Abstract
The formation of peptides from amino acids is one of the processes associated with life. Because of the dominant role of translation in extant biology, peptide‐forming processes that are RNA induced are of particular interest. We have previously reported the formation of phosphoramidate‐linked peptido RNAs as the products of spontaneous condensation reactions between ribonucleotides and free amino acids in aqueous solution. We now asked whether four‐helix bundle (4HB) DNA or RNA folding motifs with a single‐ or double‐nucleotide gap next to a 5’‐phosphate can act as reaction sites for phosphoramidate formation. For glycine, this was found to be the case, whereas phenylalanine and tryptophan showed accelerated formation of peptides without a covalent link to the nucleic acid. Free peptides with up to 11 tryptophan or phenylalanine residues were found in precipitates forming in the presence of gap‐containing DNA or RNA 4HBs. Control experiments using motifs with just a nick or primer alone did not have the same effect. Because folded structures with a gap in a double helix are likely products of hybridization of strands formed in statistically controlled oligomerization reactions, our results are interesting in the context of prebiotic scenarios. Independent of a putative role in evolution, our findings suggest that for some aromatic amino acids an RNA‐induced pathway for oligomerization exists that does not have a discernable link to translation.
Keywords: amino acids, DNA, peptides, prebiotic chemistry, RNA
Folded nucleic acids with no more than 65 base pairs were found to act as amino acid polymerase for aromatic amino acids. Both DNA and RNA 4‐helix bundles showed this effect, which was most pronounced for tryptophan.
Introduction
Proteins and peptides are a central class of biomolecules. Polypeptides are the backbone of enzymatic activity in the cell, but also function as receptors and structural units of living organisms. It is therefore interesting to ask how peptides arose during prebiotic evolution. [1] The synthesis of encoded proteins occurs through translation, but the origin of translation is a difficult problem, [2] and so is the origin of the genetic code. [3] Before the translational machinery came into existence, there may have been simpler ways in which RNA induced the formation of peptides.
What level of control ribonucleotides or oligonucleotides can exert over peptide formation is largely unknown. Because at its core, the ribosome is a folded RNA structure, [4] it is reasonable to assume that simpler assemblies of nucleic acid strands can also catalyze, or at least control, peptide syntheses. A number of hypothesis on the origin of RNA‐directed peptide formation have been proposed,[ 5 , 6 , 7 , 8 , 9 ] and RNA‐based structures have been identified that catalyze aminoacylation of RNA,[ 10 , 11 , 12 ] but how ribosomes emerged is unclear. [13] Experiments with much smaller structures that have a more reasonable probability of being formed spontaneously may shed light on how RNA could have induced peptide formation during an early stage of evolution. This includes experiments on peptide formation without a direct link to translation and the genetic code.
Peptide formation under plausibly prebiotic conditions has been the subject of extensive research,[ 14 , 15 , 16 , 17 , 18 ] but many of the studies in the literature use conditions that are not compatible with other, more labile biomolecules, such as RNA. Reports on assays involving amino acids and ribonucleotides exist, though, and many of these assays have produced interesting results. Condensation reactions with carbodiimides in mixtures of organic solvent and water were found to produce aminoacyl adenylates, [19] which, in turn, can act as starting materials in polycondensations.[ 20 , 21 ] But, aminoacyl adenylates are labile intermediates of present‐day aminoacylation of tRNAs, and are difficult to isolate in pure form.[ 22 , 23 ] Whether such labile species could have formed in sufficient quantities to serve as starting materials in a prebiotic setting is unclear.
The formation of very short peptides from amino acids upon evaporation and subsequent heating in the presence of adenosine 5’‐triphosphate (ATP) has been reported, but the role of the triphosphate was not elucidated and adenosine 5’‐monophosphate (AMP) gave similar effects. [24] A difficulty with those heating‐based condensation is also that the heat will denature folded nucleic acid structures, so that it is difficult to envision how the underlying reactions could have operated in an “RNA‐world setting” [25] that requires folded structures.
We recently observed that amino acids and ribonucleotides condense into phosphoramidate‐linked species dubbed “peptido RNAs” when allowed to react in cold, homogeneous aqueous solution containing a condensing agent and an organocatalyst. [26] The formation of peptido RNA is a ribonucleotide‐promoted process, meaning that in the presence of a ribonucleotide component, peptide chain growth is faster than background oligomerization of the amino acid. [27] The phosphoramidate‐based pathway is not limited to reactions with water‐soluble carbodiimides, like EDC, as condensation agent. It was also found for prebiotically more plausible activation agents like cyanamide or carbonyl diimidazole. [27] The formation of the peptide portion is primed by a capture step that generates the phosphoramidate link to the 5’‐phosphate of a mononucleotide or RNA chain, [28] and the experimental conditions give products in several reaction channels for all 80 combinations of a proteinogenic amino acid and a ribonucleotide (A/C/G/U). [29] Once captured on the 5’‐phosphate, amino acids or peptides can react at their C‐terminus in template‐directed fashion. [30] This includes reactions that are single nucleotide translations. [31] These findings are encouraging, but do not rule out that other forms of RNA‐induced peptide formation exist that do not involve covalently linked species.
Ribosome are nanostructures 20–30 nm in diameter composed of RNA and proteins. [32] They contain channels and cavities. When searching for simpler catalysts for (unencoded) peptide formation, it is interesting to ask whether processes leading to peptide formation exist that are induced by much smaller folded nucleic acids offering binding sites for amino acids or peptides.
