Significance
Evolutionarily conserved protein kinase R (PKR) is a powerful mediator of antiviral responses. Enigmatically, PKR activity is disabled by porcine reproductive and respiratory syndrome virus (PRRSV) during infection, while the virus still allows the phosphorylation of its substrate eIF2α. Here, we show that this selective targeting of PKR signaling down-regulates cellular inflammatory responses. Mechanistically, PRRSV takes advantage of virus-induced stress granules (SGs) and reprograms the antiviral SGs into a proviral platform by utilizing viral replicase protein nsp1β to co-opt G3BP1 to inhibit PKR activation. These findings shed light on the mechanisms of PRRSV-mediated suppression of cellular inflammation, which might help in rationale design of vaccine, and reveal a unique strategy for viral evasion of PKR and SG restriction.
Keywords: PKR, stress granules, PRRSV, inflammation, nsp1β
Abstract
Protein kinase R (PKR) is a critical host restriction factor against invading viral pathogens. However, this molecule is inactivated in the cells infected with porcine reproductive and respiratory syndrome virus (PRRSV), an economically devastating pathogen to the world swine industry. Here, we report that this event is to suppress cellular inflammation and is mediated by the viral replicase protein nsp1β. We show that nsp1β is a stress-responsive protein, enters virus-induced stress granules (SGs) during infection, and repurposes SGs into a proviral platform, where it co-opts the SG core component G3BP1 to interact with PKR in a regulated manner. RNA interference silencing of G3BP1 or mutation of specific nsp1β residues (VS19GG) can abolish the antagonization of PKR activation. The viral mutant carrying the corresponding mutations induces elevated level of PKR phosphorylation and pronounced production of inflammatory cytokines (e.g., tumor necrosis factor-α, interleukin [IL]-6, and IL-8), whereas small-interfering RNA knockdown of PKR or treatment with C16, a PKR inhibitor, blocks this effect. Thus, PRRSV has evolved a unique strategy to evade PKR restriction to suppress host inflammatory responses.
Eukaryotic cells employ an amazingly wide variety of intracellular defenses to restrict invading viral pathogens. At the virus–host interface, one such critical host player is the protein kinase R (PKR), a 551-amino acid-long serine threonine kinase (1, 2). This protein was initially discovered as a key component of interferon (IFN)-stimulated genes (3, 4) and later identified as a pattern-recognition receptor that specifically recognizes double-stranded RNAs (dsRNA) (5, 6), a common intermediate produced during infections of both DNA and RNA viruses. Structurally, PKR is composed of two tandem evolutionarily conserved dsRNA-binding motifs (DRBMs) in the N terminus, an effector/catalytic kinase domain (KD) in the C terminus, and a middle flexible linker region connecting the two domains (2, 6, 7). Binding of DRBMs to viral dsRNAs or 5′ triphosphate RNAs induces conformational changes of PKR, leading to autophosphorylation of its KD (8, 9). Alternatively, PKR can be activated via heterodimerization by an endogenous dsRNA binding protein, namely PKR-activating protein (PACT) (8).
The activated PKR phosphorylates the α-subunit of eukaryotic translation initiation factor 2 (eIF2α) to induce general translation arrest and to stimulate formation of stress granules (SGs) (7, 10), a kind of nonmembrane bound cytoplasmic aggregate that mainly include stalled translation initiation complexes, together with nucleating factors like Ras GTPase-activating protein-binding protein 1 (G3BP1), T cell intracytoplasmic antigen 1 (TIA1), and many others (11, 12). Noticeably, SGs are increasingly emerging as a critical signaling platform for antiviral immunity and a key regulator of PKR activation (10, 13). Additionally, the activated PKR critically mediates the induction of interferons and modulates cellular inflammatory responses to build up an antiviral state by inducing sustained activation of NF-κB via activating IκB kinase (IKK) and others (14–16).
To overcome PKR restriction, many viruses have evolved various strategies (2). They specify viral products to either interact directly with PKR (17–19), or sequester and degrade dsRNA (20, 21), or target host PKR regulators, such as ADAR1, P58IPK, and PACT (22–24). The focus of this report is the interplay of PKR signaling with porcine reproductive and respiratory syndrome virus (PRRSV), an economically devastating swine pathogen and a positive-stranded RNA virus in the family Arteriviridae of the order Nidovirales (25, 26). PRRSV has remained a major threat to the worldwide swine production ever since its first emergence in late 1980s (27). One of the most prominent features of PRRSV is subversion of host immunity, as manifested by crippled induction of interferon and inflammation, leading to inefficient cytotoxic T cell responses (26, 28, 29). Consequently, the currently available PRRSV vaccines are unable to induce sterilizing immunity (30), and the viral persistence in turn facilitates recombination among different strains and allows selection of immune escape mutants in the field (30, 31).
Interestingly, the activity of PKR is disabled in PRRSV-infected cells while the virus still allows the phosphorylation of its substrate eIF2α (32–34). We have previously reported that the eIF2α phosphorylation is conducive to utilization of ATF4 for viral replication (34). Here, we show that the primary purpose of this selective targeting of PKR signaling is to suppress the cellular inflammatory responses. Strategically, PRRSV repurposes antiviral SGs into a proviral platform to overcome the PKR restriction. The details are described below.
Results
Identification of PRRSV Replicase Protein nsp1β as a Critical Viral Antagonist of PKR.
To understand the temporal dynamics of PKR phosphorylation, we infected MARC-145 cells, an in vitro supporting cell line, and primary porcine alveolar macrophages (PAMs), the major in vivo target, with PRRSV strain JXwn06. Despite the presence of large amounts of dsRNAs (Fig. 1A and B), the PKR phosphorylation remained at basal level throughout the infection in both cell types (Fig. 1C and D). Moreover, the infected cells were able to inhibit poly(I:C)-induced PKR phosphorylation (Fig. 1E and F, lane 4), whereas treatment with poly(I:C) alone resulted in a sharp increase of phosphorylated PKR (p-PKR) (Fig. 1E and F, lane 2), indicating that the inability of PRRSV to activate PKR is not due to a defect of host cells, but rather attributed to a mechanism exerted by the virus itself.
Fig. 1.
The PRRSV replicase nsp1β is a critically viral antagonist of PKR. (A and B) Production of dsRNAs in MARC-145 and PAMs infected with PRRSV strain JXwn06 at an MOI of 0.1. The cells were stained with antibodies to dsRNA and to PRRSV N protein at 18 hpi and examined by a Nikon A1 confocal microscope, and the images are representative of at three independent experiments. Oil objective: 100×; zoom in 1× (A) or 2× (B). (C and D) Western blot analysis of PKR phosphorylation in PRRSV-infected MARC-145 cells and PAMs with antibodies to phosphorylated-PKR (p-PKR), PKR, β-actin, and N protein. Poly(I:C) (1.5 µg/mL) as a positive control was used to treat MARC-145 cells for 12 h or PAMs for 6 h. (E and F) Effect of PRRSV infection on poly(I:C)-induced PKR phosphorylation. MARC-145 and PAMs were infected or mock-infected with PRRSV at an MOI of 0.1, and at 24 (MARC-145) or 12 hpi (PAMs), the cells were treated with poly(I:C) at a concentration of 1.5 µg/mL for 12 or 6 h before being collected for Western blot analyses. (G) Dose-dependent effect of nsp1β on PKR phosphorylation. HEK-293FT cells seeded in six-well plates were transfected to express increasing amount of HA-nsp1β. At 24 h posttransfection, the cells were treated with poly(I:C) (1.5 µg/mL) for 12 h before being collected for Western blot analysis. The relative band density of p-PKR was normalized to the total PKR and then the loading control β-actin, and then compared to the corresponding mock control, and expressed as p-PKR(+/−).
