Abstract
Background
Neuropathic pain symptoms and signs of increased pain sensitization in osteoarthritis (OA) patients may explain persistent pain after total joint replacement (TJR). Therefore, identifying genetic markers associated with pain sensitization and neuropathic‐like pain phenotypes could be clinically important in identifying targets for early intervention.
Methods
We performed a genome‐wide gene‐based association study (GWGAS) using pressure pain detection thresholds (PPTs) from distal pain‐free sites (anterior tibia), a measure of distal sensitization, and from proximal pain‐affected sites (lateral joint line), a measure of local sensitization, in 320 knee OA participants from the Knee Pain and related health in the Community (KPIC) cohort. We next performed gene‐based fixed‐effects meta‐analysis of PPTs and a neuropathic‐like pain phenotype using genome‐wide association study (GWAS) data from KPIC and from an independent cohort of 613 post‐TJR participants, respectively.
Results
The most significant genes associated with distal and local sensitization were OR5B3 and BRDT, respectively. We also found previously identified neuropathic pain‐associated genes—KCNA1, MTOR, ADORA1 and SCN3B—associated with PPT at the anterior tibia and an inflammatory pain gene—PTAFR—associated with PPT at the lateral joint line. Meta‐analysis results of anterior tibia and neuropathic‐like pain phenotypes revealed genes associated with bone morphogenesis, neuro‐inflammation, obesity, type 2 diabetes, cardiovascular disease and cognitive function.
Conclusions
Overall, our results suggest that different biological processes might be involved in distal and local sensitization, and common genetic mechanisms might be implicated in distal sensitization and neuropathic‐like pain. Future studies are needed to replicate these findings.
Significance
To the best of our knowledge, this is the first GWAS for pain sensitization and the first gene‐based meta‐analysis of pain sensitization and neuropathic‐like pain. Higher pain sensitization and neuropathic pain symptoms are associated with persistent pain after surgery hence, identifying genetic biomarkers and molecular pathways associated with these traits is clinically relevant.
1. INTRODUCTION
It is estimated that pain is not adequately controlled in 4 of 10 people that suffer from knee osteoarthritis (OA) (Neogi, 2013). Knee pain hugely contributes to disability (Guccione et al., 1994; March & Bagga, 2004). Although OA pain has traditionally been associated with peripheral pain mechanisms due to joint damage and inflammation, the presence of radiographic joint pathology does not always explain the severity of pain (Finan et al., 2013; Valdes et al., 2012). Many people with OA suffer from pain that they describe as numbness, electric shocks, or a burning, itching sensation that can initiate spontaneously and in the absence of a painful stimulus, suggestive of a neuropathic source (den Boer et al., 2019; Cavalli et al., 2019). A role of the central nervous system (CNS) in augmenting nociceptive processing has been described in OA, which is known as pain sensitization and displays similar characteristics to neuropathic pain (Chappell et al., 2009; Petersen et al., 2015; Soni et al., 2019).
Quantitative sensory testing (QST) can be used to quantify alterations in pain sensitivity. Reduced pressure pain detection thresholds (PPTs) at the affected site may indicate the presence of peripheral sensitization, while reduced PPTs at a distal pain‐free side is suggestive of altered pain processing in the CNS and central pain augmentation (Croft et al., 2005; Graven‐Nielsen & Arendt‐Nielsen, 2002; Suokas et al., 2012). Magnetic resonance imaging‐detected inflammation in knee OA, and not the severity of radiographic features, is associated with the development and worsening of local pressure pain sensitivity in the knee (Neogi et al., 2016), suggesting that inflammation is a potential mechanism underlying local sensitization. Neuropathic pain symptoms on the painDETECT questionnaire have been associated with signs of central sensitization on QST, such as associations with PPTs at sites distant from the affected joints, suggesting that painDETECT may reflect central pain processing in patients with knee OA (Hochman et al., 2013; Moreton et al., 2015; Moss et al., 2018).
Genetic variants implicated in pain sensitivity have arisen from candidate gene studies. These included amino acid change variants in the catechol‐O‐methyltransferase (COMT) (van Meurs et al., 2009), the voltage‐gated sodium channel Nav1.7 (SCN9A) (Reimann et al., 2010) and the transient receptor potential cation channel, subfamily V, member 1 (TRPV1) (Valdes et al., 2011). However, there is a gap in our knowledge regarding the genes and molecular pathways influencing pain sensitization in knee OA. Evidence indicates that signs of increased pain sensitization might be a barrier to treatment response. Indeed, widespread hyperalgesia assessed by PPTs predicts poor outcomes to arthroplasty (Lundblad et al., 2008; Petersen et al., 2016; Rakel et al., 2012; Wylde et al., 2015). Furthermore, knee OA patients with neuropathic pain symptoms identified using the painDETECT questionnaire are most at risk of developing chronic postoperative pain after total knee replacement (Kurien et al., 2018). Therefore, understanding the underlying mechanisms involved in pain sensitization and neuropathic‐like pain may promote better profiling and diagnosis of pain patients and development of new regimes for mechanism‐based therapy. Thus, we completed two GWASs with PPTs from distal and affected sites to identify genetic variants for distal and local sensitization, respectively, using data from the Knee Pain and related health In the Community (KPIC) cohort (Fernandes et al., 2017). We then completed a genome‐wide gene‐based association analysis (GWGAS) and gene‐set analysis on both PPT phenotypes separately to explore the genes and underlying genetic mechanisms of distal and local sensitization in OA pain. Using existing GWAS data from an independent Nottingham cohort study of neuropathic‐like pain (Warner, Walsh, et al., 2017), we performed a GWGAS to identify genes related to the neuropathic‐like pain phenotype. GWGAS may have higher power to identify the causal variants of complex diseases compared to GWAS because it considers the joint effect of several single nucleotide polymorphisms within a single gene (Chung et al., 2019; Kang et al., 2013). As pain sensitization is suggested to display similar characteristics to neuropathic‐like pain, we performed two separate gene‐based meta‐analyses by combining our neuropathic‐like pain GWGAS findings with our findings from distal and local sensitization GWGAS to identify genes common in distal sensitization and neuropathic‐like pain, as well as local sensitization and neuropathic‐like pain, respectively.
2. METHODS
The study design is outlined in Figure 1. We used data from two independent study cohorts. Participants from both cohorts had similar demographic characteristics (age, sex and BMI). We used a three‐stage design for the identification of any potential associations between genetic variants and two pain sensitivity phenotypes (distal sensitization and local sensitization), as well as genes common in pain sensitivity and neuropathic‐like pain phenotypes. First, two GWASs with PPTs from both distal and affected sites were run to identify genetic variants for distal and local sensitization, respectively, using data from the KPIC cohort. Second, three GWGASs were run to identify the genes associated with the two pain sensitivity phenotypes and a neuropathic pain‐like phenotype using the GWASs outputs from KPIC and existing GWAS data from a Nottingham cohort. Third, we conducted two meta‐analyses of our GWGAS findings to identify genes common in pain sensitization (distal and local) and neuropathic‐like pain.
FIGURE 1.
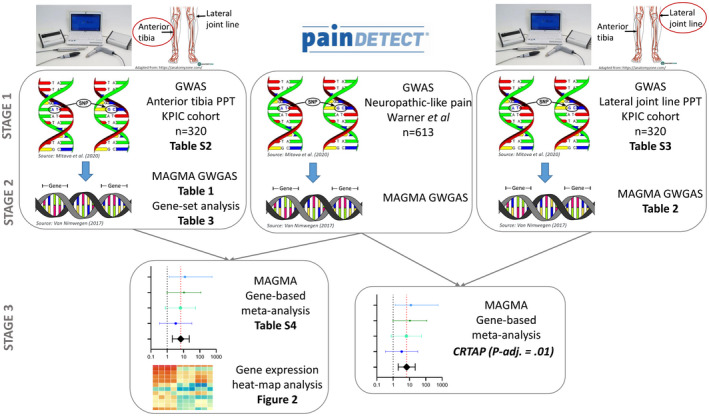
Study design. A three‐stage design for identification of associations between genetic variants and two pain sensitivity phenotypes: distal sensitization [anterior tibia pressure pain threshold (PPT)] and local sensitization (lateral joint line PPT), as well as genes common in pain sensitivity and neuropathic‐like pain (painDETECT questionnaire) phenotypes. First, two GWASs with PPTs from both distal and affected sites were run to identify genetic variants for distal and local sensitization, respectively, using data from the KPIC cohort (Tables S2 and S3). Second, three GWGASs and gene‐set analyses were run to identify the genes associated with the two pain sensitivity phenotypes and a neuropathic pain‐like phenotype using the GWASs outputs from KPIC and existing GWAS data from a Nottingham cohort (Tables 1, 2, 3). Third, two meta‐analyses of our GWGAS findings were conducted to identify genes common in pain sensitization (distal and local) and neuropathic‐like pain (Table S4) and a heat map analysis of the common genes identified from the distal sensitization/neuropathic‐like pain meta‐analysis was performed (Figure 2)
2.1. Participants
2.1.1. KPIC cohort
Baseline data from a subgroup of the KPIC (n = 320) that undertook clinical assessments including knee radiographs, knee ultrasound, quantitative sensory testing, muscle strength, balance, gait analysis and biomarker sampling and for which we had genetic information and no missing data on anterior tibia and lateral joint line PPTs was used (Fernandes et al., 2017) (Table S1). KPIC is a prospective community‐based cohort of men and women aged 40 years or over within the East Midlands region (UK).
2.1.2. Nottingham post‐total joint replacement (TJR) cohort
Participants were recruited post‐TJR for OA (n = 613) from secondary care in the Nottinghamshire area (Warner, Walsh, et al., 2017). Demographic characteristics of this cohort are presented in Table S1.
