Abstract
The molecular complexity displayed in acute myeloid leukemia (AML) hinders patient stratification and treatment decisions. Previous studies support the utility of using specific gene panels for this purpose. Focusing on two salient features of AML, the production of reactive oxygen species (ROS) by NADPH oxidases (NOX) and metabolism, we aimed to identify a gene panel that could improve patient stratification. A pairwise comparison of AML versus healthy gene expression revealed the downregulation of four members of the NOX2 complex including CYBB (coding for NOX2) in AML patients. We analyzed the expression of 941 genes related to metabolism and found 28 genes with expression correlated to CYBB. This panel of 29 genes (29G) effectively divides AML samples according to their prognostic group. The robustness of 29G was confirmed by 6 AML cohort datasets with a total of 1821 patients (overall accuracies of 85%, 78%, 80%, 75%, 59% and 83%). An expression index (EI) was developed according to the expression of the selected discriminatory genes. Overall Survival (OS) was higher for low 29G expression index patients than for the high 29G expression index group, which was confirmed in three different datasets with a total of 1069 patients. Moreover, 29G can dissect intermediate‐prognosis patients in four clusters with different OS, which could improve the current AML stratification scheme. In summary, we have found a gene signature (29G) that can be used for AML classification and for OS prediction. Our results confirm NOX and metabolism as suitable therapeutic targets in AML.
1. INTRODUCTION
Acute myeloid leukemia (AML) is the most prevalent leukemia in adults, with an incidence between 3 and 5 cases per 100 000 individuals, and median age at diagnostic around 67. 1 , 2 AML is characterized by the clonal proliferation of immature myeloid progenitors in bone marrow, peripheral blood and other tissues. The classical French‐American‐British (FAB) classification recognized eight different AML subtypes (M0 to M7) according to morphological features. 3 Evolution of AML progenitors is triggered by the accumulation of multiple genetic, epigenetic and cytogenetic alterations, with the possibility of the existence of several competing clones. Indeed, mutations in more than 250 genes can be found in AML patients, 23 of these genes are repetitively mutated. Mutations can be functionally grouped as transcription factor fusions, the NPM1 gene, tumor suppressor genes, DNA methylation‐related genes, signaling genes, chromatin‐modifying genes, myeloid transcription factor genes, cohesin complex genes, and spliceosome complex genes. 4 This provides a complex molecular landscape that hinders patient stratification and treatment decisions. Bearing this in mind, in 2010 a panel of experts on behalf of the European LeukemiaNet (ELN) provided recommendations for AML stratification (ELN2010), 5 which have been revised in 2017 (ELN2017). 4 Patients are classified into three categories: favorable, intermediate and adverse. 6 The 5‐year survival rates of patients below 60, are 64%, 42%, and 20% for each category respectively; while in patients over 60 survival is decreased to 37%, 16% and 6%. 7 The presence of cytogenetic alterations and the occurrence of certain mutations (NPM1, CEBPA, FLT3, RUNX1, TP53 and ASXL1) are taken into consideration for stratification. 4 However, it is convenient to highlight that around 45% of AML patients present a normal karyotype and that some recurrent mutations (DNMT3A, IDH1, IDH2) have not yet been assigned to a specific risk group. 4 Previous reports support the use of unbiased transcriptomic approaches for AML diagnosis. 8 , 9 , 10 , 11 Said strategy, which does not rely on the presence of genetic alterations, can assist in patient stratification and management. 12 Targeting these analyses to biological processes important in AML, such as metabolism, may improve prognosis and unveil novel therapeutic targets.
Metabolism rewiring is an important hallmark of cancer. 13 Many tumor cells respond to the pattern described by Otto Warburg almost a century ago and use aerobic glycolysis as the main source of energy. 14 AML blasts are glycolytic, 15 representing a paradigmatic example of aerobic glycolysis, also known as the Warburg effect. 14 Enhanced glycolysis has been associated with drug resistance, 16 and a more aggressive leukemic phenotype in AML. 17 In contrast, AML leukemic stem cells (LSCs) rely on oxidative phosphorylation (OXPHOS). 18 Despite this general view, AML blasts are distinguished by their metabolic plasticity, and their capacity to switch from glycolysis to OXPHOS. 19 Targeting metabolism is a promising strategy that might have soon an important clinical impact in the treatment of cancer, 20 , 21 including AML. 22
Reactive oxygen species (ROS) can promote leukemic cell proliferation 23 , 24 while inducing DNA damage and genome instability, contributing to tumor transformation. 25 The NADPH oxidase family (NOX) stands out among the cellular sources contributing to oxidative stress in tumor cells. 23 , 26 Some reports suggest the importance of NOX for the regulation of energetic metabolism. 27 , 28 , 29 AML cells exhibit high levels of ROS, which has been attributed to excessive NOX2 activity. 30 This enhanced NOX2‐driven ROS production supports AML cell proliferation through the activation of the glycolytic pathway. 31 Additionally, it seems that inhibition of NOX2 may also alter lipid and nitrogen metabolism. 32 This evidence supports the relevance of NOX2 in the control of metabolism homeostasis in AML.
Bearing the foregoing in mind, here we have compared gene expression between bone marrow samples from AML patients and healthy donors. Among more than 600 differentially expressed genes (DEGs), we found four genes encoding NOX2 subunits to be downregulated in AML samples, including CYBB (coding for NOX2). By analyzing the expression of 941 genes associated with metabolism in groups with different CYBB levels, we found 28 genes with expression correlated to CYBB. This panel of 29 genes (29G) can predict AML prognosis with high accuracy. Predictions were consistent across 6 different AML cohorts with a total of 1821 patients, demonstrating the robustness of 29G. Moreover, a low 29G expression index (EI) was linked to a higher Overall Survival (OS) which was confirmed by three independent datasets with a total of 1069 patients. Finally, 29G can dissect four clusters with different OS within the intermediate prognosis group. All these features make 29G a useful tool for AML prognosis, which could complement the current ELN scheme, for better management and therapeutics decisions.
