Summary
Background
Individuals with Lynch syndrome are at high risk for colorectal cancer (CRC). Regular colonoscopies have proven to decrease CRC incidence and mortality. However, colonoscopy is burdensome and interval CRCs still occur. Hence, an accurate, less‐invasive screening method that guides the timing of colonoscopy would be of important value.
Aim
To outline the performance of non‐endoscopic screening modalities for Lynch‐associated CRC and adenomas.
Methods
Systematic literature search in MEDLINE and EMBASE to identify studies investigating imaging techniques and biomarkers for detection of CRC and adenomas in Lynch syndrome. The QUADAS‐2 tool was used for the quality assessment of included studies.
Results
Seven of 1332 screened articles fulfilled the inclusion criteria. Two studies evaluated either CT colonography or MR colonography; both techniques were unable to detect CRC and (advanced) adenomas <10 mm. The other five studies evaluated plasma methylated‐SEPTIN9, faecal immunochemical test (FIT), faecal tumour DNA markers (BAT‐26, hMLH1, p53, D9S171, APC, D9S162, IFNA and DCC) and faecal microbiome as screening modalities. Sensitivity for CRC varied from 33% (BAT‐26) to 70% (methylated‐SEPTIN9) to 91% (hMLH1). High specificity (94–100%) for CRC and/or adenomas was observed for methylated‐SEPTIN9, FIT and BAT‐26. Desulfovibrio was enriched in the stool of patients having adenomas. However, all these studies were characterised by small populations, high/unclear risk of bias and/or low prevalence of adenomas.
Conclusions
Imaging techniques are unsuitable for colon surveillance in Lynch syndrome, whereas biomarkers are understudied. Having outlined biomarker research in Lynch‐associated and sporadic CRC/adenomas, we believe that these non‐invasive markers may hold potential (whether or not combined) for this population. As they could be of great value, (pre‐)clinical studies in this field should be prioritised.