2.2. Phenotypes
2.2.1. Neuropathic‐like pain
This phenotype has been described extensively elsewhere (Warner, van Meurs, et al., 2017). Briefly, neuropathic‐like pain was measured with the painDETECT (Freynhagen et al., 2006), a self‐report seven‐item questionnaire with scores ranging from 0 to 39 developed to discriminate between nociceptive and neuropathic pain. The questionnaire asks about the intensity, pattern and quality of their knee pain, for example persistent with slight fluctuations, burning, tingling or sudden, with Likert scale and tick‐box questions. Individuals were classified as having possible or probable neuropathic‐like pain if they scored >12 according to the validated cut‐offs for diagnosis by Freynhagen et al. (2006) making up 109 possible or probable neuropathic pain cases versus 504 controls (Warner, van Meurs, et al., 2017). The dichotomised value for the classification of participants to neuropathic‐like pain versus controls was used for the GWAS and the results from this analysis can be found elsewhere (Warner, van Meurs, et al., 2017).
2.2.2. Pressure pain detection thresholds (PPT)
PPTs (in kPa) were measured in triplicate with an algometer (Somedic AB, Sweden) that is connected to a computer (HP ProBook 4520s). Pressure was administered manually at a progressively increasing rate (standardized rate set at 30 kPa/s) by a trained researcher through the algometer, a circular rubber‐coated pressure probe (1 cm2). Subjects were instructed to push a button when the sensation changed from pressure to pain and the algometer was immediately taken off the skin. Among other areas, PPT was applied to the lateral tibiofemoral joint line adjacent to the patellar ligament of the index knee and the anterior tibia 5 cm distal to the tibial tuberosity of the contralateral knee (Fernandes et al., 2017). PPT has been shown to be a reproducible measure of nerve sensitivity throughout localized, distal and remote sites and it is part of QST, which is used to quantify pain perception. PPT at sites away from the affected joint has been interpreted as an index of central sensitization (i.e. increased pain perception in areas away from the knee) (Arendt‐Nielsen et al., 2010; Pavlaković & Petzke, 2010). PPT at an affected joint may indicate a combination of peripheral and central sensitization. Results from a systematic review and meta‐analysis revealed that people with OA have lower PPTs both at the affected joint and at areas away from the joint when compared to controls (Suokas et al., 2012). The mean of PPT values for each site were computed from all three PPT rounds for analysis. Log transformation resulted in more symmetrical and less skewed distributions of the PPT values. Therefore, the log‐transformed variables of PPT values were used for all further analyses.
2.3. GWAS
For all cohort study participants, genomic DNA was extracted from peripheral blood leukocytes. Genotype data were analysed using the Illumina Global screening array Inc Basic BioIT (Illumina). Quality Control (QC) checks and genotyping were done both at the sample and single nucleotide polymorphism (SNP) level and have been described in detail elsewhere (Warner, Walsh, et al., 2017; Zeggini et al., 2012). PLINK software (version 1.07) was used to analyse GWAS data from this array (Purcell et al., 2007). Two GWASs were conducted to identify associated SNPs and genes with the two distal and local sensitization phenotypes, respectively. The GWAS output contains information about the SNPs’ location in the genome, a regression coefficient beta (β) and a test statistic that indicate the level of association of the genetic variants with the phenotype along with a p value to determine significance.
2.4. Functional annotation
To gain insights into the functions of the identified genes, we next tested the probability of these genes to map into specific biological pathways as defined by the Kyoto Encyclopaedia of Genes and Genomes (KEGG) or Reactome databases using the Database for Annotation, Visualisation and Integrated Discovery (DAVID) (Dennis et al., 2003) and Reactome online platforms, respectively. The gene list was composed of genes corresponding to all SNPs with a p‐value of p < 0.0001 in the GWAS analysis.
2.5. Gene‐based association analysis
Subsequently, our GWAS results were used for GWGAS using Multi‐marker Analysis of GenoMic Annotation (MAGMA) (de Leeuw et al., 2015). MAGMA takes as input the p‐values derived from the GWAS and annotates SNPs to known protein‐coding genes to estimate aggregate associations based on all SNPs in a gene (linkage disequilibrium) accounting for multi‐marker effects, by applying multiple regression analyses. MAGMA then uses Fisher's test to compute p‐values to test the association between a gene and the phenotype and does not assign a positive or negative coefficient to a gene‐based association. It differs from functional annotation as it provides a statistical gene‐based test, whereas functional annotation methods map individually significant SNPs to genes. The 1,000 Genomes Project (phase 1, release 3) was used as a reference panel to calculate linkage disequilibrium between genomic variants (Auton et al., 2015). A gene‐based analysis was performed for each phenotype using the results from our GWAS for local and distal sensitization and existing GWAS data for neuropathic‐like pain. The NCBI 37.3 build was used to obtain the SNPs that were attributed to each gene (de Leeuw et al., 2015). A total of 14,428, 14,428 and 14,722 protein‐coding genes were assessed for an association with the distal sensitization phenotype, the local sensitization phenotype and the neuropathic‐like pain phenotype, respectively. Benjamin–Hochberg (B–H) correction was used to determine significance.
2.6. Gene‐based meta‐analysis
Our GWGAS resulted in different genes for anterior tibia PPT and lateral joint line PPT. For anterior tibia PPT, we identified genes that have been previously related to central mechanisms of pain, whereas one of the lateral joint line PPT genes found, has been previously associated with peripheral pain mechanisms. Accordingly, we conducted two separate fixed‐effects gene meta‐analyses combining our GWGAS neuropathic‐like pain results with GWGAS results from anterior tibia PPT and lateral joint line PPT to identify genes common in neuropathic‐like pain and distal sensitization as well as neuropathic‐like pain and local sensitization, respectively. The Z‐scores for each gene across the two cohorts, KPIC and Nottingham post‐TJR, were combined using the weighted‐Z method. According to this method, the weights are computed as the inverse of the squared standard error of the effect size estimate for each cohort, resulting in different weights for each cohort according to their power (Lipták, 1958; Whitlock, 2005). B–H correction was used to determine significance.
2.7. Pathway over‐representation analysis
Significant results from the GWGAS were entered in FUMA for gene‐set/GWAS catalogue over‐representation analysis with MAGMA (de Leeuw et al., 2015; Watanabe et al., 2017). We identified statistically significant overrepresented pathways for the anterior tibia PPT phenotype after adjusting for false detection rate (FDR) with B–H correction method.
3. RESULTS
3.1. Stage 1: GWAS
The 10 ‘top‐hits’ (i.e. genes with highest p‐values that survived correction for multiple testing) of the adjusted GWAS on anterior tibia and lateral joint line PPT phenotypes are shown in Tables S2 and S3, respectively. The total genotyping rate was 0.99 with 700,078 variants passing filtering and QC and each tested for association with both phenotypes. The genomic inflation factor lambda (based on median chi‐square) was low for both phenotypes (λ = 1.07, for anterior tibia PPT and λ = 1.00, for lateral joint line PPT). After adjusting for multiple testing, 554,759 SNPs remained in the analysis. The results of GWAS on neuropathic‐like pain are described elsewhere (Warner, van Meurs, et al., 2017).
3.2. Stage 2: Gene‐based association analysis
As knee OA is a complex, polygenic disease, we next decided to perform gene‐based association analysis, which has been suggested to be more powerful at unravelling associations compared to GWAS, as it accounts for the correlations among SNPs within a single gene (Chung et al., 2019). The ‘top’ results, in terms of statistical significance (i.e. genes with highest p‐values that survived correction for multiple testing) as well as biological relevance, of the gene‐association analyses on anterior tibia and lateral tibiofemoral joint line PPT can be seen in Tables 1 and 2, respectively. Several of the genes identified for anterior tibia PPT phenotype are traditional pain‐related genes and members of the ‘sensory perception’ gene set according to the Gene Ontology definition (GO: 0007600) (i.e. KCNA1, MTOR, ADORA1 and SCN3B). For lateral joint line PPT, we found a single gene, PTAFR, member of the ‘sensory perception’ gene set.
TABLE 1.
The results of interest from the MAGMA gene‐based association analysis of anterior tibia PPT phenotype
| Gene symbol | CHR. | Start position | Stop position | SNPS | Z statistic | Gene p‐value | Adjusted p * |
|---|---|---|---|---|---|---|---|
| OR5B3 | 11 | 58169938 | 58170882 | 1 | 9.48 | 1.22E‐21 | 1.75E‐17 |
| WNT9A | 1 | 228109165 | 228135676 | 1 | 8.59 | 4.36E‐18 | 3.14E‐14 |
| DNAH7 | 6 | 29364416 | 29365448 | 2 | 7.04 | 9.47E‐13 | 2.48E‐09 |
| IFNGR1 | 6 | 139456249 | 139501946 | 2 | 7.01 | 1.04E‐12 | 2.48E‐09 |
| HECA | 11 | 64863587 | 64879332 | 2 | 7.06 | 1.20E‐12 | 2.48E‐09 |
| AIM2 | 1 | 159028790 | 159046685 | 3 | 6.98 | 1.48E‐12 | 2.68E‐09 |
| POLR3E | 16 | 22308696 | 22346424 | 3 | 5.69 | 6.41E‐09 | 4.87E‐06 |
| IL10RA | 11 | 117857106 | 117872198 | 3 | 5.33 | 4.95E‐08 | 2.98E‐05 |
| HRH2 | 5 | 175084847 | 175136239 | 6 | 5.04 | 2.37E‐07 | 9.23E‐05 |
| ROPN1 | 3 | 123687862 | 123711017 | 1 | 4.96 | 3.46E‐07 | 1.28E‐04 |
| KCTD11 | 18 | 13218729 | 13652753 | 93 | 4.78 | 8.70E‐07 | 2.68E‐04 |
| CCDC14 | 3 | 123616152 | 123680255 | 4 | 4.78 | 8.96E‐07 | 2.69E‐04 |
| KCNA1 | 12 | 5019073 | 5027422 | 1 | 4.32 | 7.98E‐06 | 1.52E‐03 |
| MTOR | 1 | 11166588 | 11322614 | 6 | 3.98 | 3.40E‐05 | 4.42E‐03 |
| HRH1 | 10 | 38383264 | 38412280 | 1 | 3.90 | 4.84E‐05 | 5.82E‐03 |
| CACNB2 | 10 | 18429373 | 18830688 | 85 | 3.89 | 5.10E‐05 | 6.08E‐03 |
| TNFAIP3 | 6 | 138663930 | 138790381 | 11 | 3.71 | 1.05E‐04 | 1.02E‐02 |
| ADORA1 | 1 | 203096833 | 203136533 | 9 | 3.39 | 3.56E‐04 | 2.17E‐02 |
| SCN3B | 11 | 123499895 | 123525315 | 6 | 3.20 | 6.87E‐04 | 3.43E‐02 |
| TAOK3 | 12 | 118587606 | 118810750 | 8 | 3.08 | 1.04E‐03 | 4.51E‐02 |
| CACNA2D3 | 3 | 54156620 | 55108584 | 156 | 3.03 | 1.23E‐03 | 4.97E‐02 |
Adjusted p values after applying B–H correction to control for multiple testing FDR.