We subsequently screened the viral proteins for their ability to block PKR activity by transfection, and the replicase protein nsp1β was found to be a critical antagonist (SI Appendix, Fig. S1). Ectopic expression of this protein led to strong inhibition of poly(I:C)-induced PKR phosphorylation, and a dose-dependent activity was evident (Fig. 1G).
Nsp1β Is Sorted into PRRSV-Induced SGs During Infection.
To understand the mechanism of how nsp1β executes its function, we investigated the temporality of nsp1β subcellular location during infection. The time-course studies revealed that nsp1β exhibited a dynamic localization property (Fig. 2A). It emerged around 7 h postinfection (hpi) in the cytoplasm with a diffusive distribution pattern, and then quickly shifted to discrete puncta around 9 hpi, followed by accumulation in the nucleus around 12 hpi (Fig. 2A). At 24 hpi, nsp1β was distributed mostly in the nucleus, but with a substantial portion in the cytoplasm as strikingly discrete puncta (Fig. 2A). Surprisingly, the nsp1β-containing puncta did not colocalize with the viral core replicase proteins nsp2 or nsp9 (Fig. 2B), suggesting that it is not recruited to the viral replication and transcription complex, but rather somewhere else.
Fig. 2.
Nsp1β is sorted into PRRSV-induced SGs during infection. (A) Time-course analysis of G3BP1 and nsp1β distribution. MARC-145 cells on coverslips were infected with PRRSV strain JXwn06 at an MOI of 0.1. At the indicated time points, the cells were stained with antibodies to the indicated protein. (B) Colocalization analysis of nsp1β with nsp2 or nsp9 at 24 hpi. (C) Colocalization analysis of nsp1β with G3BP1 or TIA1 at 24 hpi. (D) Colocalization analysis of G3BP1 with nsp2, nsp9, or dsRNA at 24 hpi. (E) Colocalization analysis of G3BP1 with PRRSV RNAs. MARC-145 cells were infected with PRRSV at an MOI of 0.1, and at 24 hpi, the viral negative- and positive-strand RNA were detected by the RNAscope in situ hybridization, and SGs were stained against G3BP1. The images were acquired by Nikon A1 confocal microscope and processed by imageJ. Oil objective: 100×; zoom in 2×.
The cytoplasmic staining pattern of nsp1β is reminiscent of that of SGs. In line with this hypothesis, PRRSV is known to be capable of inducing stable SGs during infection (33, 35). As predicted, we observed a perfect colocalization relationship between nsp1β and the SG markers G3BP1 and TIA1 in the cytoplasm (Fig. 2C). The SGs emerged at around 8 to 9 hpi and were maintained throughout the infection (Fig. 2A). Meanwhile, the expression and localization dynamics of nsp1β coincided with the SG kinetics (Fig. 2A). However, PRRSV-induced SGs did not colocalize well with the viral replication and transcription complex (Fig. 2D), virus-induced dsRNAs (Fig. 2D), or viral RNAs (Fig. 2E). Thus, the findings reveal a previously unrecognized localization for cytoplasmic nsp1β during infection.
PRRSV nsp1β Is a Stress-Responsive Protein and Colocalizes with PKR in SGs During Infection.
We next investigated how nsp1β enters SG during infection. Since PRRSV prefers to induce a stress environment (e.g., induction of unfolded protein responses, phosphorylation of eIF2α, and so forth) conducive to replication (34), we hypothesized that nsp1β might be a stress-responsive protein. If this is the case, then exposure of nsp1β in an artificial stress condition should allow it to enter SG. Indeed, treatment of MARC-145 cells with dithiothreitol (DTT), an endoplasmic reticulum (ER) stress inducer, readily led to relocation of ectopically expressed nsp1β to SGs (Fig. 3A, Right). In contrast, PRRSV nsp4 and nsp9 were not responsive to this stimulus (Fig. 3A). Treatment with other ER stress inducers (arsenite [Ars] and thapsigargin [TG]) obtained the same result (Fig. 3B). Thus, the sorting of nsp1β into SGs is an intrinsic property of this molecule; it does not require other viral factors, but rather depends on the cellular stress itself.
Fig. 3.
Nsp1β is a stress-responsive protein and colocalizes with PKR in SGs during infection. (A) Colocalization analysis of G3BP1 with nsp1β under condition of stress. MARC-145 cells were transfected to HA-nsp1β, HA-nsp4, or HA-nsp9 at 24 h posttransfection treated with DTT (2 mM) or mock-treated with PBS for 1 h before immunofluorescent antibody with antibodies to HA and G3BP1. (B) The same as A, except that the cells were treated with Ars (0.5 mM, 0.5 h) or TG (200 nM, 1 h). (C) Colocalization analysis of PKR with nsp1β or TIA1 in mock- or PRRSV-infected MARC-145 cells at an MOI of 0.1. At 24 h posttransfection, the cells were stained with antibodies to nsp1β, PKR, and TIA1. (D) Colocalization analysis of PKR with transiently expressed HA-nsp1β in MARC-145 cells treated with different stimuli (PBS, DTT, or Ars). The images were acquired by Nikon A1 confocal microscope and processed by imageJ. Oil objective: 100×; zoom in 2×.
PKR was also relocalized to PRRSV-induced SGs (Fig. 3C). This observation is consistent with previous reports that PKR is a stress-responsive protein and that its activation is dependent on SGs (10, 13, 36). In infected cells, PKR colocalized well with nsp1β and the SG markers, whereas in mock-infected cells it exhibited a diffusive distribution (Fig. 3C). In transfected cells, nsp1β did not colocalize with PKR, but they became colocalized once the cells were stressed with SG inducers (e.g., DTT or Ars) (Fig. 3D). Thus, these findings suggest that SGs are likely the battleground for nsp1β to counteract PKR.
Regulated Interaction between nsp1β and PKR.
It is likely that there is an interaction between nsp1β and PKR. We tested this hypothesis by coimmunoprecipitation assay (co-IP). The antibodies to PKR could readily pull down nsp1β from PRRSV-infected MARC-145 cell lysates (Fig. 4A), and the same was true under the condition of transfection (Fig. 4C, lane 5). The nsp1β-binding region was further mapped by construction of PKR truncation mutants containing only the DRBM or the KD (Fig. 4B). WT PKR and its derivatives (Flag-DRBM and Flag-KD) were expressed either individually or in combination with HA-nsp1β in HEK-293FT cells. The co-IP assay showed that the mutant Flag-DRBM retained the ability to interact with nsp1β to the level of full-length PKR (Fig. 4C, lane 6), whereas the mutant Flag-KD failed to interact (Fig. 4C, lane 7), suggesting that DRBM is the key region for binding to nsp1β.
