Summary
Grazing controls bacterial abundances and composition in many ecosystems. In marine systems, heterotrophic flagellates (HFs) are important predators. Assemblages of HFs are primarily formed by species still uncultured; therefore, many aspects of their trophic behaviour are poorly known. Here, we assessed the functional response of the whole assemblage and of four taxa grown in an unamended seawater incubation. We used fluorescently labelled bacteria to create a prey gradient of two orders of magnitude in abundance and estimated ingestion rates. Natural HFs had a half‐saturation constant of 6.7 × 105 prey ml−1, a value lower than that of cultured flagellates and within the range of marine planktonic bacterial abundances. Minorisa minuta was well adapted to low prey abundances and very efficient in ingesting bacteria. MAST‐4 and MAST‐7 were also well adapted to the typical marine abundances but less voracious. In contrast, Paraphysomonas imperforata, a typical cultured species, did not achieve ingestion rate saturation even at the highest prey concentration assayed. Our study, beside to set the basis for the fundamental differences between cultured and uncultured bacterial grazers, indicate that the examined predator taxa have different functional responses, suggesting that they occupy distinct ecological niches according to their grazing strategies and prey preferences.
Introduction
Heterotrophic flagellates (HFs) are colourless small protists that are active predators of aquatic bacteria (Sherr and Sherr, 2002). Grazing by HFs controls bacterial abundance and diversity in a wide range of ecosystem conditions, channels organic carbon to higher trophic levels, and releases inorganic nutrients that controls regenerated primary production (Fuhrman and Noble, 1995; Šimek et al., 2001; Pernthaler, 2005). In fact, HFs are central in the microbial loop concept (Azam et al., 1983). Grazing rates of natural HF assemblages are estimated using tracer techniques that follow the fate of an added bacterial surrogate or by manipulation techniques that uncouple predators and preys (Vaqué et al., 1994; Strom, 2000; Jürgens and Massana, 2008). Community grazing rates may be then used to calculate growth rates of HFs (Fenchel, 1987) and to evaluate their contribution to bacterial mortality. However, these rates average the activities of the different taxa in the community, each one perhaps having different grazing rates and prey preferences. Indeed, molecular surveys have unveiled a large diversity of marine protists assemblages (de Vargas et al., 2015; Massana et al., 2015), including HFs (Jürgens and Massana, 2008; Logares et al., 2012). Recent studies conducted mostly in freshwater systems have been dealing with characterizing the growth rates of major bacterivorous taxa (Grujcic et al., 2018; Šimek et al., 2018, 2020). So, for a better understanding of the bacterial grazing and its impact on microbial food web structure, it is still necessary to investigate the physiological parameters of the dominant marine HF taxa (Piwosz et al., 2021).
Grazing rates directly depend on prey abundance. This dependence, named functional response, has been determined in a variety of marine and freshwater predators (Weisse et al., 2016), including copepods (Henriksen et al., 2007; Isari and Saiz, 2011), dinoflagellates (Kim and Jeong, 2004; Jeong et al., 2005; Roberts et al., 2011), ciliates (Jonsson, 1986; Jürgens and Simek, 2000; Gismervik, 2005; Lu et al., 2021) and heterotrophic flagellates (Jeong et al., 2008). Functional responses can be fitted to different mathematical models (Holling, 1959), being the most popular among ecologists the equivalent to the enzyme kinetic model developed in 1913 by Leonor Michaelis and Maude Menten. This model includes two parameters, the maximum ingestion rate (IRmax), determined by the capacity of the predator to capture, handle and digest preys, and the half‐saturation constant (Ks: prey concentration that allows half IRmax), which is a proxy of the prey abundance at which the predator is adapted to live (Fenchel, 1980). Only few studies have measured the functional response of small flagellates, due to the difficulty in obtaining grazing rates, while it is more common to measure the numerical response, the relationship of growth rates and prey abundance (Jürgens and Matz, 2002). As the growth efficiency in these predators is considered constant regardless prey abundance, growth rates are proportional to ingestion rates, and the numerical and functional responses have the same form (Fenchel, 1987). Numerical response reports exist for cultured heterotrophic flagellates (Fenchel, 1982a; Eccleston‐Parry and Leadbeater, 1994; Mohapatra and Fukami, 2004; Anderson et al., 2011), but it has been suggested that these cultured species may not represent the dominant grazers in the sea, many of which are still uncultured (Massana et al., 2014). A few trophic experiments (grazing rates and prey preferences) have been done with uncultured species (Massana et al., 2009; Piwosz and Pernthaler, 2010; Meira et al., 2018), but it is still unknown if they have a fundamentally different functional response than cultured ones. There is an overall need to increase the effort on functional studies on ecologically relevant organisms, including HFs, placed in natural multispecies assemblages under in situ conditions (Caron et al., 2012; Worden et al., 2015; Weisse et al., 2016).
The aim of this study is to determine the functional responses of uncultured flagellates living in natural assemblages. We combined short‐term ingestion experiments based on counting fluorescently labelled bacteria (FLB) inside protist food vacuoles (Sherr et al., 1987), with specific FISH counts of HF taxa within mixed assemblages. This time‐consuming approach is so far the only way to provide specific ingestion rates for individual flagellate taxa. To overcome the typically low in situ abundances of HFs, we carried out this experiment in an unamended seawater incubation known to promote the growth of uncultured HFs (Massana et al., 2006a). By preparing a gradient of FLB abundance, we obtained the functional responses of the whole HF assemblage and of four distinct taxa (MAST‐4, MAST‐7, Minorisa minuta and Paraphysomonas imperforata). This is the first report of the functional response of uncultured HF taxa and highlights intrinsic features that might explain why they have not been cultured by classical approaches.
Experimental procedures
Enrichment of heterotrophic flagellates by an unamended incubation
Surface seawater from the Blanes Bay Microbial Observatory (BBMO) was taken on 16 October 2007 and transported to the laboratory in less than 2 h. Six litres of seawater were filtered by gravity through a 200‐μm nylon mesh and then through 3‐μm pore size polycarbonate filters. The resulting community (bacteria plus eukaryotes ≤ 3 μm) was incubated into a Nalgene polycarbonate bottle at in situ temperature (19°C) in the dark, to prevent the growth of phototrophic cells (Massana et al., 2006a), and sampled daily during 4 days (Fig. 1). Glutaraldehyde fixed aliquots (1% final concentration) were stained with 4,6‐diamidino‐2‐phenylindole (DAPI; 5 μg ml−1 according to Sieracki et al., 1985) and filtered on 0.2 μm (for bacteria) or 0.6 μm (for flagellates) pore size black polycarbonate filters (DHI Lab Products). Counts of heterotrophic bacteria (including archaea), Synechococcus, and phototrophic (PF) and heterotrophic flagellates (HF) were carried out in an Olympus BX61 microscope at 1000× magnification using UV irradiance (DAPI‐stained DNA signal) and blue light (chlorophyll signal) (Porter and Feig, 1980). Samples for FISH were taken daily by filtering formaldehyde fixed aliquots (3.7% final concentration) on 0.8 or 1‐μm pore size polycarbonate filters, which were then kept at −80°C. A sample for DNA extraction was taken at day 3 of the incubation by filtering 100 ml onto a 0.2‐μm pore size Durapore.
Fig. 1.
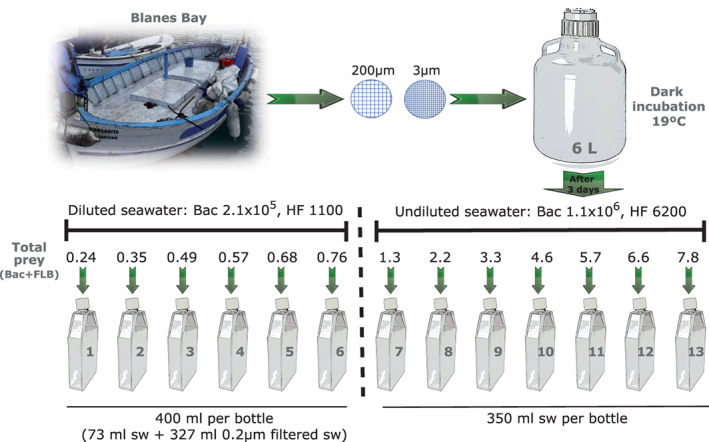
Scheme of the experimental design for assessing the functional response of natural heterotrophic flagellates. Several bottles with increasing amounts of prey (native bacteria plus added FLBs, 106 total prey) were prepared. Bacterial and HF concentration (cells ml−1) are indicated for the diluted (left) and undiluted (right) sets. A short‐term ingestion experiment was performed in each experimental bottle.