TABLE 2.
The results of interest from the MAGMA gene‐based association analysis of lateral joint line PPT phenotype
| Gene symbol | CHR. | Start position | Stop position | SNPS | Z statistic | Gene p‐value | Adjusted p * |
|---|---|---|---|---|---|---|---|
| BRDT | 1 | 92414928 | 92479985 | 4 | 6.92 | 2.24E‐12 | 3.23E‐08 |
| RFX6 | 6 | 117198376 | 117253326 | 7 | 6.59 | 2.19E‐11 | 1.05E‐07 |
| CRTAP | 3 | 33155450 | 33189265 | 8 | 6.11 | 4.84E‐10 | 1.74E‐06 |
| SUOX | 12 | 56391043 | 56399309 | 2 | 6.06 | 6.73E‐10 | 1.94E‐06 |
| CHD3 | 17 | 7788096 | 7816075 | 2 | 5.48 | 2.15E‐08 | 5.15E‐05 |
| CRAT | 9 | 131857073 | 131873070 | 2 | 5.18 | 1.10E‐07 | 1.97E‐04 |
| IKZF4 | 12 | 56401268 | 56432219 | 5 | 4.92 | 4.31E‐07 | 6.90E‐04 |
| TMEM26 | 10 | 63166401 | 63213208 | 5 | 4.36 | 6.53E‐06 | 8.95E‐03 |
| IARS | 9 | 94972489 | 95056038 | 1 | 4.35 | 6.83E‐06 | 8.95E‐03 |
| PTAFR | 1 | 28473677 | 28520447 | 1 | 4.02 | 2.89E‐05 | 2.78E‐02 |
| C12ORF10 | 12 | 53693132 | 53700965 | 1 | 3.94 | 4.09E‐05 | 3.10E‐02 |
| DAPK1 | 9 | 90112601 | 90323566 | 4 | 3.85 | 6.01E‐05 | 4.33E‐02 |
Adjusted p values after applying B–H correction to control for multiple testing FDR.
We then performed a gene‐set analysis to identify pathways related to PPT phenotypes using FDR‐adjusted significant genes as identified from the GWGAS. We found significantly overrepresented pathways for anterior tibia PPT after B–H correction (Table 3). Enrichment was seen in several metabolic‐related pathways.
TABLE 3.
Gene‐set analysis for anterior tibia PPT MAGMA FDR‐adjusted significant genes
| GeneSet | N | n | p‐value | Adjusted p | Genes |
|---|---|---|---|---|---|
| Systolic blood pressure | 793 | 26 | 1.88e‐7 | 2.52e‐4 | CPSF3L, NME7, ADORA1, WNT9A, GPR137B, CACNB2, SYNPO2L, TCF7L2, NOX4, SYT1, MYCBP2, HOXB7, INSR, RGL3, SLC8A1, COBLL1, TNS1, COL4A4, JPH2, ITPR1, HRH1, FGD5, CTNNB1, MITF, ADRB2, TBXAS1 |
| Obesity‐related traits | 756 | 25 | 2.77e‐7 | 2.52e‐4 | CSF1, AIM2, CACNB2, SLC29A3, NAV2, NOX4, IL10RA, ANO2, SYT1, FAM155A, SAMD4A, MAX, FAM189A1, CEP152, SLC8A1, MACROD2, ITPR1, NCEH1, TLL1, LRFN2, GRIK2, NXPH1, AUTS2, TBXAS1, HR |
| Night sleep phenotypes | 538 | 20 | 7.48e‐7 | 4.51e‐4 | SPTA1, WNT9A, LRIG3, SAMD4A, AEN, PMP22, ZNF830, RTTN, ZNF486, LRP1B, UBOX5, TASP1, DTD1, CACNA2D3, LRTM1, GALNTL6, NADK2, RANBP3L, EPB41L4A, HRH2 |
| Amyotrophic lateral sclerosis (sporadic) | 164 | 11 | 9.94e‐7 | 4.51e‐4 | FANK1, RYR3, ANKRD29, RNF165, MACROD2, MYO18B, TAPT1, TLL1, LRFN2, AUTS2, TBXAS1 |
| Coronary artery calcified atherosclerotic plaque score in type 2 diabetes | 26 | 5 | 5.65e‐6 | 2.05e‐3 | LRP1B, MAGI1, ZBTB49, OSBPL3, MTSS1 |
| Systemic juvenile idiopathic arthritis | 34 | 5 | 2.24e‐5 | 6.77e‐3 | KLF17, WWOX, ZNF521, TAPT1, COL12A1 |
| Body mass index | 1365 | 31 | 2.65e‐5 | 6.88e‐3 | MTOR, FAM63A, CACNB2, TCF7L2, PNLIPRP3, STK33, SFSWAP, POLR2 M, CHTF18, CYLD, MYO19, CDC27, POP4, LRP1B, COBLL1, COL4A4, MACROD2, ENTPD6, TAF4, CACNA2D3, COL25A1, FGF2, TLL1, LRFN2, COL19A1, IFNGR1, RGS17, SBDS, AUTS2, TSGA13, DMD |
| Hand grip strength | 156 | 9 | 3.21e‐5 | 7.28e‐3 | TCF7L2, SYT1, SAMD4A, SLC8A1, DNER, ITPR1, CADPS, LRFN2, AUTS2 |
The results of pathway analysis showing overrepresented pathways, before and after B–H correction.
3.3. Stage 3: Gene‐based meta‐analysis
We then conducted a fixed‐effects gene meta‐analysis of anterior tibia PPT and neuropathic‐like pain in both cohorts (KPIC and Nottingham post‐TJR) and performed heat map analysis (Table S4 and Figure 2, respectively). Results from the meta‐analysis revealed ‘top‐hits’ (i.e. genes with highest p‐values that survived correction for multiple testing) in various genes related to bone morphogenesis and neuro‐inflammation, including WNT9A, POLR3E, AIM2, HECA and IFNGR1 (Table S4). We visualized tissue specific expression patterns based on GTEx v6 RNA‐seq data (GTEx Consortium, 2015) for each significant gene as an interactive heat map plot (Figure 2).
FIGURE 2.
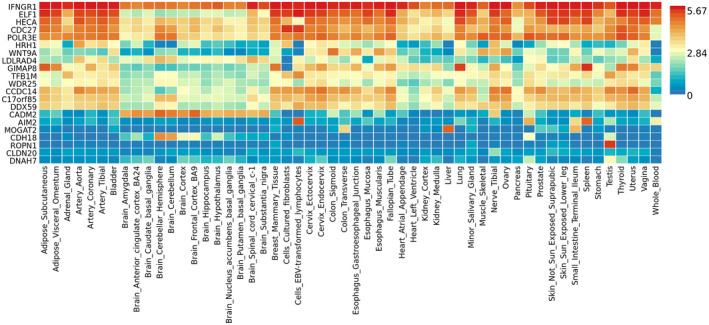
Gene expression heat‐map plot of significant genes from the meta‐analysis of anterior tibia PPT and neuropathic‐like pain in both cohorts
We also performed a fixed‐effects gene meta‐analysis of lateral joint line PPT and neuropathic‐like pain in both cohorts, which yielded a single significant gene, CRTAP at chromosome 3 (Z = 4.83, p‐adjusted = 0.01, after applying B–H correction).
4. DISCUSSION
In this study, we used a GWGAS approach to identify novel genes for pain sensitization in knee OA. The most significant genes associated with distal and local sensitization were OR5B3 and BRDT, respectively. Additionally, we identified several traditional pain‐related genes, KCNA1, MTOR, ADORA1 and SCN3B, for anterior tibia PPT phenotype and PTAFR for lateral joint line PPT phenotype. Gene‐set analysis for the anterior tibia PPT revealed enrichment of metabolic‐related pathways. We found genes involved in bone morphogenesis (CRTAP and WNT9A), inflammation and neuroinflammation (POLR3E, AIM2 and IFNGR1), metabolic disease and obesity (AIM2, DNAH7, CADM2), and cognitive function (CRTAP and CCDC14) from meta‐analysis of distal sensitization and neuropathic‐like pain traits in the two cohorts. A single gene, CRTAP, was significant after meta‐analysis of local sensitization and neuropathic‐like pain traits.
