Significance
Retinal degeneration associated with photoreceptor death represents a major source of blindness worldwide. Photoreceptors are known to have unique functional properties, sculpted to a large extent by alternative splicing. In recent years, increasing efforts have been made to understand the intricacies of the photoreceptor-specific transcriptome, with the goal of identifying new therapeutic targets to treat retinal diseases. Here, we performed a comprehensive characterization of retinal and photoreceptor-specific microexons in vertebrates. We demonstrate that this splicing program is regulated by the splicing factor srrm3 and that its misregulation results in severe photoreceptor alteration and visual impairment, similar to classic zebrafish models of human retinopathies. This work thus places SRRM3 and retina-specific microexons as players in retinal health and disease.
Keywords: alternative splicing, retinal disease model, zebrafish
Abstract
Retinal photoreceptors have a distinct transcriptomic profile compared to other neuronal subtypes, likely reflecting their unique cellular morphology and function in the detection of light stimuli by way of the ciliary outer segment. We discovered a layer of this molecular specialization by revealing that the vertebrate retina expresses the largest number of tissue-enriched microexons of all tissue types. A subset of these microexons is included exclusively in photoreceptor transcripts, particularly in genes involved in cilia biogenesis and vesicle-mediated transport. This microexon program is regulated by Srrm3, a paralog of the neural microexon regulator Srrm4. Despite the fact that both proteins positively regulate retina microexons in vitro, only Srrm3 is highly expressed in mature photoreceptors. Its deletion in zebrafish results in widespread down-regulation of microexon inclusion from early developmental stages, followed by other transcriptomic alterations, severe photoreceptor defects, and blindness. These results shed light on the transcriptomic specialization and functionality of photoreceptors, uncovering unique cell type-specific roles for Srrm3 and microexons with implications for retinal diseases.
Impaired vision is a highly heterogeneous condition affecting millions of individuals worldwide. The key feature that accounts for most visual disabilities is the primary or secondary loss of photoreceptor (PR) cells, arising from a number of different genetic and environmental causes (1). PRs, comprising rods and cones, have a unique ciliary structure named the outer segment (OS) that physically supports the phototransduction cascade, the process by which incoming light is converted into electrical signals that the brain can process (2). This ciliary function contrasts with those of other cell types, which are generally involved in sensing vibration, liquid circulation, hormones, chemicals, or temperature, and it is reflected by unique morphological characteristics. OSs are filled with densely packed and organized membranous disks that undergo rapid and extensive renewal to guarantee the proper supply of proteins, such as rhodopsin, cone opsins, and other visual pigments (3). The delivery of those components from the Golgi to the ciliary tip and back is sustained by a high-capacity vesicle-mediated transport system, carried along the ciliary axoneme (4). Consequently, multiple genes whose mutations are known to cause PR degeneration, leading to syndromic or nonsyndromic retinal ciliopathies, encode for proteins involved in OS vesicular transport (4, 5).
Alternative splicing is a pretranslational mechanism used by specialized cell types to remodel their transcriptomes to produce protein isoforms necessary for their activity. Indeed, the high degree of functional specialization of PR cells is mirrored by the unusually large fraction of genes that undergo retina-specific alternative splicing, thereby creating unique isoforms in PRs essential for different cellular properties, such as OS biogenesis (6). The unique pattern of alternative splicing in PRs has been described and is known to be at least in part driven by the action of an RNA binding protein, called Musashi 1 (MSI1) (7). Recently, MSI1 expression, combined with down-regulation of the splicing factors PTBP1 and PCBP2, has been linked to the activation of PR-specific exons (8). Considering that aberrant splicing due to mutations in splicing factors and retina-specific exons has been linked to several retinal diseases (e.g., retinitis pigmentosa) (9), a complete characterization of the retinal alternative splicing programs would be of great benefit for precision medicine.
Microexons are very short exons, ranging from 3 to 27 nt, that are evolutionarily conserved among vertebrates and enriched in neurons (10). These exons can encode as little as one amino acid and are often located on the surface of proteins, where they can modulate protein–protein interactions (10–13). The neuronal-specific Ser/Arg repetitive matrix protein 4 (SRRM4) controls the inclusion of most neuronal microexons annotated so far (10, 14). SRRM4 encodes a 39-amino acid domain (enhancer of microexons, or eMIC) at its C terminus that is necessary and sufficient for the inclusion of neural microexons (14). Unlike most other neural-enriched splicing regulators (e.g., Nova or Rbfox) (15, 16), SRRM4 is only known to act as a positive regulator through the binding of the eMIC domain to UGC motifs in the upstream intronic region near the 3′ splice sites of microexons (10). Depletion of SRRM4 leads to neurodevelopmental defects in cell cultures as well as in in vivo models (17–20). Recently, a neurally enriched, vertebrate-specific paralog of SRRM4, called SRRM3, has been reported (14, 21, 22). SRRM3 also encodes an eMIC domain, and both paralogs regulate a highly overlapping set of alternatively spliced small exons in vivo and in vitro (14, 21). Srrm3 gene-trapped (Srrm3gt/gt) mice show reduced body size and lifespan, as well as tremors and ataxia. These mice also exhibit neuronal splicing defects, which become more severe when SRRM4 expression levels are low (21). However, the roles of SRRM4 and SRRM3 in PRs remain unexplored.
Here, we report that the human retina expresses the largest program of tissue-enriched microexons among all tissue types, with a subset of those included only in PRs. Retina-enriched microexons (hereafter, RetMICs) are enriched in genes involved in cilia biogenesis and vesicle transport, as well as loci known to be associated with retinal diseases. A large fraction of RetMICs and their retina-enriched regulation date back to the last common ancestor of vertebrates, suggesting a critical functional role for this program across vertebrate species. We further identify SRRM3 as the key regulator of RetMIC inclusion in PRs. Consistently, zebrafish lacking the srrm3 eMIC domain show progressive OS shortening, PR degeneration, and visual impairment preceded by RetMIC down-regulation from early developmental stages. Together, these results demonstrate that the conserved program of retina-specific microexons regulated by Srrm3 is essential for PR functionality and vision, contributing to a novel understanding of retinal physiology in health and disease.
Results
The Human Retina Has the Highest Number of Tissue-Enriched Microexons.
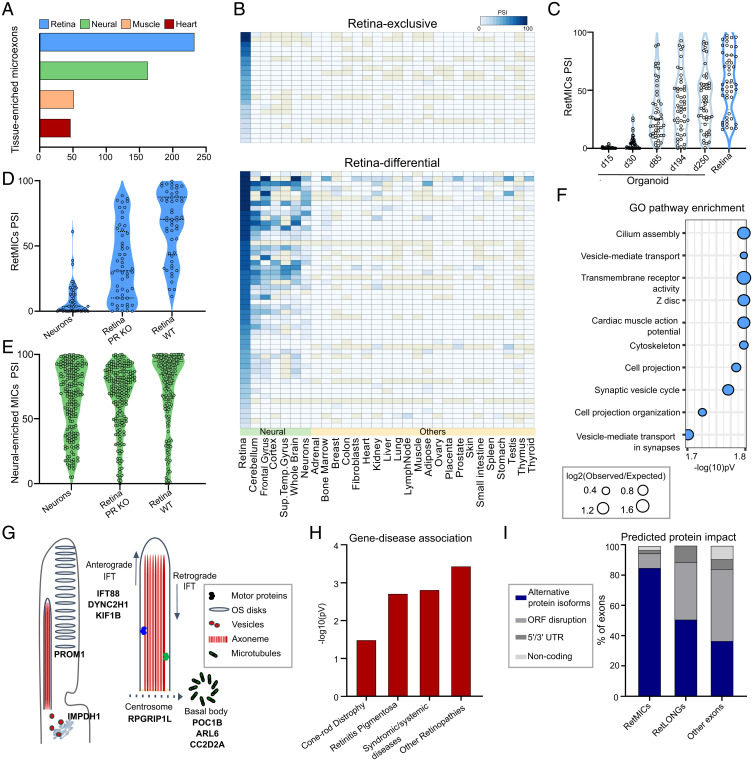
In order to systematically profile tissue-enriched microexon programs, we used data from VastDB (https://vastdb.crg.eu) (23) comprising 136 samples from different human cell/tissue types. For each tissue type, we aimed at identifying microexons with biased inclusion, which we defined as tissue-enriched (Methods and Materials). We found that the retina has the largest program of tissue-enriched microexons of all analyzed tissues (Fig. 1A). While this could be related to the particularly high fraction of neurons and low cell-type heterogeneity of the retina (PRs account for ∼60% of all retinal cells) (24), it could also indicate the presence of an additional set of microexons not shared with other, nonretinal neurons. Therefore, we implemented a retina specificity score to discriminate events specifically enriched in retina that takes into account the average inclusion level (using the metric “percent spliced in” or PSI) and its SD for a given exon in retinal, neuronal and nonneuronal samples (Methods and Materials). Using this metric, we found 75 microexons that are specifically enriched in human retinal samples (RetMICs), as well as 116 retina-enriched long exons (i.e., > 27 nt, hereafter RetLONGs) (Dataset S1). RetMICs were further subdivided into 23 microexons included exclusively in the retina (retina-exclusive) and 52 with higher inclusion in the retina compared to other neuronal samples (retina-differential) (Fig. 1B).