Detection of possible predators by clone library and FISH
DNA extraction was done by using lysozyme, proteinase K and SDS for cell lysis, phenol:chloroform:isoamyl alcohol for DNA extraction, and a Centricon‐100 (Millipore) for DNA purification (Massana et al., 2000). Two nanogram of the DNA extract was added to a PCR mixture (50 μl) containing 0.5 μM of each primer, 200 μM of dNTP, 1.5 mM MgCl2, and 1.25 units of a Taq DNA polymerase (ProOmega). We used eukaryotic 18S rDNA primers 528F (Elwood et al., 1985) and EUKR (Medlin et al., 1988) and the following PCR cycle: initial denaturation at 94°C for 3 min; 30 cycles with denaturation at 94°C for 45 s, annealing at 55°C for 1 min and extension at 72°C for 3 min; and a final extension at 72°C for 10 min. PCR products were purified with the QIAquick PCR Purification kit (QIAGEN) and cloned using the TOPO‐TA cloning kit (Invitrogen). The presence of correct insert in the bacterial clones was checked by PCR reamplification with the same primers and PCR amplicons were sequenced at the Macrogen sequencing service (Korea). Chimera detection and phylogenetic affiliation of sequences were obtained by a basic local alignment search tool (BLAST).
Oligonucleotide probes for FISH or CARD‐FISH (Table 1) were labelled at the 5′ end with the fluorescent dye CY3 or the enzyme HRP, respectively, and supplied by Thermo Electron Corporation (Waltham, MA, USA). We generally used CARD‐FISH except for three probes (NS4, CRN 02 and CET1). For FISH, filter portions were hybridized for 3 h at 46°C in the appropriate buffer (with 30% formamide) and washed at 48°C in a second buffer, following the protocol and conditions detailed elsewhere (Pernthaler et al., 2001; Massana et al., 2006b). For CARD‐FISH, we followed the protocol and conditions detailed in the study by Pernice et al. (2015). Briefly, filters with protist cells were first embedded in 1% (w/v) low‐gelling‐point agarose to minimize cell loss. Then, filter portions were hybridized overnight at 35°C, washed at 37°C, and tyramide signal amplification was done for 60 min at 46°C using Alexa 594‐labelled tyramide. A final washing step of 1 h with 1:1 ethanol : PBS at room temperature was done to remove background fluorescence. After hybridization, filters were counter‐stained with DAPI, mounted in a slide, and cells were observed by epifluorescence microscopy at 1000× under green light excitation. Cell biovolumes were calculated by measuring two dimensions (length and width) in about 100 cells of the target group stained by FISH or CARD‐FISH, and applying the prolate spheroid formula (Hillebrand et al. 1999) considering the third dimension (height) as two‐thirds of the width.
Table 1.
Probes used to visualize specific taxa within the unamended seawater incubation by FISH or CARD‐FISH.
| Probe | Sequence (5′–3′) | References | Group | Species | Cells ml−1 | Clones | Mismatches |
|---|---|---|---|---|---|---|---|
| NS1A a | ATTACCTCGATCCGCAAA | Massana et al. (2006b) | MAST‐1A | nd | 0 | ‐ | |
| NS1B a | AAC GCA AGT CTC CCC GCG | Massana et al. (2006b) | MAST‐1B | 3.5 | 0 | ‐ | |
| NS1C a | GTGTTCCCTAACCCCGAC | Massana et al. (2006b) | MAST‐1C | 3.9 | 0 | ‐ | |
| NS2 a | ATGGGCCGACCGGTCGCT | Massana et al. (2006b) | MAST‐2 | 22.5 | 10 | 0 | |
| NS4 | TACTTCGG TCTGCAAACC | Massana et al. (2002) | MAST‐4 | 372.4 | 1 | 0 | |
| NS7 a | TCATTACCATAGTACGCA | Giner et al. (2016) | MAST‐7 | 362.1 | 4 | 0–1 | |
| CRN 02 | TACTTAGCTCTCAGAACC | del Campo et al. (2013) | Chlorarachniophyta | Minorisa minuta | 782 | 6 | 0 |
| PIMP 663 a | GGACGCAGAGACCAGGTGCACA | Lim et al. (1999) | Chrysophyte | Paraphysomonas imperforata | 165.4 | 2 | 1 |
| CET1 | CAGCTCAATACGGACACC | Massana et al. (2007) | Bicosoecida | Caecitellus parvulus, C. paraparvulus | nd | 1 | 0 |
| Cafeteria a | ACAGTGCTGACACCCTGT | Massana et al. (2007) | Bicosoecida | Cafeteria burkhardae | nd | 0 | ‐ |
The abundance of targeted cells at day 3 of the incubation is shown, together with the number of clones (and mismatches) detected having the probe sequence. nd: Not determined: no positive cells were observed in the hybridization.
CARD‐FISH.
Fluorescently labelled bacteria used as prey
Brevundimonas diminuta (syn. Pseudomonas diminuta; Caulobacteraceae, α‐Proteobacteria) was obtained from the Colección Española de Cultivos Tipo (Valencia, Spain), grown in LB agar plates and used to prepare FLBs (Sherr et al., 1987). B. diminuta has already been used to prepare FLB (Vazquez‐Dominguez et al., 1999) because of their small size close to that of natural marine bacteria. Two‐week‐old colonies were scraped, diluted in carbonate–bicarbonate buffer (pH 9.5), and stained with 5‐(4,6‐dichlorotriazinyl)‐aminofluorescein (DTAF; 100 pg ml−1) for 2 h in a water bath at 60°C. Stained cells were centrifuged five times (10 min, 10 000 rpm) and resuspended in 0.2 μm‐filtered carbonate–bicarbonate buffer to prevent the transfer of leftover dye to experimental samples. Cell suspensions (average cell biovolume 0.099 μm3) were kept frozen at −20°C. Before used in the grazing experiments, the FLB solution was thawed and gently sonicated for three 10 s rounds with the microtip at 35% of power output (Dynatech sonic dismembrator, Model 300) to minimize cell clustering (Unrein et al., 2007).
Grazing experiments
Seawater in the unamended incubation at day 3 was divided in two sets (Fig. 1). One set was diluted (1 to 5.5) with filtered seawater in order to decrease the initial bacterial concentration and used to fill bottles 1 to 6 (400 ml each). The second set remained undiluted and was used to fill bottles 7 to 13 (350 ml each). Bottles were acclimated in a large container for 2–4 h at in situ temperature (19°C), and then increasing amounts of FLB were added to the bottles, at tracer concentrations in the first bottle (~15% of total bacteria) and becoming the main bacterial prey in the other bottles (~600% in the last bottle). Instead replicating the same prey concentration, we decided to obtain more points along the prey gradient, a recommended strategy in a regression analysis (Montagnes and Berges, 2004). Aliquots for DAPI‐stained microbial counts (bacteria and small protists) and for FISH analyses (only small protists) were taken immediately after the addition of FLB and after 40 min of incubation. Fixation was done with an equal volume of diluted fixative to reduce cell egestion (Sieracki et al., 1987), reaching the same final concentration as detailed before. The incubation time (40 min) was chosen based on a previous time series that showed a plateau in the number of ingested FLB at 45 min (Unrein et al., 2007), and it is much shorter than the half‐life of bacteria in food vacuoles reported in several heterotrophic nanoflagellates (Shannon et al., 2007). Grazing of the HF assemblage was estimated by counting FLB inside colourless flagellates in the DAPI‐stained samples. Grazing of specific predators was assessed counting FLB inside FISH or CARD‐FISH positive cells, by combining green light excitation (FISH or CARD‐FISH signal of the predator) and blue light excitation (FLB detection). The mean number of cells counted per sample was 325 in HFs, 299 in M. minuta, 109 in MAST‐4, 279 in MAST‐7 and 84 in P. imperforata. The number of FLB per cell was multiplied by the ratio of total prey (native bacteria plus FLB, obtained by separate DAPI counts) to FLB, to obtain the preys ingested at time 0 (I0) and at 40 min (I40). Ingestion rates (IRs: preys predator−1 h−1) were calculated according to:
Data for IR and prey abundance were fitted by iteration to the hyperbolic Michaelis–Menten equation:
where IRmax is the maximum ingestion rate, N is the prey concentration (prey ml−1) and Ks is the half‐saturation constant (prey ml−1). To fit this model, we used the R package ‘drc’ (analysis of dose–response curves) (Ritz et al., 2015) with functions ‘drm’ and ‘nls’. Results were visualized using the ggplot2 package for R (Gómez‐Rubio, 2017). Growth efficiency was calculated as the percentage of protist biovolume produced (μ × predator biovolume) to the bacteria biovolume ingested (IRmax × prey biovolume).