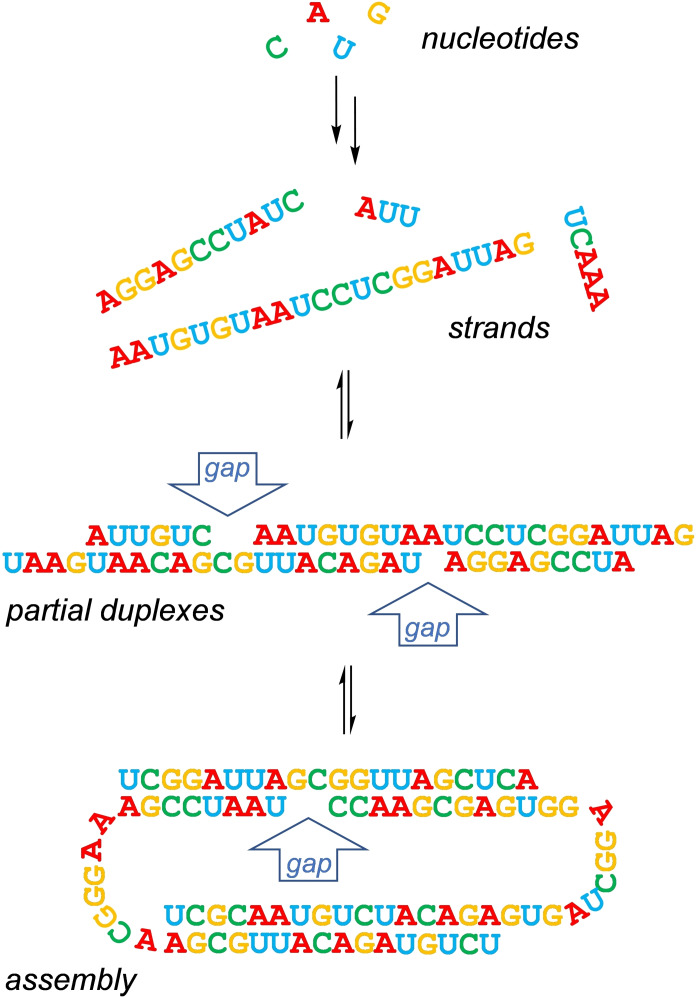
The simplest binding sites may consist of gaps in a linear duplex, which result from hybridization of oligonucleotide strands with partial complementarity (Figure 1). Gaps may also be found in three‐dimensionally folded structures of modest size assembled from several strands. If oligomerization is statistically controlled, gap‐containing structures are more likely to assemble than structures with perfect complementarity of all strands.
Figure 1.
Scenario that can lead to the formation of nucleic acid structures with gaps in a helix. Mononucleotides oligomerize to strands, which hybridize to imperfect duplexes.
Gaps in linear duplexes, triplexes, and small DNA nanostructures have previously been found to be good binding sites for nucleobases, nucleosides and nucleotides.[ 33 , 34 , 35 , 36 , 37 ] This prompted us to ask whether a DNA or RNA structure can induce or control peptide chain growth, if only in a modest, unencoded way. We reasoned that if peptide chain growth was to occur in the confinements of a cavity, interactions between nucleotides and the growing chain (or simple steric effects) would affect the outcome of the growth‐process in a fashion that depended more strongly on amino acid structure than oligomerizations in solvent. [29] We opted for folded structure with a more stable cavity than that of a linear duplex with a gap in one strand, which is prone to close by bulging out the unpaired nucleotide.[ 37 , 38 ]
We were interested in studying both amino acids believed to have been formed easily through abiotic processes, such as glycine, and aromatic amino acids, such as phenylalanine and tryptophan. The latter are also proteinogenic amino acids, but are not believed to have been encoded early, based on the proposed genealogy of the 64 codons of the genetic code.[ 39 , 40 ] It is not unlikely that they were present in prebiotic mixtures, though. Phenylalanine has been found in the product mixture of the Miller experiment. [41] There are also other syntheses of phenylalanine and tyrosine under plausible prebiotic earth conditions. [42] Tryptophan is too labile to survive under intense UV irradiation, [43] but photodissociation of complexes with other amino acids may have played a role in the origin of homochirality. [44] Perhaps more importantly, tryptophan has been found in rock samples on the Atlantic ocean floor, where it is believed to have been formed by abiotic processes catalyzed by saponite clays. [45] This mineral‐catalyzed, abiotic synthesis through a Friedel–Crafts reaction [46] complements earlier synthetic proposals, including one involving hydroxymethylation of indole with formaldehyde, followed by substitution with HCN, hydrolysis, reductive carboxylation and transamination. [47]
Aromatic amino acids formed by abiotic syntheses can react to form peptides in the absence of enzymes. Dimerization and oligomerization of amino acids, including the formation of the dipeptide Trp–Trp from tryptophan upon irradiation with high energy charged particles simulating cosmic radiation, has been demonstrated experimentally. [48] Unfortunately, such harsh reaction conditions are incompatible with many biological processes. Instead, processes occurring under milder conditions may have filled the evolutionary gap between abiotically synthesized amino acids (and the useful peptides derived from them) and the biosyntheses of extant biology that became necessary once the initial source of the amino acids was depleted. [47]
Because peptides containing tryptophan are biologically useful, it is not surprising that the biosynthetic pathway producing tryptophan is ancient. The operon of the corresponding genes is believed to have been present in the common ancestor of bacteria and archaea. [48] Still, even this ancient biosynthesis required translation, placing it at a later stage than the presumed early use of amino acids produced abiotically. What this use was and how it was driven by other biomolecules has not been explored in detail. As in earlier work on the possible role of tryptophan in the origin of life, [50] it is interesting to focus on nucleic acids in this context. This is why we decided to study the oligonucleotide assemblies mentioned above for a possible role in inducing or controlling oligomerization of aromatic amino acids.
When we initiated the project, we expected to find accelerated formation of peptido RNAs in the confinements of oligonucleotide assemblies with gaps. Instead, we chanced upon an unexpected effect that we report upon here. Free peptides, rather than the nucleotide‐bound phosphoramidates expected,[ 26 , 29 ] were formed in macroscopic quantities from phenylalanine and tryptophan when a DNA or RNA assembly with a single‐ or double‐nucleotide gap in one of its helices was present at a micromolar concentration in a condensation buffer containing EDC. [26] This unencoded form of accelerated peptide formation is probably unrelated to translation, but may have provided a valuable biomaterial in prebiotic settings.
Results
The design of our folded nucleic acid structures was chosen to produce a putative reaction site adjacent to the 5’‐phosphate of an RNA primer, from which a peptido RNA was expected to form. The reaction site was to be of sufficient size to accommodate even the largest of the amino acids (tryptophan). The indole moiety of tryptophan is similar in size to the purine ring of adenine and guanine, so that leaving out one or two purine nucleotides in one strand of a helix was expected to produce a suitable gap. This led to the structures shown in Figure 2. Based on our experience with nucleoside and nucleotide binding,[ 37 , 38 ] we selected a four‐helix bundle (4HB) structure as our core motif. The 4HB is a small, well folding structure, in which gaps in one strand of a helix can exist as stable cavities that do not collapse, due to the rigidifying effect of the neighboring helices, avoiding transitions between open and closed forms that would otherwise complicate the interpretation of data. Further, the adjacent helices provide steric shielding that allows placing a cavity at a solvent‐exposed, outer position or at more sheltered interior position.