2. METHODS
2.1. Datasets
Gene expression levels from 1821 healthy and AML bone marrow donors were publicly available obtained from the Gene Expression Omnibus database. GSE15061 33 was selected as the training dataset while GSE14468, 34 GSE10358, 35 GSE68833 36 and GSE165656 37 were used for validation (Figure S1 and Table S1). Gene expression data of one extra validation dataset (phs001657.v1.p1) were obtained from Tyner et al. 38 (Figure S1 and Table S1).
2.2. Canonical Biplot, Linear discriminant analysis (LDA) and Hierarchical k‐means clustering
Discriminant functions of LDA and canonical axes of Canonical Biplot were estimated as a linear combination of the 29 genes so that the ratio of between‐group variance to within‐group variance was maximized. Prognostic group discrimination abilities associated with the expression of the 29G were graphically visualized in reduced dimensional space generated by discriminant axes. LDA was extended with Canonical Biplot analysis 39 , 40 that enables the simultaneous representation of the three prognosis groups and the 29 genes, so that the relevance of each individual 29G gene to the differences among prognosis groups could be visualized (described in detail in Data S1).
Hierarchical k‐means cluster was used to determine 4 non‐overlapping segments for GSE10358 and phs001657.v1.p1 intermediate samples based on 29G expression data. Each data point belongs to one group only.
2.3. Expression index groups generation
The was computed as a linear combination of gene expression values ( for each ‐sample ), as follows:
where denotes the overall accuracy of each ‐discriminant function ( for LD1 and for LD2), denotes the discriminant coefficients of each ‐selected gene (, see results) in the corresponding ‐discriminant function and refers to the gene expression values of ‐gene in ‐sample.
AML patients were separated into Low‐Index or High‐Index groups via ROC curve analysis conducted with an optimal EI cut‐off and sensibility and specificity properties established to maximize the Youden index.
2.4. Statistical analysis of clinical factors
For each continuous variable analyzed, the median and interquartile range or mean and standard deviation (SD) were calculated depending on the variable distribution. For categorical variables, frequencies and percentages were reported. Statistical significance (p‐value <.05) of differences observed between groups was determined via Student's t‐test (for quantitative parametric variables); Mann–Whitney U and Kruskal–Wallis tests (for quantitative nonparametric variables) or Chi‐square test (for categorical variables). The linear relationship between two continuous variables was studied using Pearson's correlation. The Kaplan–Meier method was used to construct OS and EFS curves, and the log‐rank test was used to assess the statistical significance of the survival curves between groups (p‐value <.05).
Bioinformatics analyses were executed in R 41 (version 3.5.2; http://www.r-project.org) and Bioconductor (http://www.bioconductor.org) software tools. All described computations were implemented using affy, GenomicFeatures, limma, MultBiplotR, MASS, pROC, survminer and factorextra. Figures were generated via geneplotter, ggplot and plotly R‐packages.
3. RESULTS
3.1. CYBB is downregulated in AML patients and correlates with FAB classification, RUNX1‐RUNX1T1 translocation, as well as IDH1 , IDH2 , FLT3‐TKD and N‐RAS mutations
To identify differentially expressed genes in AML which hold prognostic value we chose a public data cohort (GSE15061) 33 as a training set that included information from AML and healthy samples (n = 449). Box plots and density plots (Figure S2) ensured sample normalization. Among more than 600 statistically significant DEGs identified, four members of the NOX2 complex were downregulated in AML: NCF1 (coding for p47 phox ), NCF2 (p67 phox ), NCF4 (p40 phox ), and CYBB (NOX2 or gp91 phox catalytic subunit), with fold change values of 5.14, 4.59, 2.26 and 2.45 respectively. No differences were found in the expression of CYBA (p22phox) (data not shown), an essential protein not only for NOX2 complex but also for other NADPH oxidases.
The four NOX2 complex subunits perform a similar trend to inferior expression pattern if compared to healthy donors suggesting a coordinated action, and consequently an altered activity of NOX2 complex in AML patients (Figure S3). We decided to characterize the samples based on the catalytic subunit CYBB as a reference for the activity of the complex.
CYBB expression denoted wide variability among AML patients (Figure 1A), which allowed the arrangement of AML samples into three groups: High‐CYBB (n = 64), Medium‐CYBB (n = 234) and Low‐CYBB (n = 78) (Figure 1A). We next questioned the effect of CYBB expression on the clinical characteristics of samples. To test this, an AML data cohort (GSE14468) 34 was used where clinical features are registered (Table S2). No significant variation was found in gender, age, prognosis, karyotype alteration or CEBPA, NPM1, FLT3‐ITD, K‐RAS and EVI1 variables (p‐value >.05). However, the frequency of RUNX1‐RUNX1T1 translocation and mutation of IDH1 and IDH2 increased in Low‐CYBB samples (Figure 1B). Conversely, N‐RAS is more frequent in patients whose CYBB expression levels are high, while FLT3‐TKD presents the same trend, although not reaching statistical significance (Figure 1C). In regards to FAB classification, 3 75% of High‐CYBB cases belong to FAB M4 and M5, whereas almost 60% of Medium‐CYBB and 80% of Low‐CYBB are described as FAB M1 and M2 (Figure 1D). These results suggest CYBB expression levels are associated with FAB classification, RUNX1‐RUNX1T1 translocation, and mutations of IDH1, IDH2, FLT3‐TKD and N‐RAS.
FIGURE 1.