Fig. 1.
Characterization and identification of the human RetMIC program. (A) Number of tissue-enriched microexons by tissue type in humans. Only the four tissues with the highest number of tissue-enriched microexons are depicted. (B) Heatmap showing RetMIC inclusion in different human tissues. RetMICs are divided into retina-exclusive (inclusion only in retina samples) and retina-enriched (biased inclusion in retina compared to neural samples). Each row corresponds to a different microexon. Inclusion levels were obtained from VastDB (23). (C) Inclusion levels (PSIs) of RetMICs in cone-rich human organoids (SRP056957) and whole retina. Developing organoid time points: day 15 (d15), day 30 (d30), day 85 (d85), day 194 (d194), and day 250 (d250). Data plotted are averaged PSIs of three biological replicates; events with NA values due to insufficient read coverage were omitted. (D and E) Violin plots depicting inclusion levels of mouse RetMICs (D) and neural-enriched microexons (neural MICs) (E) in hippocampal neurons, WT and Aipl1 KO retinae (data from SRP068974). Events with insufficient read coverage were omitted. (F) Top 10 enriched GO terms for human RetMIC-containing genes. P values are false-discovery rate (FDR)-adjusted. (G) Schematic representation of the OS and localization of selected RetMIC genes; IFT, intraflagellar transport. (H) Enrichment of RetMIC-containing genes among loci associated with different retinal diseases. P values from hypergeometric tests. Complete inputs and results are provided in Dataset S3. (I) Predicted protein impact of different exon types as annotated in VastDB.
Although the mammalian retina is primarily composed of PRs, it also contains other neural cell types, such as bipolar, ganglion, horizontal, and amacrine cells. To determine whether RetMICs are mainly PR-specific, we analyzed public RNA-sequencing (RNA-seq) data from cone-rich human retinal organoids, developed from human embryonic stem cells and closely mimicking functional cones with mature OS (25). These data showed that RetMIC inclusion strongly increases during organoid development and, consequently, PR maturation (Fig. 1C), and shows a positive association with the expression of various PR differentiation markers (SI Appendix, Fig. S1 A and B). Moreover, reanalysis of an RNA-seq dataset of retinae from Aipl1 knockout (KO) mice, which leads to specific PR degeneration (7), revealed that the vast majority (73.5%) of mouse RetMICs (see below) exhibit a substantial reduction of inclusion (ΔPSI < −15) in Aipl1 KO mice retina compared to the control (Fig. 1D and SI Appendix, Fig. S1C). This contrasts with neural-enriched microexons, which did not significantly change their inclusion patterns (Fig. 1E and SI Appendix, Fig. S1D). RetMICs also had no or low inclusion in mouse hippocampal neurons, further supporting their PR enrichment (Fig. 1D and SI Appendix, Fig. S1C).
Human RetMICs Are Enriched in Cilia-Related Genes and Associated with Retinal Diseases.
Gene ontology (GO) analysis revealed that genes containing RetMICs are enriched for functions important for PR homeostasis, such as cilium assembly and vesicle-mediated intracellular transport (Fig. 1F and Dataset S2). We identified RetMIC-containing genes that localize to the cilia basal body and transition zone (POC1B, ARL6, CC2D2A) (26–28), where they control ciliogenesis and vesicle anchoring to the OS microtubule axoneme (Fig. 1G). Others regulate OS disks morphogenesis (PROM1) (29), PR homeostasis (IMPDH1) (30), are involved in anterograde and retrograde transport (IFT88, DYNC2H1, KIF1B) (31, 32), or are part of the centrosomal complex (RPGRIP1L) (33) (Fig. 1G). This contrasts with the GO analysis for genes harboring neural-enriched microexons, which were also enriched in functions related to vesicle-mediated transport and neural development, but not to cilia biogenesis (Dataset S2). Remarkably, RetMIC-containing genes were significantly enriched among loci associated with multiple retinal diseases (Fig. 1H and Dataset S3)—including retinitis pigmentosa, cone-rod dystrophies, and Bardet-Biedl syndrome—pointing to a potential role for RetMICs in vision. In line with this hypothesis, at least two individual RetMICs have been previously linked to visual impairment [arl6 (34) and DYNC2H1 (35)].
RetMICs Generate Alternative Protein Isoforms with Remodeled Structures.
Similar to neural microexons (10), we found that RetMICs were less likely to disrupt open reading frames than RetLONGs and other cassette exons (Fig. 1I). Moreover, a larger fraction of RetMICs overlapped annotated PFAM or PROSITE protein domains (36, 37) (46% of RetMICs vs. 35% of RetLONGs) (Dataset S4). Together, these observations suggest that RetMICs may serve to modulate the activity of protein domains and protein–protein interactions, rather than affecting gene expression through nonsense-mediated decay or by causing gross alterations to protein folding. To perform a more comprehensive analysis of the structural impact of RetMIC inclusion, we compiled and examined all available PDB structures of human RetMIC-containing genes, as well as high-confidence structural models from ModBase (38) and Interactome3D (39) (SI Appendix, Fig. S2 and Dataset S5). Of 32 protein structures containing the microexon insertion site, the vast majority (96%) of those sites occurred in solvent accessible regions, namely regions with relative solvent accessibility ≥20% (40). Most insertion sites were mapped to unstructured loops (72%) rather than within structured helices (20%), sheets (4%), or turns (4%), further suggesting that RetMICs generally serve to modify protein surfaces instead of impacting overall protein folding.
Through our structural analysis, we identified interesting examples of RetMICs in ubiquitous vesicular trafficking-related proteins that are likely to affect their functionality. For example, a highly retina-specific and evolutionarily conserved 3-nt microexon (VastID: HsaEX0015816) is present in the clathrin heavy chain (CLTC) gene, a key protein in charge of packaging cargo into clathrin-coated vesicles at the plasma membrane (SI Appendix, Fig. S3 A and B). This RetMIC occurs within the N-terminal β-propeller domain, which mediates interactions with multiple clathrin-box motif-containing adapter proteins that compete for binding to the same surface (41). Upon inclusion, the microexon introduces a single negatively charged aspartic acid residue into the otherwise hydrophobic clathrin-box binding groove of the β-propeller, thereby modifying the electrostatic landscape of this interaction surface and likely affecting its interaction with adapters (SI Appendix, Fig. S3 C–E). Another interesting example is the motor protein myosin-VI (MYO6), which colocalizes with clathrin-coated vesicles and plays important roles in ciliary vesicular transport (42). MYO6 harbors a conserved 9-nt retina-enriched microexon (VastID: HsaEX0041235) in the catalytic motor domain, through which myosin binds to and catalyzes movement along the actin filament through its ATPase activity (SI Appendix, Fig S4 A and B). The microexon is located within a loop termed “insert-1” that is not shared with other myosins and has been shown to modulate nucleotide binding (SI Appendix, Fig. S4 C and D) (43). This insert is thought to explain some of the unique kinetic properties of myosin-VI relative to other myosins, which may be further fine-tuned in PRs by the inclusion of the RetMIC, especially given its highly acidic sequence (Glu-Asp-Glu) that might affect the binding of negatively charged ATP. Another recent study identified a retina-enriched microexon (VastID: HsaEX0021304) within a different motor protein, dynein cytoplasmic 2 heavy chain 1 (DYNC2H1), which, from its position within an ATP-binding domain, also appears likely to affect conformational dynamics during microtubule binding and ATP hydrolysis (44).