1. INTRODUCTION
Lynch syndrome is the most common hereditary colorectal cancer (CRC) syndrome, accounting for approximately 2%‐4% of all CRC cases. 1 The syndrome is characterised by early onset CRC and accelerated carcinogenesis, 2 which occurs due to mutated DNA mismatch repair genes. 3 As a result, individuals with Lynch syndrome have a cumulative lifetime CRC risk of 15%–70% at age 70 4 and are at increased risk of metachronous CRC. 5 , 6 Therefore, the European Society of Gastrointestinal Endoscopy guideline recommends surveillance colonoscopy every 2 years for individuals with Lynch syndrome, starting at the age of 25 for MLH1 and MSH2 and age of 35 for MSH6 and PMS2 mutation carriers. 4
Although colonoscopic surveillance is effective in reducing the incidence, morbidity and mortality of CRC in this high‐risk population, 7 it also has limitations. First, individuals experience both colonoscopy and the required bowel preparation as burdensome. 8 This negative perception is a predictor of non‐compliance with regular colonoscopic surveillance in high‐risk patients, 9 and hence needs attention as compliance rates in individuals with Lynch syndrome are sub‐optimal. 10 Another drawback of colonoscopy is the small risk for serious complications such as bleeding and perforation. Furthermore, the procedure is resource intensive and costly, so a surveillance program involving frequent colonoscopies puts pressure on healthcare systems. 11
Despite regular (1–3 yearly) colonoscopies, up to 15% of individuals with Lynch syndrome still develop CRC. 12 These interval carcinomas might arise from missed or incompletely resected adenomas during colonoscopy or from the progression of normal mucosa to carcinoma within the recommended surveillance interval. 12
The above limitations underline the need for an accurate, less‐invasive screening modality for CRC and adenomas in individuals with Lynch syndrome, which could guide the timing of colonoscopy. An accurate, less‐invasive screening method would reduce (1) the number of unnecessary colonoscopies and (2) potentially, the interval carcinoma rate by selecting individuals who require a colonoscopy at a shorter time interval, resulting in improved prognosis. For colorectal neoplasia in an average‐risk population, such methods (primarily biomarkers and imaging techniques) are studied extensively, with promising results and some (i.e. FIT and Cologuard) are even being implemented for population screening strategies. 13 , 14 In contrast, no study to date has provided an overview of potential alternative, less‐invasive surveillance modalities for individuals with a genetically determined elevated lifetime risk of CRC (including Lynch syndrome). Such data are valuable because particularly this population would benefit substantially from an accurate, minimally invasive surveillance test.
Given this knowledge gap, the aim of this systematic review was to outline non‐endoscopic screening modalities for CRC and adenomas in individuals with Lynch syndrome.
2. MATERIALS AND METHODS
The protocol of this systematic review was registered in the PROSPERO database (registration number: CRD42020215405). The review was performed in accordance with the protocol and the Preferred Reporting Items for Systematic Reviews and Meta‐analysis (PRISMA) statement. 15
A systematic electronic literature search was performed in July 2020 using the MEDLINE (via PubMed) and Embase databases. An update of the search was conducted in February 2021 to gather recently published articles. The search was developed by the first author in cooperation with an information specialist and included search terms representing the following components: Lynch syndrome, colorectal neoplasia, surveillance and non‐endoscopic test. All types of non‐endoscopic tests were evaluated, which included both imaging and (bio‐ or tumour) markers such as proteins (e.g. faecal haemoglobin), DNA, RNA, metabolites (e.g. volatile organic compounds) and gut microbiome composition. The full search strategy can be found in Table S1. No language or publication date restrictions were applied.
Titles and abstracts of articles identified were screened using the Rayyan web application 16 by two independent researchers (EvL and NdB) to determine eligibility for full‐text evaluation. Discrepancies were discussed, and if needed, a third reviewer (DR) was consulted for advice. The same method was applied during the second, full‐text phase of the study selection process. Articles were deemed relevant if data consisted of (a) individuals with Lynch syndrome, that is proven MMR mutation or those fulfilling the Amsterdam I or II criteria; (b) CRC and/or adenomas (all types) and (c) a non‐endoscopic screening modality for CRC and/or adenomas. Non‐original studies, single case reports, duplicates and articles written in a language other than Dutch or English were excluded. Reference lists of included studies and of relevant reviews identified during the search were reviewed for additional eligible studies, as were articles that had cited an included record. Data extraction from the included studies was performed by EvL and verified by NdB. If additional, unpublished information was necessary for inclusion or analysis, the authors were contacted to request the data.
Two independent reviewers (EvL and NdB) evaluated the quality of included studies using the Quality Assessment of Diagnostic Accuracy Studies‐2 (QUADAS‐2) tool, 17 with a third researcher (DR) being consulted in case of any disagreement.
3. RESULTS
The selection process of the records yielded by the systematic literature search is outlined in a PRISMA flow diagram (Figure 1). In total, 1332 articles were screened for eligibility, of which 58 were selected for full‐text evaluation. This second phase resulted in seven studies being included for analysis. 18 , 19 , 20 , 21 , 22 , 23 , 24 One of these was only published as an abstract. 21 We decided to include this study as the results were described in sufficient detail and the methodology was provided upon request by the first author (by referring to two high‐quality studies 25 , 26 ).
FIGURE 1.
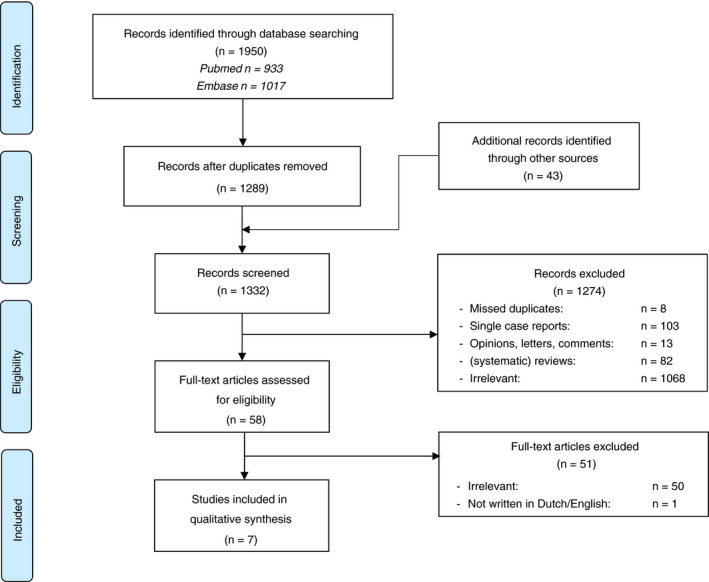
Flow diagram of the study selection process
Study characteristics of included records are provided in Table 1. A summary of the quality assessment using the QUADAS‐2 tool is detailed in Table 2 and Figure 2. Five of seven studies were deemed at high or unclear risk of bias and had concerns regarding applicability. Each study investigated a different screening method for CRC and/or adenomas in individuals with Lynch syndrome, consisting of imaging techniques (n = 2), blood markers (n = 1) and stool markers (n = 4).
TABLE 1.
Study characteristics of included records
| Study | Study design a | Surveillance test/marker | No. participants | Gender (F/M) | Age (years) | MMR mutation (n) b | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|---|---|---|
| Imaging | Lim et al. 22 | Prospective, single centre | MRC | 30 | 17/13 | Median 48 (range 22–70) |
|
|
|
| Renkonen et al. 23 | Prospective, single centre | CTC | 78 | 40/38 | Median 41 (range 20–70) |
|
|
||
| Blood marker | Hitchins et al. 19 | Retrospective | Methylated SEPTIN9 |
20 cases (CRC) 34 controls |
Unknown |
|
Cases/controls:
|
CRC: 70% | CRC: 100% |
| Stool markers | Digby et al. 18 , d | Prospective, multi‐centre | FIT | 17 | 7/10 | Median 56 (range 33–65) | Unknown | Adenomas:
|
High‐risk adenomas
f
:
|
| Laken et al. 21 , d | Prospective | BAT‐26 | 44 | Unknown | Unknown | Unknown |
CRC: 33% Adenomas: unknown |
CRC + adenomas: 100% | |
| Koshiji et al. 20 | Prospective | hMLH1, p53, D9S171, APC, D9S162, IFNA, DCC c | 11 | 4/7 | Mean 56 (range 36–75) | Unknown | CRC:
|
Unknown | |
| Yan et al. 24 | Prospective, multi‐centre | Microbiome profile (metagenomics and meta‐transcriptomics) | 100 | 56/44 | Median 50 (range 21–89) |
|
Unknown | Unknown |
Abbreviations: CTC, computed tomographic colonography; CRC, colorectal cancer; FIT, faecal immunochemical test; MMR, mismatch repair; MRC, magnetic resonance colonography; SD, standard deviation.
Some authors did not describe whether their study was single‐ or multi‐centre.
All participants, except those in the study of Kosiji et al., had a proven mutation in one of the MMR genes. The article of Lim et al. contained a miscalculation of MMR status; correct numbers were obtained by contacting the first author.
Located on the following chromosomes: 3p21.3 (hMLH1), 17p13.2‐17p12 (p53), 9p21 (D9S171), 5q15‐5q23.1 (APC), 9p22‐21 (D9S162), 9p22 (IFNA), 18q21.3 (DCC).
Additional information was provided by the first author.
Sensitivity and specificity were radiologist dependent, therefore a range is provided.
Defined as ≥3 adenomas or any ≥10 mm.
TABLE 2.
Quality of included studies using the quality assessment of diagnostic accuracy studies‐2 (QUADAS‐2) tool
| Risk of bias | Applicability concerns | ||||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Patient selection | Index test | Reference standard | |
| Lim et al. 22 |