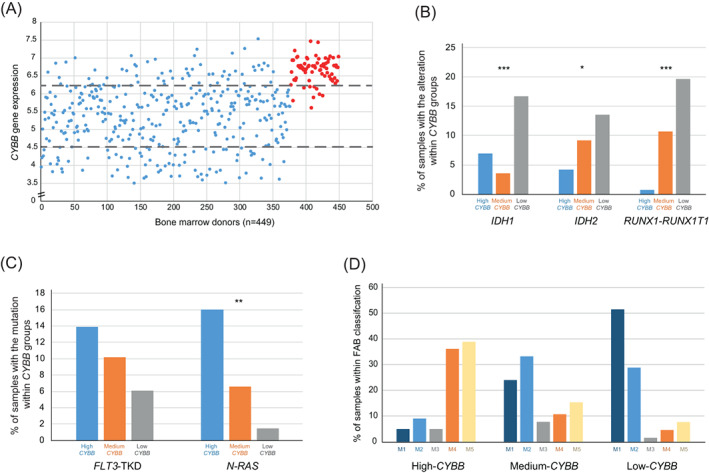
FAB classification, RUNX1‐RUNX1T1 translocation and IDH1, IDH2, FLT3‐TKD and N‐RAS mutations are associated with CYBB in AML patients. (A) CYBB expression levels in bone marrow cells ‐ AML individuals (blue dots) and healthy donors (red dots) ‐ extracted from GSE15061 are shown. Distribution of CYBB‐level groups are also illustrated as a result of cut‐off points drawn in gray: 10th percentile of CYBB expression in healthy donors (upper line) and 25th percentile of AML CYBB expression (lower line). (B) Bar plot representation showing the percentage of GSE14468 samples positive for the mutations IDH1 and IDH2 as well as RUNX1‐RUNX1T1 translocation in each of the CYBB groups. (C) Bar plot representing percentage of GSE14468 samples positive for N‐RAS and FLT3‐TKD mutations in each of the CYBB groups. (D) Bar plot showing percentage of GSE14468 samples classified in each FAB class within CYBB groups. (*) represents p‐value <.05; (**) p‐value <.01 and (***) p‐value <.001 in Chi‐Square test [Color figure can be viewed at wileyonlinelibrary.com]
3.2. 28 metabolic genes show a linked expression with CYBB in AML patients
Given the potential relevance of NOX2 in the regulation of metabolism in AML, 30 , 31 , 32 an analysis of the differential expression of 941 metabolic genes was performed on the GSE15061 dataset. Following the mentioned strategy for group generation (Figure 1A), differentially expressed genes in CYBB, NCF1, NCF2 and NCF4 expression groups were evaluated. High versus Low group comparison for CYBB, NCF1 and NCF2 genes reported mostly shared genes (Figure S4). Indeed, CYBB expression groups comparison resulted in the shortest list of genes and, potentially the one with the greatest clinical applicability. 10 genes were differentially expressed (p‐value <.05) between High‐CYBB and Medium‐CYBB and 35 genes were distinctly expressed (p‐value <.05) in High‐CYBB vs. Low‐CYBB. Of the 35 DEGs identified, 6 genes were excluded since their expression was also altered upon the comparison of the High‐CYBB group with the healthy group. The remaining 29 genes (29G) (Figure S5) are implicated in immune response and metabolic processes such as NADPH oxidase activity, glucose metabolism, OXPHOS, fatty acid biosynthesis or key metabolism regulation routes such as the p53 pathway (Figure S6). In particular, NCF2, HK3, IFI30 and FBP1 appeared even more downregulated than CYBB in Low‐CYBB versus High‐CYBB groups (FC values of 19.6, 14.0, 9.2 and 6.8, respectively). Discriminant analysis corroborated the ability of the 29G signature to separate CYBB groups (Figure S7). Moreover, 29G‐pairwise correlation showed a direct relationship between all genes except PXDN which presented an inverse association (Figure 2A). Said correlation was confirmed in validation datasets (Figure S8). Altogether, we have obtained a set of 28 genes whose expression is correlated with CYBB, which could reflect a common system of regulation.
FIGURE 2.
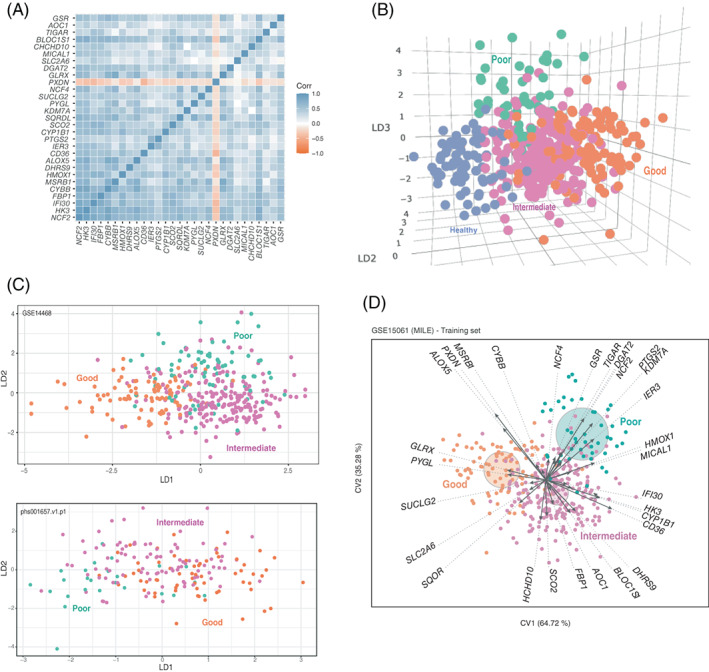
29G is a correlated gene signature that separates AML patients on the basis of prognosis. (A) Pearson's correlation pairwise coefficients (from −1 to 1) of 29G GSE15061 samples were plotted. (B) Dot representation of GSE15061 samples in the three LDA coordinates. LD1, LD2 and LD3 correspond to the discriminant functions obtained in the LDA. (C) Scatter plot data from LDA prognosis group separation is shown for two validation datasets: GSE14468 and phs001657.v1.p1. LD1 and LD2 correspond to the discriminant functions obtained in the LDA. (D) Canonical Biplot representation of the different prognosis samples (good: orange filled dots; intermediate: pink filled dots; and poor: green filled dots) of GSE15061 on the axes 1–2. Discriminant genes (arrows) and confidence circles of each prognosis subtype distribution are plotted based on univariate Student t‐tests to perform post hoc analysis of each gene [Color figure can be viewed at wileyonlinelibrary.com]
3.3. The 29 correlated genes constitute a signature for AML patient prognosis
Multiple gene signatures have shown potential for AML prognosis. 8 , 9 , 10 , 11 Therefore, we tested the prognostic value of 29G. Discriminant analyses showed that 29G efficiently separates healthy and AML samples according to their prognostic group (Figures 2B and S9). The first discriminant axis, LD1, was able to separate good from intermediate samples (p‐value <.001), intermediate from poor samples (p‐value <.001) and good from poor samples (p‐value <.001). The second discriminant dimension, LD2, could separate intermediate from poor (p‐value <.001) and good from poor (p‐value <.001) but was not effective in distinguishing between good and intermediate samples (p‐value >.05).