Results
The grazing experiment was done with a natural community incubated for 3 days in the dark, in which predators from higher trophic levels had been filtered out and the growth of bacterivorous HFs was promoted. During this unamended incubation in situ bacterial abundance (5.9 × 105 cells ml−1) increased to 1.1 × 106 cells ml−1 and in situ HF abundance (804 cells ml−1) increased to 6200 cells ml−1, being in exponential growth at the moment of the experiment (Fig. 2A). The cell abundance of photosynthetic flagellates and Synechococcus decreased continuously during the incubation (Fig. 2A), as a result of the inhibition of the photosynthesis during the dark incubation and perhaps of grazing mortality. Short‐term ingestion experiments with the HF‐enriched assemblage were prepared along a gradient of prey abundance (native bacteria plus FLB) covering almost two orders of magnitude (105–107 preys ml−1) in 13 bottles (Fig. 1). To cover properly this gradient, some of the conditions were prepared with diluted samples. The gradient covers the natural marine bacterial abundance, typically around 106 cells ml−1. This experimental set‐up allowed to measure grazing rates at different prey abundances and therefore estimate the functional responses of the whole natural assemblage and of specific heterotrophic flagellate taxa.
Fig. 2.
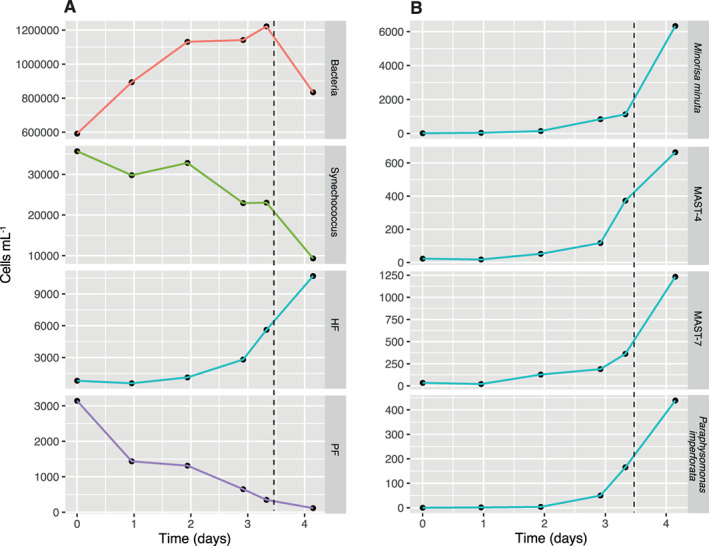
Temporal dynamics of microbial components in the unamended incubation.
A. Changes in cell abundance of bacteria, Synechococcus, heterotrophic flagellates (HF) and phototrophic flagellates (PF) obtained by epifluorescence microscopy after DAPI staining.
B. Cell abundances estimated by FISH for Minorisa minuta, MAST‐7, MAST‐4 and Paraphysomonas imperforata. Dashed lines represent the moment of the grazing experiment.
In order to characterize the protist species composition of the assemblage used for the grazing experiment (mostly formed by HFs cells, as shown by microscopic inspections), we did first a simple clone library with a sample taken at the same time of the grazing experiments (day 3). This clone library yielded 44 sequences distributed in 15 phylogenetic groups, being most of them highly similar to environmental sequences from previous marine surveys (Massana et al., 2004). More than half of the clones affiliated to uncultured MAST (Marine Stramenopiles) with similarities with the closest match in GenBank generally at the range 99%–100%: 10 clones to MAST‐2, 5 to MAST‐8, 4 to MAST‐7 and MAST‐12, and a single one to MAST‐4. Another three clones affiliated to uncultured MALV (Marine Alveolates) I and II (similarities above 99%). Six clones were almost identical to Minorisa minuta, an heterotrophic flagellate basal to Chlorarachniophyceae and cultured in oligotrophic conditions (del Campo et al., 2013). Five clones affiliated to chrysophytes, two highly similar to Paraphysomonas imperforata and P. foraminifera, one belonging to the cultured clade C and two other to the uncultured clades H and I. The single bicosoecida clone was moderately related to Caecitellus sp. (96.5%). The remaining groups (Haptophyte, Fungi, Cercozoa, and Bolidomonas) presented only one clone.
We then applied a battery of FISH and CARD‐FISH probes to directly quantify the presence of these taxa (and others) in our experimental sample. We used 10 FISH probes from uncultured and cultured heterotrophic flagellates available at the moment (Table 1). Four groups appeared at a reasonable abundance at the day 3 of the experiment, which made them suitable for grazing inspections. M. minuta was the most abundant flagellate of the four (782 cells ml−1), followed by MAST‐4 (372 cells ml−1), MAST‐7 (362 cells ml−1), and P. imperforata (165 cells ml−1). Probe sequences matched perfectly the sequences retrieved in the clone libraries (probes NS4, CRN 02) or with a single mismatch (NS7 and PIMP 663), indicating that these probes targeted properly these populations. Some probes gave none or very little signal, consistent with the absence of the corresponding taxa in clone libraries: MAST‐1A, ‐1B and ‐1C and Cafeteria burkhardae. Finally, two groups gave inconsistent results, despite the probes matched perfectly the sequences from the sample: MAST‐2, the group dominant at the clone library was detected in low cell abundance (22.5 cells ml−1), whereas Caecitellus spp. was not detected at all. After the selection of the four candidates for grazing analysis, they were counted during the unamended incubation to estimate their growth dynamics. The four taxa were in exponential growth at the moment of the experiment (Fig. 2B). The derived growth rates for these taxa ranged from 0.7 to 1.9 day−1 (Table 2).
Table 2.
A summary of specific functional parameters for each HF taxa: growth rates (μ, d−1), biovolume (Size, μm3, mean values), maximum ingestion rates (IRmax, prey cell−1 h−1), and half‐saturation constant (Ks, prey ml−1, 106).
| Organisms | μ | Size (μm3) | IRmax | Ks | IRexp | IRGE40 |
|---|---|---|---|---|---|---|
| HF | 0.71 | 8.6 | 2.3 | 0.67 | 1.5 | 6.4 |
| Minorisa minuta | 1.54 | 6.3 | 5.3 | 0.62 | 3.4 | 10.2 |
| MAST‐4 | 0.90 | 3.3 | 1.0 | 0.87 | 0.6 | 3.1 |
| MAST‐7 | 0.86 | 9.0 | 2.0 | 0.97 | 1.1 | 8.14 |
| P. imperforata | 1.95 | 21.2 | 2.1 | 6.10 | 0.3 | 43.5 |
From the functional response, the ingestion rate of the day of the experiment was estimated (IRexp). The last column (IRGE40) shows ingestion rates needed to explain the observed growth rates with a growth efficiency of 40%.
In the grazing experiments, we calculated the ingestion rate in the different bottles having a gradient of prey abundance, allowing the delineation of the functional response of the grazer (Table 2). Images of FISH‐stained cells with ingested FLBs are exemplified in Fig. 3 for the four taxa analysed. We did first this analysis for the community of heterotrophic flagellates (Fig. 4A), just by inspecting DAPI‐filters without FISH hybridization. The fit of the Michaelis–Menten curve to the ingestion data yielded a maximum ingestion rate IRmax of 2.3 prey HF−1 h−1 and a half‐saturation constant Ks of 6.7 × 105 prey ml−1. Then, we estimated the functional response of four different heterotrophic flagellate taxa. M. minuta presented an IRmax more than double the community rates (Fig. 4B), and a Ks only slightly lower. Both parameters were estimated with a high significance. For the MAST‐4, the IRmax was rather low, only 1.0 prey HF−1 h−1 and the Ks was slightly higher than that of the HF community, although in this fit the Ks parameter was not significant in the model (Fig. 4C). The situation of the MAST‐7 was similar, with an IRmax similar to the HF community and a Ks also slightly higher than the HF community (Fig. 4D). In this case, the significance of the Ks value is low, close to the 0.01 level. Finally, the curve of P. imperforata was different from the other four, as there was not clear sign of saturation of the ingestion rates along the prey concentration used in this experiment (Fig. 4E). Even though the parameters estimated were not significant, the Ks shown for this species was one order of magnitude higher than that of all the other taxa.