|

|

|

|

|

|

|
| Renkonen et al. 23 |

|

|

|

|

|

|

|
| Hitchins et al. 19 |

|
? | ? |

|
? |

|

|
| Digby et al. 18 |

|

|

|

|

|

|

|
| Laken et al. 21 | ? | ? | ? | ? | ? |

|

|
| Koshiji et al. 20 |

|
? | ? |

|

|

|

|
| Yan et al. 24 |

|
? |

|

|

|

|

|
Note:
 = low risk,
= low risk, = high risk, ? = unclear risk.
= high risk, ? = unclear risk.
Review Question: Test population = patients with Lynch syndrome (proven mutation mismatch repair genes), Index test = non‐endoscopic test, Reference standard = histology of tissue obtained with colonoscopy or if applicable with subsequent surgery, Target condition = colorectal carcinoma and colorectal adenoma, Setting = hospital/medical centre, Intended use of the index test = surveillance, Patient presentation = follow‐up according to a surveillance program, Prior testing = not applicable.
FIGURE 2.
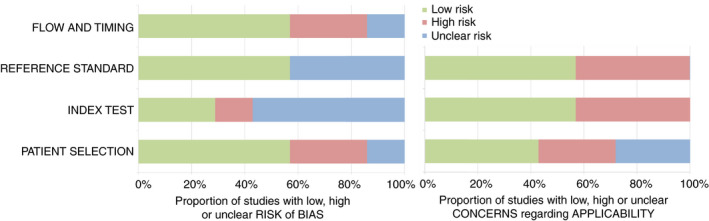
Risk of bias and applicability concerns using the quality assessment of diagnostic accuracy studies‐2 (QUADAS‐2) tool
3.1. Imaging
Two prospective studies reported on the use of imaging techniques for surveillance: one evaluated magnetic resonance colonography (MRC), 22 the other computed tomographic colonography (CTC). 23 The procedures were performed after standard bowel preparation, with MRC/CTC undertaken prior to but on the same day as colonoscopy. For MRC and CTC, optimal colon distension was achieved by rectal administration of water or room air, respectively. Intravenous scopolamine was routinely used.
MRC was evaluated in 30 individuals, using a 1.5 T MRI and intravenous contrast. 22 Three‐dimensional T2‐weighed images were acquired, which were interpreted by two independent radiologists using 2D and 3D modes. Eighty‐three percent of scans were rated as adequate to good quality. With MRC a total of three lesions, all adenocarcinomas (70, 36 and 17 mm), in three separate patients were detected by both radiologists (one in a poor‐quality scan). However, MRC failed to detect any lesion smaller than 10 mm, including a 3 mm adenocarcinoma and two tubulovillous adenomas (4 and 3 mm). As such, the sensitivity of MRC for detecting all‐sized CRC and adenomas was reported to be 50%. Whether an association existed between the quality of scan and the miss‐rate was not described. In this study, individuals’ perception of MRC and colonoscopy (performed under sedation) was also compared, using a visual analogue scale. MRC was associated with less reported discomfort than colonoscopy (20% vs 68%, P = 0.003), whereas mean inconvenience scores were equal for both entities.
CTC performance was assessed in 78 individuals with Lynch syndrome by Renkonen et al. (using a 4‐row multidetector scanner). 23 This generated 2D images in supine and, if necessary, in prone position, which were analysed by two independent radiologists. Of all colorectal segments studied, 19% (88/468) were regarded as technically suboptimal. Nevertheless, both CRCs (both ≥10 mm) were detected by each radiologist. A total of three large adenomas (≥10 mm), 1 medium‐sized adenoma (6–9 mm), 11 small adenomas (2–5 mm) and 20 small hyperplastic polyps were missed by one or both radiologists. Hence, per‐patient sensitivity of CTC for detecting all‐sized lesions was 25% (radiologist A) and 29% (radiologist B), whereas specificity was 82% and 76%, respectively. Specific performance for all‐sized neoplastic lesions (so without including hyperplastic polyps) was not reported. Per‐patient sensitivity for CRC and adenomas ≥10 mm was 60% (radiologist A) and 100% (radiologist B), whereas specificity was 96% and 96%. No significant difference in performance was observed between individuals receiving (35%) and those not receiving intravenous contrast.
Although an advantage of MRC/CTC is the possibility to detect extra‐colonic pathology, no relevant abnormalities were found except for lymphadenopathy in one patient with CRC. 23
To summarise, in these two small studies, neither MRC (n = 3 neoplastic lesions <10 mm at colonoscopy) nor CTC (n = 12 neoplastic lesions <10 mm at colonoscopy) were able to detect CRC and (advanced) adenomas <10 mm in individuals with Lynch syndrome.
3.2. Blood marker
One study investigated a blood‐based marker, that is methylated SEPTIN9 (mSEPT9), as a screening method for CRC in individuals with Lynch syndrome. 19 In this retrospective case–control study, the authors found that hypermethylation of the SEPTIN9‐gene was present in 97% (36/37) and 90% (18/20) of Lynch‐associated CRC and advanced adenoma specimens, respectively. As a next step, plasma samples of individuals with Lynch syndrome with or without CRC were assessed for detectability of mSEPT9 as circulating tumour DNA, using the Epi proColon 2.0 CE assay adapted for 1 mL plasma (1/1 PCR algorithm). Controls were diagnosed with Lynch syndrome but had no history of CRC/adenomas and did not develop CRC within 2 years after the blood draw. CRC cases were categorised into two groups. The first group had drawn blood after a colonoscopy‐based CRC diagnosis (“post‐diagnosis cases,” n = 20). The second group consisted of samples collected within 1 year (median 103 days, range 20–328 days) prior to CRC diagnosis (“pre‐diagnosis cases,” n = 18). These samples were evaluated to investigate whether mSEPT9 is detectable before the usual CRC diagnosis and thus could serve as an interval test between colonoscopies. In this study, the diagnostic performance of mSEPT9 for colorectal adenomas was not evaluated.
A valid mSEPT9 test result was obtained in 20/20 post‐ and 17/18 pre‐diagnosis cases. Of the 20 post‐diagnosis patients, 14 (with CRC stages I – IV) tested positive for mSEPT9. Hence, sensitivity for detecting CRC with plasma‐based mSEPT9 was 70%. Contrarily, only three pre‐diagnosis patients demonstrated a positive test. These were among five patients with samples collected up to approximately 2 months (median 49 days, IQR 28–57 days) prior to colonoscopy‐based diagnosis of CRC stage I–III, resulting in a 60% sensitivity of mSEPT9 to detect CRC approximately 2 months prior to usual diagnosis. All valid plasma samples from controls (31/34) were negative for mSEPT9, so specificity was reported to be 100%.
3.3. Stool markers
Stool‐based markers for surveillance of individuals with Lynch syndrome were explored prospectively in four studies and included the following: FIT, 18 tumour DNA markers (i.e. BAT‐26 21 and hMLH1, p53, D9S171, APC, D9S162, IFNA, DCC 20 ) and microbiome profile. 24