3.4. 29 gene‐prognosis efficacy validation
To validate our findings, we used five additional independent datasets including 862 samples classified according to ELN2010 (GSE10358, 35 GSE14468, 34 GSE68833 36 ) and 510 samples classified according to ELN2017 (phs001657.v1.p1 38 and GSE165656 37 ), that corroborated the efficacy of 29G to separate samples into prognosis groups, with overall accuracies of 78%, 80%, 75%, 59.2% and 82.7% respectively (Figure 2C and Figure S10). Of note, individuals with acute promyelocytic leukemia (APL) – an example of a good prognosis ‐ showed the greatest pattern of separation, indicating a differential 29G expression profile (first panel of Figure S10).
To further validate our gene panel, we compared its performance with two different panels previously reported. 9 , 11 The percentages of correct classification were very similar among the three gene panels, which strongly supports the robustness of 29G (illustrated for training dataset in Figure S11 and described for validation datasets in Table S3). We noticed that the accuracy of 29G classifying patients belonging to the poor prognosis group tended to be higher than with the other gene panels (76.1% of correct classification with 29G compared to 60.9 and 54.3 for Li et al. 9 and Ng et al. 11 respectively, in the training dataset).
As it can be analyzed from Figures 2D and S12, some genes of 29G excelled especially on determining prognosis. IFI30, CD36, HK3 and CYP1B1 appear downregulated in samples with a good prognosis; in contrast, PXDN and ALOX5 exhibit high expression levels in the same samples. Regarding intermediate prognosis samples, low expression levels of PXDN and ALOX5 on par with upregulation of CD36, BLOC1S1, FBP1 and SCO2 are characteristic. Finally, decreased SQOR and augmented DAGT2 denote a poor patient prognosis.
3.5. Low expression index of the 29 genes is linked with higher OS and EFS
We next investigated whether 29G expression was linked to the Overall Survival (OS) and Event Free Survival (EFS) of AML patients. To test this, we used 260 samples from GSE10358, where OS and EFS information is available. 36
We elaborated an expression index (EI) described as a linear combination of the expression of 29G to distinguish AML patients in terms of OS or EFS. A Receiver Operating Characteristic (ROC) curve was computed to establish the optimal cut‐off point of the diagnostic EI to distinguish between Low‐ and High‐risk survival samples. The cut‐off point was registered as −4.307, which showed a sensitivity of 0.826, a specificity of 0.887 and an area under the ROC curve (AUC) of 0.916 (95% CI: 0.879–0.953) (Figure 3A). EI sample segregation reported 83 cases in the Low‐Index value group (median OS: 34.4 48.0 months) with the remaining 177 cases in the High‐Index value group (median OS: 17.0 33.0 months). Survival curves derived from EI groups showed considerably significant differences in OS (Figure 3B) and EFS (Figure 3C) between Low‐ and High‐Index groups of AML samples (p‐value <.001 in both cases).
FIGURE 3.
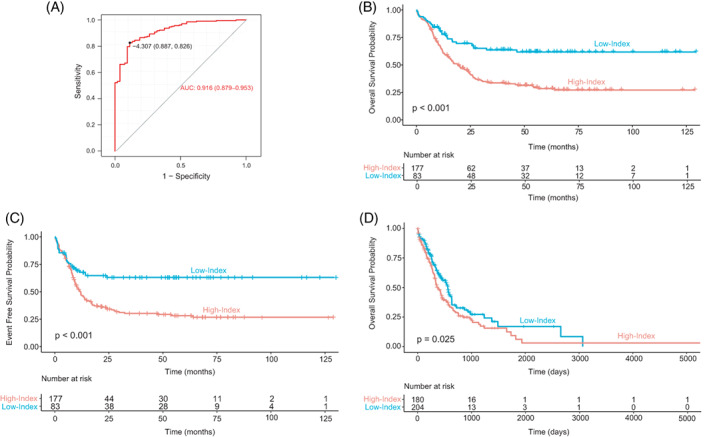
Patients with a low expression index exhibit greater OS. (A) Representation of ROC curve assessment for establishing an appropriate prognosis discrimination cut‐off point for EI in training dataset GSE10358. (B) Kaplan–Meier overall survival (OS) curves of EI groups (High‐Index and Low‐Index) in 260 samples from the GSE10358 dataset. (C) Kaplan–Meier event‐free survival (EFS) curve representation of EI groups (High‐Index and Low‐Index) from 260 samples of the GSE10358 dataset. (D) Kaplan–Meier overall survival (OS) curves of EI groups (High‐Index and Low‐Index) in 429 samples from the phs001657.v1.p1dataset [Color figure can be viewed at wileyonlinelibrary.com]
OS data corresponding to 384 samples from phs001657.v1.p1 was used for validation. EI sample segregation reported 204 cases in the Low‐Index value group (median OS: 329.5 385.3 days) with the remaining 180 cases in the High‐Index value group (median OS: 319 390 days). The Low‐Index group of patients have significantly better survival (Figure 3D), demonstrating that such an EI could reliably report survival, and reinforcing the relevance of 29G. Consistent results were found in the GSE14468 dataset (data not shown).
In view of the increased population with high CYBB levels observed in samples M4 and M5, we wonder whether 29G would be equally prognostic in different FAB groups. Therefore, we segregated GSE10358 into two groups based on their FAB classification: M0 to M3 formed a group while M4 and M5 grouped together. Replicating what was seen in the whole population, Low‐Index samples demonstrated longer survival, further the differences in survival between Low‐Index and High‐Index are magnified for patients in the M4 and M5 groups (Figure S13). The same results were observed in the GSE14468 dataset (data not shown).
Similarly, we tested if 29G was useful for OS prediction within young and older patients by establishing 60 years old as the cut‐off point for constituting two groups. Although no differences were found when using phs001657.v1.p1 (data not shown), 29G could distinguish OS in young and older patients of the GSE10358 dataset (Figure S14), supporting the interest of further analyzing this issue in the future.