The most significant gene associated with distal sensitization was OR5B3. OR5B3 is an olfactory receptor gene involved in the production of G‐protein coupled transmembrane receptors, which enable the detection and transmission of olfactory stimuli. A recent study revealed that knockdown of cathepsin S (CTSS) upregulates almost all olfactory receptor family genes and vice versa, in cell overexpressing CTSS and an increase of the expression level of olfactory receptor 5B3 protein was observed when cells were treated with a CTSS inhibitor (Chen et al., 2021). It has been shown that the release of CTSS from microglial cells causes neuropathic pain (Clark & Malcangio, 2012) that could be reversed by intraspinal injection of CTSS inhibitor. Consistent with our findings, these results suggest a potential mechanistic role of OR5B3 in neuropathic pain. BRDT was the most significant gene associated with local sensitization and is a member of the bromodomain and extra‐terminal (BET) family of proteins. BET proteins bind to acetylated lysine residues in the histones of nucleosomal chromatin and function either as co‐activators or co‐repressors of gene expression (Filippakopoulos & Knapp, 2014), where they regulate the expression of key oncogenes, anti‐apoptotic proteins and many immunity‐associated genes and pathways (Wang et al., 2021), which is consistent with an underlying role of inflammation in local sensitization.
Genes previously associated with neuropathic pain and signs of central sensitization—KCNA1, MTOR, ADORA1 and SCN3B—were identified from GWGAS of anterior tibia PPT, while an inflammatory pain gene—PTAFR—was found from GWGAS of lateral joint line. These findings are consistent with a role of joint damage and inflammation in local sensitization, as well as altered pain processing in the CNS in distal sensitization, and thus are biologically plausible. Specifically, KCNA1 encodes for the potassium voltage‐gated channel subfamily A member 1, which is a key player in the perception of mechanical stimuli (Hao et al., 2013). ADORA1 that codes for adenosine A1 receptor is suggested to have anti‐nociceptive properties, following surgery (Gan & Habib, 2007), and to reduce thermal hyperalgesia and mechanical allodynia in animal models of neuropathic pain (Gong et al., 2010; Wu et al., 2005). MTOR that codes for the mammalian target of rapamycin has a role in inflammatory‐ and opioid‐induced hyperalgesia (Xu et al., 2011, 2014) and acts as a regulator of neuroplasticity in the CNS (Hoeffer & Klann, 2010; Jaworski & Sheng, 2006). SCN3B encodes for the b3 subunit of voltage‐gated sodium channel (Nav) (Morgan et al., 2000) and is implicated in neuropathic pain (Casula et al., 2004; Lopez‐Santiago et al., 2006; Pertin et al., 2005). Finally, PTAFR encodes for a member of the G‐protein coupled receptor 1 family of proteins for platelet‐activating factor and is involved in the perception and maintenance of neuropathic pain by regulating the production of pro‐inflammatory cytokines in the dorsal root ganglion (Okubo et al., 2012; Shindou et al., 2017; Tsuda et al., 2011).
The results of pathway analysis for anterior tibia PPT revealed overrepresented pathways associated with systolic blood pressure, obesity‐related traits, night sleep phenotypes, amyotrophic lateral sclerosis, type 2 diabetes, systemic juvenile idiopathic arthritis, body mass index and hand grip strength. These traits are often comorbid with OA and have been previously associated with pain from OA (Swain et al., 2020). For instance, cardiovascular disease and OA share similar underlying disease mechanisms (Fernandes & Valdes, 2015). Obesity is a major risk factor for OA and shares common genetic variations with OA (Panoutsopoulou et al., 2014). Disturbed sleep has been associated with increased pain in OA (Doherty & Smith, 1993; Smith et al., 2009), while pain has been suggested to increase the risk of developing frailty (low hand grip strength) in OA (Valdes & Stocks, 2018; Veronese et al., 2017).
Meta‐analysis of distal sensitization and neuropathic‐like pain revealed ‘top‐hits’ in genes involved in bone morphogenesis, CRTAP (Morello et al., 2006) and WNT9A (Regard et al., 2012). A recent GWAS identified WNT9A as a robust novel genetic marker for hand OA (Boer et al., 2021). We identified several inflammation‐related genes, POLR3E (Chiu et al., 2009), AIM2 (Hornung et al., 2009), IFNGR1 (van de Wetering et al., 2010), LDLRAD4 (Nakano et al., 2014) and HRH1 (Dong et al., 2014). Increased accumulation of pro‐inflammatory factors in the joint accompanying synovitis, is highly correlated to OA pain (Berenbaum, 2013). Despite the role of POLR3E, AIM2, IFNGR1 in inflammation, evidence suggests that these genes are also involved in neuroinflammation, neurodegeneration and neuroplasticity, processes related to central pain facilitation and neuropathic pain (Ji et al., 2018; Latremoliere & Woolf, 2009; Myers et al., 2006). For example, POLR3E participates in pre‐mRNA splicing and transcription and aged mice showed reduced expression of POLR3E (Kohman et al., 2011). Improper RNA splicing can result in abnormal translation of RNA and is associated with many age‐related diseases including Alzheimer's disease (Meshorer & Soreq, 2002). Furthermore, AIM2 is involved in neuroinflammation and neurodegeneration (Cox et al., 2015; Wu et al., 2017), as well as in neuronal plasticity and memory (Chen et al., 2019; Wu et al., 2016). Moreover, deletion of IFNGR1 results in complete abrogation of neuroinflammation and nigrostriatal degeneration, suggesting a role of this gene in neuroinflammation and neurodegeneration (Strickland et al., 2018). We also identified genes involved in obesity and adiposity, AIM2 (Gong et al., 2019), IFNGR1 (Locke et al., 2015), DNAH7 (Söhle et al., 2012) and CADM2 (Graff et al., 2017), as well as type 2 diabetes and cardiovascular disease, DNAH7 (Vujkovic et al., 2020), DDX59 (van der Harst & Verweij, 2018), WDR25 (Perry et al., 2014) and CLDN20 (Heid et al., 2010). Additionally, we found genes associated with intelligence, educational attainment, cognitive performance and psychological traits, CRTAP (Hall et al., 2018), CCDC14 (Hill et al., 2019), TFB1 M, ROPN1 (Lee et al., 2018), HRH1 (Shan et al., 2015) and CADM2 (Okbay et al., 2016). There is strong epidemiological evidence of a link between cognitive function, depression/anxiety and pain in people with arthritis (James & Ferguson, 2019).
We do not find significant associations with some of the candidate genes previously reported to be associated with a pain phenotype, such as COMT, SCN9A and TRPV1, in our GWGAS. This can be partly explained due to different analysis methods. In candidate gene studies, genes are selected a priori and only very small regions of the genome are investigated at a time, meaning that important genes may be overlooked using this method (Warner & Valdes, 2017). Furthermore, a lack of reproducibility of SNPs in candidate genes in GWAS meta‐analyses has been previously shown. For example, candidate COMT SNPs were not reproduced in a GWAS meta‐analysis of chronic widespread pain (Peters et al., 2013). In addition, although COMT has been extensively studied in relation to pain, results from candidate gene studies are not consistent (Hagen et al., 2006; Hocking et al., 2010; Nicholl et al., 2010). Rare and drastic mutations in the SCN9A gene that explain different types of congenital insensitivity to pain have been identified (Cox et al., 2006). Nevertheless, in addition to these rare‐causing mutations, it is known that the genetic risk for chronic pain is due to common variations with small effect size (Mogil, 2012). TRPV1 has been associated with heat pain sensitivity, and thus it is not necessarily implicated in mechanical pain transduction. Indeed, it was shown that TrpV1 neurons are selectively tuned nociceptors that mediate responses to thermal but not mechanical pain (Mishra & Hoon, 2010).
Limitations of the current study include the small sample sizes of the individual cohorts used that are not sufficient to power genome‐wide significant results. We tried to overcome this by running gene‐based meta‐analysis of the two cohorts. Findings from GWAS are generally prone to type 1 errors (Bacanu et al., 2000). This was addressed by adjusting results for FDR with B–H (Benjamini, 2010). Another limitation with the use of GWAS is the possibility of inflated effect sizes (Ioannidis, 2008). We performed GWGAS and gene‐set analysis that may have higher power to identify the causal variants of complex diseases, as it takes into consideration the correlations among SNPs within a single gene (Kang et al., 2013). Gene‐based tests are designed to identify genes containing multiple risk variants that individually are weakly associated with a univariate trait (Chung et al., 2019). There is a possibility that distal PPTs are reflective of central sensitization mechanisms but can also reflect features of peripheral (inflammatory) mechanisms and while neuropathic pain is presumed to result from abnormal neuronal activity of the somatosensory nervous system, glial cell dysfunction may also contribute (Ji et al., 2013). Indeed, distal and local PPTs were correlated in our cohort (r = 0.72, p < 0.001) and from our meta‐analysis of distal sensitization and neuropathic‐like pain, we found some inflammation‐related genes. However, these genes are also associated with neuroinflammation, neurodegeneration and neuroplasticity, processes related to central pain processing and neuropathic‐like pain (Ji et al., 2018; Latremoliere & Woolf, 2009; Myers et al., 2006). In addition, painDETECT scores apart from being associated with signs of central sensitization (e.g. reduced PPTs) (Hochman et al., 2013) can also reflect nerve damage (Sumitani et al., 2016). Although the majority of the patients in the KPIC cohort had unilateral OA (75.19%), 33 patients had bilateral OA and thus, for these patients the contralateral tibia could be an OA affected area.
To conclude, our results suggest that different biological processes might be involved in distal and local sensitization, while common genetic mechanisms might be implicated in distal sensitization and neuropathic‐like pain. Further research is needed to confirm these findings and to explore whether other measures of altered pain processing (temporal summation and conditioned pain modulation) demonstrate similar results.
CONFLICTS OF INTEREST
None to declare.
AUTHOR CONTRIBUTIONS
AK is the primary author and all other authors are secondary. AMV is the main supervisor and is leading this project. All authors discussed the results and commented on the manuscript.
Supporting information
Table S1‐S4
ACKNOWLEDGEMENTS
We are grateful to the study participants that took part in these studies.