Figure 2.
Four‐helix structures assembled from three DNA or four RNA strands and one 5’‐phosphorylated RNA primer (1p). a) Cartoon of a 4HB fold with a gap in one of its helices. b) The DNA motifs had a two‐nucleotide gap (Iag) or a single‐nucleotide gap, where an A or a G nucleotide is left out (Ia/Ig). Control motifs 1fa/g contain an unphosphorylated primer (1), and control motif (In) is without a gap, that is, the 5’‐terminus of the primer is directly adjacent to the 3’‐terminus of the red‐colored staple strand. c) Related RNA folding motifs containing a two‐nucleotide gap (IIag), a single‐nucleotide gap (IIa), a two‐nucleotide gap with unphosphorylated primer (IIf), or no gap at all (IIn). Tilted lines at the end of a sequence indicate the 5’‐terminus. The expansions below the sequence maps show enlargements of the regions where gaps were created. The letter P indicates the phosphate group at the 5’‐terminus of the primer. An asterisk indicates an unphosphorylated primer. See the Supporting Information for numbering of individual strands.
The initial experiments were performed with DNA motifs of general structure I, but we also established an RNA 4HB motif, relying on strands of a length that can reasonably be expected to have formed in a prebiotic setting.[ 51 , 52 ] Six different structures consisting of DNA were employed. These included In, without a gap adjacent to the primer, Ia and Ig with a single‐nucleotide gap, and Iag with a two‐nucleotide gap, respectively. Motif Ia has an A nucleotide missing, Ig has a G nucleotide missing, whereas in Iag the gap is where formerly the dinucleotide AG was found. Hexamer 1p was employed as the 5’‐phosphorylated strand serving as primer for peptido RNA formation. [27] It was chosen to be as short as possible, while still being reliably anchored in the 4HB, in order to facilitate monitoring by MALDI‐TOF mass spectrometry. Because phosphate‐terminated strands are the products of the oligomerization of nucleoside 5’‐monophosphates, a phosphorylated primer was considered a plausible choice.[ 26 , 52 ] For control assays, motifs Ifa and Ifg were designed, which lack a 5’‐phosphate, featuring a primer with 5’‐hydroxy terminus instead. Unlike the DNA motifs with their 100mer scaffold strand, RNA motifs (general structure II) contained a 62mer and a 38mer as longest strands to make them prebiotically plausible structures, assembled from strands of a length accessible by oligomerization. Motifs II contained the same phosphorylated primer (1p) as the DNA motifs.
Assembly of the motifs was induced by briefly heating equimolar mixtures of the strands in HEPES buffer containing MgCl2 to 85 °C and slow cooling to 4 °C. The assemblies were separated from any remaining oligonucleotides by spin filtration with 30 kDa molecular weight cut‐off filters. Successful assembly was confirmed by polyacrylamide gel electrophoresis (Figure 3), and quantification relied on UV absorption. Yields were in the range of 54–95 % and are reported for each motif in the Supporting Information. While for DNA the procedure was similar to that of earlier work on 4HBs, [37] folding of the RNA motifs required optimization to minimize hydrolysis and to reduce formation of alternative structures. Among the parameters that were optimized were the strand concentration during folding, which was lowered from 10 to 5 μM to avoid concatenated oligomers, shortening of the annealing time from 16 to 2 h, and lowering the magnesium concentration to 10 mM to reduce decomposition during the hot phase of the assembly process. Results from the optimization study are shown in Figure S2a in the Supporting Information.
Figure 3.
Representative results of gel electrophoresis analyses confirming the folding of 4HB motifs. A) Photograph of a 10 % native PAGE gel of DNA assembly Ig, with the component strands in the control lanes, indicated as colored lines above each lane. A color code for strands identical to that in Figure 2 was used. B) Image of a 10 % native PAGE gel for RNA motif IIa and its components. Staining used the dye “Stains All” in either case. Please see the Supporting Information for further details.
We then asked whether single‐nucleotide gaps in the 4HB motifs can act as binding sites for an aromatic amino acid. We chose tryptophan as putative ligand because the indole ring system of its side chain should fit well into gaps left by purines deleted from the base stacks. We assumed that binding in the pocket would place the amino acid in proximity to the terminal phosphate of the primer, favoring the formation of a phosphoramidate linkage. We used equilibrium filtration to measure binding. [35] In the event, a dissociation constant (K d) of 100 μM was measured for the complex of tryptophan with Ia, and the corresponding complex with Ig gave a K d of 90 μM (Figure 4), thus confirming that either motif acts as a binding partner for the aromatic amino acid.
Figure 4.
Four‐helix bundle motifs with a single‐nucleotide gap bind an aromatic amino acid: Cartoon showing the binding event and UV spectra from equilibrium filtration assays with tryptophan (40 μM) in Tris⋅EDTA buffer in the presence or absence of a DNA binding motif (40 μM). The red spectrum shows the absorbance in the eluate when incubated with Ig, yielding a K d value of 100 μM, and the blue spectrum shows the tryptophan absorption after incubation with Ia and filtration, giving a K d value of 90 μM. The black spectrum is for the control filtration with tryptophan but without DNA assemblies.
Encouraged by this finding, we then proceeded to peptide‐forming assays using phenylalanine or tryptophan. Solutions of the folded motifs were incubated at 0 °C with the respective amino acid in aqueous condensation buffer, pH 7.5, containing 1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide hydrochloride (EDC) as condensing agent, MgCl2 as salt and 1‐ethylimidazole (EtIm) as organocatalyst.[ 26 , 27 , 28 ] These are non‐denaturing conditions, initially developed for primer extension, [53] that is, a type of reaction that relies on Watson–Crick base pairing as the molecular recognition principle.