RetMICs Are Evolutionarily Conserved and Enriched in Cilia-Related Genes in Vertebrates.
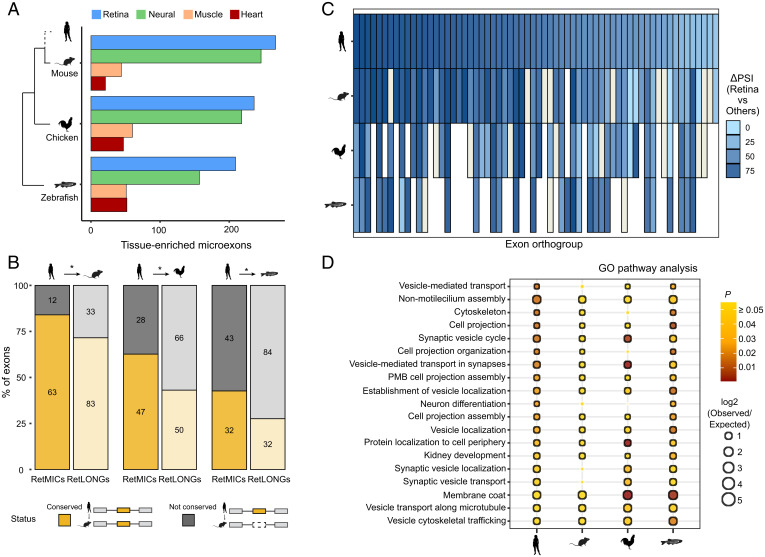
To investigate the evolutionary conservation of RetMICs across vertebrate species, we used publicly available RNA-seq samples for mouse, chicken, and zebrafish (23), as well as zebrafish adult retina samples that we generated for this study. Similar to humans, retina was the tissue with the largest tissue-enriched microexon program in all species (Fig. 2A). Applying the retina specificity score to define RetMICs in mouse, chicken, and zebrafish, we identified 63 mouse, 75 chicken, and 72 zebrafish RetMICs (Dataset S1). As in mammals, zebrafish RetMICs were highly PR-specific, as shown by RNA-seq comparisons between FACS-sorted PRs and non-PR neurons (SI Appendix, Fig. S5). To evaluate RetMIC conservation from both the genomic and regulatory perspective, we first derived exon orthologies among all selected vertebrate species using ExOrthist (45) (SI Appendix, Methods). RetMICs showed significantly higher levels of genomic conservation compared to RetLONGs (Fig. 2B), with 84%, 62%, and 42% of human RetMICs conserved in mouse, chicken, and zebrafish, respectively. To evaluate regulatory conservation, we next investigated if genomically conserved RetMICs had retina-enriched inclusion. Preferential inclusion in the retina compared to the other tissues was observed for the majority of the human RetMIC orthologs (Fig. 2C), with few exceptions represented by exons characterized by broader, neural-enriched inclusion (Fig. 2C and SI Appendix, Fig. S6A). Overall, these results suggest an ancestral role of the RetMIC program in the vertebrate retina. To assess whether RetMICs impact similar biological processes across vertebrates, we then performed comparative GO enrichment analyses. To avoid biases coming from the different genome annotations, we transferred human GO annotations to the mouse, chicken, and zebrafish gene orthologs (SI Appendix, Methods). As for humans, cilium organization and vesicle transport appeared as enriched categories across the studied species (Fig. 2D), in contrast to RetLONGs (SI Appendix, Fig. S6B and Dataset S2). In summary, these results show that a program of RetMICs modulating PR ciliogenesis and vesicle transport was likely already present in the last common ancestor of jawed vertebrates.
Fig. 2.
Evolutionary study of RetMICs. (A) Number of tissue-enriched microexons (as in Fig. 1A) in mouse, chicken, and zebrafish. Only the four tissues with the highest number of tissue-enriched microexons are represented. (B) Genomic conservation of human RetMICs (full color, left bar) and human RetLONGs (transparent color, right bar) in mouse, chicken, and zebrafish. Human RetMICs/RetLONGs are considered genomically conserved in another species when they belong to an exon orthogroup including at least one exon from that species. Asterisks indicate significant differences between RetMICs and RetLONGs conservation (P < 0.05, hypergeometric test). (C) Heatmap showing the bias in retina inclusion for genomically conserved human RetMICs (top row) and their respective orthologs in mouse, chicken, and zebrafish. Each column corresponds to a different RetMIC, and the color represents the ΔPSI between the average of the retina samples and of all the other tissues, with darker blue reflecting greater retina inclusion bias. In case of multiple orthologs, only the one with the highest ΔPSI (retina − others) was plotted. Blanks and ivory rectangles indicate missing orthologs and missing ΔPSI values due to lack of read coverage, respectively. (D) Dot plot representing functional enrichment of RetMIC-containing genes across species. The functional enrichment of genes containing RetMICs was separately tested for each of the species, and significant categories in at least two species (FDR-adjusted P ≤ 0.05) were plotted. The color reflects the adjusted P value, with yellow color depicting P ≥ 0.05. The size of the dots is proportional to the log2 of the observed vs. expected ratio (O/E), and black borders around the categories highlight log2 O/E ≥ 1. PMD, plasma membrane bounded.
SRRM3 and SRRM4 Regulate RetMIC Inclusion In Vitro.
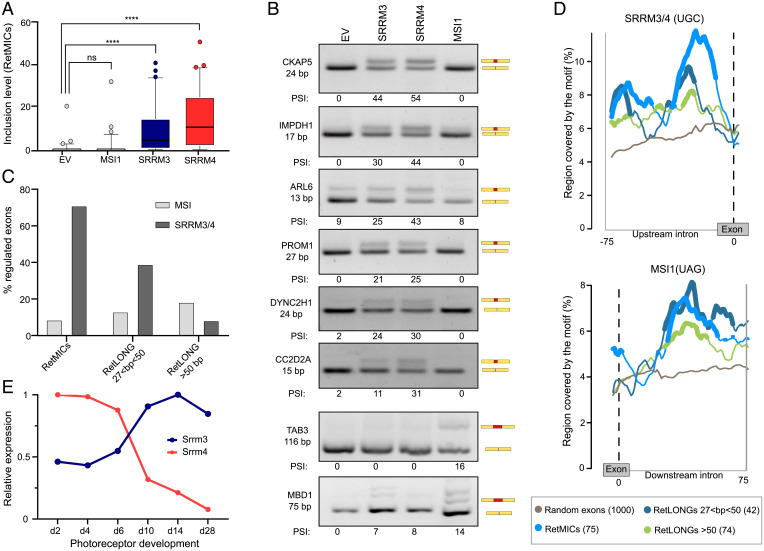
We next looked into the regulation of RetMICs. First, we focused on MSI1, a splicing factor that is highly expressed in PRs and can promote the inclusion of retina-enriched exons through direct binding to UAG motifs in their downstream introns (7). However, stable ectopic expression of MSI1 for 24 h in HEK293 cells (SI Appendix, Fig. S7A) revealed that RetMICs were largely not responsive to this regulator (Fig. 3 A and B), in contrast to some RetLONGs and known MSI1 targets (Fig. 3B). Next, we tested the effect of SRRM3 and SRRM4 ectopic expression (SI Appendix, Fig. S7 B and C). Similar to neural-enriched exons (10, 14), expression of either gene was sufficient to promote the inclusion of most RetMICs (Fig. 3 A and B). Similar results were obtained using previously published data from ectopic expression of MSI1 and SRRM3/4 in different neural and nonneural cell lines (8, 46). Altogether, 71% of human RetMICs responded substantially (ΔPSI ≥ 15) to SRRM3/4 expression in at least one experiment, in contrast to 9% for MSI1 (Dataset S6). Moreover, SRRM3/4 and MSI1 showed the opposite exon length preference: whereas SRRM3/4 enhanced mostly short retina-specific exons, MSI1 regulated a larger fraction of long exons than of short exons (Fig. 3C). Analysis of the surrounding intronic sequences revealed a significant enrichment of SRRM3/4-binding UGC motifs upstream of RetMICs and, to a lesser extent, of short (28 to 50 nt) RetLONGs, as compared to longer RetLONGs or a set of random control exons (Fig. 3D and SI Appendix, Fig. S8A). In contrast, both RetMICs and RetLONGs were enriched for MSI1-binding UAG motifs in the downstream intron compared to control exons (Fig. 3D and SI Appendix, Fig. S8B). Taken together, these results suggest that SRRM3/4 activity is sufficient to promote the inclusion of the majority of RetMICs, although MSI1 may be able to further modulate their inclusion levels. Interestingly, we also found a significant enrichment for known binding motifs for the neural-enriched factors Nova, Rbfox, and Elavl in the upstream introns of RetMICs (SI Appendix, Fig. S8 C–E), where their binding is expected to cause exon down-regulation (47). This is in contrast with neural microexons, which showed enrichment for inclusion-enhancing Nova and Rbfox binding motifs in the downstream introns (SI Appendix, Fig. S8 C–E), as previously reported (11, 48). These neuronal-enriched splicing factors are known to have very low expression in PRs (SI Appendix, Fig. S8F) (49, 50), suggesting a differential role for these proteins in the regulation of RetMICs and neural microexons.