Fig. 4.
Regulated interaction between nsp1β and PKR. (A) Interaction of nsp1β with PKR in infected cells. MARC-145 cells were mock-infected or infected with PRRSV at an MOI of 0.1. At 36 hpi, the cells were harvested for co-IP analysis with rabbit antibodies to PKR, followed by Western blot with antibodies to PKR, nsp1β and GAPDH. A rabbit isotype antibody used as a control. (B) Diagram of PKR truncation mutants. The individual domains of PKR were tagged with either Flag or TF at the N terminus. (C) Interaction of nsp1β with PKR in transfected cells. HEK-293FT cells were transfected to coexpress HA-nsp1β and Flag-PKR or its derivatives. At 24 h posttransfection, the co-IP assay was performed with rabbit antibodies to Flag, followed by Western blot with mouse antibodies to Flag, HA, or GAPDH. (D) Interaction of nsp1β with PKR in vitro. GST and GST-nsp1β were bacterially expressed purified with glutathione beads and used to pull down TF, TF-PKR, TF-DRBM, and TF-KD from bacterial lysates, followed by Western blot analysis with antibodies to TF. A ponceau staining shows the input. (E) MARC-145 cells were transfected to express Flag-PKR, Flag-DRBM, and Flag-KD either individually or in combination with HA-nsp1β. At 24 h posttransfection, the cells were stained with antibodies to the epitope tags. The images were acquired by Nikon A1 confocal microscope and processed by ImageJ. Oil objective: 60×; zoom in 3×. Arrows indicate co-localization regions.
To test whether the interaction requires any eukaryotic factor, we carried out the in vitro binding assay. The relevant proteins were expressed in a bacterial system. Specifically, PRRSV nsp1β was expressed as a GST fusion protein, whereas PKR or its derivatives (DRBM and KD) were fused to the C terminus of bacterial chaperone trigger factor (TF) to promote protein solubility (Fig. 4B). GST-nsp1β was purified with glutathione beads to pull down PKR or derivatives from bacterial lysates, whereas GST alone served as a negative control (Fig. 4D, lanes 5 to 8). To our surprise, GST-nsp1β was unable to pull down the full-length PKR (TF-PKR) (Fig. 4D, lane 10), but the TF-DRBM mutant was abundantly recovered (Fig. 4D, lane 11). This result is in a stark contrast with that from the above co-IP assay using mammalian cells (Fig. 4C), suggesting that a conformational change of PKR is likely necessary for the interaction. Consistent with this notion, nsp1β showed partial colocalization with Flag-DRBM in the cytoplasm, but not with the full-length Flag-PKR or Flag-KD in transfected cells (Fig. 4E). Together, these results suggest that the nsp1β-PKR interaction is regulated and facilitated by cellular factors during infection.
The SG Component G3BP1 Promotes the nsp1β-PKR Interaction.
As nsp1β was sorted into SGs during PRRSV infection, the induction of SG or its core components, such as G3BP1 and TIA1, might promote the nsp1β–PKR interaction. In agreement with this idea, treatment with the SG inhibitor cycloheximide (CHX) led to dramatically reduced pulldown of nsp1β by antibodies to PKR (Fig. 5A). In the RNA interference (RNAi) assay, knockdown of G3BP1 significantly crippled the nsp1β–PKR interaction in PRRSV-infected MARC-145 cells (Fig. 5B), whereas the effect of TIA1 depletion was less prominent (Fig. 5C). In the cotransfection condition, we found that the DTT treatment significantly enhanced the nsp1β–PKR interaction, as demonstrated by co-IP assay (Fig. 5D). Thus, the above results suggest that SG formation plays an important role in promoting the nsp1β–PKR interaction and that G3BP1 is a critical player.
Fig. 5.
The SG formation promotes the nsp1β-PKR interaction. (A) MARC-145 cells were mock-infected with DMEM or infected with PRRSV at an MOI of 0.1. At 24 hpi, the cells were treated with CHX (50 μM) for 3 h or mock-treated with DMSO. The co-IP assay was carried out with rabbit anti-PKR, followed by Western blot with rabbit anti-PKR, mouse anti-nsp1β and anti-GAPDH. (B) The same as A, except that the cells were first transfected with siRNA-G3BP1 (siG3BP1) and siRNA-NC (siNC), followed by PRRSV infection at 24 h posttransfection. A rabbit antibody was used for isotype control. (C) The same as B, except for siRNA-TIA1 (siTIA1) and siRNA-NC. (D) HEK-293FT cells were transfected to coexpress nsp1β and PKR. After 24 h posttransfection, the cells were treated with PBS or DTT for 1 h. The co-IP assay were performed with rabbit anti-Flag, followed by Western blot with mouse antibodies to Flag, HA, or GAPDH. Quantitative analysis is shown (Right). The relative band density of immunoprecipitated nsp1β in the treated group was measured by fold-changes compared to that in the untreated group after being individually normalized against immunoprecipitated PKR and then nsp1β and GAPDH in the corresponding input panel. Statistical analysis was performed by two-tailed Student’s t test, and error bars indicate means ± SDs. Asterisks (*) indicate the statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001; NS, no significance.
G3BP1 has been shown to interact with PKR (13). Thus, it is possible that it also interacts with nsp1β to provide a bridging function. Indeed, the antibodies to G3BP1 could pull down nsp1β in both infection and transfection conditions (Fig. 6A and B). In contrast, the antibodies to TIA1 could pull down nsp1β only in an infection condition, but poorly in a transfection condition (Fig. 6B and C). We investigated whether the interaction is correlated with SG status. As shown in SI Appendix, Fig. S2, overexpression of G3BP1 tended to induce SG formation in transfected cells, an observation that is consistent with a previous report (37), and a colocalization relationship with nsp1β could be observed when coexpressed. In contrast, TIA1 failed to do so, suggesting that the nsp1β–TIA1 interaction is more dependent on SG formation. Moreover, knockdown of G3BP1 significantly decreased the interaction of nsp1β with TIA1 in the infection condition (SI Appendix, Fig. S3A), but depletion of TIA1 did not affect much the nsp1β–G3BP1 interaction (SI Appendix, Fig. S3B). Thus, the nsp1β–TIA1 interaction is more contingent on G3BP1, highlighting a critical role of G3BP1 in the interaction.
Fig. 6.
G3BP1 promotes the interaction between nsp1β and PKR. (A) MARC-145 cells were mock-infected with DMEM or infected with PRRSV. At 24 hpi, the co-IP assay was performed with antibodies to G3BP1, followed by Western blot analysis. A rabbit isotype antibody was used as a control. (B) Co-IP analysis of the interaction between HA-nsp1β and Flag-G3BP1 or Flag-TIA1 in cotransfected cells. A pCMV-Flag empty vector used as a control. (C) The same as A, except that rabbit anti-TIA1 was used for co-IP analysis. (D) Interaction of nsp1β with G3BP1 in vitro. GST and GST-nsp1β were bacterially expressed purified with glutathione beads, and incubated with increasing amount of His6-G3BP1 purified from E. coli BL-21 cells. After three washes, the beads were resuspended in sample buffer prior to analysis by SDS/PAGE and Western blot with antibodies to the His tag. A Ponceau staining was performed to shown the input. (E) The same as above, except that GST-G3BP1–containing beads were used to pull down TF, TF-PKR, TF-DRBM, or TF-KD from bacterial lysates. (F) The same as above, except that the GST-nsp1β–containing beads were used to pull down bacterially expressed PKR and its derivatives in the presence or absence of purified His-G3BP1. Red asterisks refer to the target protein.