The diagnostic accuracy of the faecal immunochemical test (FIT), which quantifies faecal haemoglobin concentration, was studied by Digby et al. in individuals at increased risk of CRC. 18 Of the 593 individuals in this study, 19 had Lynch syndrome (of whom 17 had complete colonoscopy). FIT performance data of this specific population were provided by the first author for the purpose of the current review. In the study, faecal samples were collected within 4 weeks prior to colonoscopy and analysed using an OC‐Sensor with a limit of detection of 2 μg Hb/g faeces. Different cut‐offs for a positive FIT were assessed, that is 2, 4 and 10 μg Hb/g faeces. Of the 17 individuals with Lynch syndrome who underwent complete colonoscopy, 1/17 had high‐risk adenoma (defined as ≥3 adenomas or any ≥10 mm), 1/17 had low‐risk adenoma and 0/17 CRC. At all three FIT cut‐offs, the single patient with high‐risk adenoma tested positive, whereas the single patient with low‐risk adenoma tested negative. However, increasing the threshold from 2 to either 4 or 10 μg Hb/g faeces resulted in fewer false‐positive cases (3 vs 1) and subsequently improved specificity of FIT for high‐risk adenomas (81% [95% CI 54–96%] vs 94% [95% CI 70–100%]).
Another faecal marker that was investigated in individuals with Lynch syndrome concerned altered DNA, that is deletions within BAT‐26 (a microsatellite instability marker), which is exfoliated into a stool by neoplastic tissue. 21 After sequence‐specific purification (to isolate human DNA from stool) and amplification, BAT‐26 analysis was performed using gel electrophoresis. 25 Mutated BAT‐26 was found in the faeces of 2/44 participants, of whom one had CRC and the other advanced adenomas. 21 Two patients with CRC had a false‐negative test, while in one of them BAT‐26 mutations were present in the tumour tissue. Hence, the sensitivity of faecal BAT‐26 to detect CRC in individuals with Lynch syndrome was 33%. Sensitivity for adenomas could not be determined, since the number of missed adenomas was not described. Nonetheless, all 40 individuals with Lynch syndrome and no CRC or adenomas at colonoscopy had a negative faecal BAT‐26 assay, so specificity was 100%.
DNA shed in stool by tumour cells was also studied by Koshiji et al. 20 They measured the loss of heterozygosity (LOH) by assaying seven chromosomal location markers (i.e. hMLH1, p53, D9S171, APC, D9S162, IFNA and DCC) in faeces of 11 patients with microsatellite instable CRC and likely to have Lynch syndrome according to the Amsterdam I criteria. These markers were used to measure LOH and not microsatellite instability; both are signature features of chromosomal instability though different conditions. Mismatch repair gene status was not reported in this study, nor was the performance for colorectal adenomas. Three methods for DNA extraction were tested in their ability to provide sufficient DNA with a low level of colonocyte DNA contamination. Applying one of these methods, LOH scores found in stool matched those determined in corresponding tumour tissue in each patient. LOH at hMLH1 was found in the faeces (and thus the neoplastic tissue) of 10/11 patients included. Moreover, LOH at DCC was observed in 9/11 patients, at IFNA in 8/11, at D9S162 in 6/11, at APC in 4/11, at D9S171 in 4/11 and at p53 in 4/11. On the other hand, 2/15 controls without cancer also showed LOH of faecal D9S162. The authors were contacted, unsuccessfully, to clarify whether LOH of the other chromosomal location markers was detected in stool samples of controls and whether these controls were (suspected of) having Lynch syndrome.
The last study included in this systematic review is the study of Yan et al., in which faecal microbiome composition was examined in hundred individuals with Lynch syndrome. 24 Stool specimens and colon biopsies of 56 patients with adenomas (all ≤10 mm in size) and of 31 individuals without adenomas were collected. No patients with CRC were included in this study. Stool samples were assayed by metagenomics and metatranscriptomics to yield taxonomic and functional profiles, whereas biopsies were analysed using 16s rRNA gene sequencing. In general, the microbial composition of stool samples accurately reflected the colonic (mucosal) microbiome, illustrating low within‐subject variation. Strikingly, the bacterium Desulfovibrio was enriched both in stool samples and in mucosal biopsies of patients with adenomas. On the contrary, 9% of all unique species in the cohort, among which were Clostridiaceae, showed depleted abundances in patients with adenomas. However, no single taxon was found to be consistently predictive of adenoma carriage. Nevertheless, adenoma carriage could best be predicted by microbial transcripts (using random forest classifiers), although weakly with an area under the curve of 0.63 (with approximately similar sensitivity and specificity rates).
4. DISCUSSION
In this systematic review, we outlined non‐endoscopic screening modalities for CRC and adenomas in individuals with Lynch syndrome. Seven studies were identified, which evaluated the following screening methods: MRC, CTC, plasma mSEPT9, FIT, faecal tumour DNA markers (BAT‐26, hMLH1, p53, D9S171, APC, D9S162, IFNA and DCC) and faecal microbiome profile.
In the two imaging studies, CRC and (advanced) adenomas of <10 mm observed at colonoscopy could not be detected with either CTC or MRC. 22 , 23 Considering the accelerated adenoma‐carcinoma sequence, these techniques are therefore unsuitable for surveillance of individuals with Lynch syndrome. In other high‐ and average‐risk populations, polyps <10 mm (including adenomas) are also frequently missed with MRC or CTC, but sensitivity may improve when advanced technology and thin slice thickness are used. 27 , 28 Nonetheless, the burden for the patient (bowel preparation, hospital visit, rectal tube insertion) and the exposure to ionising radiation in CTC remain important drawbacks of these methods.
Biomarkers are most practical for colon surveillance in Lynch syndrome. Promising outcomes were observed in average‐risk populations for CRC. 13 However, we could not draw firm conclusions with regard to (blood‐ and stool‐based) biomarkers for Lynch‐associated CRC and adenomas, since only five studies were conducted in this field, which were small (with thus limited cases of CRC/adenomas) and/or had high or unclear risk of bias. Sensitivity varied from 33% (faecal tumour BAT‐26), to 70% (plasma tumour mSEPT9) to 91% (faecal tumour hMLH1). High specificity (94%–100%) was observed for mSEPT9, FIT and BAT‐26.