Due to differences in solubility, phenylalanine was used at a concentration of 100 mM, tryptophan at a concentration of 40 mM, and tyrosine at a concentration of 4 mM. In our earlier work, tyrosine proved to be the amino acid with the lowest yield in phosphoramidate‐forming reactions among the 20 proteinogenic amino acids. [29] Therefore, it came as no surprise that assays with this aromatic amino acid did not produce detectable levels of peptidic products in our current study (Figure S14), and tyrosine was not pursued further.
Based on our former results,[ 26 , 27 , 28 , 29 ] we expected the formation of peptido RNAs, with a phosphoramidate link between the peptide chain and the 5’‐phosphate of the primer. Further, we expected that such RNA‐bound peptido RNAs would remain water soluble, as the molecular weight of the (very soluble) 4HB motifs should far exceed that of the peptide chain. When the reaction tubes were visually inspected, colorless precipitates were observed after several days at 0 °C, though, both for tryptophan and for phenylalanine as amino acid (Figure 5). The extent to which precipitation occurred depended on the structure of the 4HB motif and the amino acid employed.
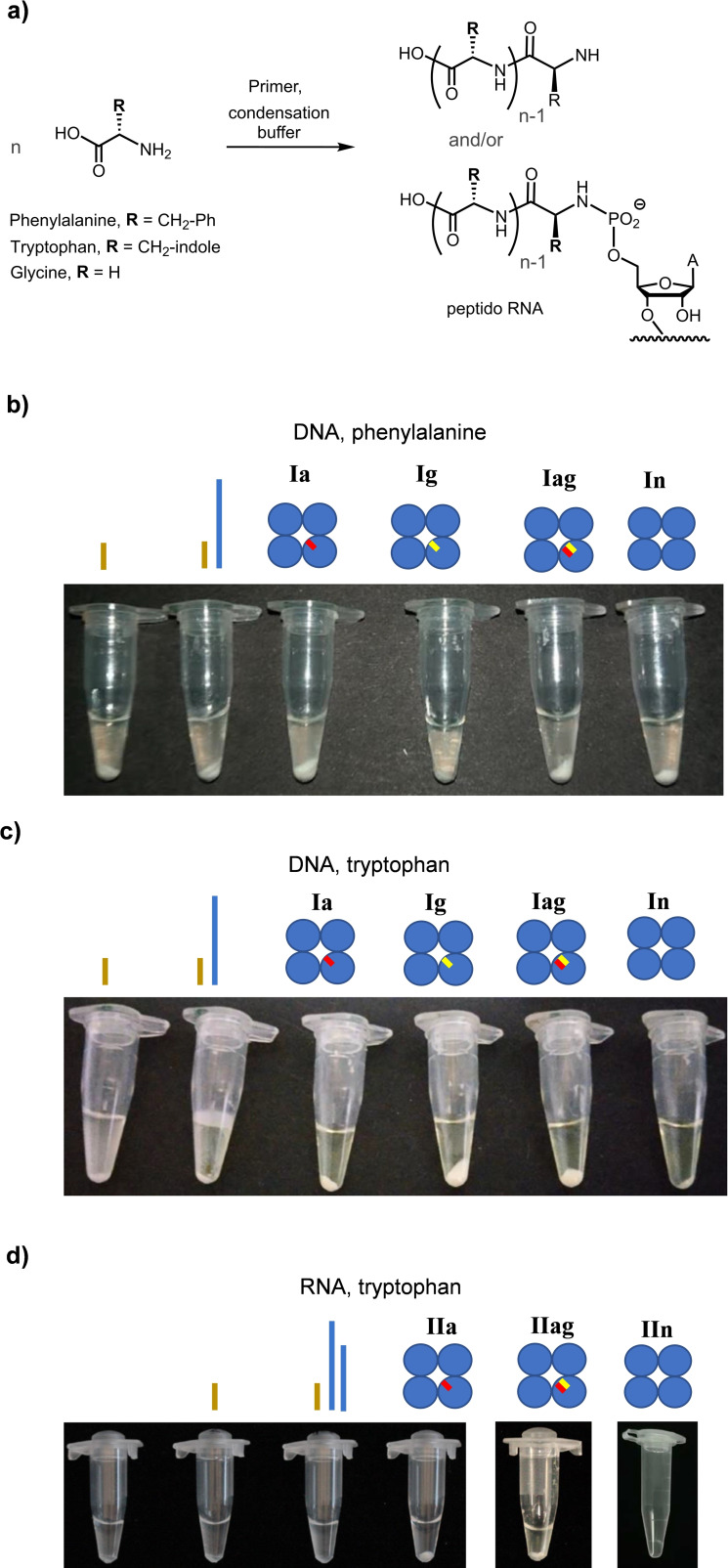
Figure 5.
Results of reactions of aromatic amino acids in aqueous condensation buffer in the presence or absence of nucleic acids. The formation of precipitates is discernable in photographs of tubes containing the reaction mixtures, taken after centrifugation, with the nucleic acids in the respective assay mixture indicated above each photograph in cartoon format. The 4HBs are shown as four blue circles with gaps indicated by yellow/red bars. a) Reaction scheme showing both peptido RNA and free peptides as possible products. b) Assays with phenylalanine in the presence or absence of 4HBs. c) Assay results for tryptophan with or without DNA motifs. d) Photographs of tubes from assays with tryptophan and RNA components. The left‐most tube is without primer. Differences in volumes are due to sampling during the course of some assays. Conditions: 100 mM phenylalanine or 40 mM tryptophan, 0.1 M EDC, 0.5 M HEPES, 125 mM 1‐ethylimidazole, 80 mM MgCl2, 50 μM primer, pH 7.5, 0 °C, 11 days.
For phenylalanine, precipitates were observed in either of the samples. Even when an assay was performed without a primer, some insoluble product was found. However, the most voluminous precipitate was found in the presence of Ia (Figure 5b). We had noticed some insoluble peptido RNAs for phenylalanine in earlier studies, [26] but those had been assays with high millimolar concentrations of ribonucleotide, not 50 μM concentrations of folding motifs containing a single terminal phosphate group. Because the extent of peptide formation depended on the nucleic acids in the solution, a reaction other than peptido RNA formation appeared to have occurred.
For tryptophan, for which the binding site in the 4HBs was designed, the dependence of the extent of precipitation on the nucleic acids in solution was even more pronounced than for phenylalanine. In the absence of a template strand, or when the DNA 4HB without a gap (In) was present, hardly any insoluble products were detectable (Figure 5c). On the other hand, the most extensive precipitation among the DNA‐containing assays was found when Ig with its single‐nucleotide gap was in the reaction mixture.