We performed an in vitro binding assay to investigate whether G3BP1 is sufficient to bridge the nsp1β–PKR interaction. Specifically, the His6-tagged G3BP1 was bacterially expressed, purified, and then incubated with GST-nsp1β beads overnight at 4 °C. GST alone did not bind to G3BP1 (Fig. 6D, lanes 5 to 8), whereas GST-nsp1β was able to pull down His6-G3BP1 in a dose-dependent manner (Fig. 6D, lanes 9 to 12). As expected, the purified GST-G3BP1 was able to pull down TF-PKR and TF-DRBM (Fig. 6E, lanes 10 and 11). Thus, G3BP1 can bind directly to both nsp1β and PKR. In the presence of purified His6-G3BP1, GST-nsp1β could readily pull down TF-PKR (Fig. 6F, lane 10), suggesting the formation of a triple complex. Thus, G3BP1 serves as a direct adaptor to promote the nsp1β–PKR interaction.
G3BP1 Contributes to nsp1β-Mediated Inhibition of PKR Activity.
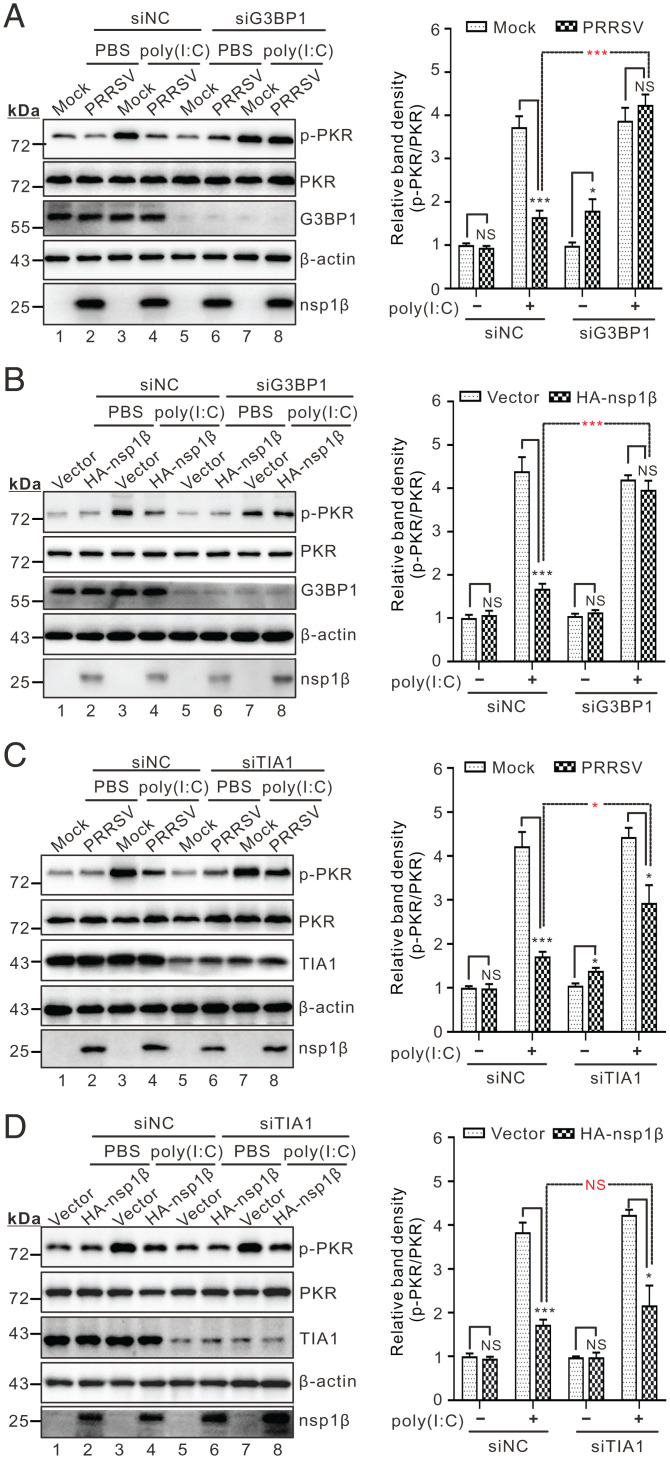
We next tested the effect of RNAi silencing of G3BP1 on PKR phosphorylation during infection in the presence or absence of poly(I:C) stimulation. When G3BP1 was knocked down, PRRSV failed to inhibit poly(I:C)-induced PKR phosphorylation (Fig. 7A, lane 8). This was also true in transfected cells expressing nsp1β (Fig. 7B, lane 8). In contrast, TIA1 knockdown had only a mild influence on nsp1β-mediated inhibition of PKR phosphorylation (Fig. 7C, lane 8), especially in the transfection condition (Fig. 7D, lane 8). These differing effects between the two molecules, however, cannot be attributed to a differential effect on the SG formation, as knockdown of either G3BP1 or TIA1 equally reduced the diameter and number of SGs (SI Appendix, Fig. S4). As a control, the small-interfering RNA (siRNA)-negative control had no effect (Fig. 7A–D, line 4). Together, these data suggest that G3BP1 is a critical host factor required for nsp1β to restrict PKR.
Fig. 7.
G3BP1 contributes to nsp1β-mediated inhibition of PKR activation. (A) MARC-145 cells were transfected with siRNAs targeting G3BP1 or scrambled siRNA (siNC). At 36 h posttransfection, the cells were infected with PRRSV strain JXwn06 at an MOI of 0.1 for 36 h before being treated with poly(I:C) for 12 h. The cells were then subject to Western blot analysis with antibodies to the indicated proteins. (B) The same as A, except cells were transfected to express HA-nsp1β. (C and D) The same as A and B, except that siRNAs were used to target TIA1. The signals of protein bands were determined by ImageJ. The relative abundance of p-PKR in each treatment was expressed as fold-changes compared to the intensity of p-PKR in the first lane after being normalized against PKR and then β-actin in the same lane. Statistical analysis was performed by two-tailed Student’s t test, and error bars indicate means ± SDs. Asterisks (*) indicate the statistical significance: *P < 0.05; ***P < 0.001; NS, no significance.
Identification of Critical Residues of nsp1β for Antagonizing PKR Activity.