Fig. 3.
Regulation of RetMICs by MSI1, SRRM3, and SRRM4. (A) Inclusion levels (using the PSI metric) of RetMICs in HEK293 cells ectopically expressing SRRM3, SRRM4, and MSI1. EV, empty vector. PSIs are quantified using vast-tools on RNA-seq data from cells 24 h postinduction with 1 µg/mL doxycycline; ****P < 0.0001 Wilcoxon test; ns: not significant. (B) RT-PCR assays showing the inclusion of RetMICs (CKAP5, ARL6, IMPDH1, PROM1, DYNC2H1, and CC2D2A), a RetLONG (MBD1), and a known MSI1-dependent exon (TAB3) in HEK293 cell lines upon ectopic expression of SRRM3, SRRM4, and MSI1. PSI levels quantified using ImageJ are shown below each gel. (C) Percent of retina-enriched exons by length group that showed substantial up-regulation (ΔPSI > 15) upon SRRM3/4 (dark gray) or MSI1 (gray) expression in at least one experiment. (D) RNA maps of SRRM3/4 and MSI1 associated binding motifs in the regions surrounding retina-enriched exons by length group and 1,000 random exons used as a control set. For simplicity, only the relevant upstream (SRRM3/4) or downstream (MSI1) introns are shown (full maps shown in SI Appendix, Fig. S8). Regions with a significant difference (FDR < 0.05) in motif coverage in the tested exon group with respect to the random exons are marked by thicker lines. Sliding window = 27 bp. (E) Srrm3 and Srrm4 gene expression levels (using the cRPKM metric) across mouse developing rods (data from VastDB). Expression levels are normalized to the stage with the highest cRPKM value for each gene.
srrm3 Is Necessary for RetMIC Inclusion in Zebrafish.
Unlike in most other types of neurons, Srrm4 has been shown to be lowly expressed in adult PRs (7). Indeed, analysis of Srrm4 expression in mouse developing rods revealed a sharp down-regulation of its mRNA levels over time (Fig. 3E). Interestingly, Srrm3 displays the opposite pattern, with increasing expression during PR maturation (Fig. 3E). This switch in expression from Srrm4 to Srrm3 in mature PRs suggests that, while both Srrm4 and Srrm3 can induce RetMICs inclusion in vitro, Srrm3 may be primarily responsible for RetMIC inclusion in mature PRs in vivo. To evaluate this possibility and investigate the physiological roles of these regulators and their targets, we used the CRISPR/Cas9 system to generate zebrafish mutant lines for srrm3 and srrm4 by targeting the eMIC domain (14) (Methods and Materials and SI Appendix, Fig. S9 A and B). To probe the functional impact of the srrm3 and srrm4 mutations that we generated, we overexpressed the mutated and WT zebrafish sequences in human HEK293 cells and assessed their ability to drive ectopic microexon inclusion (14). As predicted, in contrast to the WT versions, the mutated srrm3 and srrm4 proteins failed to promote the inclusion of all tested microexons (SI Appendix, Fig. S9C).
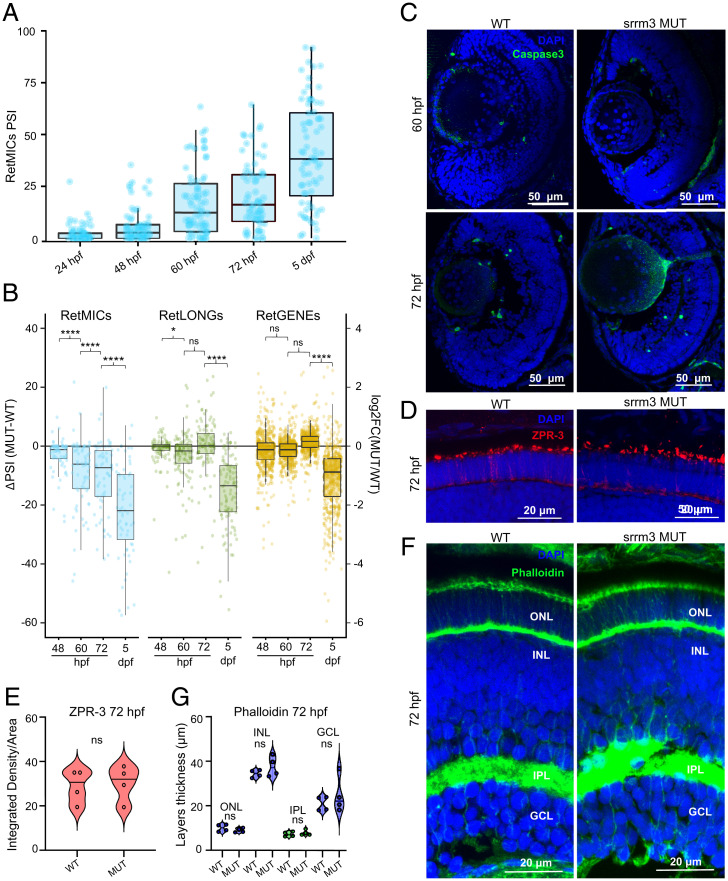
While fish homozygous for the srrm4 mutation (srrm4 MUT) did not display any evident phenotype, including changes in size or survival rate, srrm3 MUT and double homozygous mutant (DMUT) larvae died between 10 and 15 days postfertilization (dpf) (Fig. 4A). We hypothesized that this early mortality was linked to visual impairment, as reported for other zebrafish models of blindness in which the mutant fish are unable to forage for food and die of starvation after exhaustion of the yolk sac around the same days of development (51). If this were the case, we would expect that, in dark conditions, all genotypes resulting from a heterozygous (HET) cross would be equally affected, resulting in genotype ratios consistent with the Mendelian expectation. In line with this, in contrast to the specific depletion of homozygous mutants observed in control light conditions, the fish that survived at 13 dpf in the dark showed no homozygous depletion (26% WT, 39% HET, and 35% MUT) (Fig. 4B). These results thus indicate that, whereas all genotypes are equally likely to die in darkness, only MUT fish are more likely to do so in light conditions, pointing to a visual impairment upon srrm3 depletion. Therefore, to directly monitor visual performance, we performed an optokinetic response (OKR) test at 10 dpf, which measures reflexive eye movements elicited by a rotating visual pattern, confirming that srrm3 MUT fish display a severe decrease in visual performance compared to WT siblings (SI Appendix, Fig. S10 and Dataset S7).
Fig. 4.
srrm3 depletion in zebrafish causes early lethality and RetMIC down-regulation. (A) Genotype distribution for surviving larvae at different time points from in-crosses of srrm4+/−, srrm3+/−, or srrm4−/−;srrm3+/− (double mutants) fish. (B) Genotype distribution for surviving larvae at 13 dpf from a srrm3+/− in-cross in dark and control light conditions. For both A and B, the number of animals per genotype and time-point is indicated in the plots. (C) Enriched biological process GO terms for genes down-regulated in srrm3 homozygous mutants (MUT) eyes at 5 dpf [log2FC(MUT/WT) ≤ −1.5]. GO terms are grouped by ClueGO into three networks according to their GO groups. GO groups are highlighted using three different arbitrary colors, as listed in Dataset S9. P values are corrected with Bonferroni step down. (D) Change in inclusion levels [ΔPSI (MUT-WT)] quantified using vast-tools for all exons shorter than 300 bp with sufficient read coverage in WT and srrm3 MUT eyes at 5 dpf. Blue/green dots correspond to RetMICs and RetLONGs, respectively. (E) Density plots for ΔPSI distributions of RetMICs, RetLONGs, and other alternative exons (10 < PSI < 90 in WT and MUT at 5 dpf) (P = 8.0e-5; Wilcoxon rank-sum test).