When RNA motifs were added, the effect of the nucleic acids on the formation of insoluble products was pronounced (Figure 5d). Assays without a 4HB did not lead to visible precipitation. The same was found for IIn without nucleotide gap. Assembly IIa with a single‐nucleotide gap gave the most clearly visible solid, whereas IIag with its two‐nucleotide gap induced a more modest, but still significant level of precipitation. Taken together, these observations showed that the extent of product formation from the two aromatic amino acid depends strongly on whether a folded nucleic acid motif with a gap is present or not.
To shed light on the unexpected findings, we analyzed the products of the assays by MALDI‐TOF mass spectrometry. When a solid had formed, the supernatant and the homogenized mixture were analyzed separately. For the soluble components in the supernatant, spectra were acquired in negative mode, using trihydroxyacetophenone as matrix. [54] The precipitate‐containing samples were analyzed in positive mode, using 4‐chloro‐α‐cyanocinnamic acid as matrix. [55] We also performed control assays with glycine as a small, well water‐soluble amino acid. The latter did not show any precipitates at the end of the reaction time (11 days).
Moreover, mass spectra indicated that, with glycine, the peptido RNA pathway was active, forming covalently linked species with oligoglycines up to eight residues in length being detected (Figure 6a, b). In contrast, spectra from assays with phenylalanine or tryptophan showed little or no product in the peptido RNA channel, but significant peaks in the mass range of free peptides (Figures 6 and S5–S12). For DNA motif Ig, this is shown in Figure 6c, where no peptido RNA peak is discernible and Figure 6d, where a peak pattern up to a heptamer is observed for free peptides. Likewise, assays with RNA motif IIa gave no covalently linked species in the peptido RNA channel (Figure 6e), but an oligomer pattern for free peptides up to a decamer (Figure 6f). For Ia and IIag and phenylalanine, up to undecamers were detectable in mass spectra in the peptide channel (Figures 6h and S6). In addition, even at an early time point (48 h), more precipitate was observed in the presence of RNA motif IIag than in the control assay, when phenylalanine was allowed to react (Figure S4a).
Figure 6.
Representative mass spectra of the products of peptide‐forming reactions in the presence of 50 μM 4HB motifs after 11 days in 0.5 M HEPES buffer, pH 7.5, 80 mM MgCl2, 0 °C. Either the solution phase containing the primer‐based peptido RNA products or the precipitates were analyzed by MALDI‐TOF MS in negative or positive mode, respectively. Which product channel was analyzed is indicated on each spectrum. a) Peptido RNA peaks from the assay with glycine in the presence of Ig, b) peptido RNA channel for the assay with glycine and IIa c) Peptido RNA channel for tryptophan and DNA 4HB Ig, and d) peptide channel for the same assay. e) Peptido RNA channel for tryptophan and RNA assembly IIa, and f) peptide channel for the same experiment. g) Peptido RNA channel for phenylalanine and RNA assembly IIa, and h) peptide channel for the same experiment. Assays with glycine were with 400 mM amino acid, 150 mM ethylimidazole, 400 mM EDC; assays with aromatic amino acids were with 100 mM phenylalanine or 40 mM tryptophan; 125 mM EtIm and 100 mM EDC.
To obtain more than a qualitative impression and to corroborate the shift from phosphoramidate‐linked peptido RNAs to free peptides, the results of assays were analyzed in more detail for tryptophan. This included measuring the longest chain detectable for both primer‐bound and free peptides at a signal‐to‐noise cut‐off of 2 : 1 in MALDI spectra, the maximum in the peak pattern of free peptide chains, and the percentage of amino acid residues found in the precipitate relative to the amino acid quantity in the assay (Table 1).
Table 1.
Results from peptide‐forming assays with tryptophan and 4HB.[a]
|
Nucleic acids[b] |
Longest peptido RNA found |
Most intense peptide peak |
Longest free peptide detected |
Trp in precipitate [%][c] |
Molar ratio of nucleic acid strands to Trp in precipitate[d] |
|---|---|---|---|---|---|
|
Primer |
|
|
|
|
|
|
1p |
2 |
4 |
4 |
1 |
1 : 5 |
|
DNA |
|
|
|
|
|
|
1p/3a |
2 |
3 |
3 |
3 |
1 : 25 |
|
In |
1 |
5 |
5 |
3 |
1 : 22 |
|
Ia |
1 |
5 |
7 |
17 |
1 : 138 |
|
Ig |
2 |
4 |
7 |
53 |
1 : 420 |
|
Iag |
3 |
4 |
7 |
22 |
1 : 176 |
|
Ifa |
<1[e] |
4 |
7 |
21 |
1 : 163 |
|
Ifg |
<1[e] |
4 |
6 |
54 |
1 : 432 |
|
RNA |
|
|
|
|
|
|
1p/7/8 |
<1[e] |
4 |
5 |
–[f] |
–[f] |
|
IIn |
<1[e] |
<1[e] |
<1[e] |
–[f] |
–[f] |
|
IIf |
<1[e] |
5 |
7 |
15 (34)[g] |
1 : 117 (250)[g] |
|
IIa |
<1[e] |
6 |
10 |
23[h] (32)[g] |
1 : 180[h] (425)[g] |
|
IIag |
<1[e] |
6 |
12 |
(31)[g] |
(500)[g] |
[a] Conditions: 40 mM tryptophan, 50 μM nucleic acid strands, in 40 μL buffer containing 0.5 M HEPES, pH 7.5, 80 mM MgCl2, 125 mM EtIm and 100 mM EDC, 0 °C; after 11 days. [b] 3 a and 7/8 are scaffold strands. See the Supporting Information for full sequences. [c] Amount of tryptophan precipitated, as detected by UV. [d] Molar ratio between nucleic acid strands and amino acid residues found in precipitated peptides. [e] No signal outside matrix‐dominated region of the spectrum (>500 Da). [f] No precipitate. [g] Calculated from remaining Trp content of solution. [h] Assay performed at half the scale (20 μL solution).
