autophagy. 2013; 9:951–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Di Malta C., Cinque L., Settembre C.. Transcriptional regulation of autophagy: mechanisms and diseases. Front. Cell Dev. Biol. 2019; 7:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li L., Friedrichsen H.J., Andrews S., Picaud S., Volpon L., Ngeow K., Berridge G., Fischer R., Borden K.L.B., Filippakopoulos P.et al.. A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat. Commun. 2018; 9:2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., Ferguson S.M.. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012; 5:ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shin H.J.R., Kim H., Oh S., Lee J.G., Kee M., Ko H.J., Kweon M.N., Won K.J., Baek S.H.. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016; 534:533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Y.S., Shin H.R., Kim D., Baek S.A., Choi S.A., Ahn H., Shamim A., Kim J., Kim I.S., Kim K.K.et al.. Pontin arginine methylation by CARM1 is crucial for epigenetic regulation of autophagy. Nat. Commun. 2020; 11:6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P., Choi K., Bendall S., Friedman N., Pe’Er D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 2016; 34:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weng G., Kim J., Won K.J.. VeTra: a tool for trajectory inference based on RNA velocity. Bioinformatics. 2021; 37:3609–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pratapa A., Jalihal A.P., Law J.N., Bharadwaj A., Murali T.M.. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods. 2020; 17:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J., Jakobsen S.T., Natarajan K.N., Won K.. TENET: gene network reconstruction using transfer entropy reveals key regulatory factors from single cell transcriptomic data. Nucleic Acids Res. 2020; 49:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim D., Nam H.J., Lee W., Yim H.Y., Ahn J.Y., Park S.W., Shin H.J.R., Yu R., Won K.J., Bae J.S.et al.. PKCα-LSD1-NF-κB-Signaling cascade is crucial for epigenetic control of the inflammatory response. Mol. Cell. 2018; 69:398–411. [DOI] [PubMed] [Google Scholar]
- 29. Huang M., Wang J., Torre E., Dueck H., Shaffer S., Bonasio R., Murray J.I., Raj A., Li M., Zhang N.R.. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods. 2018; 15:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gene Ontology Consortium Gene ontology consortium: going forward. Nucleic Acids Res. 2015; 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999; 27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A.et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C.. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017; 14:979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huggins C.J., Mayekar M.K., Martin N., Saylor K.L., Gonit M., Jailwala P., Kasoji M., Haines D.C., Quiñones O.A., Johnson P.F.. C/EBPγ is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol. Cell. Biol. 2015; 36:693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 37. Langmead B., Salzberg S.L.Langmead . Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2013; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. B’Chir W., Maurin A.C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., Bruhat A.. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013; 41:7683–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y.Y., Slotine J.J., Barabási A.L.. Controllability of complex networks. Nature. 2011; 473:167–173. [DOI] [PubMed] [Google Scholar]
- 41. Kim J., Park S.-M., Cho K.-H.. Discovery of a kernel for controlling biomolecular regulatory networks. Sci. Rep. 2013; 3:2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jauhiainen A., Thomsen C., Strömbom L., Grundevik P., Andersson C., Danielsson A., Andersson M.K., Nerman O., Rörkvist L., Ståhlberg A.et al.. Distinct cytoplasmic and nuclear functions of the stress induced protein DDIT3/CHOP/GADD153. PLoS One. 2012; 7:e33208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang C., Santofimia-Castaño P., Iovanna J.. NUPR1: a critical regulator of the antioxidant system. Cancers. 2021; 13:3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cohen D.M., Won K.-J., Nguyen N., Lazar M.A., Chen C.S., Steger D.J.. ATF4 licenses C/EBPβ activity in human mesenchymal stem cells primed for adipogenesis. Elife. 2015; 4:e06821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Efeyan A., Comb W.C., Sabatini D.M.. Nutrient-sensing mechanisms and pathways. Nature. 2015; 517:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luecken M.D., Büttner M., Chaichoompu K., Danese A., Interlandi M., Mueller M.F., Strobl D.C., Zappia L., Dugas M., Colomé-Tatché M.et al.. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods. 2021; 19:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng W., Cha J., Yuan J., Haraguchi H., Bartos A., Leishman E., Viollet B., Bradshaw H.B., Hirota Y., Dey S.K.. p53 coordinates decidual sestrin 2/AMPK/mTORC1 signaling to govern parturition timing. J. Clin. Invest. 2016; 126:2941–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tracy K., Dibling B.C., Spike B.T., Knabb J.R., Schumacker P., Macleod K.F.. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 2007; 27:6229–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S., Sun X., Yoon G., Kang Y., Zhong W.et al.. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014; 516:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNAseq (GSE189710) and bulk RNAseq (GSE199923) data can be downloaded from Gene Expression Omnibus database.