To investigate the effect of the srrm3 depletion in the retina at the molecular level, we next examined gene expression and splicing changes by enucleating the eyes of WT and srrm3 MUT fish at 5 dpf and performing RNA-seq. Several genes crucial for PR functionality showed reduced expression in srrm3 MUT eyes (e.g., rhodopsin: log2FC(MUT/WT) = −3.72) (Dataset S8), and GO analysis of down-regulated genes further revealed a strong enrichment for visual function and phototransduction (Fig. 4C and Dataset S9). Quantification of alternative splicing events using vast-tools revealed multiple misregulated exons, 53% (225 of 409) of which corresponded to microexons. Moreover, although both RetMICs and RetLONGs showed global down-regulation in mutant eyes (Fig. 4D and Dataset S10), RetMICs exhibited significantly larger decreases in inclusion levels than RetLONGs (P = 8.0e-5; Wilcoxon Rank-Sum test) (Fig. 4E).
srrm3 Is Necessary for OS Maintenance and Visual Function in Zebrafish.
To follow up on these observations pointing at major visual defects, we then investigated the retinal morphology of the mutants. We performed immunostaining of frozen retinal sections of 5- and 10-dpf larvae using: 1) ZPR-3, a PR marker commonly used to stain rhodopsin in the OS (52), and 2) ZPR-1 (Arrestin3), a PR-specific antigen expressed in red and green double cones (52). ZPR-3 staining at 5 dpf revealed that OSs appear severely shortened and disorganized in srrm3 MUT larvae, with different spots corresponding to rhodopsin mislocalization throughout the outer nuclear layer (ONL) (Fig. 5 A–C and Dataset S11). Moreover, we observed a significant decrease of the ONL thickness in mutant retinae already at 5 dpf (Fig. 5G and Dataset S11), a common signature of PR degeneration. Indeed, at 10 dpf only a few spots of rhodopsin were detected in the mutants, while the ONL disappeared completely (Fig. 5 D and E). ZPR-1 staining confirmed that cones also displayed disorganized OS and progressive degeneration at 5 and 10 dpf (SI Appendix, Fig. S11 A–G and Dataset S11). In contrast, histological examination of single srrm4 MUT retinae revealed no morphological changes compared to WT fish (SI Appendix, Fig. S11 C and F and Dataset S11). The DMUT retinal phenotype at 5 dpf was similar to that of srrm3 MUT but stronger (SI Appendix, Fig. S11 H and I). Beyond the ONL, at 5 dpf the srrm3 MUT inner nuclear layer, inner plexiform layer, and ganglion cell layer displayed no thickness alterations compared to WT siblings, suggesting a PR-specific phenotype (Fig. 5G and Dataset S11). Finally, staining of Müller glial cells revealed no obvious differences in cell morphology between genotypes (SI Appendix, Fig. S11J).
Fig. 5.
PR degeneration and visual impairment in srrm3 mutants. (A–F) ZPR-3 staining in WT (A and D) and srrm3 homozygous mutants (MUT) (B and E) siblings at 5 and 10 dpf. Arrows show rhodopsin mislocalization. n = 5 for 5 dpf and n = 3 for 10 dpf fish. Quantifications of the ZPR-3+ area are provided for 5 dpf (C) and 10 dpf (F). P values from unpaired t tests. (G) Phalloidin staining in WT and srrm3 MUT retinae. The violin plot shows a thickness analysis for different retina layers: inner nuclear layer (INL), inner plexiform layer (IPL), and ganglion cell layer (GCL); n = 6. P values from unpaired t tests. (H–J) Electron microscopy images show absence of OSs or a dramatic OS length decrease in eyes of both srrm3 MUT (I) and DMUT (J) compared to WT ones (H) at 5 dpf; n = 2 for all the genotypes. Further magnification in the Lower panels (H–J) revealed smaller mitochondria (asterisks indicate mitochondria in fission process) and enlarged interphotoreceptor space (ips), quantified as mean of mitochondria area per field and interphotoreceptor space area normalized on field area, represented as fold-change (FC) in K and L, respectively. P values from one-way ANOVA tests with Tukey post hoc analyses. (Scale bars, 2 μm, Upper, and 1 μm, Lower.) n ≥ 4 eyes per genotype; n ≥ 13 fields per genotype were analyzed. (M) Caspase3 staining and associated quantifications in 5 dpf retina sections. n = 4 for each genotype. (N) Averaged ERG b-wave amplitudes from WT, srrm3 HET, and srrm3 MUT, or srrm3 MUT and DMUT at 5 dpf upon different light stimuli (1%, 10%, and 100%). All recordings were done in two independent experiments. n = 20 for WT, n = 34 for srrm3 HET, n = 17 for srrm3 MUT (N), n = 22 for srrm3 MUT (O), and n = 15 for DMUT. P values from one-way ANOVA tests. Error bars correspond to the SEM. Significance code for all tests: ****P = 0; ***, 0 < P < 0.001; **, 0.001 ≤ P < 0.01; *, 0.01 ≤ P < 0.05.
We next used electron microscopy to analyze OS ultrastructure. This analysis confirmed that OSs were dramatically shortened and disorganized at 5 dpf in both srrm3 MUT and DMUT eyes (Fig. 5 H–J), indicating that the ZPR-1 and ZPR-3 signals observed by immunofluorescence largely corresponded to mislocalized proteins. These results also revealed smaller mitochondria in both mutants (Fig. 5 H–J), with a decrease in the total mitochondria area per field with respect to WT retinae (Fig. 5K and Dataset S12). In particular, the mitochondria appeared more fragmented in mutant eyes, indicating an increase in the fission process (asterisks in Fig. 5 I and J, Lower panels), which represents an early event prior to PR cell death (53, 54). This phenotype was more severe in DMUT eyes, showing increased numbers of mitochondria, which are smaller and even more fragmented, probably due to a higher rate of fission (SI Appendix, Fig. S11K and Dataset S12). Mutant retinae also exhibited an enlargement of the interphotoreceptor space, particularly in DMUT eyes (ips in Fig. 5 I, J, and L, Lower, and Dataset S12). This interphotoreceptor space enlargement has been reported to occur due to the accumulation of extracellular vesicles (55) and is commonly observed in animal models of PR degeneration caused by alterations in ciliary trafficking (56, 57), suggesting the presence of compromised PR cells. To monitor PR cell death, we stained retinal sections from WT and srrm3 MUT larvae at 5 dpf with an anticaspase3 (active caspase3) antibody. The number of caspase3+ cells was significantly increased in MUT animals compared to WT siblings, with the signal being specifically localized to the PR layer (Fig. 5M and Dataset S11).
Altogether, these results point to strong defects in retinal function already at 5 dpf. To directly monitor visual performance at this stage, we did electroretinography (ERG) recordings (Dataset S13). ERG measures field potential changes of the whole retina induced by light (58), resulting in characteristic waveforms defining the neurons contributing to the response. At 5 dpf, zebrafish photoresponse is dominated by cones while rods are not mature until around 15 dpf (59). Thus, negative ERG a-wave representing photoresponse is masked by the positive b-wave, which mirrors ON-bipolar cell response (58). We observed that srrm3 MUT fish have a striking decrease in b-wave amplitude upon light stimulation with respect to HET and WT siblings, indicating a strong visual impairment (Fig. 5N and SI Appendix, Fig. S12A). DMUT fish performed even worse than srrm3 MUT fish (Fig. 5N and SI Appendix, Fig. S12B), supporting a minor but significant overlapping role for srrm4. Nevertheless, single srrm4 MUT fish did not display any significant visual impairment compared to WT siblings (SI Appendix, Fig. S12 C and D).
Early Missplicing of RetMICs Precedes PR Degeneration and Other Transcriptomic Alterations.