Faeces can be considered an obvious matrix for colorectal neoplasia biomarker discovery research since it reflects the intestinal metabolic and microbial state. While research from the United Kingdom demonstrated low patient acceptability with stool sample collection, 29 a larger and more recent study from the Netherlands showed great willingness. 30 Candidate faecal markers for surveillance of individuals with Lynch syndrome may include haemoglobin, tumour DNA and RNA, gut microbiome composition and metabolomics (see next paragraphs).
Faecal haemoglobin is used in national bowel screening programs and is an attractive marker since analysis (performed with FIT) is inexpensive, easy and fast. Efficacy of FIT in individuals at increased risk of neoplasia is highly dependent on the cut‐off of haemoglobin concentration: sensitivity improves significantly, although, at the expense of specificity, when low cut‐offs (i.e. ≤10 μg Hb/g faeces, corresponding to 50 ng Hb/mL buffer) are used, predominantly through enhanced detection of adenomas. 18 , 31 , 32 A threshold as low as 2 μg Hb/g faeces resulted in a sensitivity of 78% for high‐risk adenomas and 42% for low‐risk adenomas (specificity 62% and unknown, respectively). 18 Of note, the threshold in national bowel screening programs varies between 15 and 180 μg Hb/g faeces. 33 , 34 , 35 Future studies should assess the performance of FIT in a large Lynch syndrome cohort, including strategies that potentially increase detection of both CRC and adenomas, such as low cut‐offs, repeated FIT sampling 36 , 37 or sampling at shorter intervals (e.g. annual).
Exfoliated tumour DNA is another emerging faecal marker. The yet only FDA‐approved stool DNA test, Cologuard, outperformed FIT (thresholds of 50, 75 and 100 ng Hb/mL) in average‐risk individuals in terms of sensitivity (for both CRC and premalignant lesions). 38 , 39 This might be explained by the fact that DNA, in contrast to blood, is released into the colorectal lumen continuously rather than intermittently. 40 Of note, specificity was roughly equal (FIT: 94.7%, Cologuard: 94.1%) using a low FIT threshold of 50 ng Hb/ml, whereas slightly higher for FIT using a threshold of 100 ng Hb/ml (FIT: 98.0%, Cologuard: 94.1%). 39 For population screening purposes, multipanel faecal DNA assays (such as Cologuard, which tests for KRAS mutations, NDRG4 and BMP3 hypermethylation, and haemoglobin) are more effective than single targets. 41 This could be attributed to the genetically heterogeneous nature of sporadic colorectal neoplasia, with no single mutation found to be expressed across all lesions to date. 42 Nonetheless, DNA shedding in Lynch syndrome‐associated neoplasms might be more homogeneous as it is postulated these lesions develop along particular pathways, related to the underlying gene defect. 43 Based on the molecular pathways of colorectal tumorigenesis that have been identified in individuals with Lynch syndrome, a list of potential DNA markers worth further clinical research was recently created (among which are mutated KRAS, APC, CTNNB1 and TP53, and hypermethylated IGF2, NEUROG1, CDKN2A and CRABP1). 44 To the best of our knowledge, whether hypermethylation of NDRG4 and/or BMP3 (tested for with Cologuard) is present in Lynch‐associated neoplastic lesions is unknown, hence should be examined. Although stool DNA tests may hold promise for non‐invasive colon surveillance of individuals with Lynch syndrome, remaining challenges include the high degree of time and expertise required for analysis, the necessity of whole stool samples (associated with lower patient acceptability) and high costs. Hence, these tests are unsuitable for implementation in clinical practice at this moment.
Other candidates, but yet less studied stool‐based markers involve tumour RNA (especially microRNA), proteins and gut microbiome composition. 13 , 45 , 46 It would be interesting to further study the bacterium Desulfovibrio as a biomarker in Lynch syndrome since it was enriched in stool samples of patients with Lynch syndrome having adenomas (as described in the results of the current review). 24 This phenomenon has been observed in patients with CRC as well and might be explained by the potential DNA‐damaging and pro‐inflammatory role of Desulfovibrio. 24 , 47 , 48 , 49
Next to markers in stool, blood‐based markers may have potential for surveillance of individuals with Lynch syndrome, especially circulating tumour RNA and DNA. Methylated SEPTIN9 (mSEPT9) is such a DNA marker and is incorporated in an FDA‐approved test for sporadic colorectal neoplasms (Epi proColon 2.0 CE). 45 Hypermethylation of the SEPTIN9 gene was shown to be present in 97% and 90% of Lynch‐associated CRCs and advanced adenomas as well, respectively. 19 A recent meta‐analysis demonstrated a pooled sensitivity and specificity of mSEPT9 for CRC of 78% and 84%, respectively, when using the 1/3 PCR algorithm. 50 Likewise, rates were 70% and 100% in the single study assessing mSEPT9 (1/1 algorithm) in individuals with Lynch syndrome, as described in the results of the current review, 19 but validation studies are needed. In addition, mSEPT9 and FIT performance in an average‐risk population were similar in terms of sensitivity, while sensitivity increased substantially to 89% (specificity 79%) when these tests were combined. 51 Nevertheless, the detection rate seems to be stage‐dependent according to the meta‐analysis (ranging from 60% to 93% for stage I and IV cancer respectively) 50 and unsatisfactory for adenomas. 52 , 53 , 54 , 55 , 56 This might be explained by marker release into blood being dependent on the degree of vascular invasion, which in turn is dependent on the phase of tumorigenesis. 52 Nonetheless, recent developments (e.g. analysis of additional parameters as nucleosomes or fragment size) may overcome this problem and could therefore lead to a suitable DNA marker for individuals with Lynch syndrome, 45 hence further research in this field is warranted. Circulating microRNAs may be another solution since they have the potential to detect adenomas according to preliminary studies. 45 , 57 In this context miR‐29a, miR‐21 and the miR‐17‐92 cluster are the most promising microRNAs. The next steps would be to determine the most beneficial microRNA or microRNA‐panel and detection methodology and to perform large, prospective validation studies, also in individuals with Lynch syndrome.