3.6. 29G complements ELN prognosis classification
Patients included within the intermediate group present a very variable response to therapy and a high degree of relapse, which makes risk‐stratification and treatment of this group enormously challenging. 42 In fact, some authors propose dividing the intermediate group into different subgroups. 43 Bearing this in mind we decided to analyze whether 29G could differentiate OS within the intermediate group. Application of a hierarchical k‐means clustering technique based on 29G expression to intermediate samples resulted in a separation into four differentiated clusters (Figure 4A). Notably, said clusters showed differences in OS and EFS Kaplan–Meier curves (Figure 4B, C). These results were corroborated in the phs001657.v1.p1 validation dataset, where 29G can also dissect four different OS groups within the intermediate group (Figure 4D). Therefore, 29G could complement ELN2017 classification by stratifying patients within the intermediate group.
FIGURE 4.
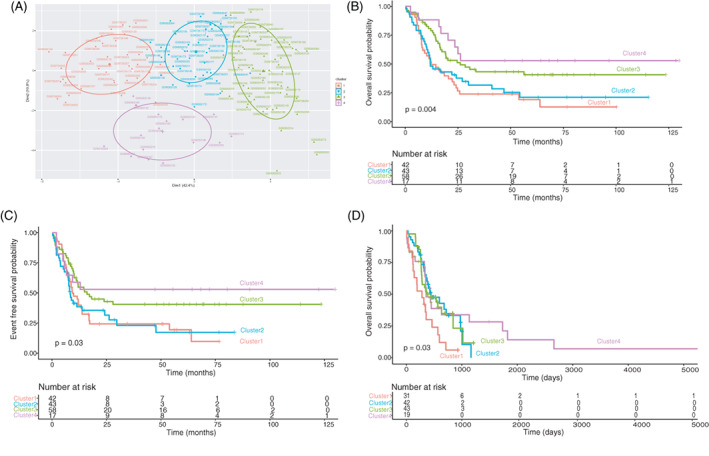
29G based cluster formation in intermediate samples generates four clusters that show differences in OS and EFS. The ability of 29G to discriminate different groups among the intermediate prognosis group was tested on GSE10358 samples (n = 160) and phs001657.v1.p1 (n = 135). (A) PCA representation of the four clusters generated by hkmeans emanated from 29G for intermediate samples of GSE10358 dataset. (B) Kaplan–Meier overall survival (OS) curves of the clusters shown in panel A. Time is expressed in months. (C) Kaplan–Meier event free survival (EFS) curves of the clusters shown in panel A. Time is expressed in months. (D) Kaplan–Meier overall survival (OS) curves of the clusters constituted from intermediate samples of phs001657.v1.p1 following the same procedure as in the training. Time is expressed in days [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
Given the complexity of AML, a great effort has been made by the ELN experts to provide a reliable system of patient classification, which is broadly accepted. 4 , 5 Screening of cytogenetic alterations and some recurrent mutations (NPM1, CEBPA, FLT3, RUNX1, TP53 and ASXL1) are the main factors in the latest version of this scheme. 4 However, a considerable percentage of AML patients present a normal karyotype, and the relevance in the prognosis of some recurrent mutations (DNMT3A, IDH1, IDH2) is yet unknown. 4 High throughput DNA sequencing techniques hold the potential to uncover gene panels with prognostic value for AML. 8 , 9 , 10 , 11 Such an approach could be applied indiscriminately to all patients, including those with a normal karyotype and not bearing mutations, and therefore it could be an interesting complement to ELN guidelines.
ROS has evolved from being non‐desirable by‐products of cellular reactions to their consolidated role as essential secondary messengers. In this progress, NADPH oxidases emerge from the dark as the only cellular system whose main responsibility is ROS production. 44 NOX2, the first described isoform, stands out in the hematopoietic system for its immune function, though it may also be involved in hematopoietic differentiation, 45 and leukemia. 46 , 47
Here we have focused on two distinctive features of AML cells, the relevance of NOX2 as a source of ROS, 30 and the importance of metabolism rewiring for leukemic cells. 22 We show that AML blast cells present a reduced expression of CYBB compared with their sane counterparts. This notion is in agreement with a recent publication. 48 The use of specific gene signatures for the classification of AML is supported by previous studies. 8 , 9 , 11 Here, we present a list of 29 genes sharing no commonalities with those previous panels, whose expression effectively differentiates AML prognosis groups. The performance of 29G and that of the gene panels described by Li et al. 9 and Ng et al. 11 was very similar in six different datasets, further validating our results.
Even more important than the ability of 29G for patient classification, is its predictive value regarding patient survival. By studying the expression data of 29G, we provide a predictive function (29G EI) to calculate the OS of AML patients, in which a low 29G EI correlates with a better prognosis. Besides the possibility of applying 29G to all patients regardless of whether they show genetic alterations or not, a great asset of 29G is its potential to dissect patients belonging to the intermediate group according to their OS. Risk stratification and treatment decisions within the intermediate group are enormously challenging, 42 and 29G provides the possibility of improving the stratification of these patients. All these features make 29G an interesting tool that could effectively complement the current ELN classification system.
In contrast with some previous reports, 8 , 9 but in line with Ng et al,11 the genes belonging to 29G reflect a genetic program related to the regulation of a biological process important for AML cells. 29G are implicated in immune response and metabolic processes such as NADPH oxidase activity, glucose metabolism, oxidative phosphorylation, fatty acid biosynthesis or key metabolism regulation routes such as the p53 pathway. Thus, besides the predictive value of our panel, 29G also highlights NOX2 as well as metabolism as a suitable therapeutic target. In line with this notion, it has been shown before that the use of a NOX2 inhibitor reduces relapse risk in AML patients. 49 Moreover, it seems that the success of arsenic in the treatment of APL depends on NOX activity. 50 , 51 Interestingly our results show that APL patients display a differential 29G expression profile. We surmise that metabolic rewiring may vary among AML prognosis groups, presenting a possibility to design specific treatments based on the NOX2‐metabolism axis.
Furthermore, our results reinforce the relevance of NOX2 in the control of AML metabolism, as outlined by other authors. 27 , 31 , 52 Our data suggest that NOX2 could induce glycolytic metabolism in AML by increasing glucose transport (SLC2A6), glycolytic rate (HK3), glucose mobilization from glycogen (PGYL), and gluconeogenesis (FBP1). Our analysis did not show the correlation between CYBB and lactate dehydrogenase, however, we found a strong correlation between CYBB with several genes involved in mitochondrial metabolism (SCO2, SQOR, SUCLG2, CHCHD10), suggesting that CYBB would promote the complete oxidation of glucose in AML cells.
