Abstract
In the current era of T cell–based immunotherapies, it is crucial to understand which types of MHC-presented T cell antigens are produced by tumor cells. In addition to linear peptide antigens, chimeric peptides are generated through proteasome-catalyzed peptide splicing (PCPS). Whether such spliced peptides are abundantly presented by MHC is highly disputed because of disagreement in computational analyses of mass spectrometry data of MHC-eluted peptides. Moreover, such mass spectrometric analyses cannot elucidate how much spliced peptides contribute to the pool of immunogenic antigens. In this Perspective, we explain the significance of knowing the contribution of spliced peptides for accurate analyses of peptidomes on one hand, and to serve as a potential source of targetable tumor antigens on the other hand. Toward a strategy for mass spectrometry independent estimation of the contribution of PCPS to the immunopeptidome, we first reviewed methodologies to identify MHC-presented spliced peptide antigens expressed by tumors. Data from these identifications allowed us to compile three independent datasets containing 103, 74, and 83 confirmed T cell antigens from cancer patients. Only 3.9%, 1.4%, and between 0% and 7.2% of these truly immunogenic antigens are produced by PCPS, therefore providing a marginal contribution to the pool of immunogenic tumor antigens. We conclude that spliced peptides will not serve as a comprehensive source to expand the number of targetable antigens for immunotherapies.
Keywords: PCPS, spliced antigens, tumor antigens
The presentation of antigenic peptides by MHC class I (MHC-I; HLA class I in humans) is of major importance for the initiation and execution of CD8+ T cell–mediated immune responses. Until the beginning of this century, MHC-I-presented peptides were thought to arise from linearly degraded proteins. This perspective changed when, in 2004, the first spliced antigen was identified. Spliced antigens consist of two joined peptide fragments which are not linearly encoded, for example, with an intervening sequence in the natural protein. Since then, five more spliced peptides have been characterized (1–6).
Currently, there are two essential but unsolved issues related to proteasome-catalyzed peptide splicing (PCPS). First, the existence of spliced peptides raises questions on the reliability of current mass spectrometry (MS)-based identification of MHC-I-presented peptides (immunopeptidome), because the data may contain a high error rate when spliced peptides significantly contribute to the immunopeptidome. But it is yet unknown what the contribution of spliced peptides to the immunopeptidome is.
Different analysis workflows and validation methods of MS data of MHC-I-derived peptides divide the field into groups convinced of a limited versus a large contribution of spliced peptides to the peptidome, varying from close to 0% to up to 34% (7–17). This vast disagreement is reinforced in the absence of a different, widely accepted, or more objective method to determine this percentage.
Secondly, the existence of spliced peptides is of great interest to the oncoimmunology community, since spliced peptides potentially serve as new immunotherapeutic targets (18). But, since the contribution of these peptides to the immunopeptidome is unknown, it is unclear whether they represent a significant new source of tumor antigens. Additionally, the immunogenicity of peptides identified by MS usually remains unknown, while this information is crucial to draw (patho)physiologically relevant conclusions. Thus, preferably, an alternative method should not just focus on reliable identification of spliced peptides in the MHC-I peptidome but also provide information on the contribution of such peptides to immune responses.
In this Perspective, we introduce spliced peptide generation, discuss the debate in the immunopeptidomics community, and elaborate on the potential relevancy in the field of tumor immunology. More importantly, we summarize the various methods used to identify individual T cell antigens, including spliced peptides, and highlight the undeniable biases that accompany each of these methods. Finally, based on the most unbiased approaches to date, we make a substantiated estimation of the involvement of spliced peptides in antitumor immune responses, and discuss how these findings relate to the debate in the immunopeptidomics field.
Spliced Peptides and Immunopeptidomics
Proteasomal Activity Generates both Regular and Spliced (Antigenic) Peptides.