Kouraki, A. , Doherty, M. , Fernandes, G. S. , Zhang, W. , Walsh, D. A. , Kelly, A. , & Valdes, A. M. (2022). Different genes may be involved in distal and local sensitization: A genome‐wide gene‐based association study and meta‐analysis. European Journal of Pain, 26, 740–753. 10.1002/ejp.1902
Funding information
This work was part of a PhD project funded by the National Institute of Health Research via the NIHR Nottingham Biomedical Research Centre. The KPIC was supported financially by the Versus Arthritis Pain Centre; and Arthritis Research UK Centre for Sport Exercise and Osteoarthritis.
REFERENCES
- Arendt‐Nielsen, L. , Nie, H. , Laursen, M. B. , Laursen, B. S. , Madeleine, P. , Simonsen, O. H. , & Graven‐Nielsen, T. (2010). Sensitization in patients with painful knee osteoarthritis. Pain, 149(3), 573–581. 10.1016/j.pain.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Auton, A. , Abecasis, G. R. , Altshuler, D. M. , Durbin, R. M. , Abecasis, G. R. , Bentley, D. R. , Chakravarti, A. , Clark, A. G. , Donnelly, P. , Eichler, E. E. , Flicek, P. , Gabriel, S. B. , Gibbs, R. A. , Green, E. D. , Hurles, M. E. , Knoppers, B. M. , Korbel, J. O. , Lander, E. S. , Lee, C. , … Abecasis, G. R. (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacanu, S.‐A. , Devlin, B. , & Roeder, K. (2000). The power of genomic control. The American Journal of Human Genetics, 66(6), 1933–1944. 10.1086/302929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. (2010). Discovering the false discovery rate. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 72(4), 405–416. 10.1111/j.1467-9868.2010.00746.x [DOI] [Google Scholar]
- Berenbaum, F. (2013). Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthritis Cartilage, 21(1), 16–21. 10.1016/j.joca.2012.11.012 [DOI] [PubMed] [Google Scholar]
- Boer, C. G. , Yau, M. S. , Rice, S. J. , Coutinho de Almeida, R. , Cheung, K. , Styrkarsdottir, U. , Southam, L. , Broer, L. , Wilkinson, J. M. , Uitterlinden, A. G. , Zeggini, E. , Felson, D. , Loughlin, J. , Young, M. , Capellini, T. D. , Meulenbelt, I. , & van Meurs, J. B. J. (2021). Genome‐wide association of phenotypes based on clustering patterns of hand osteoarthritis identify WNT9A as novel osteoarthritis gene. Annals of the Rheumatic Diseases, 80(3), 367–375. 10.1136/annrheumdis-2020-217834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casula, M. A. , Facer, P. , Powell, A. J. , Kinghorn, I. J. , Plumpton, C. , Tate, S. N. , Bountra, C. , Birch, R. , & Anand, P. (2004). Expression of the sodium channel β3 subunit in injured human sensory neurons. NeuroReport, 15(10), 1629–1632. 10.1097/01.wnr.0000134927.02776.ae [DOI] [PubMed] [Google Scholar]
- Cavalli, E. , Mammana, S. , Nicoletti, F. , Bramanti, P. , & Mazzon, E. (2019). The neuropathic pain: an overview of the current treatment and future therapeutic approaches. International Journal of Immunopathology and Pharmacology, 33, 2058738419838383.– 10.1177/2058738419838383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell, A. S. , Ossanna, M. J. , Liu‐Seifert, H. , Iyengar, S. , Skljarevski, V. , Li, L. C. , Bennett, R. M. , & Collins, H. (2009). Duloxetine, a centrally acting analgesic, in the treatment of patients with osteoarthritis knee pain: A 13‐week, randomized, placebo‐controlled trial. Pain, 146(3). 10.1016/j.pain.2009.06.024 [DOI] [PubMed] [Google Scholar]
- Chen, J. , Shu, S. , Chen, Y. , Liu, Z. , Yu, L. , Yang, L. , Xu, Y. , & Zhang, M. (2019). AIM2 deletion promotes neuroplasticity and spatial memory of mice. Brain Research Bulletin, 152, 85–94. 10.1016/j.brainresbull.2019.07.011 [DOI] [PubMed] [Google Scholar]
- Chen, S.‐J. , Chen, L.‐H. , Yeh, Y.‐M. , Lin, C.‐C. , Lin, P.‐C. , Huang, H.‐W. , Shen, M.‐R. , Lin, B.‐W. , Lee, J.‐C. , Lee, C.‐C. , Lee, Y.‐F. , Chiang, H.‐C. , & Chang, J.‐Y. (2021). Targeting lysosomal cysteine protease cathepsin S reveals immunomodulatory therapeutic strategy for oxaliplatin‐induced peripheral neuropathy. Theranostics, 11(10), 4672–4687. 10.7150/thno.54793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y. H. , Macmillan, J. B. , & Chen, Z. J. (2009). RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG‐I pathway. Cell, 138(3), 576–591. 10.1016/j.cell.2009.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J. , Jun, G. R. , Dupuis, J. , & Farrer, L. A. (2019). Comparison of methods for multivariate gene‐based association tests for complex diseases using common variants. European Journal of Human Genetics, 27(5), 811–823. 10.1038/s41431-018-0327-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. K. , & Malcangio, M. (2012). Microglial signalling mechanisms: Cathepsin S and Fractalkine. Experimental Neurology, 234(2), 283–292. 10.1016/j.expneurol.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Cox, D. J. , Field, R. H. , Williams, D. G. , Baran, M. , Bowie, A. G. , Cunningham, C. , & Dunne, A. (2015). DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia, 63(5), 812–825. 10.1002/glia.22786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. J. , Reimann, F. , Nicholas, A. K. , Thornton, G. , Roberts, E. , Springell, K. , Karbani, G. , Jafri, H. , Mannan, J. , Raashid, Y. , Al‐Gazali, L. , Hamamy, H. , Valente, E. M. , Gorman, S. , Williams, R. , McHale, D. P. , Wood, J. N. , Gribble, F. M. , & Woods, C. G. (2006). An SCN9A channelopathy causes congenital inability to experience pain. Nature, 444(7121), 894–898. 10.1038/nature05413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft, P. , Jordan, K. , & Jinks, C. (2005). “Pain elsewhere” and the impact of knee pain in older people. Arthritis and Rheumatism, 52(8), 2350–2354. 10.1002/art.21218 [DOI] [PubMed] [Google Scholar]
- de Leeuw, C. A. , Mooij, J. M. , Heskes, T. , & Posthuma, D. (2015). MAGMA: generalized gene‐set analysis of GWAS data. PLoS Computational Biology, 11(4), e1004219. 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boer, C. , Dries, L. , Terluin, B. , van der Wouden, J. C. , Blankenstein, A. H. , van Wilgen, C. P. , Lucassen, P. , & van der Horst, H. E. (2019). Central sensitization in chronic pain and medically unexplained symptom research: A systematic review of definitions, operationalizations and measurement instruments. Journal of Psychosomatic Research, 117, 32–40. 10.1016/j.jpsychores.2018.12.010 [DOI] [PubMed] [Google Scholar]
- Dennis, G. , Sherman, B. T. , Hosack, D. A. , Yang, J. , Gao, W. , Lane, H. C. , & Lempicki, R. A. (2003). DAVID: Database for annotation, visualization, and integrated discovery. Genome Biology, 4(9), R60. 10.1186/gb-2003-4-9-r60 [DOI] [PubMed] [Google Scholar]
- Doherty, M. , & Smith, J. (1993). Elusive ‘alpha‐delta’ sleep in fibromyalgia and osteoarthritis. Annals of the Rheumatic Diseases, 52(3), 245. 10.1136/ard.52.3.245-b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Zhang, W. , Zeng, X. , Hu, G. , Zhang, H. , He, S. , & Zhang, S. (2014). Histamine induces upregulated expression of histamine receptors and increases release of inflammatory mediators from microglia. Molecular Neurobiology, 49(3), 1487–1500. 10.1007/s12035-014-8697-6 [DOI] [PubMed] [Google Scholar]
- Fernandes, G. S. , Sarmanova, A. , Warner, S. , Harvey, H. , Akin‐Akinyosoye, K. , Richardson, H. , Frowd, N. , Marshall, L. , Stocks, J. , Hall, M. , Valdes, A. M. , Walsh, D. , Zhang, W. , & Doherty, M. (2017). Knee pain and related health in the community study (KPIC): a cohort study protocol. BMC Musculoskeletal Disorders, 18(1), 404. 10.1186/s12891-017-1761-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, G. S. , & Valdes, A. M. (2015). Cardiovascular disease and osteoarthritis: common pathways and patient outcomes. European Journal of Clinical Investigation, 45(4), 405–414. 10.1111/eci.12413 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos, P. , & Knapp, S. (2014). Targeting bromodomains: epigenetic readers of lysine acetylation. Nature Reviews Drug Discovery, 13(5), 337–356. 10.1038/nrd4286 [DOI] [PubMed] [Google Scholar]
- Finan, P. H. , Buenaver, L. F. , Bounds, S. C. , Hussain, S. , Park, R. J. , Haque, U. J. , Campbell, C. M. , Haythornthwaite, J. A. , Edwards, R. R. , & Smith, M. T. (2013). Discordance between pain and radiographic severity in knee osteoarthritis: findings from quantitative sensory testing of central sensitization. Arthritis and Rheumatism, 65(2), 363–372. 10.1002/art.34646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freynhagen, R. , Baron, R. , Gockel, U. , & Tölle, T. R. (2006). painDETECT: a new screening questionnaire to identify neuropathic components in patients with back pain. Current Medical Research and Opinion, 22(10), 1911–1920. 10.1185/030079906x132488 [DOI] [PubMed] [Google Scholar]