The correlation between the expression of CYBB and the p53 pathway genes (SCO2, TIGAR), also called our attention. The activation of the p53 pathway could also contribute to enhancing OXPHOS metabolism in the High‐CYBB AML group. 53 We can surmise that these patients would show a high level of intracellular ROS, derived from NOX2 complex activity and mitochondrial metabolism. The enhancement in the expression of enzymes involved in redox homeostasis, especially with glutathione reduction (GLRX, GSR) and the p53 target TIGAR 54 could allow AML cells to cope with a high level of oxidative stress.
The implications of coexisting molecular alterations and their effects on disease classification and treatment are fundamental to developing more effective combination therapies. In the same line, we analyzed whether CYBB levels were correlated with any of the recurrent genetic alterations observed in AML. Constitutive activation mutations in FLT3 receptor (FLT3‐ITD and FLT3‐TKD) 55 and RAS 56 occur frequently in AML. Both FLT3 and RAS mutations increase NADPH oxidase ROS production in AML. 56 Interestingly, our results show a correlation between high CYBB levels and mutations in FLT3‐TKD and RAS. FLT3 and RAS activating mutations would lead to high ROS production through the NOX2 complex, which may confer an advantage of increased proliferation with respect to other AML clones.
Mutations in IDH1/2 and the RUNX1‐RUNX1T1 chromosomal rearrangements also appear with a high frequency in AML. 4 Our results show an inverse correlation between these genetic alterations and CYBB levels. On the other hand, it seems that low CYBB expression is linked to a less differentiated phenotype, as it could be expected, given the requirement of NOX2 for the innate immunity mediated by terminal differentiated myeloid cells. In agreement with this notion it has been described that NOX2 complex is highly expressed in M4/M5 but not in M1/M2 AML samples. 57 , 58 This could be explained by the requirement of ROS for triggering myeloid differentiation. 59 , 60 The RUNX1‐RUNX1T1 translocation is found in 40% of the M2 FAB group. 61 Therefore, it seems that a less differentiated phenotype correlates with both a low CYBB expression and with the occurrence of RUNX1‐RUNX1T1.
In summary, here we present a panel of metabolic genes correlated to CYBB (29G) as a tool for AML stratification and survival prediction, indicating metabolism as a therapeutic target. Integrating all genetic abnormalities into a prognostic scheme is becoming increasingly difficult due to the vast array of such alterations and their numerous possible combinations. The use of gene panels in prognosis could be easily implemented into clinical practice and holds the possibility to improve AML risk stratification. The robustness of the 29G signature we describe is supported by the analyses of six different AML cohorts with a total of 1821 samples, suggesting a great potential for future clinical applications.
CONFLICT OF INTEREST
The authors declare no potential competing interests.
AUTHOR CONTRIBUTIONS
Carla Ijurko, Nerea González‐García and Ángel Hernández‐Hernández conception and design. Carla Ijurko and Nerea González‐García collected and analyzed data. Nerea González‐García and Purificación Galindo‐Villardón provided statistical support. Carla Ijurko and Ángel Hernández‐Hernández provided resources. Carla Ijurko and Ángel Hernández‐Hernández wrote the manuscript. All authors reviewed and edited the manuscript.
Supporting information
Data S1. Supporting Information.
ACKNOWLEDGMENTS
We thank L. Stockdale for reviewing the English version of this manuscript.
Ijurko C, González‐García N, Galindo‐Villardón P, Hernández‐Hernández Á. A 29‐gene signature associated with NOX2 discriminates acute myeloid leukemia prognosis and survival. Am J Hematol. 2022;97(4):448-457. doi: 10.1002/ajh.26477
Funding information This work was supported by the Regional Government of Castile & Leon (SA077P20) and by the Spanish Government (PID2020‐117692RB‐I00) grants received by Dr Hernández‐Hernández. Carla Ijurko was the recipient of pre‐doctoral fellowships from the Regional Government of Castile and Leon, Spain and ERDF funds.
DATA AVAILABILITY STATEMENT
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/. Gene expression data of one extra validation dataset (phs001657.v1.p1) were obtained from Tyner et al. 38
REFERENCES
- 1. Khwaja A, Bjorkholm M, Gale RE, et al. Acute myeloid leukaemia. Nat Rev Dis Primers. 2016;2:16010. doi: 10.1038/nrdp.2016.10 [DOI] [PubMed] [Google Scholar]
- 2. O'Donnell MR, Abboud CN, Altman J, et al. Acute myeloid leukemia. JNCCN J Natl Compr Cancer Netw. 2012;10(8):984‐1021. doi: 10.6004/jnccn.2012.0103 [DOI] [PubMed] [Google Scholar]
- 3. Canaani J, Beohou E, Labopin M, et al. Impact of FAB classification on predicting outcome in acute myeloid leukemia, not otherwise specified, patients undergoing allogeneic stem cell transplantation in CR1: an analysis of 1690 patients from the acute leukemia working party of EBMT. Am J Hematol. 2017;92(4):344‐350. doi: 10.1002/ajh.24640 [DOI] [PubMed] [Google Scholar]
- 4. Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424‐447. doi: 10.1182/blood-2016-08-733196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453‐474. doi: 10.1182/blood-2009-07-235358 [DOI] [PubMed] [Google Scholar]
- 6. Estey EH. Acute myeloid leukemia: 2019 update on risk‐stratification and management. Am J Hematol. 2018;93(10):1267‐1291. doi: 10.1002/ajh.25214 [DOI] [PubMed] [Google Scholar]
- 7. Herold T, Rothenberg‐Thurley M, Grunwald VV, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34(12):3161‐3172. doi: 10.1038/s41375-020-0806-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullinger L, Dohner K, Bair E, et al. New England journal medicine. N Engl J Med. 2004;351(4):2605‐2615. [Google Scholar]