We set out to map the key residues of nsp1β required for suppressing PKR activity. By triple alanine scanning, a total of 30 nsp1β mutants were engineered. These mutants were transiently expressed in MARC-145 cells and screened for their ability to enter SG under the DTT treatment. One nsp1β mutant (VSW19AAA) with the substitutions at positions 19 to 21 (VSW) failed to be localized to SG (SI Appendix, Fig. S5). Replacement with glycine residue (VSW19GGG) did not affect the phenotype (Fig. 8A). Further delineation by making substitution mutants (V19G, S20G, W21G, VS19GG, and SW20GG) revealed that the residues at positions 19 and 20 worked in coordination (Fig. 8A and SI Appendix, Fig. S5). When tested in a co-IP assay, both nsp1β VS19GG and VSW19GGG failed to interact with either G3BP1 (Fig. 8B) or PKR (Fig. 8C). Consequently, they lost the capability to inhibit poly(I:C)-induced PKR phosphorylation (Fig. 8D, lanes 3 and 4). As a control, the mutant GGK16AAA retained the ability to interact with PKR and could block PKR phosphorylation (Fig. 8B–D). Thus, the residues at positions 19 to 20 (VS) are necessary for nsp1β function.
Fig. 8.
Identification of critical residue required for nsp1β to counteract PKR activity. (A) Effect of point mutations on nsp1β sorting into SGs in transfected cells treated with either PBS (Left) or DTT (2 mM, Right) for another 1 h. (B and C) Co-IP analysis of the interaction of Flag-G3BP1 or Flag-PKR with HA-nsp1β or its derivatives in cotransfected HEK-293FT cells. (D) Inhibitory effect of nsp1β mutations on poly(I:C)-induced PKR activation. HEK-293FT cells were transfected to express HA-nsp1β or its mutants for 24 h, and then treated with poly(I:C) for 12 h prior to Western blot analysis with antibodies to the indicated proteins. The relative band density of p-PKR was expressed as fold-changes compared to the empty vector control (lane 6) after being normalized against total PKR and β-actin in the corresponding lane. Statistical analysis was performed by two-tailed Student’s t test, and error bars indicate means ± SDs. Asterisks (*) indicate the statistical significance: ***P < 0.001; NS, no significance.
The PRRSV Mutant Carrying the nsp1β Mutation VS19GG Induces Elevated Level of PKR Phosphorylation.
We next introduced the corresponding nsp1β point mutations into the infectious cDNA clone of PRRSV strain JXwn06 in a DNA-launched system by site-directed mutagenesis to generate three viral mutants: VSW19GGG, VS19GG, and GGK16AAA. The cDNA clone plasmids were transfected into MARC-145 cells for virus recovery. Only two mutants (VS19GG and GGK16AAA) were successfully rescued. The mutant GGK16AAA displayed a growth property similar to the parental virus in both MARC-145 cells and PAMs, whereas the growth of VS19GG was reduced (Fig. 9A and B). Similar to that in the transfection condition, the interaction of nsp1β with G3BP1 and PKR were almost diminished in VS19GG infected cells (Fig. 9C and D). This was accompanied by a concurrent elevation of PKR phosphorylation in both cell types (Fig. 9E and F), as opposed to both WT and the mutant nsp1β GGK16AAA (SI Appendix, Fig. S6A and B). Thus, nsp1β is responsible for inhibiting PKR activation during infection.
Fig. 9.
Characterization of the nsp1β mutant viruses. (A and B) MARC-145 cells or PAMs were infected with WT PRRSV or the mutants at an MOI of 0.1. At indicated time points, the whole-cell lysates were collected for virus titration. (C and D) Co-IP analysis of the interaction of G3BP1 or PKR with nsp1β or its mutants in infected MARC-145 cells at an MOI of 0.1. (E and F) Time-course study of the mutant VS19GG to induce PKR activation in MARC-145 cells and PAMs infected at an MOI of 0.1. Poly(I:C) (1.5 µg/mL) treatment served as a positive control. (G and H) The same as A and B, except that the cytokine mRNA was measured by quantitative PCR (qPCR). The relative mRNA abundance of TNF-α was normalized against GAPDH and then compared to the mock-infected group. Statistical analysis was performed by two-tailed Student’s t test, and error bars indicate means ± SDs. Asterisks (*) indicate the statistical significance: **P < 0.01; ***P < 0.001; NS, no significance.
The Mutant nsp1β VS19GG Induces Inflammatory Responses via G3BP1-PKR Axis.
To investigate the biological significance, we measured the production of interferons and inflammatory cytokines by RT-qPCR in PRRSV-infected cells. In all treatments, the CT values for IFN-α mRNA were near or over 35 cycles (SI Appendix, Fig. S6C), and therefore the fold-changes are not reliable. In contrast, production of IFN-β mRNA was highly inducible (SI Appendix, Fig. S6D). As compared with WT virus, there was a slight up-regulation of IFN-β mRNA, but for both mutants (SI Appendix, Fig. S6E and F), suggesting that the mutational effect is less likely due to PKR activation. On the other side, despite a reduced viral growth, the mutant VS19GG induced significantly higher level of inflammatory cytokines, such as tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-8, in both MARC-145 cells (Fig. 9G and SI Appendix, Fig. S6G and I) and primary PAMs (Fig. 9H and SI Appendix, Fig. S6H and J).
The correlation of increased PKR phosphorylation with inflammation induction was further investigated in a series of experiments. In the first assay, we treated virus-infected cells with C16, a specific PKR inhibitor. The treatment significantly reduced the level of TNF-α and IL-8 in nsp1β VS19GG-infected cells (Fig. 10A and B), coinciding with the decreased level of PKR phosphorylation (Fig. 10C, lane 11). Similar results were obtained in infected PAMs (Fig. 10D). In the second assay, we applied a genetics approach. RNAi knockdown of PKR greatly down-regulated the production of TNF-α and IL-8 in the MARC-145 cells infected with nsp1β VS19GG, but not with WT PRRSV (Fig. 10E–G). Similar results were obtained by RNAi knockdown of G3BP1 (Fig. 10H and SI Appendix, Fig. S7). Interestingly, knockdown of G3BP1 resulted in an increased replication of the mutant VS19GG, but not WT PRRSV or GGK16AAA (Fig. 10I), suggesting that, in addition to the role in PKR activation, G3BP1 itself performs additional antiviral roles (38, 39) (see more in Discussion). In the third assay, we tested the ability of VS19GG to respond to poly(I:C) treatment in infected cells. Compared with WT and GGK16AAA, the mutant VS19GG failed to inhibit poly(I:C)-stimulated inflammatory responses and PKR phosphorylation (Fig. 10A–D, lane 7). Similar results were obtained in the transfection condition (SI Appendix, Fig. S8). Taken together, we conclude that the increased phosphorylation of PKR is closely linked to induction of inflammatory cytokines in VS19GG-infected cells and that PRRSV nsp1β is a critical viral factor to restrict PKR activity.
Fig. 10.
The mutant nsp1β VS19GG induces inflammatory responses via PKR signaling. MARC-145 cells were mock-infected with DMEM or infected with indicated viruses at an MOI of 0.1 and harvested at 48 hpi before being treated with poly(I:C) for 12 h or C16 for 6 h. (A and B) Quantitative analysis of the relative mRNA abundance of TNF-α and IL-8 via qPCR that was normalized against GAPDH and then compared to the mock-infected, PBS treated group. (C) Western blot analysis of PKR activation. (D) The same as A, except that PAMs were used. (E–G) Effect of PKR knockdown on virus-induced cytokines production. MARC-145 cells were transfected with siRNA targeting PKR or scramble siRNA for 24 h, and then infected with indicated viruses at an MOI of 0.1 for 48 h. (E) Western blot analysis of the PKR knockdown effect. (F and G) show the relative mRNA abundance of TNF-α and IL-8 via qPCR that was normalized against GAPDH and then compared to the mock-infected, siNC treated group. (H) Knockdown effect of G3BP1 on TNF-α production. The measurement is the same as F. (I) Knockdown effect of G3BP1 or in combination with TIA1 on virus replication. At 24 h posttransfection, MARC-145 cells were infected with indicated viruses and the virus total titer was determined by end-point dilution assay. Statistical analysis was performed by two-tailed Student’s t test, and error bars indicate means ± SDs. Asterisks (*) indicate the statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001; NS, no significance.