- Gan, T. J. , & Habib, A. S. (2007). Adenosine as a non‐opioid analgesic in the perioperative setting. Anesthesia and Analgesia, 105(2), 487–494. 10.1213/01.ane.0000267260.00384.d9 [DOI] [PubMed] [Google Scholar]
- Gong, Q.‐J. , Li, Y.‐Y. , Xin, W.‐J. , Wei, X.‐H. , Cui, Y. , Wang, J. , Liu, Y. , Liu, C.‐C. , Li, Y.‐Y. , & Liu, X.‐G. (2010). Differential effects of adenosine A1 receptor on pain‐related behavior in normal and nerve‐injured rats. Brain Research, 1361, 23–30. 10.1016/j.brainres.2010.09.034 [DOI] [PubMed] [Google Scholar]
- Gong, Z. , Zhang, X. , Su, K. , Jiang, R. , Sun, Z. , Chen, W. , Forno, E. , Goetzman, E. S. , Wang, J. , Dong, H. H. , Dutta, P. , & Muzumdar, R. (2019). Deficiency in AIM2 induces inflammation and adipogenesis in white adipose tissue leading to obesity and insulin resistance. Diabetologia, 62(12), 2325–2339. 10.1007/s00125-019-04983-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff, M. , Scott, R. A. , Justice, A. E. , Young, K. L. , Feitosa, M. F. , Barata, L. , Winkler, T. W. , Chu, A. Y. , Mahajan, A. , Hadley, D. , Xue, L. , Workalemahu, T. , Heard‐Costa, N. L. , den Hoed, M. , Ahluwalia, T. S. , Qi, Q. , Ngwa, J. S. , Renström, F. , Quaye, L. , … Kilpeläinen, T. O. (2017). Genome‐wide physical activity interactions in adiposity – A meta‐analysis of 200,452 adults. PLoS Genetics, 13(4), e1006528. 10.1371/journal.pgen.1006528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graven‐Nielsen, T. , & Arendt‐Nielsen, L. (2002). Peripheral and central sensitization in musculoskeletal pain disorders: an experimental approach. Current Rheumatology Reports, 4(4), 313–321. 10.1007/s11926-002-0040-y [DOI] [PubMed] [Google Scholar]
- GTEx Consortium . (2015). The Genotype‐Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science, 348(6235), 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione, A. A. , Felson, D. T. , Anderson, J. J. , Anthony, J. M. , Zhang, Y. , Wilson, P. W. , Kelly‐Hayes, M. , Wolf, P. A. , Kreger, B. E. , & Kannel, W. B. (1994). The effects of specific medical conditions on the functional limitations of elders in the Framingham Study. American Journal of Public Health, 84(3), 351–358. 10.2105/ajph.84.3.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, K. , Pettersen, E. , Stovner, L. J. , Skorpen, F. , & Zwart, J.‐A. (2006). No association between chronic musculoskeletal complaints and Val158Met polymorphism in the Catechol‐O‐methyltransferase gene. The HUNT study. BMC Musculoskeletal Disorders, 7(1), 40. 10.1186/1471-2474-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, L. S. , Adams, M. J. , Arnau‐Soler, A. , Clarke, T.‐K. , Howard, D. M. , Zeng, Y. , Davies, G. , Hagenaars, S. P. , Maria Fernandez‐Pujals, A. , Gibson, J. , Wigmore, E. M. , Boutin, T. S. , Hayward, C. , Scotland, G. , Porteous, D. J. , Deary, I. J. , Thomson, P. A. , Haley, C. S. , & McIntosh, A. M. (2018). Genome‐wide meta‐analyses of stratified depression in Generation Scotland and UK Biobank. Translational Psychiatry, 8(1), 9. 10.1038/s41398-017-0034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J. , Padilla, F. , Dandonneau, M. , Lavebratt, C. , Lesage, F. , Noël, J. , & Delmas, P. (2013). Kv1.1 channels act as mechanical brake in the senses of touch and pain. Neuron, 77(5), 899–914. 10.1016/j.neuron.2012.12.035 [DOI] [PubMed] [Google Scholar]
- Heid, I. M. , Jackson, A. U. , Randall, J. C. , Winkler, T. W. , Qi, L. U. , Steinthorsdottir, V. , Thorleifsson, G. , Zillikens, M. C. , Speliotes, E. K. , Mägi, R. , Workalemahu, T. , White, C. C. , Bouatia‐Naji, N. , Harris, T. B. , Berndt, S. I. , Ingelsson, E. , Willer, C. J. , Weedon, M. N. , Luan, J. , … Lindgren, C. M. (2010). Meta‐analysis identifies 13 new loci associated with waist‐hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nature Genetics, 42(11), 949–960. 10.1038/ng.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. D. , Marioni, R. E. , Maghzian, O. , Ritchie, S. J. , Hagenaars, S. P. , McIntosh, A. M. , Gale, C. R. , Davies, G. , & Deary, I. J. (2019). A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Molecular Psychiatry, 24(2), 169–181. 10.1038/s41380-017-0001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman, J. R. , Davis, A. M. , Elkayam, J. , Gagliese, L. , & Hawker, G. A. (2013). Neuropathic pain symptoms on the modified painDETECT correlate with signs of central sensitization in knee osteoarthritis. Osteoarthritis and Cartilage, 21(9), 1236–1242. 10.1016/j.joca.2013.06.023 [DOI] [PubMed] [Google Scholar]
- Hocking, L. J. , Smith, B. H. , Jones, G. T. , Reid, D. M. , Strachan, D. P. , & Macfarlane, G. J. (2010). Genetic variation in the beta2‐adrenergic receptor but not catecholamine‐O‐methyltransferase predisposes to chronic pain: results from the 1958 British Birth Cohort Study. Pain, 149(1), 143–151. 10.1016/j.pain.2010.01.023 [DOI] [PubMed] [Google Scholar]
- Hoeffer, C. A. , & Klann, E. (2010). mTOR signaling: at the crossroads of plasticity, memory and disease. Trends in Neurosciences, 33(2), 67–75. 10.1016/j.tins.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, V. , Ablasser, A. , Charrel‐Dennis, M. , Bauernfeind, F. , Horvath, G. , Caffrey, D. R. , Latz, E. , & Fitzgerald, K. A. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase‐1‐activating inflammasome with ASC. Nature, 458(7237), 514–518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis, J. P. (2008). Why most discovered true associations are inflated. Epidemiology, 640–648. 10.1097/EDE.0b013e31818131e7 [DOI] [PubMed] [Google Scholar]
- James, R. J. E. , & Ferguson, E. (2019). The dynamic relationship between pain, depression and cognitive function in a sample of newly diagnosed arthritic adults: a cross‐lagged panel model. Psychological Medicine 50(10), 1663–1671. 10.1017/S0033291719001673 [DOI] [PubMed] [Google Scholar]
- Jaworski, J. , & Sheng, M. (2006). The growing role of mTOR in neuronal development and plasticity. Molecular Neurobiology, 34(3), 205–219. 10.1385/mn:34:3:205 [DOI] [PubMed] [Google Scholar]
- Ji, R. R. , Berta, T. , & Nedergaard, M. (2013). Glia and pain: is chronic pain a gliopathy? Pain, 154(Supplement 1), S10–s28. 10.1016/j.pain.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, R.‐R. , Nackley, A. , Huh, Y. , Terrando, N. , & Maixner, W. (2018). Neuroinflammation and central sensitization in chronic and widespread pain. Anesthesiology, 129(2), 343–366. 10.1097/ALN.0000000000002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, G. , Jiang, B. , & Cui, Y. (2013). Gene‐based Genomewide Association analysis: A comparison study. Current Genomics, 14(4), 250–255. 10.2174/13892029113149990001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohman, R. A. , Rodriguez‐Zas, S. L. , Southey, B. R. , Kelley, K. W. , Dantzer, R. , & Rhodes, J. S. (2011). Voluntary wheel running reverses age‐induced changes in hippocampal gene expression. PLoS One, 6(8), e22654. 10.1371/journal.pone.0022654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurien, T. , Arendt‐Nielsen, L. , Petersen, K. K. , Graven‐Nielsen, T. , & Scammell, B. E. (2018). Preoperative neuropathic pain‐like symptoms and central pain mechanisms in knee osteoarthritis predicts poor outcome 6 months after total knee replacement surgery. The Journal of Pain: Official Journal of the American Pain Society, 19(11), 1329–1341. 10.1016/j.jpain.2018.05.011 [DOI] [PubMed] [Google Scholar]
- Latremoliere, A. , & Woolf, C. J. (2009). Central sensitization: a generator of pain hypersensitivity by central neural plasticity. The Journal of Pain, 10(9), 895–926. 10.1016/j.jpain.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. J. , Wedow, R. , Okbay, A. , Kong, E. , Maghzian, O. , Zacher, M. , Nguyen‐Viet, T. A. , Bowers, P. , Sidorenko, J. , Karlsson Linnér, R. , Fontana, M. A. , Kundu, T. , Lee, C. , Li, H. , Li, R. , Royer, R. , Timshel, P. N. , Walters, R. K. , Willoughby, E. A. , … Cesarini, D. (2018). Gene discovery and polygenic prediction from a genome‐wide association study of educational attainment in 1.1 million individuals. Nature Genetics, 50(8), 1112–1121. 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipták, T. (1958). On the combination of independent tests. Magyar Tud Akad Mat Kutato Int Kozl, 3, 171–197. [Google Scholar]
- Locke, A. E. , Kahali, B. , Berndt, S. I. , Justice, A. E. , Pers, T. H. , Day, F. R. , Powell, C. , Vedantam, S. , Buchkovich, M. L. , Yang, J. , Croteau‐Chonka, D. C. , Esko, T. , Fall, T. , Ferreira, T. , Gustafsson, S. , Kutalik, Z. , Luan, J. , Mägi, R. , Randall, J. C. , … Speliotes, E. K. (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature, 518(7538), 197–206. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Santiago, L. F. , Pertin, M. , Morisod, X. , Chen, C. , Hong, S. , Wiley, J. , & Isom, L. L. (2006). Sodium channel beta2 subunits regulate tetrodotoxin‐sensitive sodium channels in small dorsal root ganglion neurons and modulate the response to pain. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience, 26(30), 7984–7994. 10.1523/jneurosci.2211-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, H. , Kreicbergs, A. , & Jansson, K.‐å (2008). Prediction of persistent pain after total knee replacement for osteoarthritis. The Journal of Bone and Joint Surgery. British Volume, 90‐B(2), 166–171. 10.1302/0301-620X.90B2.19640 [DOI] [PubMed] [Google Scholar]