Fig. 3.
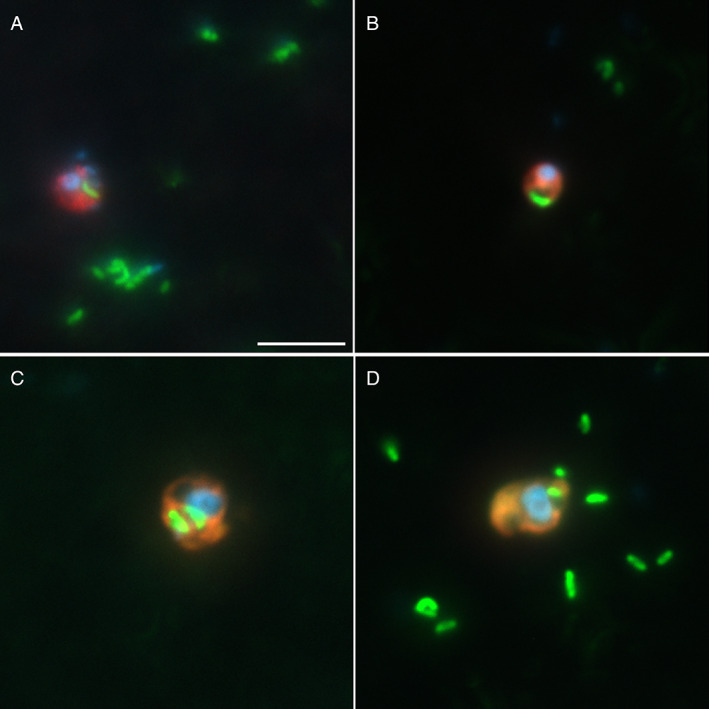
Epifluorescence micrographs of cells with ingested FLBs for Minorisa minuta (A), MAST‐4 (B), MAST‐7 (C) and Paraphysomonas imperforata (D). Each image is an overlay of three pictures of the same cell observed under UV radiation (showing the blue nucleus after DAPI staining), green light (red cytoplasm after CARD‐FISH) and blue light excitation (FLB detection). Scale bar is 5 μm and applies to all figures.
Fig. 4.
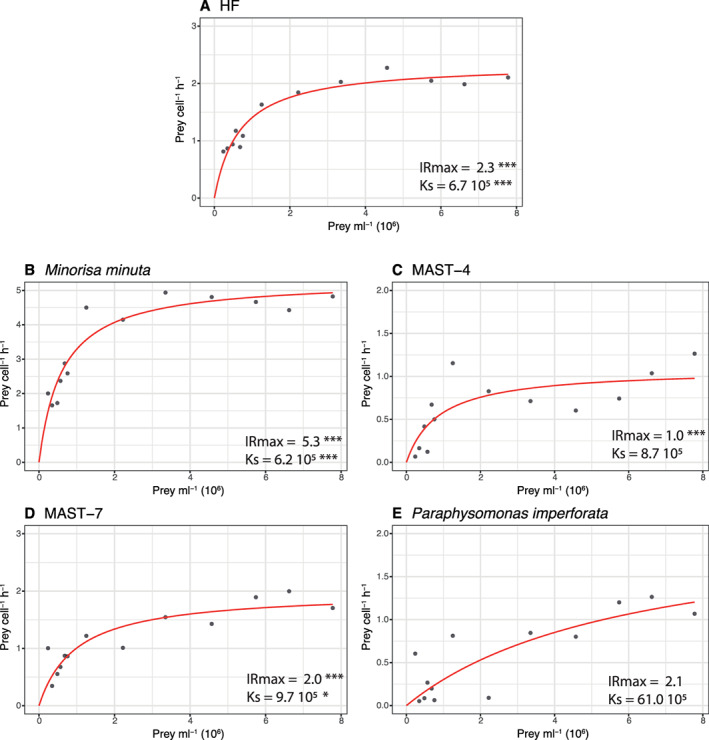
Functional responses (relationship of ingestion rates and prey abundance) of the natural community of heterotrophic flagellates (A), Minorisa minuta (B), MAST‐4 (C), MAST‐7 (D) and Paraphysomonas imperforata (E). Ks = half‐saturation constant (prey ml−1) and IRmax = maximum ingestion rate (prey cell−1 h−1). Significance of the estimate in the fit: ‘***’(P ≤ 0.001), ‘**’(P ≤ 0.01), ‘*’(P ≤ 0.05), ‘.’(P ≤ 0.1), ‘’(P ≤ 1).
We then evaluated the distribution of ingested FLBs (estimated at time 40 subtracting time 0) within individual cells for the whole community and for the four individual taxa, considering only the six bottles with highest prey abundance (Fig. 5). When considering the maximal number of FLBs ingested, the number was highest for HF (one cell had 15 FLBs), and relatively similar in the other cases: seven in Minorisa, five in MAST‐4, 8 in MAST‐7, and seven in Paraphysomonas. Except in one case, the majority of cells appeared with any FLB ingested. The percentage of cells without ingestion was highest in Paraphysomonas (82%), followed by MAST‐4 (about 70%) and MAST‐7 and the HF assemblage (about 60%). As a striking contrast, Minorisa presented only about 20% of cells without ingestion. When observing then the ingested cells, in the case of MAST‐4 most cells presented only one FLB ingested, while several had two FLBs. In the case of MAST‐7 a similar number of cells exhibited one or two ingested FLBs, while for Minorisa the majority of cells presented between two and four ingested FLBs. Paraphysomonas presented few cells with ingestion, but with a constant decrease until seven ingested FLBs per cell. The HF community presented a less marked profile, with many cells having between one and four ingested FLBs.
Fig. 5.
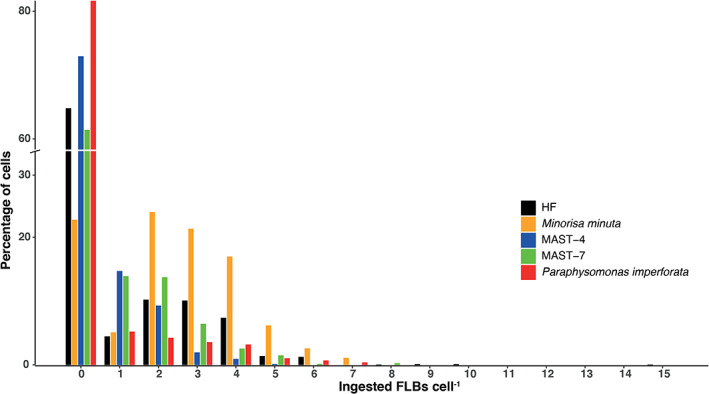
Percentage of cells within five HF groups having different number of ingested FLBs. These numbers are calculated with all cells observed per group at saturating food abundances (the last six bottles of the gradient).
Finally, we evaluated the fit between the observed growth rates and the measured ingestion rates (Table 2). From the functional response equations and the measured bacterial abundance of the day of the experiment (1.1 × 106 prey ml−1) we calculated the ingestion rate of that day and used this value to calculate the growth efficiency, which resulted in values unrealistically high. We then calculated which ingestion rate was needed to get a typical growth efficiency of 40% (Table 2).
Discussion
A main challenge in microbial ecology is to shed light on the black box approach. Indeed, for decades the abundance and activity of microbial components (such as bacteria, bacterial grazers, picoalgae, etc.), have been studied as bulk properties, ignoring the different capacities and performances of the composing species in the assemblage. A very relevant topic when investigating predation in nature is the shape of the functional or numerical responses, i.e., the changes of grazing rates or growth rates with food abundance (Weisse et al., 2016). These physiological features have been studied in cultured heterotrophic flagellate strains grazing on bacteria (summarized in Table 3). However, to our knowledge, there are no studies analysing functional responses of natural assemblages, including uncultured species. This was the motivation of our study.