Although more invasive than a stool test, patients preferred a blood test according to a German study. 58 Nevertheless, the technically complex and labour‐intensive analysis of circulating tumour RNA or DNA remains challenging. Also, aberrantly expressed nucleic acids are not necessarily colorectal neoplasia‐specific. However, for individuals predisposed to various malignancies (like in Lynch syndrome) detection of multiple (pre)cancer types at once could be valuable as they could potentially improve prognosis. Of note, a blood‐based DNA test was yet able to detect eight common malignancies (including CRC) in a recent, large trial. 59 On the contrary, overdiagnosis is a potential pitfall of such a multicancer detection test. Diagnostic and therapeutic pathways following a positive test may cause an (economic) burden for patients and healthcare systems, particularly disadvantageous when a test turns out to be false‐positive. Ideally, the test would detect the malignancies most frequently observed in Lynch syndrome (e.g. colorectal, skin, endometrial, genitourinary, breast, prostate and ovarian cancer) and be gender‐ and/or mutation‐specific. 60 Next to discovery and validation studies in a preclinical and thereafter clinical setting, the long‐term effect as well as cost‐effectiveness have to be addressed in order to establish the clinical utility of a multicancer detection test in this population.
A novel approach within the field of biomarker exploration is the analysis of volatile organic compounds (VOCs). 61 These gaseous metabolites reflect biochemical processes (like cancer growth) and emanate from a wide range of excreted materials. 62 Most studies focussed on VOCs originating from exhaled breath or faeces in the context of colorectal neoplasia, with promising outcomes regarding detection of sporadic CRC, advanced adenomas and non‐advanced polyps. 63 , 64 , 65 , 66 Combining FIT with VOCs resulted in further detection of CRC. 67 Additionally, faecal VOC profiles showed potential as a follow‐up test after polypectomy. 64 This would make faecal VOC profiles, which are rapid, easy and low‐cost, attractive for surveillance of individuals with Lynch syndrome, hence further validation studies are needed. Moreover, the optimal source of sample (e.g. faeces, exhaled breath, urine) and performance of FIT‐VOC combination remain to be elucidated in this population.
Biomarkers could complement personalised timing of colonoscopy based on mismatch repair gene variants. 68 The ideal biomarker for individuals with Lynch syndrome has to compromise between high sensitivity and high specificity for both CRC and adenomas, high patient acceptability, simple sample and data analysis, high throughput capacities and cost‐effectiveness. Both discovery and validation studies should be conducted in this population. These studies could be guided by the potential biomarkers and associated literature gaps outlined in this review and by general recommendations for how to evaluate and report new diagnostic tests for colorectal neoplasia. 69 , 70 , 71 , 72 The aberrant biology of Lynch‐associated colorectal neoplasia should be taken in mind when designing and performing such studies. Perhaps a combination of markers will be inevitable if biomarkers are going to play a role in the surveillance of this population.
In conclusion, in this systematic review we show that non‐invasive biomarkers for CRC and adenomas in individuals with Lynch syndrome, which could guide the timing of colonoscopy, are yet poorly studied. Nevertheless, plasma tumour mSEPT9, FIT, faecal tumour BAT‐26 and faecal Desulfovibrio may hold potential, along with other tumour DNA/RNA markers and gaseous metabolites. Biomarkers could be of important value for this population since colonoscopy is burdensome and interval CRCs still occur, hence, (pre‐)clinical studies in this field should be prioritised.
AUTHORSHIP
Guarantor of the article: Dewkoemar Ramsoekh.
Author contributions: DR, NdB and EvL conceived the study design and collected data. EvL drafted the manuscript. All authors critically revised the manuscript for important intellectual content. All authors approved the final version of the manuscript, including the authorship list.
Declaration of funding interests: This study was not funded.
Supporting information
Table S1
ACKNOWLEDGEMENT
We would like to thank Marijke A.E. Mol, an information specialist at the Vrije Universiteit Amsterdam, for her aid in developing and performing the systematic literature search in MEDLINE and Embase.
Declaration of personal interests: EvL and MvL have nothing to declare. NdB has served as a speaker for AbbVie and MSD and has served as a consultant and principal investigator for TEVA Pharma BV and Takeda. He has received a research grant (unrestricted) from Dr. Falk, TEVA Pharma BV, Dutch Digestive Foundation (MLDS) and Takeda. ED has endoscopic equipment on a loan from FujiFilm and Olympus and has received a research grant from FujiFilm. She has received an honourarium for a consultancy from FujiFilm, Olympus, GI Supply, CPP‐FAP, PAION and Ambu and speakers’ fees from Olympus, Roche, GI Supply, Norgine, IPSEN, PAION and FujiFilm. TdM has served as a speaker for Nutricia, Mead Johnson and Winclove. He has served as an advisory board member for Nutricia. DR has received a research grant (unrestricted) from AbbVie and is a member of the Data Safety Monitoring Board of Vivoryon Therapeutics.
van Liere ELSA, de Boer NKH, Dekker E, van Leerdam ME, de Meij TGJ & Ramsoekh D. Systematic review: non‐endoscopic surveillance for colorectal neoplasia in individuals with Lynch syndrome. Aliment Pharmacol Ther. 2022;55:778–788. doi: 10.1111/apt.16824
The Handling Editor for this article was Professor Rohit Loomba, and this uncommissioned review was accepted for publication after full peer‐review.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783‐5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edelstein DL, Axilbund J, Baxter M, et al. Rapid development of colorectal neoplasia in patients with Lynch syndrome. Clin Gastroenterol Hepatol. 2011;9:340‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044‐2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Leerdam ME, Roos VH, van Hooft JE, et al. Endoscopic management of Lynch syndrome and of familial risk of colorectal cancer: European Society of Gastrointestinal Endoscopy (ESGE) guideline. Endoscopy. 2019;51:1082‐1093. [DOI] [PubMed] [Google Scholar]