- March, L. M. , & Bagga, H. (2004). Epidemiology of osteoarthritis in Australia. Medical Journal of Australia, 180(S5). 10.5694/j.1326-5377.2004.tb05906.x [DOI] [PubMed] [Google Scholar]
- Meshorer, E. , & Soreq, H. (2002). Pre‐mRNA splicing modulations in senescence. Aging Cell, 1(1), 10–16. 10.1046/j.1474-9728.2002.00005.x [DOI] [PubMed] [Google Scholar]
- Mishra, S. K. , & Hoon, M. A. (2010). Ablation of TrpV1 neurons reveals their selective role in thermal pain sensation. Molecular and Cellular Neuroscience, 43(1), 157–163. 10.1016/j.mcn.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil, J. S. (2012). Pain genetics: past, present and future. Trends in Genetics, 28(6), 258–266. 10.1016/j.tig.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Morello, R. , Bertin, T. K. , Chen, Y. , Hicks, J. , Tonachini, L. , Monticone, M. , Castagnola, P. , Rauch, F. , Glorieux, F. H. , Vranka, J. , Bächinger, H. P. , Pace, J. M. , Schwarze, U. , Byers, P. H. , Weis, M. A. , Fernandes, R. J. , Eyre, D. R. , Yao, Z. , Boyce, B. F. , & Lee, B. (2006). CRTAP is required for prolyl 3‐ hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell, 127(2), 291–304. 10.1016/j.cell.2006.08.039 [DOI] [PubMed] [Google Scholar]
- Moreton, B. J. , Tew, V. , das Nair, R. , Wheeler, M. , Walsh, D. A. , & Lincoln, N. B. (2015). Pain phenotype in patients with knee osteoarthritis: classification and measurement properties of painDETECT and self‐report Leeds assessment of neuropathic symptoms and signs scale in a cross‐sectional study. Arthritis Care and Research, 67(4), 519–528. 10.1002/acr.22431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, K. , Stevens, E. B. , Shah, B. , Cox, P. J. , Dixon, A. K. , Lee, K. , Pinnock, R. D. , Hughes, J. , Richardson, P. J. , Mizuguchi, K. , & Jackson, A. P. (2000). beta 3: an additional auxiliary subunit of the voltage‐sensitive sodium channel that modulates channel gating with distinct kinetics. Proceedings of the National Academy of Sciences, 97(5), 2308–2313. 10.1073/pnas.030362197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, P. , Benson, H. A. E. , Will, R. , & Wright, A. (2018). Patients with knee osteoarthritis who score highly on the PainDETECT questionnaire present with multimodality hyperalgesia, increased pain, and impaired physical function. The Clinical Journal of Pain, 34(1), 15–21. 10.1097/ajp.0000000000000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, R. R. , Campana, W. M. , & Shubayev, V. I. (2006). The role of neuroinflammation in neuropathic pain: mechanisms and therapeutic targets. Drug Discovery Today, 11(1), 8–20. 10.1016/S1359-6446(05)03637-8 [DOI] [PubMed] [Google Scholar]
- Nakano, N. , Maeyama, K. , Sakata, N. , Itoh, F. , Akatsu, R. , Nakata, M. , Katsu, Y. , Ikeno, S. , Togawa, Y. , Vo Nguyen, T. T. , Watanabe, Y. , Kato, M. , & Itoh, S. (2014). C18 ORF1, a novel negative regulator of transforming growth factor‐β signaling. Journal of Biological Chemistry, 289(18), 12680–12692. 10.1074/jbc.M114.558981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neogi, T. (2013). The epidemiology and impact of pain in osteoarthritis. Osteoarthritis and Cartilage, 21(9), 1145–1153. 10.1016/j.joca.2013.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neogi, T. , Guermazi, A. , Roemer, F. , Nevitt, M. C. , Scholz, J. , Arendt‐Nielsen, L. , Woolf, C. , Niu, J. , Bradley, L. A. , Quinn, E. , & Frey Law, L. (2016). Association of joint inflammation with pain sensitization in knee osteoarthritis: The multicenter osteoarthritis study. Arthritis and Rheumatology, 68(3), 654–661. 10.1002/art.39488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholl, B. I. , Holliday, K. L. , Macfarlane, G. J. , Thomson, W. , Davies, K. A. , O'Neill, T. W. , Bartfai, G. , Boonen, S. , Casanueva, F. , Finn, J. D. , & Forti, G. ; E. M. A. S. Group . (2010). No evidence for a role of the catechol‐O‐methyltransferase pain sensitivity haplotypes in chronic widespread pain. Annals of the Rheumatic Diseases, 69(11), 2009–2012. 10.1136/ard.2009.126086 [DOI] [PubMed] [Google Scholar]
- Okbay, A. , Beauchamp, J. P. , Fontana, M. A. , Lee, J. J. , Pers, T. H. , Rietveld, C. A. , Turley, P. , Chen, G.‐B. , Emilsson, V. , Meddens, S. F. W. , Oskarsson, S. , Pickrell, J. K. , Thom, K. , Timshel, P. , de Vlaming, R. , Abdellaoui, A. , Ahluwalia, T. S. , Bacelis, J. , Baumbach, C. , … Benjamin, D. J. (2016). Genome‐wide association study identifies 74 loci associated with educational attainment. Nature, 533(7604), 539–542. 10.1038/nature17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo, M. , Yamanaka, H. , Kobayashi, K. , Kanda, H. , Dai, Y. , & Noguchi, K. (2012). Up‐regulation of platelet‐activating factor synthases and its receptor in spinal cord contribute to development of neuropathic pain following peripheral nerve injury. Molecular Pain, 8, 1744–8069‐1748‐1748. 10.1186/1744-8069-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoutsopoulou, K. , Metrustry, S. , Doherty, S. A. , Laslett, L. L. , Maciewicz, R. A. , Hart, D. J. , & Valdes, A. M. (2014). The effect of %3cem%3eFTO%3c/em%3e variation on increased osteoarthritis risk is mediated through body mass index: a mendelian randomisation study. Annals of the Rheumatic Diseases, 73(12), 2082. 10.1136/annrheumdis-2013-203772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlaković, G. , & Petzke, F. (2010). The role of quantitative sensory testing in the evaluation of musculoskeletal pain conditions. Current Rheumatology Reports, 12(6), 455–461. 10.1007/s11926-010-0131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J. R. , Day, F. , Elks, C. E. , Sulem, P. , Thompson, D. J. , Ferreira, T. , & Ong, K. K. (2014). Parent‐of‐origin‐specific allelic associations among 106 genomic loci for age at menarche. Nature, 514(7520), 92–97. 10.1038/nature13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertin, M. , Ji, R.‐R. , Berta, T. , Powell, A. J. , Karchewski, L. , Tate, S. N. , & Decosterd, I. (2005). Upregulation of the voltage‐gated sodium channel beta2 subunit in neuropathic pain models: characterization of expression in injured and non‐injured primary sensory neurons. The Journal of Neuroscience : the Official Journal of the Society for Neuroscience, 25(47), 10970–10980. 10.1523/JNEUROSCI.3066-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, M. J. , Broer, L. , Willemen, H. L. D. M. , Eiriksdottir, G. , Hocking, L. J. , Holliday, K. L. , & van Meurs, J. B. J. (2013). Genome‐wide association study meta‐analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Annals of the Rheumatic Diseases, 72(3), 427–436. 10.1136/annrheumdis-2012-201742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, K. K. , Arendt‐Nielsen, L. , Simonsen, O. , Wilder‐Smith, O. , & Laursen, M. B. (2015). Presurgical assessment of temporal summation of pain predicts the development of chronic postoperative pain 12 months after total knee replacement. Pain, 156(1), 55–61. [DOI] [PubMed] [Google Scholar]
- Petersen, K. K. , Graven‐Nielsen, T. , Simonsen, O. , Laursen, M. B. , & Arendt‐Nielsen, L. (2016). Preoperative pain mechanisms assessed by cuff algometry are associated with chronic postoperative pain relief after total knee replacement. Pain, 157(7), 1400–1406. 10.1097/j.pain.0000000000000531 [DOI] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. R. , Bender, D. , & Sham, P. C. (2007). PLINK: A tool set for whole‐genome association and population‐based linkage analyses. The American Journal of Human Genetics, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakel, B. A. , Blodgett, N. P. , Zimmerman, B. M. , Logsden‐Sackett, N. , Clark, C. , Noiseux, N. , & Sluka, K. A. (2012). Predictors of postoperative movement and resting pain following total knee replacement. Pain, 153(11), 2192–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard, J. B. , Zhong, Z. , Williams, B. O. , & Yang, Y. (2012). Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harbor Perspectives in Biology, 4(12), a007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann, F. , Cox, J. J. , Belfer, I. , Diatchenko, L. , Zaykin, D. V. , McHale, D. P. , & Woods, C. G. (2010). Pain perception is altered by a nucleotide polymorphism in %3cem%3eSCN9A%3c/em>. Proceedings of the National Academy of Sciences, 107(11), 5148. 10.1073/pnas.0913181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L. , Bao, A. M. , & Swaab, D. F. (2015). The human histaminergic system in neuropsychiatric disorders. Trends in Neurosciences, 38(3), 167–177. 10.1016/j.tins.2014.12.008 [DOI] [PubMed] [Google Scholar]