Table 3.
A report of maximum growth rates (μmax, d−1), maximum ingestion rates (IRmax, prey cell−1 h−1) and half‐saturation constants (Ks, 106 prey ml−1) for cultured flagellate species and prey types.
| Flagellate cultures | Prey | μmax | IRmax | Ks | References |
|---|---|---|---|---|---|
| Actinomonas mirabilis | Pseudomonas sp. | 6.00 | 1.4 | Fenchel (1982b) | |
| Bodo designis | Alteromonas or Shewanella | 3.84 | 3.4 | Eccleston‐Parry and Leadbeater, (1994) | |
| Aeromonas sp. | 2.88 | 8.8 | Hammond (1991) | ||
| Ciliophrys infusionum | Alteromonas or Shewanella | 1.08 | 45.0 | Eccleston‐Parry and Leadbeater (1994) | |
| Codosiga gracilius | Alteromonas or Shewanella | 1.25 | 9.7 | Eccleston‐Parry and Leadbeater (1994) | |
| Diaphanoeca grandis | Pseudomonas sp. | 2.88 | 2.4 | Andersen (1989) | |
| Jakoba libera | Aeromonas sp. | 1.92 | 5.3 | Hammond (1991) | |
| Alteromonas or Shewanella | 0.86 | 5.4 | Eccleston‐Parry and Leadbeater (1994) | ||
| Monosiga sp. | Pseudomonas sp. | 4.08 | 13.5 | Fenchel (1982a) | |
| Ochromonas sp. | Pseudomonas sp. | 4.56 | 19.0 | Fenchel (1982a) | |
| Paraphysomonas vestita | Pseudomonas sp. | 5.52 | 14.9 | Fenchel (1982a) | |
| P. imperforata | Alteromonas or Shewanella | 5.04 | 1.1 | Eccleston‐Parry and Leadbeater (1994) | |
| Aeromonas sp. | 2.88 | 4.4 | Hammond (1991) | ||
| Vibrio sp. | 5.28 | 13.0 | Edwards (1989) | ||
| Pseudoalteromonas sp. | 4.56 | 12.6 | 9.7 | Tophøj et al. (2018) | |
| Pseudoalteromonas sp. | 2.40 | 6.3 | 3.6 | Tophøj et al. (2018) | |
| Procryptobia sorokini | Pseudoalteromonas sp. | 6.00 | 11.7 | 2.8 | Tophøj et al. (2018) |
| Pseudoalteromonas sp. | 2.64 | 6.7 | 1.1 | Tophøj et al. (2018) | |
| Pseudoalteromonas sp. | 4.80 | 20.7 | 8.3 | Tophøj et al. (2018) | |
| Pleuromonas jaculans | Pseudomonas sp. | 3.84 | 38.6 | Fenchel (1982a) | |
| Pseudobodo tremulans | Pseudomonas sp. | 3.60 | 8.4 | Fenchel (1982a) | |
| Stephanoeca diplocostata | Pseudomonas sp. | 1.90 | 6.8 | Geider and Leadbeater (1988) | |
| Alteromonas or Shewanella | 0.84 | 2.3 | Eccleston‐Parry and Leadbeater (1994) | ||
| Cafeteria roenbergensis | Photobacterium angustum | 6.24 | 5.8 | Anderson et al. (2011) | |
| Vibrio vuinificus | 5.04 | 2.7 | Anderson et al. (2011) | ||
| Sphingopyxis alaskensis | 5.76 | 7.4 | Anderson et al. (2011) | ||
| Cafeteria sp. | Mixed bacterial communities | 0.98 | 8.7 | Mohapatra and Fukami (2004) | |
| Flavobacterium sp. | 0.98 | 9.1 | Mohapatra and Fukami, 2004 | ||
| Alteromonas sp. | 0.96 | 9.2 | Mohapatra and Fukami (2004) | ||
| Pseudomonas sp. | 0.96 | 9.1 | Mohapatra and Fukami (2004) | ||
| Jakoba libera | Mixed bacterial communities | 0.58 | 5.1 | Mohapatra and Fukami (2004) | |
| Flavobacterium sp. | 0.77 | 3.7 | Mohapatra and Fukami, 2004 | ||
| Alteromonas sp. | 0.10 | 9.5 | Mohapatra and Fukami (2004) | ||
| Pseudomonas sp. | 0.98 | 1.4 | Mohapatra and Fukami (2004) | ||
| Poterioochromonas malhamensis | Polynucleobacter | 1.01 | 18.2 | Boenigk et al. (2006) | |
| Listonella pelagia | 1.70 | 1.5 | Boenigk et al. (2006) | ||
| Spumella sp. | Polynucleobacter | 1.99 | 20.5 | Boenigk et al. (2006) | |
| 2.30 | 22.0 | Pfandl and Boenigk (2006) | |||
| Listonella pelagia | 2.40 | 1.2 | Boenigk et al. (2006) | ||
| 2.81 | 1.2 | Pfandl and Boenigk (2006) | |||
| Mixed bacteria | 3.86 | 72.8 | 2.4 | Jürgens (1995) | |
| Isolated bacteria | 5.40 | 64.6 | Jürgens (1995) | ||
| Bodo sp. | Mixed bacteria | 4.39 | 3.9 | Jürgens (1995) | |
| Oxyrrhis marina | FLB | 71.3 | 1.1 | Jeong et al. (2008) | |
| Gyrodinium cf. guttula | FLB | 23.2 | 1.3 | Jeong et al. (2008) | |
| Pfiesteria piscicida | FLB | 13.7 | 0.8 | Jeong et al. (2008) |
Our grazing experiments were based in short‐term incubations adding FLBs, which were then counted inside protist food vacuoles by epifluorescence. This technique has been highly used (Jezbera et al., 2005; Ng and Liu, 2016; Šimek and Sirova, 2019) and is the best to calculate specific ingestion rates for taxonomic classes of protists. Major limitations of this approach are the negative selection against fluorochrome labelled and heat‐killed bacteria (Landry et al., 1991; Fu et al., 2003; Massana et al., 2009), and statistical problems in obtaining reliable counts of ingested FLBs at low predator densities (McManus and Okubo, 1991). The first limitation can be solved by using alive monospecific bacteria as food and CARD‐FISH detection in food vacuoles (Jezbera et al., 2005; Massana et al., 2009). In the later paper, we found that MAST‐4 ingested alive bacteria two to three times faster than FLBs. However, using alive bacteria would have added an extra layer of complexity to our experiment. At any rate, the underestimation because using FLB would be similar along the prey gradient, therefore not affecting the shape of the functional response. To minimize the problem of low predator densities, we did our grazing experiment with a sample enriched in HF. As expected (Massana et al., 2006a), our unamended seawater incubation selected for heterotrophic flagellates abundant in situ, many of them being uncultured MAST taxa, but also including cultured species like M. minuta, which had been cultured mimicking natural conditions (del Campo et al., 2013). Luckily enough, some typical cultured HF also developed in our incubation, highlighting a very interesting and contrasting behaviour.
The growth rate (Table 2) during the incubation of the whole HF assemblage (0.71 day−1) was similar to previous incubations studies (between 0.66 and 1.25 day−1) (Massana et al., 2006a), and faster than typical growth rates of natural assemblages, 0.05–0.50 day−1 (Jürgens and Massana, 2008; Piwosz and Pernthaler, 2010), in accordance with the promotion of growth in the incubation. Moreover, each flagellate taxa presented a slightly higher growth rate than the community rate, particularly for M. minuta (1.54 day−1). Specific growth rates measured here were similar to those from previous studies: M. minuta (1.56 day−1; del Campo et al., 2013) and MAST‐4 (0.62 day−1 on average; Massana et al., 2006a). Moreover, growth rates of cultured species in the laboratory were remarkably larger (up to 10 times) than the rates reported in our unamended incubations.