- 9. Li Z, Herold T, He C, et al. Identification of a 24‐gene prognostic signature that improves the european LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31(9):1172‐1181. doi: 10.1200/JCO.2012.44.3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marcucci G, Yan P, Maharry K, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven‐gene score. J Clin Oncol. 2014;32(6):548‐556. doi: 10.1200/JCO.2013.50.6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng SWK, Mitchell A, Kennedy JA, et al. A 17‐gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433‐437. doi: 10.1038/nature20598 [DOI] [PubMed] [Google Scholar]
- 12. Warnat‐Herresthal S, Perrakis K, Taschler B, et al. Scalable prediction of acute myeloid leukemia using high‐dimensional machine learning and blood transcriptomics. iScience. 2020;23(1):100780. doi: 10.1016/j.isci.2019.100780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27‐47. doi: 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309‐314. doi: 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 15. Herst PM, Howman RA, Neeson PJ, Berridge MV, Ritchie DS. The level of glycolytic metabolism in acute myeloid leukemia blasts at diagnosis is prognostic for clinical outcome. J Leukoc Biol. 2011;89(1):51‐55. doi: 10.1189/jlb.0710417 [DOI] [PubMed] [Google Scholar]
- 16. Song K, Li M, Xu X, et al. Resistance to chemotherapy is associated with altered glucose metabolism in acute myeloid leukemia. Oncol Lett. 2016;12(1):334‐342. doi: 10.3892/ol.2016.4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watson A, Riffelmacher T, Stranks A, et al. Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Dis. 2015;1(1):1‐10. doi: 10.1038/cddiscovery.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Culp‐Hill R, D'Alessandro A, Pietras EM. Extinguishing the embers: targeting AML metabolism. Trends Mol Med. 2020;27(4):332‐344. doi: 10.1016/j.molmed.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poulain L, Sujobert P, Zylbersztejn F, et al. High mTORC1 activity drives glycolysis addiction and sensitivity to G6PD inhibition in acute myeloid leukemia cells. Leukemia. 2017;31(11):2326‐2335. doi: 10.1038/leu.2017.81 [DOI] [PubMed] [Google Scholar]
- 20. Gentric G, Mieulet V, Mechta‐Grigoriou F. Heterogeneity in cancer metabolism: new concepts in an old field. Antioxid Redox Signal. 2017;26(9):462‐485. doi: 10.1089/ars.2016.6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 2013;104(3):275‐281. doi: 10.1111/cas.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Castro I, Sampaio‐Marques B, Ludovico P. Targeting metabolic reprogramming in acute myeloid leukemia. Cell. 2019;8(9):967. doi: 10.3390/cells8090967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchez‐Sanchez B, Gutierrez‐Herrero S, Lopez‐Ruano G, et al. NADPH oxidases as therapeutic targets in chronic myeloid leukemia. Clin Cancer Res. 2014;20(15):4014‐4025. doi: 10.1158/1078-0432.CCR-13-3044 [DOI] [PubMed] [Google Scholar]
- 24. Pérez‐Fernández A, López‐Ruano G, Prieto‐Bermejo R, et al. SHP1 and SHP2 inhibition enhances the pro‐differentiative effect of phorbol esters: an alternative approach against acute myeloid leukemia. J Exp Clin Cancer Res. 2019;38(1):80. doi: 10.1186/s13046-019-1097-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panieri E, Santoro M M. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death & Disease. 2016;7:(6):e2253–e2253. doi: 10.1038/cddis.2016.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Naughton R, Quiney C, Turner SD, Cotter TG. Bcr‐Abl‐mediated redox regulation of the PI3K/AKT pathway. Leukemia. 2009;23(8):1432‐1440. doi: 10.1038/leu.2009.49 [DOI] [PubMed] [Google Scholar]
- 27. Lu W, Hu Y, Chen G, et al. Novel role of NOX in supporting aerobic glycolysis in cancer cells with mitochondrial dysfunction and as a potential target for cancer therapy. PLoS Biol. 2012;10(5):e1001326. doi: 10.1371/journal.pbio.1001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prata C, Maraldi T, Fiorentini D, Zambonin L, Hakim G, Landi L. Nox‐generated ROS modulate glucose uptake in a leukaemic cell line. Free Radic Res. 2008;42(5):405‐414. doi: 10.1080/10715760802047344 [DOI] [PubMed] [Google Scholar]
- 29. Shanmugasundaram K, Nayak BK, Friedrichs WE, Kaushik D, Rodriguez R, Block K. NOX4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance. Nat Commun. 2017;8(1):1‐15. doi: 10.1038/s41467-017-01106-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hole PS, Zabkiewicz J, Munje C, et al. Overproduction of NOX‐derived ROS in AML promotes proliferation and is associated with defective oxidative stress signaling. Blood. 2013;122(9):3322‐3330. doi: 10.1182/blood-2013-04-491944 [DOI] [PubMed] [Google Scholar]
- 31. Robinson AJ, Hopkins GL, Rastogi N, et al. Reactive oxygen species drive proliferation in acute myeloid leukemia via the glycolytic regulator PFKFB3. Cancer Res. 2020;80(5):937‐949. doi: 10.1158/0008-5472.CAN-19-1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinson AJ, Davies S, Darley RL, Tonks A. Reactive oxygen species rewires metabolic activity in acute myeloid leukemia. Front Oncol. 2021;11:632623. doi: 10.3389/fonc.2021.632623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mills KI, Kohlmann A, Williams PM, et al. Microarray‐based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood. 2009;114(5):1063‐1072. doi: 10.1182/blood-2008-10-187203 [DOI] [PubMed] [Google Scholar]
- 34. Wouters BJ, Löwenberg B, Erpelinck‐Verschueren CAJ, van Putten WLJ, Valk PJM, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088‐3091. doi: 10.1182/blood-2008-09-179895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomasson MH, Xiang Z, Walgren R, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111(9):4797‐4808. doi: 10.1182/blood-2007-09-113027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Genomic and Epigenomic landscapes of adult De novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059‐2074. doi: 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abbas HA, Mohanty V, Wang R, et al. Decoupling lineage‐associated genes in acute myeloid leukemia reveals inflammatory and metabolic signatures associated with outcomes. Front Oncol. 2021;11:705627. doi: 10.3389/fonc.2021.705627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tyner J, Tognon C, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526‐531. doi: 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vicente‐Villardón JL. Una alternativa a los métodos factoriales clásicos basada en una generalización de los métodos biplot.[PhD Thesis]. Universidad de Salamanca; 1992. [Google Scholar]