Discussion
The host restriction factor PKR is a crucial sensor of virus infections and plays an important role in modulating cellular innate immunity, including protein synthesis, SG formation, induction of interferons, and inflammatory responses (5, 10). However, this molecule is inactivated in PRRSV-infected cells with yet-known purposes (32). On the other hand, PRRSV is notoriously capable of subverting host innate immunity and has evolved strategies to evade inflammation and induce little production of TNF-α, IL-6, and IL-1β, leading to a weak activation of adaptive immunity (26, 29). In this study, we established a link of PKR inactivation to PRRSV suppression of inflammation and provide evidence that PRRSV repurposes SG and subverts its antiviral function to evade PKR, thereby downregulating cellular inflammatory responses. We propose a model for PRRSV manipulation of SGs to evade PKR (SI Appendix, Fig. S9).
Repurposing SG into a Proviral Platform.
Cytoplasmic SGs are increasingly emerging as a critical immune signaling platform to detect and restrict invading viral pathogens (10, 11). As so, many viral sensors—such as MDA5, RIG1, and PKR—are recruited to SGs for activation to amount antiviral responses (11, 40). To evade this antiviral immunity, viruses have evolved various mechanisms to control SG assembly (10, 41, 42). The key strategies include encoding gene products to cleave critical SG components, such as G3BP1 (e.g., picornaviruses and porcine epidemic diarrhea virus) (43–45), to prevent PKR activation such as by indirect masking or degrading the dsRNA (e.g., coronavirus, reovirus, adenovirus, and so forth) (21, 46–48), or to redirect SG factors to viral replication complexes (e.g., alphaviruses, flaviviruses, and so forth) (42, 49, 50).
Contrary to the trend, PRRSV is one of the very few examples (e.g., Newcastle disease virus, infectious bursal disease virus, and so forth) that instead induce formation of stable SGs (33, 35, 51, 52). Our results here reveal that such SGs serve as a critical platform for PRRSV to counteract PKR activation. Mechanistically, the viral replicase protein nsp1β took advantage of SG to coopt G3BP1 to interact with PKR. Unexpectedly, the nsp1β–PKR interaction is regulated. This is evidenced by the results from the in vitro binding assay, which showed that nsp1β bound efficiently to PKR dsRBMs, but poorly to the full-length form, unless in the presence of purified G3BP1 (Fig. 6F), suggesting that a conformational change of PKR is necessary for efficient interaction with nsp1β. It is conceivable that binding of G3BP1 to PKR facilitates exposure of the nsp1β-binding sites within DRBMs, allowing formation of nsp1β–G3BP1–PKR triplex. This event likely leads to a structural alteration of PKR, which may in some manner prevent the ligands (e.g., dsRNA, PACT, and so forth) from binding to dsRBMs, disable induction of PKR KD, or produce an alternative, inactive dimer configuration. The detailed mechanisms are not clear and await to be discovered in the future.
The proviral function for PRRSV of SG assembly is twofold. First, the SG formation is conducive to nsp1β cooption of G3BP1 to disable PKR. Impairment of SG assembly via G3BP1 knockdown or CHX treatment all reduced the ability of nsp1β to interact with PKR and the capacity to counteract PKR activation (Figs. 5 and 7), whereas the SG inducers promote nsp1β function (Fig. 5). When the interaction with PKR was blocked by mutating specific residues of nsp1β (VS19GG) (Fig. 8), the PKR signaling was activated, accompanied by a pronounced cellular inflammatory response (Fig. 9G and H). Second, SGs likely provide a place for nsp1β to disarm G3BP1. Our results suggest that G3BP1 itself may have additional antiviral activities (38, 39). This is evidenced by the fact that knockdown of G3BP1 did not have an effect on WT virus or the mutant GGK16AAA, but instead increased replication of the mutant VS19GG (Fig. 10). This seemingly confusing result may be explained by WT nsp1β normally inhibiting the antiviral function of G3BP1 via interaction, whereas the mutant nsp1β VS19GG fails to do so due to loss of interaction (Fig. 8B and C). This notion is in line with the observation that the mutant VS19GG had a reduced growth rate in both cell types (Fig. 9A and B). Certainly, this is a different story from PKR antagonization and warrants further exploration in the future. Overall, these results suggest a strategy of two birds (G3BP1 and PKR) with one stone (nsp1β).
Insight into the Inflammation Regulation by PRRSV.
Our results also unexpectedly revealed a regulatory mechanism for PRRSV-mediated suppression of host cellular inflammatory responses. PRRSV is a rather “sneaky” virus, and the infections often lead to low level production of inflammatory cytokines in the early infection (26, 28, 29). Concomitantly, induction of cellular immunity is delayed, usually 1 mo following infection, and it is maintained at a low level, contributing to viral persistence in vivo (26, 30). Despite the identification of several inflammation-inhibitory viral proteins (e.g., nsp1α, nsp4, and nsp11), the detailed mechanisms have remained poorly defined (28, 29). On the other hand, PKR is a powerful host restriction factor and possesses multiple functions in antiviral immunity, including restricting the cross-species transmission of several viruses (5, 53, 54). Enigmatically, this molecule is shut off by PRRSV. By taking advantage of a nsp1β mutant, we showed that the selective targeting of PKR signaling is to suppress cellular inflammation. Despite slightly reduced virus growth, the viral mutant induced an elevated level of PKR phosphorylation and pronounced production of inflammatory cytokines (e.g., TNF-α, IL-6, and IL-8) (Fig. 9A and B). In contrast, either siRNA knockdown of PKR or treatment with PKR inhibitor C16 could block this effect (Fig. 10). Thus, our results suggest a strategy of how PRRSV regulates cellular inflammatory responses.
Expansion of PRRSV nsp1β Functions.
PRRSV nsp1β is a multifunctional protein (55–58), and it displays a dynamic distribution during PRRSV infection (Fig. 2A). It accumulates mainly in the cytoplasm in early infection and then mostly in the nucleus, with the cytoplasmic fraction as discrete puncta. It is now becoming clear that major function of nuclear nsp1β is to cause cellular nuclear mRNA retention (56), and interestingly, loss of this nuclear location cripples the ability of nsp1β to antagonize IFN-α (57). On the other side, the function of cytoplasmic nsp1β has remained less characterized, except for its function as a protease to cleave off itself from viral polyprotein precursor and as a transactivator for frameshift expression of nsp2-TF and nsp2N (55, 58). Unexpectedly, our studies discovered a localization of nsp1β in the cytoplasm and showed that the new mission for nsp1β redistribution into SGs is to counteract PKR restriction to down-regulate cellular inflammation and also likely to disarm G3BP1. Mutation of specific residues within nsp1β could reverse this effect. Thus, our findings establish nsp1β as a potential target for PRRSV vaccine development through modulating cellular inflammatory responses.