The maximal ingestion rate of M. minuta was several times higher than that of MAST‐4 and MAST‐7 (Fig. 4). Using FLB as prey surrogates and the concentration range assayed, M. minuta results to be very efficient and well adapted to low prey abundances. The low feeding rates for MAST‐4 measured here are consistent with previous estimates, also obtained with FLB, of ingestion rates of 1.0–1.5 bacteria predator−1 h−1 (Massana et al., 2009). These rates are about half the ingestion rates of the whole HF assemblage, even though a high expression of genes involved in phagocytosis has been reported for MAST‐4 (Labarre et al., 2020). It has been inferred before that MAST‐4 biases against heat‐killed FLB, preferring alive bacteria in good physiological state (Massana et al., 2009), but this likely holds true for most HF taxa in the assemblage (Landry et al., 1991; Fu et al., 2003). Another explanation for lower ingestion rates in MAST‐4 could be that it is adapted to graze on a specific prey, as small Pelagibacter ubique detected within MAST‐4 cells by single‐cell sequencing (Martinez‐Garcia et al., 2012), therefore potentially occupying a different ecological niche than M. minuta. Finally, P. imperforata represents a very contrasting case, consistent with the fact that it is easily cultured feeding on large bacteria at very high densities (Lim et al., 1999). Studies with Paraphysomonas species have shown different results, including a strain with relatively low Ks, 1.1 × 106 bacteria ml−1 (Eccleston‐Parry and Leadbeater, 1994) and another that ceased to multiply at prey abundances below ~2 × 106 cells ml−1 (Ishigaki and Seligh, 2001). Our data seemed to agree with the later case, since P. imperforata is the taxa that grows faster but did not achieve ingestion rate saturation even at the highest prey concentrations assayed (Fig. 4). With these data, we should expect a high Ks, similar to other cultured flagellates (Table 3).
The growth efficiencies calculated from the measured ingestion rates and the observed growth rates were unrealistically high, above 100% in all cases and extremely high in P. imperforata, something vitally impossible and out of the range of 30%–60% in previous estimates (Snyder and Hoch, 1996; Zubkov and Sleigh, 2000). This inconsistency is caused by underestimation of the ingestion rates, which can be explained by several reasons: a negative selection against FLBs as previously commented (Massana et al., 2009), an incubation time in ingestion experiments being too close to the plateau (Unrein et al., 2007), or the predation on alternative preys, such as Synechococcus and phototrophic flagellates that in fact decreased during the incubation, which could contribute to biomass ingestion and therefore to the growth rates. Our calculations of the ingestion rates needed to explain the growth observed with a GE of 40% provided very realistic estimates, within the range of 2–20 bacteria prey−1 h−1 measured for in situ HF assemblages (Jürgens and Massana, 2008), while P. imperforata displayed values similar to other cultured flagellates (Table 3).
The most remarkable finding of this work was that the Ks of the functional responses for the whole community and for three flagellate taxa were in the narrow range of 6.2–9.7 × 105 prey ml−1 (Fig. 4). MAST‐4 (Rodriguez‐Martinez et al., 2012), MAST‐7 (Giner et al., 2016) and M. minuta (del Campo et al., 2013) represent heterotrophic flagellates that are widely distributed and abundant in natural marine assemblages (Mangot et al., 2018). Interestingly, our data indicate that they are very well adapted to the bacterial abundances of marine planktonic environments, typically around 106 bacteria ml−1 (Fuhrman and Hagström, 2008). For instance, in the oligotrophic coastal system sampled here (Blanes Bay, NW Mediterranean), the averaged bacterial concentration during the last 20 years (monthly sampling) was 0.90 × 106 bacteria ml−1, with a typical bacterial cell size of 0.06 μm3 (Gasol et al., 1995). In contrast, the Ks of cultured heterotrophic flagellates is typically at least one order of magnitude higher, ranging from 0.1 to 4.5 × 107 bacteria ml−1 with only one exception, Pfiesteria piscicida (Table 3). These higher Ks of cultured flagellates are the expected values for organisms that grow efficiently in rich media, and at the same time establish an obvious limitation for their development at the prevailing low in situ bacterial abundances. The results shown here for P. imperforata agree with this scenario, since this species exhibited a low grazing capacity and no food saturation in the range of prey abundance tested.
In conclusion, we have shown here that the assemblage of heterotrophic flagellates derived from a marine coastal station (BBMO) presents a functional response with a Ks of 6.7 × 105 bacteria ml−1, which is much lower than the Ks of typical cultured flagellates. This indicates that cells of the community are generally well adapted to in situ marine bacterial abundance. Inside this mixed community, there are taxa with different functional responses, therefore delineating ecological niches, perhaps with different predation strategies and prey preferences. We have shown that Minorisa minuta is well adapted to low prey abundances (Ks of 6.2 × 105 bacteria ml−1) and is very efficient in ingesting bacteria (IRmax 5.3 prey h−1). MAST‐4 is less voracious but is also well adapted to typical planktonic bacterial abundances (Ks of 8.7 × 105 bacteria ml−1). MAST‐7 has a IRmax similar to the natural HF community and a Ks slightly higher. In contrast, Paraphysomonas imperforata is food limited all along the prey gradient tested, suggesting a general poor performance in natural marine planktonic environments. Our study sets the basis for the fundamental differences between cultured and uncultured bacterial grazers.
Acknowledgements
Funding has been provided by ESTRAMAR (CTM2004‐12631/MAR, MEC), FLAME (CGL2010‐16304, MICINN) and ALLFLAGS (CTM2016‐75083‐R, MINECO) Spanish projects to R.M. and by a F.P.I. fellowship from the Spanish Ministry of Education and Science to R.R.M. The authors thank R. Terrado, G. Caló and M. Vila for help in FISH analyses, V. Balagué for molecular help and E. Sainz for statistical advices.
Contributor Information
Raquel Rodríguez‐Martínez, Email: raquelrmcs@gmail.com.
Ramon Massana, Email: ramonm@icm.csic.es.
References
- Andersen, P. (1989) Functional biology of the Choanoflagellate Diaphanoeca grandis Ellis. Mar Microb Food Webs 3: 35–50. [Google Scholar]
- Anderson, R. , Kjelleberg, S. , McDougald, D. , and Jürgens, K. (2011) Species‐specific patterns in the vulnerability of carbon‐starved bacteria to protist grazing. Aquat Microb Ecol 64: 105–116. [Google Scholar]
- Azam, F. , Fenchel, T. , Field, J.G. , Gray, J.S. , Meyer‐Reil, L.A. , and Thingstad, F. (1983) The ecological role of water‐column microbes in the sea *. Mar Ecol Prog Ser 10: 257–263. [Google Scholar]
- Boenigk, J. , Pfandl, K. , and Hansen, P. (2006) Exploring strategies for nanoflagellates living in a “wet desert.”. Aquat Microb Ecol 44: 71–83. [Google Scholar]
- del Campo, J. , Not, F. , Forn, I. , Sieracki, M.E. , and Massana, R. (2013) Taming the smallest predators of the oceans. ISME J 7: 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, D. , Countway, P.D. , Jones, A.C. , Kim, D.Y. , and Schnetzer, A. (2012) Marine protistan diversity. Ann Rev Mar Sci 4: 467–493. [DOI] [PubMed] [Google Scholar]
- Eccleston‐Parry, J.D. , and Leadbeater, B.S.C. (1994) A comparison of the growth kinetics of six marine heterotrophic nanoflagellates fed with one bacterial species. Mar Ecol Prog Ser 105: 167–177. [Google Scholar]
- Edwards, A. (1989) Heterotrophic chrysophytes; their role in energy and carbon turnover in the sea. Ph D Thesis, Univeristy Birmingham.