The quantification of tryptophan by UV absorption to calculate the latter used re‐dissolved precipitate or measured the depletion from the assay solution. It is clear from the data that a more than tenfold (and sometimes almost 100‐fold) increase in the amount of precipitated peptides was induced in the case of active 4HB motifs, when compared to the control reaction with primer 1p alone. Further, the results obtained with motifs containing an unphosphorylated primer (Ifa, Ifg and IIf) are similar to those for their phosphorylated counterparts, confirming that the 5’‐phosphate does not have a role in peptide‐forming process. Because 800 equivalents of tryptophan (40 mM) over the 4HB motifs (50 μM) were used, the most favorable cases formally correspond to hundreds of turnovers in terms of additional peptide bond forming events.
While the number of peptide bonds formed increased significantly for gap‐containing motifs, there was a more modest effect on the length of peptide chains. This suggests that there are multiple chain initiation events, and that their number increases when the gap is provided as a binding/reaction site. The maximum length may be limited by the rate of chain growth and the availability of remaining amino acids and condensation agent or both. Among the RNA assemblies, the two‐nucleotide motif IIag gave the highest peptide yield, whereas the single‐nucleotide gap 4HBs Ig and Ifg are the most efficient motifs in the DNA series. This is probably due to differences in local structure, as RNA prefers A‐type helices, whereas DNA usually forms B‐type helices. The differences in scaffold length (uninterrupted for DNA and two strands for RNA) might also play a role.
Discussion
These results are interesting for several reasons. Firstly, our hypothesis that a binding pocket next to the 5’‐phosphate of the primer would accelerate the formation of phosphoramidate‐linked peptido RNAs proved incorrect. For phenylalanine and tryptophan the peptido RNA pathway is not the preferred mode of reaction. Rather, at the 50 μM concentration of 5’‐phosphate groups of our oligoribonucleotide assemblies (much lower than the 200 mM concentration of nucleotides used earlier)[ 27 , 28 ] they prefer to react via a second pathway. This second pathway produces free peptides, not covalently linked peptido RNAs. The results with glycine indicate that this is not due to inaccessibility of the phosphate group in the activation reaction or some other inhibitory effect of the four‐helix bundle motifs. Rather, the positioning of bound tryptophans does not seem to be favorable for a reaction with the phosphate.
It is interesting to ask what causes the acceleration of peptide formation. One likely cause is increased local concentration. If two or more aromatic amino acids are found in or near the gap of a 4HB, they may be able to react with each other more readily than would be the case when reactions are the consequence of diffusional encounters only. This then leads to the question how a single‐nucleotide gap can accommodate two aromatic amino acids or more. We suspect that the cavity caused by the missing nucleotide is larger than assumed based on a perfectly regular Watson‐Crick double helix. Folding can lead to local distortion of helices where they are not interlocked through bridging segments, [56] as indicated by the bulging in Figure 7.
Figure 7.
Proposed reaction‐pathway for formation of free oligopeptides in the hydrophobic interior of a folded 4HB structure. Blue discs represent aromatic amino acids.
This effect may not only lead to a larger intra‐motif cavity next to the gap, but the gap itself may also be larger than in an unrestrained helix. The strain imposed by folding may be most easily released where a strand is nicked or interrupted. That a larger binding site can be advantageous is indicated by the activity of folding motifs with double nucleotide gaps, particularly in the case of RNA (IIag). If the bending distortion of the helices away from each other where they are not directly linked leads to a cavity that can accommodate a short oligopeptide, the scenario of Figure 7 is not unlikely.
Another effect that may contribute to the acceleration of oligomerizations is modulation of the pK a of the reacting groups. The interior of the gap should be more lipophilic than free solvent, which can be expected to decrease the extent of protonation of the α‐amino group acting as nucleophile in the coupling reaction, making it more reactive. That the protection state (and thus the protonation level) of the α‐amino substituent of amino acids undergoing carbodiimide‐induced coupling can affect oligomerization has previously been observed by Cavadore, who supplemented reaction mixtures with N‐acylated derivatives to initiate oligomerization. [14]
Additionally, the chemical microenvironment of the binding pocket may affect the activation chemistry, by increasing or modulating concentrations of the condensing agent or the organocatalyst at the reaction site. Results from exploratory assays confirm the notion that, in a modest way, our folded nucleic acid structures with gaps in one helix act as amino acid polymerase. One such exploratory experiment involved assays with increasing concentrations of IIag (Figure S13). When 10 μM IIag was added to an otherwise identical reaction mixture containing 40 mM tryptophan, no precipitation was found at the end of the assay (11 days). At a concentration of 25 μM of the RNA 4HB, precipitation did set in, and 50 μM IIag gave the significant quantity of oligotryptophans mentioned above. So, the amount of insoluble tryptophan oligomers depends on the quantity of 4HB in the solution, as expected if the latter acts a catalyst.
The second exploratory experiment that strengthens the hypothesis of a process favored by proximity of tryptophan molecules in the interior of four‐helix bundles employed a protected dipeptide as inhibitor. For this, IIag was treated with 1 mM of the N‐acetylated tryptophan dimer ester Ac‐TrpTrp‐OEt, followed by addition of the usual reaction mixture to induce oligomerization of tryptophan. The results of this and the control assay without the dipeptide are shown in Figure 8. Whereas the assay with the two‐nucleotide gap motif gave peptides up to the dodecamer, no peptides beyond traces of tetramer were detected in the presence of Ac‐TrpTrp‐OEt. Apparently the lipophilic, protected dipeptide can block oligomerization quite efficiently.
Figure 8.
Mass spectrometric results from assays with 50 μM motif IIag a) in the absence and b) in the presence of dipeptide 1 mM Ac‐WW‐OEt as inhibitor. Conditions: 40 mM tryptophan, 0.5 M HEPES, pH 7.5, 80 mM MgCl2, 125 mM EtIm, 100 mM EDC, 0 °C, 11 days; with detection in positive mode. The gray triangles indicate peptides having reacted with EDC, most probably because of the reduced supramolecular shielding in the two‐nucleotide gap RNA motif.