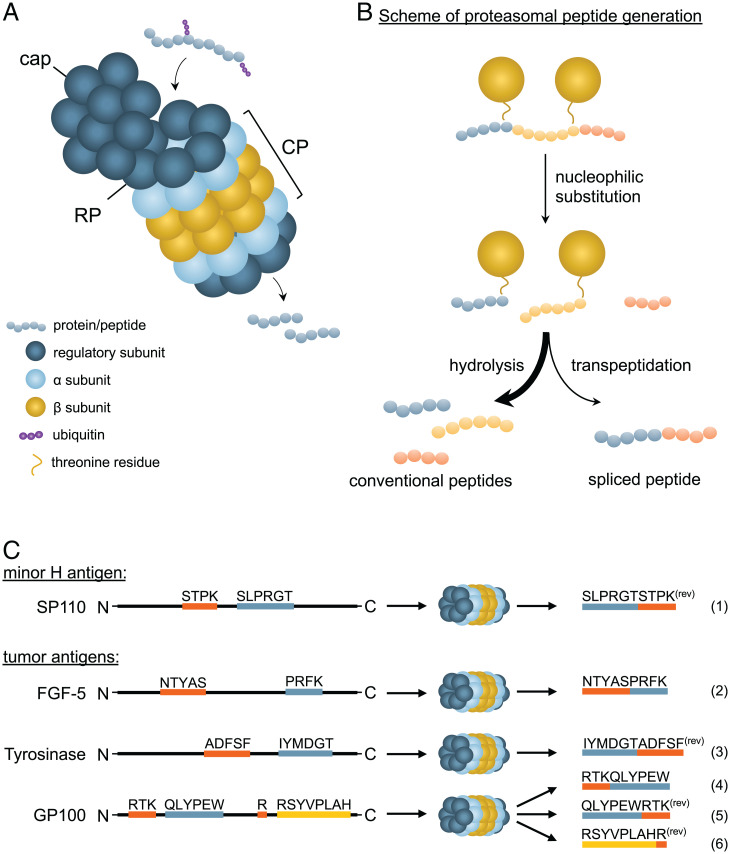
The generation and MHC-I presentation of peptides conventionally requires proteasomal cleavage of full-length proteins in the cytoplasm (Fig. 1A). Produced peptides are then transported into the endoplasmic reticulum (ER) to be loaded in the peptide-binding groove of MHC-I by the so-called peptide loading complex. Peptide splicing takes place during proteasomal processing. The proteasome consists of one or two S19 regulatory particles (RPs) that unite with an S20 catalytic particle (CP) to establish a fully operating proteasome. The RP captures ubiquitinated proteins before facilitating their translocation into the CP, where they will be catalytically processed. The cylinder-shaped CP is formed by two β-rings (β1 and β7), with subunits β1, β2, and β5 responsible for the hydrolysis of peptides, positioned between two α-rings (α1 and α7). These α-rings function as a gateway to limit entrance of substrates by the capping of one or both sides by specific proteasome regulators (as extensively reviewed by Schmidt and Finley (19) (Fig. 1A). The β1, β2, and β5 subunits contain reactive N-terminal threonine residues that cleave proteins by a process of nucleophilic substitution and hydrolysis before dispersing the ensuing peptide products into the cytosol (Fig. 1 A and B) (20).
Fig. 1.
Proteasomal activity generates both regular and spliced (antigenic) peptides. (A) The proteasome consists of one or two RPs, a cap and two rings consisting of seven α/β subunits each that form the CP. Ubiquitinated proteins or polypeptides are captured by the RP and processed by the CP into peptides. (B) Proteins are cleaved by means of nucleophilic substitution; reactive threonines attack the carbonyl carbon of scissile amino acid bonds, resulting in separate peptides bound to the β subunits. Conventional peptides are formed through hydrolysis, while spliced peptides are generated by transpeptidation of a donor peptide. (C) Six tumor-expressed cis-spliced antigens have been identified thus far, of which four are ligated in reverse order (rev). The antigen derived from SP110 is a minor H antigen, whereas the other five are tumor-associated antigens. Three of the latter group are derived from GP100.
The proteasome readily digests proteins to peptides ranging from 2 amino acids (aa) to 24 aa in length (21). During PCPS, the proteasome not only degrades proteins into smaller peptides but also ligates peptides together through a process called transpeptidation (Fig. 1B). Instead of hydrolysis, the free amino group of adjacent peptides reacts with the C terminus of the threonine-bound peptide, which, due to a lower efficiency, still yields fewer spliced compared to linear nonspliced peptides (22). Such spliced peptides are then composed of fragments that originate from the same (cis-splicing) or distinct (trans-splicing) parental proteins. In addition, reverse cis-spliced peptides may be generated when fragments are ligated in the reverse order from how they appear from N terminus to C terminus in the parental protein. Up to this day, six cis-spliced antigens have been defined, of which four are ligated in reverse order (Fig. 1C) (1–6). Although trans-splicing occurs in vitro, the occurrence and significance of trans-splicing in cells is disputable (8, 23).