Although srrm3 is mainly known to regulate short exons, it is conceivable that the phenotypes observed at 5 dpf could be caused by other molecular alterations. Therefore, we investigated the onset of molecular and morphological alterations at earlier time points. First, we determined the expression dynamics of srrm3 and srrm4 by qPCR assays on eyes from 24 hours post fertilization (hpf) to 20 dpf larvae. Although both genes had comparable expression levels at 24 and 48 hpf, srrm3 showed much higher expression than srrm4 from 5 dpf onward (SI Appendix, Fig. S13A), in line with the pattern observed in mouse PRs (Fig. 3E). In addition, we did not observe compensatory up-regulation of srrm4 in srrm3 MUT eyes (SI Appendix, Fig. S13B). Second, we performed RNA-seq of WT eyes at 24, 48, 60, and 72 hpf. These data revealed that RetMICs start being progressively included from 48 to 60 hpf (Fig. 6A and Dataset S10), around the time when the OSs start differentiating in zebrafish (60). This inclusion pattern was further validated by RT-PCR assays for several representative RetMICs (SI Appendix, Fig. S13C).
Fig. 6.
Transcriptomic and morphological alterations of srrm3 mutants throughout development. (A) Inclusion levels (PSIs) of RetMICs in WT zebrafish heads at 24 hpf and eyes at 48 hpf, 60 hpf, 72 hpf, and 5 dpf. (B) Box plots showing the distribution of changes in inclusion levels for RetMICs and RetLONGs [ΔPSI (MUT-WT)] (left y axis) and in expression levels for RetGENEs [log2FC(MUT/WT)] (right y axis) in WT and srrm3 MUT eyes at different time points. Only exons with sufficient read coverage across all conditions are shown. P values from paired one-tailed Wilcoxon tests. (C) Caspase3 staining for WT and srrm3 MUT retinae at 60 hpf and 72 hpf; n = 4 for all genotypes and conditions. (D and E) ZPR-3 staining (D) and positive area quantification (E) for WT and srrm3 MUT retinae at 72 hpf; n = 4. P values from unpaired t tests. (F and G) Phalloidin staining (F) and thickness analysis for different retinal layers (G) for WT and srrm3 MUT retinae. n = 4. P values from unpaired t tests.
Next, we performed RNA-seq of eyes from the corresponding srrm3 MUT siblings at 48, 60, and 72 hpf and investigated the timing of misregulation of microexons and longer exons, as well as of gene expression (Datasets S8 and S10). Importantly, whereas more RetMIC skipping was observed in srrm3 MUT respect WT fish as early as 48 hpf (Fig. 6B and SI Appendix, Fig. S13C), and it increased progressively over time up to 5 dpf (Fig. 6B and SI Appendix, Fig. S13C), neither RetLONGs nor retina-specific genes (RetGENEs) (Methods and Materials) showed global down-regulation of inclusion/expression until 5 dpf (Fig. 6B and Dataset S10). A similar overall pattern was observed for all microexons, long exons, and genes that were found to be down-regulated in srrm3 MUT eyes at 5 dpf (Fig. 4 C and D and Dataset S10), among which only microexons showed a general down-regulation before 5 dpf (SI Appendix, Fig. S14A and Dataset S10). Moreover, differential gene-expression analyses at each time point revealed much fewer down-regulated genes [−log2FC(MUT/WT) < 1.5] in MUT eyes at 48, 60, and 72 hpf than at 5 dpf (Dataset S8), and gene set enrichment analysis (GSEA) showed no enrichment for PR-related categories among those genes (SI Appendix, Fig. S14B and Dataset S14). This pattern was confirmed through qPCR assays for some representative PR genes (SI Appendix, Fig. S14C).
These observations thus indicate that microexon misregulation precedes the broad, transcriptome-wide changes found at 5 dpf, which are likely caused indirectly by the observed PR malformations and death (Fig. 5M). Consistently, GSEA revealed a significant enrichment in apoptosis-related categories among up-regulated genes exclusively at 5 dpf (SI Appendix, Fig. S14B), and no apoptotic cells were detected by caspase3 staining on 60 and 72 hpf retinae (Fig. 6C). Moreover, we found no alterations in retinal layering or OS morphologies in srrm3 MUT larvae compared to WT siblings at either 60 hpf (SI Appendix, Fig. S15) or 72 hpf (Fig. 6 D–G and Dataset S11). Finally, injection of mRNA encoding only the eMIC domain of human SRRM3 (10, 14) into srrm3 MUT zebrafish embryos rescued the inclusion of tested representative microexons at 5 dpf (SI Appendix, Fig. S16A) and restored PR morphology and ONL thickness, as shown by Phalloidin staining (SI Appendix, Fig. S16B and Dataset S11). Altogether, these results point to a direct and specific role of microexons in causing the described phenotypes in srrm3 MUT zebrafish.
Discussion
Alternative splicing is an essential mechanism for generating molecular and functional diversity across cell and tissue types. In the context of the nervous system, which shows a particularly high degree of tissue-enriched alternative splicing, PRs differ in their transcriptomic profiles from other neuronal subtypes (50), likely as a reflection of their unique cellular morphology and function. Our results reveal a program of retina-enriched microexons that further contribute to this molecular specialization. These microexons are included in PRs in addition to neural microexons, which are shared with other neuronal types, and both microexosn programs have distinct and common regulatory and functional properties.
We demonstrate that the ectopic expression of either Srrm3 or Srrm4 is sufficient to drive the inclusion of most RetMICs in non-PR cells, as was shown for neural microexons (10, 21). However, we show that only Srrm3 is highly expressed in mature PRs and that the inclusion of most RetMICs depends mainly on this paralog in vivo. Nevertheless, we also found evidence for a minor but significant redundant role for srrm4 in the retina, since double mutant fish displayed stronger visual impairment and histological defects than single srrm3 mutants. Given the substantial levels of both Srrm3 and Srrm4 expression in other neuronal types (17, 21), these results thus raise the question of why RetMICs, particularly the retina-exclusive ones, have low or no inclusion in non-PR neurons. At least two nonmutually exclusive hypotheses may explain this pattern. First, although we found that the PR-specific splicing factor MSI1 on its own is not sufficient to drive the inclusion of most RetMICs, it is possible that MSI1 is necessary for RetMIC splicing in PRs in vivo, acting synergistically with SRRM3. In line with this hypothesis, we found a significant enrichment for MSI1 binding motifs in the downstream introns of both RetMICs and longer retina-enriched exons, a binding location that is expected to promote exon inclusion (7). Second, we found that RetMICs, in contrast to neural microexons, have enrichment for known binding motifs of Nova, Rbfox, and Elavl families, in positions expected to repress their inclusion (47). Given that these factors have very low expression in PRs but high in other neuronal cells (49, 50), it is plausible that these splicing factors could act as negative regulators of RetMICs in non-PR neurons, allowing for their inclusion only in PRs. Further research should evaluate these hypotheses to provide a more complete mechanistic understanding of the unique inclusion profile of RetMICs promoted by Srrm3.
At the functional level, we found that, similar to neural microexons (10), RetMICs can remodel protein structures of genes enriched for vesicle-mediated transport and related functions. However, we also revealed an enrichment for genes involved in cilium assembly, which was not observed for neural microexons. Altogether, these functions suggest that RetMICs enable a unique proteome specialization that is necessary for proper development and functioning of the OS, the highly modified cilium of PRs. Given the unusually high demand for vesicle formation, transport, and recycling in the OS (4), the specific modifications introduced by RetMICs in otherwise ubiquitous trafficking and ciliary machinery may help meet this high demand by promoting interactions with PR-specific substrates and facilitating unique catalytic properties not required (or even detrimental) in other cell types. In line with this idea, depletion of srrm3 caused severe malformation of the OS and vesicle accumulation, leading to PR degeneration and impaired visual function. Importantly, in homozygous srrm3 mutant larvae, OSs are generated but not maintained. This phenotype is similar to that described for various zebrafish mutants of terminal effector genes involved in intraflagellar transport (61), where mislocalization of visual pigments is associated with OS disappearance and PR degeneration. Furthermore, mutations in other genes necessary at different stages of ciliogenesis or vesicle trafficking (29, 57, 62) result in PR phenotypic alterations characterized by vesicle accumulation and dysmorphic OSs. Remarkably, the nearly complete loss of the OS that we report for srrm3 places this microexon regulator among the genes with the strongest mutant PR phenotypes reported so far in zebrafish, with features matching the ones observed in human retinitis pigmentosa (63).