- Shindou, H. , Shiraishi, S. , Tokuoka, S. M. , Takahashi, Y. , Harayama, T. , Abe, T. , & Shimizu, T. (2017). Relief from neuropathic pain by blocking of the platelet‐activating factor‐pain loop. The FASEB Journal, 31(7), 2973–2980. 10.1096/fj.201601183R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. T. , Quartana, P. J. , Okonkwo, R. M. , & Nasir, A. (2009). Mechanisms by which sleep disturbance contributes to osteoarthritis pain: a conceptual model. Current Pain and Headache Reports, 13(6), 447–454. [DOI] [PubMed] [Google Scholar]
- Söhle, J. , Machuy, N. , Smailbegovic, E. , Holtzmann, U. , Grönniger, E. , Wenck, H. , & Winnefeld, M. (2012). Identification of new genes involved in human adipogenesis and fat storage. PLoS One, 7(2), e31193. 10.1371/journal.pone.0031193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, A. , Wanigasekera, V. , Mezue, M. , Cooper, C. , Javaid, M. K. , Price, A. J. , & Tracey, I. (2019). Central Sensitization in Knee Osteoarthritis: Relating Presurgical Brainstem Neuroimaging and PainDETECT‐Based Patient Stratification to Arthroplasty Outcome. Arthritis and Rheumatology, 71(4), 550–560. 10.1002/art.40749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland, M. R. , Koller, E. J. , Deng, D. Z. , Ceballos‐Diaz, C. , Golde, T. E. , & Chakrabarty, P. (2018). Ifngr1 and Stat1 mediated canonical Ifn‐γ signaling drives nigrostriatal degeneration. Neurobiology of Disease, 110, 133–141. 10.1016/j.nbd.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitani, M. , Ogata, T. , Natori, A. , Hozumi, J. , Shimojo, N. , Kida, K. , & Yamauchi, T. (2016). Poor efficacy of the phosphorylated high‐molecular‐weight neurofilament heavy subunit serum level, a biomarker of axonal damage, as a marker of chemotherapy‐induced peripheral neuropathy. Biomedical Reports, 4(6), 758–760. 10.3892/br.2016.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suokas, A. K. , Walsh, D. A. , McWilliams, D. F. , Condon, L. , Moreton, B. , Wylde, V. , & Zhang, W. (2012). Quantitative sensory testing in painful osteoarthritis: a systematic review and meta‐analysis. Osteoarthritis and Cartilage, 20(10), 1075–1085. 10.1016/j.joca.2012.06.009 [DOI] [PubMed] [Google Scholar]
- Swain, S. , Sarmanova, A. , Coupland, C. , Doherty, M. , & Zhang, W. (2020). Comorbidities in osteoarthritis: A systematic review and meta‐analysis of observational studies. Arthritis Care and Research, 72(7), 991–1000. 10.1002/acr.24008 [DOI] [PubMed] [Google Scholar]
- Tsuda, M. , Tozaki‐Saitoh, H. , & Inoue, K. (2011). Platelet‐activating factor and pain. Biological and Pharmaceutical Bulletin, 34(8), 1159–1162. 10.1248/bpb.34.1159 [DOI] [PubMed] [Google Scholar]
- Valdes, A. M. , De Wilde, G. , Doherty, S. A. , Lories, R. J. , Vaughn, F. L. , Laslett, L. L. , & Doherty, M. (2011). The Ile585Val %3cem%3eTRPV1%3c/em%3e variant is involved in risk of painful knee osteoarthritis. Annals of the Rheumatic Diseases, 70(9), 1556. 10.1136/ard.2010.148122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes, A. M. , Doherty, S. A. , Zhang, W. , Muir, K. R. , Maciewicz, R. A. , & Doherty, M. (2012). Inverse relationship between preoperative radiographic severity and postoperative pain in patients with osteoarthritis who have undergone total joint arthroplasty. Seminars in Arthritis and Rheumatism, 41(4), 568–575. 10.1016/j.semarthrit.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Valdes, A. M. , & Stocks, J. (2018). Osteoarthritis and ageing. EMJ, 3(1), 116–123. [Google Scholar]
- van de Wetering, D. , de Paus, R. A. , van Dissel, J. T. , & van de Vosse, E. (2010). Functional analysis of naturally occurring amino acid substitutions in human IFN‐gammaR1. Molecular Immunology, 47(5), 1023–1030. 10.1016/j.molimm.2009.11.016 [DOI] [PubMed] [Google Scholar]
- van der Harst, P. , & Verweij, N. (2018). Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circulation Research, 122(3), 433–443. 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meurs, J. B. J. , Uitterlinden, A. G. , Stolk, L. , Kerkhof, H. J. M. , Hofman, A. , Pols, H. A. P. , & Bierma‐Zeinstra, S. M. A. (2009). A functional polymorphism in the catechol‐O‐methyltransferase gene is associated with osteoarthritis‐related pain. Arthritis and Rheumatism, 60(2), 628–629. 10.1002/art.24175 [DOI] [PubMed] [Google Scholar]
- Veronese, N. , Maggi, S. , Trevisan, C. , Noale, M. , De Rui, M. , Bolzetta, F. , & Perissinotto, E. (2017). Pain increases the risk of developing frailty in older adults with osteoarthritis. Pain Medicine, 18(3), 414–427. [DOI] [PubMed] [Google Scholar]
- Vujkovic, M. , Keaton, J. M. , Lynch, J. A. , Miller, D. R. , Zhou, J. , Tcheandjieu, C. , & Saleheen, D. (2020). Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi‐ancestry meta‐analysis. Nature Genetics, 52(7), 680–691. 10.1038/s41588-020-0637-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Wu, R. , Tang, D. , & Kang, R. (2021). The BET family in immunity and disease. Signal Transduction and Targeted Therapy, 6(1). 10.1038/s41392-020-00384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, S. C. , & Valdes, A. M. (2017). Genetic association studies in osteoarthritis: Is it fairytale? Current Opinion in Rheumatology, 29(1), 103–109. [DOI] [PubMed] [Google Scholar]
- Warner, S. C. , van Meurs, J. B. , Schiphof, D. , Bierma‐Zeinstra, S. M. , Hofman, A. , Uitterlinden, A. G. , & Valdes, A. M. (2017). Genome‐wide association scan of neuropathic pain symptoms post total joint replacement highlights a variant in the protein‐kinase C gene. European Journal of Human Genetics, 25(4), 446–451. 10.1038/ejhg.2016.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, S. C. , Walsh, D. A. , Laslett, L. L. , Maciewicz, R. A. , Soni, A. , Hart, D. J. , & Valdes, A. M. (2017). Pain in knee osteoarthritis is associated with variation in the neurokinin 1/substance P receptor (TACR1) gene. European Journal of Pain, 21(7), 1277–1284. 10.1002/ejp.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K. , Taskesen, E. , van Bochoven, A. , & Posthuma, D. (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock, M. C. (2005). Combining probability from independent tests: the weighted Z‐method is superior to Fisher's approach. Journal of Evolutionary Biology, 18(5), 1368–1373. 10.1111/j.1420-9101.2005.00917.x [DOI] [PubMed] [Google Scholar]
- Wu, P. J. , Hung, Y. F. , Liu, H. Y. , & Hsueh, Y. P. (2017). Deletion of the inflammasome sensor Aim2 mitigates Aβ deposition and microglial activation but increases inflammatory cytokine expression in an Alzheimer disease mouse model. NeuroImmunoModulation, 24(1), 29–39. 10.1159/000477092 [DOI] [PubMed] [Google Scholar]
- Wu, P. J. , Liu, H. Y. , Huang, T. N. , & Hsueh, Y. P. (2016). AIM 2 inflammasomes regulate neuronal morphology and influence anxiety and memory in mice. Scientific Reports, 6, 32405. 10.1038/srep32405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W.‐P. , Hao, J.‐X. , Halldner, L. , Lövdahl, C. , DeLander, G. E. , Wiesenfeld‐Hallin, Z. , & Xu, X.‐J. (2005). Increased nociceptive response in mice lacking the adenosine A1 receptor. Pain, 113(3), 395–404. 10.1016/j.pain.2004.11.020 [DOI] [PubMed] [Google Scholar]
- Wylde, V. , Sayers, A. , Lenguerrand, E. , Gooberman‐Hill, R. , Pyke, M. , Beswick, A. D. , Dieppe, P. , & Blom, A. W. (2015). Preoperative widespread pain sensitization and chronic pain after hip and knee replacement. Pain, 156(1), 47–54. 10.1016/j.pain.0000000000000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. T. , Zhao, J. Y. , Zhao, X. , Ligons, D. , Tiwari, V. , Atianjoh, F. E. , & Tao, Y. X. (2014). Opioid receptor‐triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. The Journal of Clinical Investigation, 124(2), 592–603. 10.1172/jci70236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Fitzsimmons, B. , Steinauer, J. , O'Neill, A. , Newton, A. C. , Hua, X. Y. , & Yaksh, T. L. (2011). Spinal phosphinositide 3‐kinase‐Akt‐mammalian target of rapamycin signaling cascades in inflammation‐induced hyperalgesia. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience, 31(6), 2113–2124. 10.1523/jneurosci.2139-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini, E. , Panoutsopoulou, K. , Southam, L. , Rayner, N. W. , Day‐Williams, A. G. , Lopes, M. C. , & Loughlin, J. (2012). Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome‐wide association study. Lancet, 380(9844), 815–823. 10.1016/s0140-6736(12)60681-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S4