It is also interesting to ask how RNA‐accelerated formation of insoluble oligopeptides could have benefitted an primitive self‐replicating system in what is referred to as the “RNA world”. [25] Although hypotheses in this context necessarily remain highly speculative, we note that peptide precipitates that can redissolve upon dilution and warming may encapsulate other functional biomolecules, and may thus provide a protective coating during a time when pools have evaporated and labile molecules would otherwise be exposed to the elements. If, for example, such peptide precipitates prevented the degradation of RNA during the dry season of seasonal cycles, as proposed for gel phases by others, [57] peptide formation induced by simple RNA motifs could indeed have been useful. Such a protective effect may be tested experimentally.
One should always be cautious when proposing prebiotic scenarios. From the point of view of extant biology, it might seem unreasonable to assume that aromatic amino acids were present at sufficiently high concentrations during a phase of prebiotic evolution loosely described as the “RNA world”, to undergo the reactions described here. Today's biosynthesis of tryptophan is long and complex, and the aromatic amino acids are known to have appeared late in the genetic code.[ 39 , 40 ] From a chemical point of view, though, it is not unreasonable to assume that the known abiotic syntheses of tryptophan could have produced concentration as high as those found in the oceanic lithosphere.[ 45 , 46 ]
Indoles are resonance‐stabilized aromatic compounds that can form readily under a range of chemical conditions, as known from the Fischer indole synthesis, the Reissert indole synthesis, the Bartoli indole synthesis, the Japp–Klingemann reaction, the Leimgruber–Batcho indole synthesis, the Madelung synthesis, the Nenitzescu indole synthesis, and the Baeyer–Emmerling indole synthesis, to name just a few. Much like purines, for which the biosynthesis is long and complicated, but the abiotic synthesis is simple,[ 58 , 59 ] indole derivatives like tryptophan may have come from abiotic sources during an early phase of evolution. As pointed out by Miller, abiotic syntheses may have preceded the complicated biosyntheses of present‐day metabolism. [47] If so, the results presented here show how aromatic amino acids could have oligomerized in reactions catalyzed by oligonucleotide assemblies.
Conclusions
In conclusion, we report that gaps in DNA or RNA helices can have a significant effect on the rate of peptide formation. Acceleration was observed for two aromatic amino acids, phenylalanine and tryptophan, with the latter giving the more significant effect in terms of product formation over the background reaction. Unlike the phosphoramidate‐based peptido RNAs previously observed, where amino acids were reacted with stoichiometric amounts of ribonucleotides, the products of the nucleic‐acid‐induced pathway reported here are free peptides, not covalently linked species. Peptide chain growth depends on the size of the gap in the oligonucleotide assembly and the type of nucleic acid forming the assembly. When employed at millimolar concentration, both aromatic amino acids oligomerized to form precipitates in aqueous mixtures containing micromolar concentrations of the four‐helix bundle motifs. The effect of our nucleic acid assemblies on the formation of precipitates is somewhat reminiscent of precipitations resulting from a degradation of RNA in complexes with proteins, [60] and may be functionally significant. Cavities that accelerate the oligomerization of other amino acids (other than the aromatic amino acids studied here) may exist in the structure space of oligonucleotide assemblies, making the search for such structures an interesting endeavor.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
The authors thank D. Göhringer for technical assistance and S. G. Lorenz and E. Kervio for discussions. Funded by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project‐ID 364653263 – TRR 235 (CRC 235) and Volkswagen Foundation (Life? initiative, grant Az 92 768). Open Access funding enabled and organized by Projekt DEAL.
O. Doppleb, R. J. Schwarz, M. Landa, C. Richert, Chem. Eur. J. 2022, 28, e202104104.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1. Frenkel-Pinter M., Samanta M., Ashkenasy G., Leman L. J., Chem. Rev. 2020, 120, 4707–4765. [DOI] [PubMed] [Google Scholar]
- 2. Crick F. H. C., Brenner S., Klug A., Pieczenik G., Orig. Life 1976, 7, 389–397. [DOI] [PubMed] [Google Scholar]
- 3. Grosjean H., Westhof E., Nucleic Acids Res. 2016, 44, 8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Noller F. H., Science 2005, 309, 1508–1514. [DOI] [PubMed] [Google Scholar]
- 5. Orgel L. E., J. Mol. Evol. 1989, 29, 465–474. [DOI] [PubMed] [Google Scholar]
- 6. Morgens D. W., J. Mol. Evol. 2013, 77, 185–196. [DOI] [PubMed] [Google Scholar]
- 7. Lacey J. C., Mullins D. W., Origins Life 1983, 13, 3–42. [DOI] [PubMed] [Google Scholar]
- 8. Tamura K., Schimmel P., Proc. Natl. Acad. Sci. USA 2003, 100, 8666–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pascal R., Boiteau L., Top. Curr. Chem. 2005, 259, 69–122. [Google Scholar]
- 10. Chumachenko N. V., Novikov Y., Yarus M., J. Am. Chem. Soc. 2009, 131, 5257–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee N., Bessho Y., Wie K., Szostak J. W., Suga H., Nat. Struct. Biol. 2000, 7, 28–33. [DOI] [PubMed] [Google Scholar]
- 12. Illangasekare M., Yarus M., Proc. Natl. Acad. Sci. USA 1999, 96, 5470–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noller F. H., Cold Spring Harbor Perspect. Biol. 2012, 4, a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cavadore J.-C., Previero A., Bull. Chem. Soc. Biol. 1969, 51, 1245–1253. [PubMed] [Google Scholar]