- Elwood, H.J. , Olsen, G.J. , and Sogin, M.L. (1985) The small‐subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata . Mol Biol Evol 2: 399–410. [DOI] [PubMed] [Google Scholar]
- Fenchel, T. (1982a) Ecology of heterotrophic microflagellates. II. Bioenergetics and growth. Mar Ecol Prog Ser 8: 225–231. [Google Scholar]
- Fenchel, T. (1987) Ecology of Protozoa: The Biology of Free‐Living Phagotrophic Protists. Berlin, Germany: Springer‐Verlag. [Google Scholar]
- Fenchel, T. (1980) Suspension feeding in ciliated protozoa: functional response and particle size selection. Microb Ecol 6: 1–11. [DOI] [PubMed] [Google Scholar]
- Fenchel, T. (1982b) The bioenergetics of a heterotrophic microflagellate. Annls Inst Ocean 58: 55–60. [Google Scholar]
- Fu, Y. , O'Kelly, C. , Sieracki, M. , and Distel, D.L. (2003) Protistan grazing analysis by flow cytometry using prey labeled by in vivo expression of fluorescent proteins. Appl Environ Microbiol 69: 6848–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman, J.A. , and Hagström, K. (2008) Bacterial and archaeal community structure and its patterns. In Microbial Ecology of the Oceans. Hoboken, NJ, USA: John Wiley & Sons, Inc., pp. 45–90. [Google Scholar]
- Fuhrman, J.A. , and Noble, R.T. (1995) Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr 40: 1236–1242. [Google Scholar]
- Gasol, J. , del Giorgio, P. , Massana, R. , and Duarte, C. (1995) Active versus inactive bacteria:size‐dependence in a coastal marine plankton community. Mar Ecol Prog Ser 128: 91–97. [Google Scholar]
- Geider, R. , and Leadbeater, B. (1988) Kinetics and energetics of growth of the marine choanoflagellate Stephanoeca diplocostata . Mar Ecol Prog Ser 47: 169–177. [Google Scholar]
- Giner, C.R. , Forn, I. , Romac, S. , Logares, R. , de Vargas, C. , and Massana, R. (2016) Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl Environ Microbiol 82: 4757–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gismervik, I. (2005) Numerical and functional responses of choreo‐ and oligotrich planktonic ciliates. Aquat Microb Ecol 40: 163–173. [Google Scholar]
- Gómez‐Rubio, V. (2017) ggplot2 ‐ elegant graphics for data analysis (2nd edition). J Stat Softw 77: 3–5. [Google Scholar]
- Grujcic, V. , Nuy, J.K. , Salcher, M.M. , Shabarova, T. , Kasalicky, V. , Boenigk, J. , et al. (2018) Cryptophyta as major bacterivores in freshwater summer plankton. ISME J 12: 1668–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S. (1991) Comparative growth characteristics of heterotrophic flagellates. M Sc Thesis, Bristol University 26–37.
- Henriksen, C. , Saiz, E. , Calbet, A. , and Hansen, B. (2007) Feeding activity and swimming patterns of Acartia grani and Oithona davisae nauplii in the presence of motile and non‐motile prey. Mar Ecol Prog Ser 331: 119–129. [Google Scholar]
- Hillebrand, H. , Dürselen, C.‐D. , Kirschtel, D. , Pollingher, U. , and Zohary, T. (1999) Biovolume calculation for pelagic and benthic microalgae. J Phycol 35: 403–424. [Google Scholar]
- Holling, C.S. (1959) The components of predation as revealed by a study of small‐mammal predation of the European pine sawfly. Can Entomol 91: 293–320. [Google Scholar]
- Isari, S. , and Saiz, E. (2011) Feeding performance of the copepod Clausocalanus lividus (frost and Fleminger, 1968). J Plankton Res 33: 715–728. [Google Scholar]
- Ishigaki, T. , and Sleigh, M. (2001) Grazing characteristics and growth efficiencies at two different temperatures for three nanoflagellates fed with vibrio bacteria at three different concentrations. Microb Ecol 41: 264–271. [DOI] [PubMed] [Google Scholar]
- Jeong, H. , Yoo, Y. , Park, J. , Song, J. , Kim, S. , Lee, S. , et al. (2005) Feeding by phototrophic red‐tide dinoflagellates: five species newly revealed and six species previously known to be mixotrophic. Aquat Microb Ecol 40: 133–150. [Google Scholar]
- Jeong, H.J. , Seong, K.A. , Du Yoo, Y. , Kim, T.H. , Kang, N.S. , Kim, S. , et al. (2008) Feeding and grazing impact by small marine heterotrophic dinoflagellates on heterotrophic bacteria. J Eukaryot Microbiol 55: 271–288. [DOI] [PubMed] [Google Scholar]
- Jezbera, J. , Hornák, K. , and Simek, K. (2005) Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol Ecol 52: 351–363. [DOI] [PubMed] [Google Scholar]
- Jonsson, P. (1986) Particle size selection, feeding rates and growth dynamics of marine planktonic oligotrichous ciliates (Ciliophora: Oligotrichina). Mar Ecol Prog Ser 33: 265–277. [Google Scholar]
- Jürgens, K. (1995) Die bedeutung heterotropher nanoflagellaten als bakterienkonsumenten sowie deren regulation durch prädation und ressourcen. PhD Thesis 1–1.
- Jürgens, K. , and Massana, R. (2008) Protistan grazing on marine bacterioplankton. In Microbial Ecology of the Oceans. Hoboken, NJ, USA: John Wiley & Sons, Inc., pp. 383–441. [Google Scholar]
- Jürgens, K. , and Matz, C. (2002) Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81: 413–434. [DOI] [PubMed] [Google Scholar]
- Jürgens, K. , and Simek, K. (2000) Functional response and particle size selection of Halteria cf. grandinella, a common freshwater oligotrichous ciliate. Aquat Microb Ecol 22: 57–68. [Google Scholar]
- Kim, J. , and Jeong, H. (2004) Feeding by the heterotrophic dinoflagellates Gyrodinium dominans and G. spirale on the red‐tide dinoflagellate Prorocentrum minimum . Mar Ecol Prog Ser 280: 85–94. [Google Scholar]
- Labarre, A. , Obiol, A. , Wilken, S. , Forn, I. , and Massana, R. (2020) Expression of genes involved in phagocytosis in uncultured heterotrophic flagellates. Limnol Oceanogr 65: 149–160. [Google Scholar]
- Landry, M.R. , Lehner‐Fournier, J.M. , Sundstrom, J. , Fagerness, V.L. , and Selph, K.E. (1991) Discrimination between living and heat‐killed prey by a marine zooflagellate, Paraphysomonas vestita (stokes). J Exp Mar Bio Ecol 146: 139–151. [Google Scholar]
- Lim, E.L. , Dennett, M.R. , and Caron, D.A. (1999) The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol Oceanogr 44: 37–51. [Google Scholar]
- Logares, R. , Audic, S. , Santini, S. , Pernice, M.C. , de Vargas, C. , and Massana, R. (2012) Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J 6: 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X. , Gao, Y. , and Weisse, T. (2021) Functional ecology of two contrasting freshwater ciliated protists in relation to temperature. J Eukaryot Microbiol 68: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangot, J.‐F. , Forn, I. , Obiol, A. , and Massana, R. (2018) Constant abundances of ubiquitous uncultured protists in the open sea assessed by automated microscopy. Environ Microbiol 20: 3876–3889. [DOI] [PubMed] [Google Scholar]
- Martinez‐Garcia, M. , Brazel, D. , Poulton, N.J. , Swan, B.K. , Gomez, M.L. , Masland, D. , et al. (2012) Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J 6: 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana, R. , Balagué, V. , Guillou, L. , and Pedrós‐Alió, C. (2004) Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol Ecol 50: 231–243. [DOI] [PubMed] [Google Scholar]
- Massana, R. , del Campo, J. , Dinter, C. , and Sommaruga, R. (2007) Crash of a population of the marine heterotrophic flagellate cafeteria roenbergensis by viral infection. Environ Microbiol 9: 2660–2669. [DOI] [PubMed] [Google Scholar]
- Massana, R. , del Campo, J. , Sieracki, M.E. , Audic, S. , and Logares, R. (2014) Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J 8: 854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana, R. , DeLong, E.F. , and Pedrós‐Alió, C. (2000) A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol 66: 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana, R. , Gobet, A. , Audic, S. , Bass, D. , Bittner, L. , Boutte, C. , et al. (2015) Marine protist diversity in European coastal waters and sediments as revealed by high‐throughput sequencing. Environ Microbiol 17: 4035–4049. [DOI] [PubMed] [Google Scholar]