In summary, the findings reported here reveal a unique paradigm of how a RNA virus repurposes antiviral cellular organelle to evade PKR restriction, provide further insight into the PRRSV-mediated regulation of host inflammatory responses, and identify a promising target for modifying current attenuated vaccines to control PRRSV infection.
Materials and Methods
Reagents.
MARC-145 and HEK-293FT cells were cultured in Dulbecco’s modified Eagle’s medium at 37 °C with 5% CO2, whereas primary PAMs from 1-mo-old SPF piglets were maintained in RPMI-1640 medium. The PRRSV strain JXwn06 (GenBank no: EF641008) was used a model organism. The commercial antibodies and chemicals are from various sources (i.e., Sigma-Aldrich, Thermo Fisher, Abcam, Proteintech). The plasmids were engineered by standard recombinant DNA procedures, and the derivatives were made by PCR-based site-directed or deletion mutagenesis. All the constructs were verified by DNA sequencing.
Quantitative PCR.
Total cellular RNAs were reversed transcribed into cDNA by FastKing RT enzyme (Tiangen), and the relative qPCR was performed with the Applied Biosystems SYBR Select Master Mix (Thermo Fisher, #4472913). The cellular GAPDH was used as the internal control.
RNAi.
For each given gene, two siRNAs were designed to target different coding regions and transfected with Lipofectamine RNAiMAX. The cell viability was assessed with CellTiter 96 AQueous One Solution Reagent (Promega), and the knockdown efficiency was assessed by Western blot. For transfection/infection assay, MARC-145 cells were transfected with indicated siRNAs for 24 to 36 h before being infected with indicated viruses.
RNAscope In Situ Hybridization.
In situ hybridization assay was employed to detect both negative and positive PRRSV RNAs in virus-infected MARC-145 cells by using the RNAscope Multiplex Fluorescent Detection Reagents v2 Kit (ACD, #323110), as described previously (34). A total of eight double-Z branched pairs were designed to detect the positive-sense genome by targeting the highly conserved N gene, whereas a total of 20 double-Z branched pairs targeting the regions of PRRSV ORF6, ORF7, and 3′ UTR to detect the negative strands. The hybridization procedure was performed according to the manufacturer’s instruction.
Immunoprecipitation.
The treated MARC-145 or HEK-293FT cells in six-well plates were harvested and lysed in Nonidet P-40 buffer (0.5% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅HCl, pH 8.0) containing protease inhibitors (Sigma, #P8340). After being clarified by centrifugation at 12,000 rpm for 20 min, the supernatants were precleared with protein A/G Sepharose beads (Santa Cruz, #sc-2003) before being incubated with 3 to 5 μL indicated antibodies and protein A&G Sepharose beads overnight at 4 °C. The beads were washed and then subject to SDS/PAGE and Western blot analysis.
Immunofluorescence.
The treated cells on coverslips were fixed with 3.7% paraformaldehyde and permeabilized with 0.2% Triton X-100 in PBS. They were then incubated with proper primary antibodies in a humid chamber, followed by secondary antibodies. Nuclear DNA was stained with DAPI (Thermo Fisher, #62248) for 10 min. The cells were imaged using a Nikon A1 confocal microscope.
Protein Purification and In Vitro Binding Assay.
Protein expression in Escherichia coli BL21 cells was induced with Isopropyl β-d-1-thiogalactopyranoside (IPTG) at a concentration of 0.1 mM at 16 °C for 24 h when the optical density at 600 nm of E. coli cells reached 0.6. GST or His-tagged fusion proteins were purified with Glutathione-Sepharose 4B beads (GE Healthcare, #17075601) or nickel beads (Sigma Aldrich, #GE17-5268-02) by following the manufacturer’s instructions. For the in vitro binding assay, GST-nsp1β, GST-G3BP1, or GST, His-G3BP1 beads were incubated with the prey protein samples in 1 mL Nonidet P-40 Buffer (0.1% Nonidet P-40, 150 mM NaCl, 10 mM Tris⋅HCl, pH 7.4) with protease inhibitor mixture and RNase overnight at 4 °C with gentle rocking. The beads were washed five times with Nonidet P-40 buffer, and the protein complexes were resolved by SDS/PAGE and then analyzed by Western blot with the indicated antibodies.
Generation and Characterization of PRRSV Mutants.
The DNA-launched infectious clone of HP-PRRSV strain JXwn06 (59) was used to generate nsp1β mutants. The verified plasmids were transfected into MARC-145 cells for virus recovery. For viral growth analysis, MARC-145 cells or PAMs in six-well plates were infected with indicated viruses at a multiplicity of infection (MOI) of 0.1. At the indicated times postinfection, the medium and cells were harvested and titrated on MARC-145 cells by using the endpoint dilution assay.
Ethics Statement.
Preparation of PAMs derived from 1-mo-old SPF pigs was performed according to the Chinese Regulations of Laboratory Animals, The Guidelines for the Care of Laboratory Animals (Ministry of Science and Technology of People’s Republic of China, GB/T 35892-2018) and Laboratory Animal Requirements of Environment and Housing Facilities (GB 14925±2010, National Laboratory Animal Standardization Technical Committee). The license number associated with this research protocol was CAU 20120611, which was approved by the Laboratory Animal Ethical Committee of China Agricultural University.
Statistical Analysis.
Statistical significance was analyzed by two-tailed unpaired Student’s t test. Significance symbols are defined as follows: NS, no significance; *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate means ± SD.