- 15. Liu R., Orgel L. E., J. Am. Chem. Soc. 1997, 119, 4791–4792. [DOI] [PubMed] [Google Scholar]
- 16. Lamy C., Lemoine C. J., Bouchu D., Goekjian P., Strazewski P., ChemBioChem 2007, 9, 710–713. [DOI] [PubMed] [Google Scholar]
- 17. Illos R. A., Bisogno F. R., Clodic G., Bolbach G., Weissbuch I., Lahav M., J. Am. Chem. Soc. 2008, 130, 8651–8659. [DOI] [PubMed] [Google Scholar]
- 18. Canavelli P., Islam S., Powner M. W., Nature 2019, 571, 546–549. [DOI] [PubMed] [Google Scholar]
- 19. Berg P., J. Biol. Chem. 1958, 233, 608–611. [PubMed] [Google Scholar]
- 20. Paecht-Horowitz M., Berger J., Katchalsky A., Nature 1970, 228, 636–639. [DOI] [PubMed] [Google Scholar]
- 21. Turk R. M., Illangasekare M., Yarus M., J. Am. Chem. Soc. 2011, 133, 6044–6050. [DOI] [PubMed] [Google Scholar]
- 22. Moriguchi T., Yanagi T., Wada T., Sekine M., J. Chem. Soc.-Perkin Trans. 1999, 1859–1865. [Google Scholar]
- 23. Gao X., Zeng Z., Xu P., Tang G., Liu Y., Zhao Y., Rapid Commun. Mass Spectrom. 2011, 25, 291–300. [DOI] [PubMed] [Google Scholar]
- 24. Hawker J. R., Oró J., J. Mol. Evol. 1981, 17, 285–294. [DOI] [PubMed] [Google Scholar]
- 25. Gilbert W., Nature 1986, 319, 618. [Google Scholar]
- 26. Jauker M., Griesser H., Richert C., Angew. Chem. 2015, 127, 14772–1477; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2015, 54, 14564–14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griesser H., Tremmel P., Kervio E., Pfeffer C., Steiner U. E., Richert C., Angew. Chem. 2017, 129, 1239–1243; [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2017, 56, 1219–1223. [Google Scholar]
- 28. Tremmel P., Griesser H., Steiner U. E., Richert C., Angew. Chem. 2019, 131, 13221–13226; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2019, 58, 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griesser H., Bechthold M., Tremmel P., Kervio E., Richert C., Angew. Chem. 2017, 129, 1244–1248; [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2017, 56, 1224–1228. [Google Scholar]
- 30. Jash B., Richert C., Chem. Sci. 2020, 11, 3487–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jash B., Tremmel P., Jovanovic D., Richert C., Nat. Chem. 2021, 13, 751–757. [DOI] [PubMed] [Google Scholar]
- 32. Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A., Science 2000, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 33. Yoshimoto K., Nishizawa S., Minagawa M., Teramae N., J. Am. Chem. Soc. 2003,125, 8982–8983. [DOI] [PubMed] [Google Scholar]
- 34. Patel M., Dutta A., Huang H., Anal. Bioanal. Chem. 2011, 400, 3035–3040. [DOI] [PubMed] [Google Scholar]
- 35. Kröner C., Röthlingshöfer M., Richert C., J. Org. Chem. 2011, 76, 2933–2936. [DOI] [PubMed] [Google Scholar]
- 36. Kröner C., Thunemann M., Vollmer S., Kinzer M., Feil R., Richert C., Angew. Chem. 2014, 125, 9352–9356; [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2014, 53, 9198–9202. [DOI] [PubMed] [Google Scholar]
- 37. Schwarz R. J., Richert C., Nanoscale 2017, 9, 7047–7054. [DOI] [PubMed] [Google Scholar]
- 38. Wolfrum M., Schwarz R.-J., Schwarz M., Kramer M., Richert C., Nanoscale 2019, 11, 14921–14928. [DOI] [PubMed] [Google Scholar]
- 39. Trifonov E. N., J. Biomol. Struc. Dynamics 2004, 22, 1–11. [DOI] [PubMed] [Google Scholar]
- 40. Kubyshkin V., Budisa N., Int. J. Mol. Sci. 2019, 20, 5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson A. P., Cleaves H. J., Dworkin J. P., Glavin D. P., Lazcano A., Bada J. L., Science 2008, 322, 404. [DOI] [PubMed] [Google Scholar]
- 42. Friedmann N., Miller S. L., Science 1969, 166, 766–767. [DOI] [PubMed] [Google Scholar]
- 43. Scappini F., Capobianco M. L., Casadei F., Zamboni R., Giorgianni P., Int. J. Astrobiol. 2007, 6, 281–289. [Google Scholar]
- 44. Fujihara A., Maeda N., Anal. Chim. Acta 2017, 979, 31–35. [DOI] [PubMed] [Google Scholar]
- 45. Ménez B., Pisapia C., Andreani M., Jamme F., Vanbellingen Q. P., Brunelle A., Richard L., Dumas P., Réfrégiers M., Nature 2018, 564, 59–63. [DOI] [PubMed] [Google Scholar]
- 46. Baross J. A., Nature 2018, 564, 42–43. [DOI] [PubMed] [Google Scholar]
- 47. Keefe A. D., Lazcano A., Miller S. L., Orig. Life Evol. Biosphere 1995, 25, 99–110. [DOI] [PubMed] [Google Scholar]
- 48. Simakov M. B., Kuzicheva E. A., Dodonova N. Y., Antropov A. E., Adv. Space Res. 1997, 19, 1063–1066. [DOI] [PubMed] [Google Scholar]
- 49. Xie G., Keyhani N. O., Bonner C. A., Jensen R. A., Microbiol. Mol. Biol. Rev. 2003, 67, 303–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Majerfeld I., Yarus M., Nucleic Acids Res. 2005, 33, 5482–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang W., Ferris J. P., J. Am. Chem. Soc. 2006, 128, 8914–8919. [DOI] [PubMed] [Google Scholar]
- 52. Joshi P. C., Aldersley P. M. F., Delano J. W., Ferris J. P., J. Am. Chem. Soc. 2009, 131, 13369–13374. [DOI] [PubMed] [Google Scholar]
- 53. Jauker M., Griesser H., Richert C., Angew. Chem. 2015, 127, 14767–14771; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2015, 54, 14559–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sarracino D., Richert C., Bioorg. Med. Chem. Lett. 1996, 6, 2543–2548. [DOI] [PubMed] [Google Scholar]
- 55. Calvano C. D., Ventura G., Palmisano F., Cataldi T. R. I., J. Mass Spectrom. 2016, 51, 841–848. [DOI] [PubMed] [Google Scholar]
- 56. Bai X., Martin T., Scheres S., Dietz H., Proc. Natl. Acad. Sci. USA 2012, 109, 20012–20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Damer B., Field A., Life 2016, 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oró J., Nature 1961, 191, 1193–1194. [DOI] [PubMed] [Google Scholar]
- 59. Roy D., Najafian K., von Ragué Schleyer P., Proc. Natl. Acad. Sci. USA 2007, 104, 17272–17277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aarum J., Cabrera C. P., Jones T. A., Rajendran S., Adiutori R., Giovannoni G., Barnes M. R., Malaspina A., Shee D., EMBO Rep. 2020, 21, e49585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.