- Massana, R. , Guillou, L. , Díez, B. , and Pedrós‐Alió, C. (2002) Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl Environ Microbiol 68: 4554–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana, R. , Guillou, L. , Terrado, R. , Forn, I. , and Pedrós‐Alió, C. (2006a) Growth of uncultured heterotrophic flagellates in unamended seawater incubations. Aquat Microb Ecol 45: 171–180. [Google Scholar]
- Massana, R. , Terrado, R. , Forn, I. , Lovejoy, C. , and Pedros‐Alio, C. (2006b) Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol 8: 1515–1522. [DOI] [PubMed] [Google Scholar]
- Massana, R. , Unrein, F. , Rodríguez‐Martínez, R. , Forn, I. , Lefort, T. , Pinhassi, J. , and Not, F. (2009) Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J 3: 588–596. [DOI] [PubMed] [Google Scholar]
- McManus, G.B. , and Okubo, A. (1991) On the use of surrogate food particles to measure protistan ingestion. Limnol Oceanogr 36: 613–617. [Google Scholar]
- Medlin, L. , Elwood, H.J. , Stickel, S. , and Sogin, M.L. (1988) The characterization of enzymatically amplified eukaryotic 16S‐like rRNA‐coding regions. Gene 71: 491–499. [DOI] [PubMed] [Google Scholar]
- Meira, B.R. , Lansac‐Toha, F.M. , Segovia, B.T. , Buosi, P.R.B. , Lansac‐Tôha, F.A. , and Velho, L.F.M. (2018) The importance of herbivory by protists in lakes of a tropical floodplain system. Aquat Ecol 52: 193–210. [Google Scholar]
- Mohapatra, B. , and Fukami, K. (2004) Comparison of the numerical grazing response of two marine heterotrophic nanoflagellates fed with different bacteria. J Sea Res 52: 99–107. [Google Scholar]
- Montagnes, D.J.S. , and Berges, J.A. (2004) Determining parameters of the numerical response. Microb Ecol 48: 139–144. [DOI] [PubMed] [Google Scholar]
- Ng, W.H.A. , and Liu, H. (2016) Diel periodicity of grazing by heterotrophic nanoflagellates influenced by prey cell properties and intrinsic grazing rhythm. J Plankton Res 38: 636–651. [Google Scholar]
- Pernice, M.C. , Forn, I. , Gomes, A. , Lara, E. , Alonso‐Sáez, L. , Arrieta, J.M. , et al. (2015) Global abundance of planktonic heterotrophic protists in the deep ocean. ISME J 9: 782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler, J. (2005) Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546. [DOI] [PubMed] [Google Scholar]
- Pernthaler, J. , Glöckner, F.‐O. , Schönhuber, W. , and Amann, R. (2001) Fluorescence in situ hybridization (FISH) with rRNA‐targeted oligonucleotide probes. Methods Microbiol 30: 207–226. [Google Scholar]
- Pfandl, K. , and Boenigk, J. (2006) Stuck in the mud: suspended sediments as a key issue for survival of chrysomonad flagellates. Aquat Microb Ecol 45: 89–99. [Google Scholar]
- Piwosz, K. , Mukherjee, I. , Salcher, M.M. , Grujčić, V. , and Šimek, K. (2021) CARD‐FISH in the sequencing era: opening a new universe of protistan ecology. Front Microbiol 12: 640066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwosz, K. , and Pernthaler, J. (2010) Seasonal population dynamics and trophic role of planktonic nanoflagellates in coastal surface waters of the southern Baltic Sea. Environ Microbiol 12: 364–377. [DOI] [PubMed] [Google Scholar]
- Porter, K.G. , and Feig, Y.S. (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943–948. [Google Scholar]
- Ritz, C. , Baty, F. , Streibig, J.C. , and Gerhard, D. (2015) Dose‐response analysis using R. PLoS One 10: e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, E.C. , Wootton, E.C. , Davidson, K. , Jeong, H.J. , Lowe, C.D. , and Montagnes, D.J.S. (2011) Feeding in the dinoflagellate Oxyrrhis marina: linking behaviour with mechanisms. J Plankton Res 33: 603–614. [Google Scholar]
- Rodriguez‐Martinez, R. , Rocap, G. , Logares, R. , Romac, S. , and Massana, R. (2012) Low evolutionary diversification in a widespread and abundant uncultured protist (MAST‐4). Mol Biol Evol 29: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Shannon, S.P. , Chrzanowski, T.H. , and Grover, J.P. (2007) Prey food quality affects flagellate ingestion rates. Microb Ecol 53: 66–73. [DOI] [PubMed] [Google Scholar]
- Sherr, B.F. , Sherr, E.B. , and Fallon, R.D. (1987) Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory †. Appl Environ Microbiol 53: 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, E.B. , and Sherr, B.F. (2002) Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81: 293–308. [DOI] [PubMed] [Google Scholar]
- Sieracki, M. , Haas, L. , Caron, D. , and Lessard, E. (1987) Effect of fixation on particle retention by microflagellates: underestimation of grazing rates. Mar Ecol Prog Ser 38: 251–258. [Google Scholar]
- Sieracki, M.E. , Johnson, P.W. , and Sieburth, J.M. (1985) Detection, enumeration, and sizing of planktonic bacteria by image‐analyzed epifluorescence microscopy. Appl Environ Microbiol 49: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek, K. , Grujčić, V. , Hahn, M.W. , Horňák, K. , Jezberová, J. , Kasalický, V. , et al. (2018) Bacterial prey food characteristics modulate community growth response of freshwater bacterivorous flagellates. Limnol Oceanogr 63: 484–502. [Google Scholar]
- Šimek, K. , Grujčić, V. , Mukherjee, I. , Kasalický, V. , Nedoma, J. , Posch, T. , et al. (2020) Cascading effects in freshwater microbial food webs by predatory Cercozoa, Katablepharidacea and ciliates feeding on aplastidic bacterivorous cryptophytes. FEMS Microbiol Ecol 96: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek, K. , Pernthaler, J. , Weinbauer, M.G. , Hornák, K. , Dolan, J.R. , Nedoma, J. , et al. (2001) Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl Environ Microbiol 67: 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek, K. , and Sirova, D. (2019) Fluorescently labeled bacteria as a tracer to reveal novel pathways of organic carbon flow in aquatic ecosystems. J Vis Exp 2019: 1–9. [DOI] [PubMed] [Google Scholar]
- Snyder, R.A. , and Hoch, M.P. (1996) Consequences of protist‐stimulated bacterial production for estimating protist growth efficiencies. Hydrobiologia 341: 113–123. [Google Scholar]
- Strom, S. (2000) Bacterivory: interactions between bacteria and their grazers. In Microbial Ecology of the Oceans, Kirchmand, D.L. (ed). New York, NY: Wiley‐Liss Inc, pp. 351–386. [Google Scholar]
- Tophøj, J. , Wollenberg, R.D. , Sondergaard, T.E. , and Eriksen, N.T. (2018) Feeding and growth of the marine heterotrophic nanoflagellates, Procryptobia sorokini and Paraphysomonas imperforata on a bacterium, Pseudoalteromonas sp. with an inducible defence against grazing. PLoS One 13: e0195935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unrein, F. , Massana, R. , Alonso‐Sáez, L. , and Gasol, J.M. (2007) Significant year‐round effect of small mixotrophic flagellates on bacterioplankton in an oligotrophic coastal system. Limnol Oceanogr 52: 456–469. [Google Scholar]
- Vaqué, D. , Gasol, J.M. , and Marrasé, C. (1994) Grazing rates on bacteria : the significance of methodology and ecological factors. Mar Ecol Prog Ser 109: 263–274. [Google Scholar]
- de Vargas, C. , Audic, S. , Henry, N. , Decelle, J. , Mahe, F. , Logares, R. , et al. (2015) Eukaryotic plankton diversity in the sunlit ocean. Science 348: 1261605. [DOI] [PubMed] [Google Scholar]
- Vazquez‐Dominguez, E. , Peters, F. , Gasol, J. , and Vaqué, D. (1999) Measuring the grazing losses of picoplankton: methodological improvements in the use of fluorescently labeled tracers combined with flow cytometry. Aquat Microb Ecol 20: 119–128. [Google Scholar]
- Weisse, T. , Anderson, R. , Arndt, H. , Calbet, A. , Hansen, P.J. , and Montagnes, D.J.S. (2016) Functional ecology of aquatic phagotrophic protists – concepts, limitations, and perspectives. Eur J Protistol 55: 50–74. [DOI] [PubMed] [Google Scholar]
- Worden, A.Z. , Follows, M.J. , Giovannoni, S.J. , Wilken, S. , Zimmerman, A.E. , and Keeling, P.J. (2015) Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347: 1257594. [DOI] [PubMed] [Google Scholar]
- Zubkov, M.V. , and Sleigh, M. (2000) Comparison of growth efficiencies of protozoa growing on bacteria deposited on surfaces and in suspension. J Eukaryot Microbiol 47: 62–69. [DOI] [PubMed] [Google Scholar]