- 5. Parry S, Win AK, Parry B, et al. Metachronous colorectal cancer risk for mismatch repair gene mutation carriers: the advantage of more extensive colon surgery. Gut. 2011;60:950‐957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engel C, Vasen HF, Seppala T, et al. No difference in colorectal cancer incidence or stage at detection by colonoscopy among 3 countries with different Lynch syndrome surveillance policies. Gastroenterology. 2018;155:1400‐9 e2. [DOI] [PubMed] [Google Scholar]
- 7. Jarvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15‐year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829‐834. [DOI] [PubMed] [Google Scholar]
- 8. Denters MJ, Schreuder M, Depla AC, et al. Patients' perception of colonoscopy: patients with inflammatory bowel disease and irritable bowel syndrome experience the largest burden. Eur J Gastroenterol Hepatol. 2013;25:964‐972. [DOI] [PubMed] [Google Scholar]
- 9. Bleiker EM, Menko FH, Taal BG, et al. Screening behavior of individuals at high risk for colorectal cancer. Gastroenterology. 2005;128:280‐287. [DOI] [PubMed] [Google Scholar]
- 10. Newton K, Green K, Lalloo F, Evans DG, Hill J. Colonoscopy screening compliance and outcomes in patients with Lynch syndrome. Color Dis. 2015;17:38‐46. [DOI] [PubMed] [Google Scholar]
- 11. Peterse EFP, Naber SK, Daly C, et al. Cost‐effectiveness of active identification and subsequent colonoscopy surveillance of Lynch syndrome cases. Clin Gastroenterol Hepatol. 2020;18:2760‐7 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahadova A, Seppala TT, Engel C, et al. The "unnatural" history of colorectal cancer in Lynch syndrome: lessons from colonoscopy surveillance. Int J Cancer. 2021;148:800‐811. [DOI] [PubMed] [Google Scholar]
- 13. Loktionov A. Biomarkers for detecting colorectal cancer non‐invasively: DNA, RNA or proteins? World J Gastrointest Oncol. 2020;12:124‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Expert Panel on Gastrointestinal I , Moreno C, Kim DH, et al. ACR appropriateness criteria([R]) colorectal cancer screening. J Am Coll Radiol. 2018;15:S56‐S68. [DOI] [PubMed] [Google Scholar]
- 15. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P . Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. BMJ. 2009;339:b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan‐a web and mobile app for systematic reviews. Syst Rev. 2016;5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529‐536. [DOI] [PubMed] [Google Scholar]
- 18. Digby J, Cleary S, Gray L, et al. Faecal haemoglobin can define risk of colorectal neoplasia at surveillance colonoscopy in patients at increased risk of colorectal cancer. United European Gastroenterol J. 2020;8:559‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hitchins MP, Vogelaar IP, Brennan K, et al. Methylated SEPTIN9 plasma test for colorectal cancer detection may be applicable to Lynch syndrome. BMJ Open Gastroenterol. 2019;6:e000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koshiji M, Yonekura Y, Saito T, Yoshioka K. Microsatellite analysis of fecal DNA for colorectal cancer detection. J Surg Oncol. 2002;80:34‐40. [DOI] [PubMed] [Google Scholar]
- 21. Laken S, Lynch H, Urbanowski J, Deters C, Shuber A, Watson P. Results of a stool BAT‐26 assay in people with HNPCC; (Abstract). 63rd Annual Meeting of The American Society of Human Genetics; San Diego; 2001.
- 22. Lim EJ, Leung C, Pitman A, et al. Magnetic resonance colonography for colorectal cancer screening in patients with Lynch syndrome gene mutation. Familial Cancer. 2010;9:555‐561. [DOI] [PubMed] [Google Scholar]
- 23. Renkonen‐Sinisalo L, Kivisaari A, Kivisaari L, Sarna S, Jarvinen HJ. Utility of computed tomographic colonography in surveillance for hereditary nonpolyposis colorectal cancer syndrome. Familial Cancer. 2007;6:135‐140. [DOI] [PubMed] [Google Scholar]
- 24. Yan Y, Drew DA, Markowitz A, et al. Structure of the mucosal and stool microbiome in Lynch syndrome. Cell Host Microbe. 2020;27:585‐600 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ahlquist DA, Skoletsky JE, Boynton KA, et al. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology. 2000;119:1219‐1227. [DOI] [PubMed] [Google Scholar]
- 26. Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Colorectal cancer study G. fecal DNA versus fecal occult blood for colorectal‐cancer screening in an average‐risk population. N Engl J Med. 2004;351:2704‐2714. [DOI] [PubMed] [Google Scholar]
- 27. Graser A, Melzer A, Lindner E, et al. Magnetic resonance colonography for the detection of colorectal neoplasia in asymptomatic adults. Gastroenterology. 2013;144:743‐50 e2. [DOI] [PubMed] [Google Scholar]
- 28. Mulhall BP, Veerappan GR, Jackson JL. Meta‐analysis: computed tomographic colonography. Ann Intern Med. 2005;142:635‐650. [DOI] [PubMed] [Google Scholar]
- 29. Lecky DM, Hawking MK, McNulty CA, group Es . Patients' perspectives on providing a stool sample to their GP: a qualitative study. Br J Gen Pract. 2014;64:e684‐e693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolte LA, Klaassen MAY, Collij V, et al. Patient attitudes towards faecal sampling for gut microbiome studies and clinical care reveal positive engagement and room for improvement. PLoS ONE. 2021;16:e0249405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Castro I, Cubiella J, Rivera C, et al. Fecal immunochemical test accuracy in familial risk colorectal cancer screening. Int J Cancer. 2014;134:367‐375. [DOI] [PubMed] [Google Scholar]
- 32. Cross AJ, Wooldrage K, Robbins EC, et al. Faecal immunochemical tests (FIT) versus colonoscopy for surveillance after screening and polypectomy: a diagnostic accuracy and cost‐effectiveness study. Gut. 2019;68:1642‐1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Jonge L, Worthington J, van Wifferen F, et al. Impact of the COVID‐19 pandemic on faecal immunochemical test‐based colorectal cancer screening programmes in Australia, Canada, and The Netherlands: a comparative modelling study. Lancet Gastroenterol Hepatol. 2021;6:304‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cancer Research United Kingdom . Bowel screening test. 2021, January 27. https://www.cancerresearchuk.org/health‐professional/screening/bowel‐screening‐evidence‐and‐resources/bowel‐screening‐test#FIT2 (accessed 2021, April 13).
- 35. Senore C, Basu P, Anttila A, et al. Performance of colorectal cancer screening in the European Union member states: data from the second European screening report. Gut. 2019;68:1232‐1244. [DOI] [PubMed] [Google Scholar]
- 36. Schreuders EH, Grobbee EJ, Nieuwenburg SAV, et al. Multiple rounds of one sample versus two sample faecal immunochemical test‐based colorectal cancer screening: a population‐based study. Lancet Gastroenterol Hepatol. 2019;4:622‐631. [DOI] [PubMed] [Google Scholar]