- 40. Díaz‐Faes AA, Costas R, Purificación Galindo M, Bordons M. Unravelling the performance of individual scholars: use of canonical Biplot analysis to explore the performance of scientists by academic rank and scientific field. J Informet. 2015;9(4):722‐733. doi: 10.1016/j.joi.2015.04.006 [DOI] [Google Scholar]
- 41. R Core Team. R: A Language and Environment for Statistical Computing . R Foundation for Statistical Computing. Austria; 2019. https://www.R-project.org/ [Google Scholar]
- 42. Moarii M, Papaemmanuil E. Classification and risk assessment in AML: integrating cytogenetics and molecular profiling. Hematology. 2017;2017(1):37‐44. doi: 10.1182/asheducation-2017.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mroźek K, Marcucci G, Nicolet D, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30(36):4515‐4523. doi: 10.1200/JCO.2012.43.4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prieto‐Bermejo R, Romo‐González M, Pérez‐Fernández A, Ijurko C, Hernández‐Hernández Á. Reactive oxygen species in haematopoiesis: Leukaemic cells take a walk on the wild side. J Exp Clin Cancer Res. 2018;37(1):1‐18. doi: 10.1186/s13046-018-0797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prieto‐Bermejo R, Romo‐González M, Pérez‐Fernández A, García‐Tuñón I, Sánchez‐Martín M, Hernández‐Hernández Á. Cyba‐deficient mice display an increase in hematopoietic stem cells and an overproduction of immunoglobulins. Haematologica. 2021;106(1):142‐153. doi: 10.3324/haematol.2019.233064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adane B, Ye H, Khan N, et al. The hematopoietic oxidase NOX2 regulates self‐renewal of leukemic stem cells. Cell Rep. 2019;27(1):238‐254.e6. doi: 10.1016/j.celrep.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reddy MM, Fernandes MS, Salgia R, Levine RL, Griffin JD, Sattler M. NADPH oxidases regulate cell growth and migration in myeloid cells transformed by oncogenic tyrosine kinases. Leukemia. 2011;25(2):281‐289. doi: 10.1038/leu.2010.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nehme A, Dakik H, Picou F, et al. Horizontal meta‐analysis identifies common deregulated genes across AML subgroups providing a robust prognostic signature. Blood Adv. 2020;4(20):5322‐5335. doi: 10.1182/bloodadvances.2020002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brune M, Castaigne S, Catalano J, et al. Improved leukemia‐free survival after postconsolidation immunotherapy with histamine dihydrochloride and interleukin‐2 in acute myeloid leukemia: results of a randomized phase 3 trial. Blood. 2006;108(1):88‐96. doi: 10.1182/blood-2005-10-4073 [DOI] [PubMed] [Google Scholar]
- 50. Wang J, Li L, Cang H, Shi G, Yi J. NADPH oxidase‐derived reactive oxygen species are responsible for the high susceptibility to arsenic cytotoxicity in acute promyelocytic leukemia cells. Leuk Res. 2008;32(3):429‐436. doi: 10.1016/j.leukres.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 51. Chou WC, Jie C, Kenedy AA, Jones RJ, Trush MA, Dang CV. Role of NADPH oxidase in arsenic‐induced reactive oxygen species formation and cytotoxicity in myeloid leukemia cells. Proc Natl Acad Sci USA. 2004;101(13):4578‐4583. doi: 10.1073/pnas.0306687101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baillet A, Hograindleur MA, El Benna J, et al. Unexpected function of the phagocyte NADPH oxidase in supporting hyperglycolysis in stimulated neutrophils: key role of 6‐phosphofructo‐2‐kinase. FASEB J. 2017;31(2):663‐673. doi: 10.1096/fj.201600720r [DOI] [PubMed] [Google Scholar]
- 53. Zhang XD, Qin ZH, Wang J. The role of p53 in cell metabolism. Acta Pharmacol Sin. 2010;31(9):1208‐1212. doi: 10.1038/aps.2010.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53‐inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107‐120. doi: 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- 55. Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33(2):299‐312. doi: 10.1038/s41375-018-0357-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sillar JR, Germon ZP, De Iuliis GN, Dun MD. The role of reactive oxygen species in acute myeloid leukaemia. Int J Mol Sci. 2019;20(23):6003. doi: 10.3390/ijms20236003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aurelius J, Thorén F, Akhiani A, et al. Monocytic AML cells inactivate antileukemic lymphocytes: role of NADPH oxidase/gp91(phox) expression and the PARP‐1/PAR pathway of apoptosis. Blood. 2012;119(24):5832‐5837. doi: 10.1182/blood-2011-11-391722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dakik H, El DM, Leclerc J, et al. Characterization of NADPH oxidase expression and activity in acute myeloid leukemia cell lines: a correlation with the differentiation status. Antioxidants. 2021;10(3):498. doi: 10.3390/antiox10030498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sardina JL, López‐Ruano G, Sánchez‐Abarca LI, et al. P22phox‐dependent NADPH oxidase activity is required for megakaryocytic differentiation. Cell Death Differ. 2010;17(12):1842‐1854. doi: 10.1038/cdd.2010.67 [DOI] [PubMed] [Google Scholar]
- 60. Ludin A, Gur‐Cohen S, Golan K, et al. Reactive oxygen species regulate hematopoietic stem cell self‐renewal, migration and development, as well as their bone marrow microenvironment. Antioxid Redox Signal. 2014;21(11):1605‐1619. doi: 10.1089/ars.2014.5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saitoh K, Miura I, Ohshima A, et al. Translocation (8;12;21)(q22.1;q24.1;q22.1): a new masked type of t(8;21)(q22;q22) in a patient with acute myeloid leukemia. Cancer Genet Cytogenet. 1997;96(2):111‐114. doi: 10.1016/S0165-4608(96)00320-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information.
Data Availability Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/. Gene expression data of one extra validation dataset (phs001657.v1.p1) were obtained from Tyner et al. 38