Detailed descriptions are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (32025035, 31772759), China Agriculture Research System of Ministry of Finance and Ministry of Agriculture and Rural Affairs (CARS-35), and China Postdoctoral Science Foundation (2021M703527).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201169119/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., Jacobs B. L., Inhibition of PKR by RNA and DNA viruses. Virus Res. 119, 100–110 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Cesaro T., Michiels T., Inhibition of PKR by viruses. Front. Microbiol. 12, 757238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metz D. H., Esteban M., Interferon inhibits viral protein synthesis in L cells infected with vaccinia virus. Nature 238, 385–388 (1972). [DOI] [PubMed] [Google Scholar]
- 4.Radetskyy R., Daher A., Gatignol A., ADAR1 and PKR, interferon stimulated genes with clashing effects on HIV-1 replication. Cytokine Growth Factor Rev. 40, 48–58 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Munir M., Berg M., The multiple faces of proteinkinase R in antiviral defense. Virulence 4, 85–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dzananovic E., McKenna S. A., Patel T. R., Viral proteins targeting host protein kinase R to evade an innate immune response: A mini review. Biotechnol. Genet. Eng. Rev. 34, 33–59 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Donnelly N., Gorman A. M., Gupta S., Samali A., The eIF2α kinases: Their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., et al. , Molecular basis for PKR activation by PACT or dsRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 10005–10010 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemaire P. A., Anderson E., Lary J., Cole J. L., Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 381, 351–360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiermann N., Haneke K., Sun Z., Stoecklin G., Ruggieri A., Dance with the devil: Stress granules and signaling in antiviral responses. Viruses 12, 984 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormick C., Khaperskyy D. A., Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 17, 647–660 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Jain S., et al. , ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reineke L. C., Kedersha N., Langereis M. A., van Kuppeveld F. J. M., Lloyd R. E., Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. MBio 6, e02486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapil P., Stohlman S. A., Hinton D. R., Bergmann C. C., PKR mediated regulation of inflammation and IL-10 during viral encephalomyelitis. J. Neuroimmunol. 270, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A., Haque J., Lacoste J., Hiscott J., Williams B. R., Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. U.S.A. 91, 6288–6292 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zamanian-Daryoush M., Mogensen T. H., DiDonato J. A., Williams B. R., NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol. Cell. Biol. 20, 1278–1290 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma N. R., Majerciak V., Kruhlak M. J., Zheng Z. M., KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. PLoS Pathog. 13, e1006677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colpitts C. C., et al. , Hepatitis C virus exploits cyclophilin A to evade PKR. eLife 9, e52237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J. J., et al. , Baculovirus protein PK2 subverts eIF2α kinase function by mimicry of its kinase domain C-lobe. Proc. Natl. Acad. Sci. U.S.A. 112, E4364–E4373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schierhorn K. L., et al. , Influenza A virus virulence depends on two amino acids in the N-terminal domain of its NS1 protein to facilitate inhibition of the RNA-dependent protein kinase PKR. J. Virol. 91, e00198-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X., et al. , Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 114, E4251–E4260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou S., et al. , Double-stranded RNA deaminase ADAR1 promotes the Zika virus replication by inhibiting the activation of protein kinase PKR. J. Biol. Chem. 294, 18168–18180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., et al. , Hantaviruses use the endogenous host factor P58IPK to combat the PKR antiviral response. PLoS Pathog. 17, e1010007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao G. R., et al. , Adenosine deaminase acting on RNA 1 associates with Orf virus OV20.0 and enhances viral replication. J. Virol. 93, e01912–e01918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snijder E. J., Kikkert M., Fang Y., Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 94, 2141–2163 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Lunney J. K., et al. , Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 4, 129–154 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Wensvoort G., et al. , Mystery swine disease in The Netherlands: The isolation of Lelystad virus. Vet. Q. 13, 121–130 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Han J., Zhou L., Ge X., Guo X., Yang H., Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 209, 30–47 (2017). [DOI] [PubMed] [Google Scholar]
- 29.An T. Q., Li J. N., Su C. M., Yoo D., Molecular and cellular mechanisms for PRRSV pathogenesis and host response to infection. Virus Res. 286, 197980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou L., Ge X., Yang H., Porcine reproductive and respiratory syndrome modified live virus vaccine: A “leaky” vaccine with debatable efficacy and safety. Vaccines (Basel) 9, 362 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu F., et al. , Phylogenetics, genomic recombination, and NSP2 polymorphic patterns of porcine reproductive and respiratory syndrome virus in China and the United States in 2014–2018. J. Virol. 94, e01813–e01819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Y., et al. , Downregulation of protein kinase PKR activation by porcine reproductive and respiratory syndrome virus at its early stage infection. Vet. Microbiol. 187, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y., et al. , Porcine reproductive and respiratory syndrome virus infection induces stress granule formation depending on protein kinase R-like endoplasmic reticulum kinase (PERK) in MARC-145 cells. Front. Cell. Infect. Microbiol. 7, 111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao P., et al. , Reprogramming the unfolded protein response for replication by porcine reproductive and respiratory syndrome virus. PLoS Pathog. 15, e1008169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catanzaro N., Meng X. J., Porcine reproductive and respiratory syndrome virus (PRRSV)-induced stress granules are associated with viral replication complexes and suppression of host translation. Virus Res. 265, 47–56 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Reineke L. C., Lloyd R. E., The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J. Virol. 89, 2575–2589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reineke L. C., Dougherty J. D., Pierre P., Lloyd R. E., Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell 23, 3499–3510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bidet K., Dadlani D., Garcia-Blanco M. A., G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10, e1004242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W., et al. , G3BP1 inhibits RNA virus replication by positively regulating RIG-I-mediated cellular antiviral response. Cell Death Dis. 10, 946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onomoto K., et al. , Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7, e43031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poblete-Durán N., Prades-Pérez Y., Vera-Otarola J., Soto-Rifo R., Valiente-Echeverría F., Who regulates whom? An overview of RNA granules and viral infections. Viruses 8, 180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai W. C., Lloyd R. E., Cytoplasmic RNA granules and viral infection. Annu. Rev. Virol. 1, 147–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., et al. , Picornavirus 2A protease regulates stress granule formation to facilitate viral translation. PLoS Pathog. 14, e1006901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun L., et al. , Porcine epidemic diarrhea virus infection induces caspase-8-mediated G3BP1 cleavage and subverts stress granules to promote viral replication. J. Virol. 95, e02344-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visser L. J., et al. , Foot-and-mouth disease virus leader protease cleaves G3BP1 and G3BP2 and inhibits stress granule formation. J. Virol. 93, e00922-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao B., et al. , Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus replication. PLoS Pathog. 17, e1008690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Y., et al. , The multi-functional reovirus σ3 protein is a virulence factor that suppresses stress granule formation and is associated with myocardial injury. PLoS Pathog. 17, e1009494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price A. M., et al. , Adenovirus prevents dsRNA formation by promoting efficient splicing of viral RNA. Nucleic Acids Res. 50, 1201–1220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cristea I. M., et al. , Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: Role for G3BP1 and G3BP2 in virus replication. J. Virol. 84, 6720–6732 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholte F. E., et al. , Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J. Virol. 89, 4457–4469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Y., et al. , Newcastle disease virus induces stable formation of bona fide stress granules to facilitate viral replication through manipulating host protein translation. FASEB J. 31, 1337–1353 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Zhao D., et al. , Critical role for G3BP1 in infectious bursal disease virus (IBDV)-induced stress granule formation and viral replication. Vet. Microbiol. 248, 108806 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Peng C., Haller S. L., Rahman M. M., McFadden G., Rothenburg S., Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc. Natl. Acad. Sci. U.S.A. 113, 3855–3860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park C., et al. , Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog. 17, e1009183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue F., et al. , The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease. J. Virol. 84, 6461–6471 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han M., Ke H., Zhang Q., Yoo D., Nuclear imprisonment of host cellular mRNA by nsp1β protein of porcine reproductive and respiratory syndrome virus. Virology 505, 42–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ke H., et al. , Type I interferon suppression-negative and host mRNA nuclear retention-negative mutation in nsp1β confers attenuation of porcine reproductive and respiratory syndrome virus in pigs. Virology 517, 177–187 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Patel A., et al. , Molecular characterization of the RNA-protein complex directing -2/-1 programmed ribosomal frameshifting during arterivirus replicase expression. J. Biol. Chem. 295, 17904–17921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song J., et al. , The nsp2 hypervariable region of porcine reproductive and respiratory syndrome virus strain JXwn06 is associated with viral cellular tropism to primary porcine alveolar macrophages. J. Virol. 93, e01436-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.