- 37. Wieten E, de Klerk CM, Lansdorp‐Vogelaar I, Bossuyt PM, Dekker E, Spaander MCW. A quarter of participants with advanced neoplasia have discordant results from 2‐sample fecal immunochemical tests for colorectal cancer screening. Clin Gastroenterol Hepatol. 2020;18:1805‐11 e1. [DOI] [PubMed] [Google Scholar]
- 38. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal‐cancer screening. N Engl J Med. 2014;370:1287‐1297. [DOI] [PubMed] [Google Scholar]
- 39. Bosch LJW, Melotte V, Mongera S, et al. Multitarget stool DNA test performance in an average‐risk colorectal cancer screening population. Am J Gastroenterol. 2019;114:1909‐1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahlquist DA, Gilbert JA. Stool markers for colorectal cancer screening: future considerations. Dig Dis. 1996;14:132‐144. [DOI] [PubMed] [Google Scholar]
- 41. Bosch LJ, Carvalho B, Fijneman RJ, et al. Molecular tests for colorectal cancer screening. Clin Colorectal Cancer. 2011;10:8‐23. [DOI] [PubMed] [Google Scholar]
- 42. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759‐767. [DOI] [PubMed] [Google Scholar]
- 43. Cerretelli G, Ager A, Arends MJ, Frayling IM. Molecular pathology of Lynch syndrome. J Pathol. 2020;250:518‐531. [DOI] [PubMed] [Google Scholar]
- 44. Lepore Signorile M, Disciglio V, Di Carlo G, Pisani A, Simone C, Ingravallo G. From genetics to Histomolecular characterization: an insight into colorectal carcinogenesis in Lynch syndrome. Int J Mol Sci. 2021;22:6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marcuello M, Vymetalkova V, Neves RPL, et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol Asp Med. 2019;69:107‐122. [DOI] [PubMed] [Google Scholar]
- 46. de Klaver W, Wisse PHA, van Wifferen F, et al. Clinical validation of a multitarget fecal immunochemical test for colorectal cancer screening: a diagnostic test accuracy study. Ann Intern Med. 2021;174:1224‐1231. [DOI] [PubMed] [Google Scholar]
- 47. Hiippala K, Jouhten H, Ronkainen A, et al. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients. 2018;10:988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early‐stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hale VL, Chen J, Johnson S, et al. Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomark Prev. 2017;26:85‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta‐analysis. Sci Rep. 2017;7:3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson DA, Barclay RL, Mergener K, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS ONE. 2014;9:e98238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ahlquist DA, Taylor WR, Mahoney DW, et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10:272‐277.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Church TR, Wandell M, Lofton‐Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grutzmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS ONE. 2008;3:e3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Potter NT, Hurban P, White MN, et al. Validation of a real‐time PCR–based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183‐1191. [DOI] [PubMed] [Google Scholar]
- 56. Tanzer M, Balluff B, Distler J, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS ONE. 2010;5:e9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roberts BS, Hardigan AA, Moore DE, et al. Discovery and validation of circulating biomarkers of colorectal adenoma by high‐depth small RNA sequencing. Clin Cancer Res. 2018;24:2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adler A, Geiger S, Keil A, et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014;14:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science. 2018;359:926‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dominguez‐Valentin M, Sampson JR, Seppala TT, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the prospective Lynch syndrome database. Genet Med. 2020;22:15‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Di Lena M, Porcelli F, Altomare DF. Volatile organic compounds as new biomarkers for colorectal cancer: a review. Color Dis. 2016;18:654‐663. [DOI] [PubMed] [Google Scholar]
- 62. Shirasu M, Touhara K. The scent of disease: volatile organic compounds of the human body related to disease and disorder. J Biochem. 2011;150:257‐266. [DOI] [PubMed] [Google Scholar]
- 63. Bosch S, Berkhout DJ, Ben Larbi I, de Meij TG, de Boer NK. Fecal volatile organic compounds for early detection of colorectal cancer: where are we now? J Cancer Res Clin Oncol. 2019;145:223‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bosch S, Bot R, Wicaksono A, et al. Early detection and follow‐up of colorectal neoplasia based on faecal volatile organic compounds. Color Dis. 2020;22:1119‐1129. [DOI] [PubMed] [Google Scholar]
- 65. Hintzen KFH, Grote J, Wintjens A, et al. Breath analysis for the detection of digestive tract malignancies: systematic review. BJS Open. 2021;5:zrab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Keulen KE, Jansen ME, Schrauwen RWM, Kolkman JJ, Siersema PD. Volatile organic compounds in breath can serve as a non‐invasive diagnostic biomarker for the detection of advanced adenomas and colorectal cancer. Aliment Pharmacol Ther. 2020;51:334‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chandrapalan S, Bosch S, Cubiella J, et al. Systematic review with meta‐analysis: volatile organic compound analysis to improve faecal immunochemical testing in the detection of colorectal cancer. Aliment Pharmacol Ther. 2021;54:14‐23. [DOI] [PubMed] [Google Scholar]
- 68. Kastrinos F, Ingram MA, Silver ER, et al. Gene‐specific variation in colorectal cancer surveillance strategies for Lynch syndrome. Gastroenterology. 2021;161:453‐462.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bossuyt PM, Irwig L, Craig J, Glasziou P. Comparative accuracy: assessing new tests against existing diagnostic pathways. BMJ. 2006;332:1089‐1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem. 2015;61:1446‐1452. [DOI] [PubMed] [Google Scholar]
- 71. Srivastava S, Wagner PD. The early detection research network: a National Infrastructure to support the discovery, development, and validation of cancer biomarkers. Cancer Epidemiol Biomark Prev. 2020;29:2401‐2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Young GP, Senore C, Mandel JS, et al. Recommendations for a step‐wise comparative approach to the evaluation of new screening tests for colorectal cancer. Cancer. 2016;122:826‐839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