Importantly, we show that RetMICs (as well as other microexons), but not RetLONGs or RetGENEs, were significantly dysregulated in srrm3 mutants at early time points, before morphological alterations or increased apoptosis were observed. Moreover, we demonstrated that morphological phenotypes in PRs can be rescued solely by injection of mRNA encoding the eMIC domain into zebrafish embryos, which is specifically responsible for microexon inclusion (14). Taken together, our results suggest that microexon misregulation is, at least to a large extent, directly responsible for the phenotypes we described in srrm3 mutant fish. However, it should be noted that, in addition to RetMICs, depletion of srrm3 also affected the inclusion of many neural-enriched microexons and other short exons in zebrafish eyes, whose misregulation likely contributes to the observed phenotypes. Nevertheless, three additional observations support that RetMICs, in particular, are likely to be directly behind at least some of the srrm3 mutant phenotypes. First, the inclusion of RetMICs was strongly misregulated in the eyes of srrm3 mutants. Second, as mentioned above, misregulation or mutation of the RetMICs in arl6 (34) and DYNC2H1 (35), respectively, have been shown to directly impact visual function, in line with the srrm3 mutant phenotypes observed here. Third, zebrafish lacking individual RetMIC-containing genes, such as cc2d2a (57) and ift88 (61), have been shown to display disorganization of the vesicle fusion machinery and strong PR degeneration. Therefore, and although some of the RetMICs highlighted here are in fact not strongly misregulated in srrm3 mutants (arl6; ΔPSI = −8) or even conserved in zebrafish (DYNC2H1), it is plausible that the multiple PR-related phenotypes caused by srrm3 mutation are at least in part caused by misregulation of RetMICs in genes with PR-related functions. However, which individual RetMICs underlie the srrm3 mutant phenotypes and through which molecular and cellular mechanisms they operate need to be further investigated. Remarkably, revealing their functional roles in PR differentiation and function would place RetMICs as candidates to underlie retinopathies without a known genetic cause, opening new avenues for the understanding of the highly complex molecular genetics of retinal diseases.
Methods and Materials
Definition of Tissue-Specific Exons and RetMICs, RetLONGs, and RetGENEs.
We downloaded exon inclusions levels and associated information from VastDB (23) for all the species included in the manuscript (human: hg38; mouse: mm10; chicken: galGal4; zebrafish: danRer10) and employed the Get_Tissue_Specific_AS.pl (64) script to derive sets of tissue-enriched microexons using the config files listed in Dataset S15. Definition of RetMICs and RetLONGs was then done based on a retina specificity score that takes the distributions of retina and neural samples into account. For RetGENEs, we used a comparable approach through the Get_Tissue_Specific_GE.pl (64) script.
Regulatory Analysis of RetMICs and RetLONGs.
HEK293 lentiviral cell lines used in this study to ectopically express SRRM3, SRRM4, or MSI1 were generated using a previously described protocol (46). pCW57.1 lentiviral vector (empty vector) was used as a negative control. Expression of the transgenes was induced using 1 µg/mL of doxycycline for 24 h. RNA was used for RT-PCR assays (primers listed in Dataset S16) and RNA-seq (mapping statistics in Dataset S17). RNA maps were done using the rna_maps function from Matt (65) for NOVA (YCAY), RBFOX (GCATG), and ELAVL (TTTNTTT) binding motifs.
Generation of srrm3 and srrm4 Zebrafish Mutant Lines.
Fish procedures were approved by the Barcelona Biomedical Research Park Institutional Animal Care and Use Ethic Committee (PRBB–IACUEC). To create zebrafish mutant lines (SI Appendix, Fig. S9), we used CRISPR/Cas9 to target the eMIC domain of srrm3 [Tg(HuC:GFP; srrm3_eMIC); GGGAATAACTGCGTGAGCGGCGG] and srrm4 [Tg(HuC:GFP; srrm4_eMIC); TGATTCTGCGGGCTTCCAGGTGG]. Mutant sequences were probed for microexon regulatory activity by transfecting them into HEK293, as previously described (14).
Immunofluorescence and Histological Retinal Analyses.
Zebrafish eyes were fixed in 4% paraformaldehyde (PFA) overnight, cryoprotected with 30% sucrose overnight, embedded in OCT, and cryosectioned. Twenty-micrometer cryosections were collected on slides. For PR marker staining, sections were incubated with primary antibodies (ZPR-1 [ZDB-ATB-081002-43] 1:400 and ZPR-3 [ZDB-ATB-081002-45] 1:200, Zebrafish International Resource Center, GS6 [Millipore]) overnight in blocking solution. Sections were then incubated with the Alexa Fluor secondary antibodies (1:1,000; Invitrogen) and counterstained with DAPI (Vector Laboratories). The Phalloidin staining was performed using a previously described protocol (66). The caspase3 assay was performed on retina sections following the protocol described above (anticaspase3; Fisher Scientific, 15889738; 1:500). Imaging analyses were done with the ImageJ tool.
Electron Microscopy.
The samples were cut on ultramicrotome Leica EM UC7 and collected on the single-slot oval grids and analyzed with an FEI electron microscope. OS, mitochondrial, and interphotoreceptor space area was determined using FEI software. The analysis was carried out on four eyes per genotype, for each genotype an n ≥ 13 fields was analyzed.
ERG and OKRs.
For ERG experiments, srrm3 MUT, srrm4 MUT and DMUT larvae as well as different control siblings were recorded at 5 dpf, as previously described (67). OKRs were recorded as described previously (68).
Supplementary Material
Acknowledgments
We thank Juan Valcárcel and Juan Ramón Martínez for scientific support and critical reading of the manuscript; Xavier Hernandez-Alias, Antonio Torres-Méndez and Alessia Indrieri for scientific discussion; Claire Lastrucci for supervision during the initial stages of the project; Xavier Henandez-Alias and Miquel Anglada-Girotto for assistance during the bioinformatic analysis; Jochen Hecht and the CRG Genomics Unit for the RNA-sequencing services; Elena Polishchuk and the Telethon Institute of Genetics and Medicine Advanced Microscopy and Imaging Facility for the Electron Microscopy services. We acknowledge the support of the Spanish Ministry of Science and Innovation through the Centro de Excelencia Severo Ochoa (CEX2020-001049-S, MCIN/AEI/10.13039/501100011033), and the Generalitat de Catalunya through the CERCA programme.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117090119/-/DCSupplemental.
Data Availability
Extended methods can be found in SI Appendix. RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE180781) (69). All other RNA-seq samples used in this study are publicly available and listed in Dataset S17. All other study data are included in the main text and supporting information.
References
- 1.Wert K. J., Lin J. H., Tsang S. H., General pathophysiology in retinal degeneration. Dev. Ophthalmol. 53, 33–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoon M., Okawa H., Della Santina L., Wong R. O. L., Functional architecture of the retina: Development and disease. Prog. Retin. Eye Res. 42, 44–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell L. J., West M. C., Jensen A. M., A high content, small molecule screen identifies candidate molecular pathways that regulate rod photoreceptor outer segment renewal. Sci. Rep. 8, 14017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sánchez-Bellver L., Toulis V., Marfany G., On the wrong track: Alterations of ciliary transport in inherited retinal dystrophies. Front. Cell Dev. Biol. 9, 623734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters A. M., Beales P. L., Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol. 26, 1039–1056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheway G., Lord J., Baralle D., Splicing in the pathogenesis, diagnosis and treatment of ciliopathies. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 194433 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Murphy D., Cieply B., Carstens R., Ramamurthy V., Stoilov P., The Musashi 1 controls the splicing of photoreceptor-specific exons in the vertebrate retina. PLoS Genet. 12, e1006256 (2016). Erratum in PLos Genet. 12, e1006432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling J. P., et al. , ASCOT identifies key regulators of neuronal subtype-specific splicing. Nat. Commun. 11, 137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M. M., Zack D. J., Alternative splicing and retinal degeneration. Clin. Genet. 84, 142–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irimia M., et al. , A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y. I., Sanchez-Pulido L., Haerty W., Ponting C. P., RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome Res. 25, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volfovsky N., Haas B. J., Salzberg S. L., Computational discovery of internal micro-exons. Genome Res. 13 (6A), 1216–1221 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis J. D., et al. , Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol. Cell 46, 884–892 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Torres-Méndez A., et al. , A novel protein domain in an ancestral splicing factor drove the evolution of neural microexons. Nat. Ecol. Evol. 3, 691–701 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Ule J., et al. , An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Zhang C., et al. , Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 22, 2550–2563 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calarco J. A., et al. , Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138, 898–910 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Quesnel-Vallières M., et al. , Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol. Cell 64, 1023–1034 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Quesnel-Vallières M., Irimia M., Cordes S. P., Blencowe B. J., Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 29, 746–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano Y., et al. , A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet. 8, e1002966 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano Y., Wiechert S., Bánfi B., Overlapping activities of two neuronal splicing factors switch the GABA effect from excitatory to inhibitory by regulating REST. Cell Rep. 27, 860–871.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonatopoulos-Pournatzis T., et al. , Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Mol. Cell 77, 1176–1192.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Tapial J., et al. , An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 27, 1759–1768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Anand M., Rao K. N., Khanna H., Cilia in photoreceptors. Methods Cell Biol. 127, 75–92 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Kim S., et al. , Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. U.S.A. 116, 10824–10833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veleri S., et al. , Ciliopathy-associated gene Cc2d2a promotes assembly of subdistal appendages on the mother centriole during cilia biogenesis. Nat. Commun. 5, 4207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roosing S., et al. ; POC1B Study Group, Disruption of the basal body protein POC1B results in autosomal-recessive cone-rod dystrophy. Am. J. Hum. Genet. 95, 131–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiens C. J., et al. , Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J. Biol. Chem. 285, 16218–16230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Z., et al. , Deletion of the transmembrane protein Prom1b in zebrafish disrupts outer-segment morphogenesis and causes photoreceptor degeneration. J. Biol. Chem. 294, 13953–13963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plana-Bonamaisó A., et al. , Post-translational regulation of retinal IMPDH1 in vivo to adjust GTP synthesis to illumination conditions. eLife 9, e56418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pazour G. J., et al. , The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157, 103–113 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama K., Katoh Y., Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 163, 155–164 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Wiegering A., Rüther U., Gerhardt C., The ciliary protein Rpgrip1l in development and disease. Dev. Biol. 442, 60–68 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Pretorius P. R., et al. , Identification and functional analysis of the vision-specific BBS3 (ARL6) long isoform. PLoS Genet. 6, e1000884 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vig A., et al. ; Genomics England Research Consortium, DYNC2H1 hypomorphic or retina-predominant variants cause nonsyndromic retinal degeneration. Genet. Med. 22, 2041–2051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mistry J., et al. , Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hulo N., et al. , The PROSITE database. Nucleic Acids Res. 34, D227–D230 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieper U., et al. , ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 42, D336–D346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosca R., Pons T., Céol A., Valencia A., Aloy P., Towards a detailed atlas of protein-protein interactions. Curr. Opin. Struct. Biol. 23, 929–940 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Savojardo C., Manfredi M., Martelli P. L., Casadio R., Solvent accessibility of residues undergoing pathogenic variations in humans: From protein structures to protein sequences. Front. Mol. Biosci. 7, 626363 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ter Haar E., Harrison S. C., Kirchhausen T., Peptide-in-groove interactions link target proteins to the β-propeller of clathrin. Proc. Natl. Acad. Sci. U.S.A. 97, 1096–1100 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buss F., Arden S. D., Lindsay M., Luzio J. P., Kendrick-Jones J., Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J. 20, 3676–3684 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ménétrey J., et al. , The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature 435, 779–785 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niekamp S., Coudray N., Zhang N., Vale R. D., Bhabha G., Coupling of ATPase activity, microtubule binding, and mechanics in the dynein motor domain. EMBO J. 38, e101414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Márquez Y., et al. , ExOrthist: A tool to infer exon orthologies at any evolutionary distance. Genome Biol. 22, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Head S. A., et al. , Silencing of SRRM4 suppresses microexon inclusion and promotes tumor growth across cancers. PLoS Biol. 19, e3001138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witten J. T., Ule J., Understanding splicing regulation through RNA splicing maps. Trends Genet. 27, 89–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita M., Yamamoto R., Mitsui K., Kanazawa H., Altered motor activity of alternative splice variants of the mammalian kinesin-3 protein KIF1B. Traffic 10, 1647–1654 (2009). [DOI] [PubMed] [Google Scholar]
- 49.PLOS Genetics Staff, Correction: The Musashi 1 controls the splicing of photoreceptor-specific exons in the vertebrate retina. PLoS Genet. 12, e1006432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weyn-Vanhentenryck S. M., et al. , Precise temporal regulation of alternative splicing during neural development. Nat. Commun. 9, 2189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stearns G., Evangelista M., Fadool J. M., Brockerhoff S. E., A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J. Neurosci. 27, 13866–13874 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hochmann S., et al. , Fgf signaling is required for photoreceptor maintenance in the adult zebrafish retina. PLoS One 7, e30365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirra S., Marfany G., Mitochondrial gymnastics in retinal cells: A resilience mechanism against oxidative stress and neurodegeneration. Adv. Exp. Med. Biol. 1185, 513–517 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Mirra S., et al. , CERKL, a retinal dystrophy gene, regulates mitochondrial function and dynamics in the mammalian retina. Neurobiol. Dis. 156, 105405 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Spencer W. J., et al. , PRCD is essential for high-fidelity photoreceptor disc formation. Proc. Natl. Acad. Sci. U.S.A. 116, 13087–13096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sukumaran S., Perkins B. D., Early defects in photoreceptor outer segment morphogenesis in zebrafish ift57, ift88 and ift172 intraflagellar transport mutants. Vision Res. 49, 479–489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ojeda Naharros I., et al. , Loss-of-function of the ciliopathy protein Cc2d2a disorganizes the vesicle fusion machinery at the periciliary membrane and indirectly affects Rab8-trafficking in zebrafish photoreceptors. PLoS Genet. 13, e1007150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makhankov Y. V., Rinner O., Neuhauss S. C. F., An inexpensive device for non-invasive electroretinography in small aquatic vertebrates. J. Neurosci. Methods 135, 205–210 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Bilotta J., Saszik S., Sutherland S. E., Rod contributions to the electroretinogram of the dark-adapted developing zebrafish. Dev. Dyn. 222, 564–570 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Crespo C., Knust E., Characterisation of maturation of photoreceptor cell subtypes during zebrafish retinal development. Biol. Open 7, bio036632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsujikawa M., Malicki J., Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 42, 703–716 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Insinna C., Baye L. M., Amsterdam A., Besharse J. C., Link B. A., Analysis of a zebrafish dync1h1 mutant reveals multiple functions for cytoplasmic dynein 1 during retinal photoreceptor development. Neural Dev. 5, 12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Soest S., Westerveld A., de Jong P. T., Bleeker-Wagemakers E. M., Bergen A. A., Retinitis pigmentosa: Defined from a molecular point of view. Surv. Ophthalmol. 43, 321–334 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Martín G., Márquez Y., Mantica F., Duque P., Irimia M., Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 22, 35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gohr A., Irimia M., Matt: Unix tools for alternative splicing analysis. Bioinformatics 35, 130–132 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Letelier J., et al. , Mutation of Vsx genes in zebrafish highlights the robustness of the retinal specification network. bioRxiv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.01.20.477122v1. [DOI] [PMC free article] [PubMed]
- 67.Zang J., Keim J., Kastenhuber E., Gesemann M., Neuhauss S. C. F., Recoverin depletion accelerates cone photoresponse recovery. Open Biol. 5, 150086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rinner O., Rick J. M., Neuhauss S. C. F., Contrast sensitivity, spatial and temporal tuning of the larval zebrafish optokinetic response. Invest. Ophthalmol. Vis. Sci. 46, 137–142 (2005). [DOI] [PubMed] [Google Scholar]
- 69.L. Ciampi, S. Head, L. Serrano, and M.l Irimia, Regulation of retina microexons. GEO Data Base. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE180781. Deposited 14 April 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Extended methods can be found in SI Appendix. RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE180781) (69). All other RNA-seq samples used in this study are publicly available and listed in Dataset S17. All other study data are included in the main text and supporting information.