Upon IFN-γ stimulation, new proteasomes are synthesized using β1i, β2i, and β5i instead of β1, β2, and β5 subunits, giving rise to the so-called immunoproteasome. The β-subunits of the immunoproteasome harbor different peptidase properties compared to the β-subunits of a constitutive proteasome, resulting in a different pool of peptides (24–26). For example, the immunosubunit β5i has a broader specificity compared to its β5 counterpart, generating more hydrophobic residue-bearing peptides that generally fit better into the MHC-I-binding cleft (27). The variety in proteasomal activity may have consequences for the generation of spliced peptides. Two spliced antigens, GP100 and FGF-5, are less efficiently produced by the immunoproteasome compared to the constitutive proteasome (2, 4, 23, 28), whereas the spliced peptide derived from SP110 is slightly more efficiently produced by the immunoproteasome (1, 28). Although likely, it is too premature to state that different catalytic properties result in the generation of a different pool of spliced peptides in vivo.
Our current knowledge regarding the requirements for PCPS to occur is limited. In vitro, proteasome-mediated cleavage of short parental peptides (14 aa to 22 aa) results in a higher proportion of unique spliced peptides in the processed peptide pool compared to when a long polypeptide source (23 aa to 47 aa) is used, which indicates that shorter substrates—such as partially degraded proteins—are more susceptible to PCPS (22). In addition to size, synthetic peptides with certain amino acid motifs were more prone to participate in a splicing reaction than others, suggesting that PCPS does not occur randomly (20, 21). Lastly, splicing reactions are more efficient for shorter intervening sequences between the fragments (23, 29).
Resolving the Contribution of Spliced Epitopes to the MHC-I Immunopeptidome Will Facilitate More-Accurate Immunopeptidome Analyses.
Since the discovery of PCPS in 2004 (2), various efforts have been undertaken to reliably identify spliced peptides at a large scale using MHC-I peptidome MS (liquid chromatography–tandem MS [LC-MS/MS]). This resulted in highly contradicting reports claiming that spliced peptides are either substantially or only minimally represented in the MHC-I-presented peptidome (7, 8, 12–14, 16, 30).
In 2016, Liepe et al. (7) showed that around 30% of antigenic peptides were produced by PCPS. In this study, MHC-I peptidome MS data were analyzed using a computational algorithm. For each protein in the annotated human proteome, all theoretical 9-mer to 12-mer cis and reverse-cis spliced peptides were computed with a maximum intervening sequence of 25 aa. Similarly, all possible 9-mer to 12-mer nonspliced linear peptides were computed. The MS/MS spectra were then aligned against this reference peptidome, which led to the identification of 966 (∼30%) and 3,417 (∼34%) spliced peptides from two Epstein–Barr virus-immortalized lymphoblastoid cell lines (EBV-LCLs), and 1,154 spliced peptides (∼29%) from primary human fibroblasts, of which the raw data were derived from Bassani-Sternberg et al. (31).
In contrast, a different computational analysis of LC-MS/MS MHC-I peptidome datasets performed by Mylonas et al. (14) revealed that less than 2 to 6% of MHC-I-binding peptides are spliced peptides. The algorithm computed the five best scoring de novo sequences for each MS/MS spectrum. Spectra without human proteome matches of these sequences were analyzed to match the proteome with one splicing event (cis or reverse cis). Interestingly, analysis of the fibroblast data (31), taking the spliced peptides by Liepe et al. (7) into account, reidentified only 6 of these 1,154 peptides as spliced peptides (14). Additionally, Rolfs et al. (12) found that a maximum of 1 to 4% of all peptides from the MHC-I immunopeptidome, MHC-II immunopeptidome, and trypsin-digested mouse islet proteins are spliced peptides. Their reanalysis of the Bassani-Sternberg fibroblast dataset reported back ∼1% of peptide sequences as cis-spliced peptides. These findings coincide with the theory that PCPS occurs rarely, based on the fact that only six spliced antigens have been reported since the discovery of PCPS (1–6, 32). In line with these results, Erhard et al. (9) reanalyzed one of the EBV-LCL datasets of Liepe et al. using Peptide-PRISM and identified 10 spliced peptides (<0.1%). Willimsky et al. (33) and Lichti (34) also concluded that spliced peptides contribute minimally to the immunopeptidome, whereas Faridi et al. (8), Kuckelkorn et al. (35), Specht et al. (36), and Paes et al. (22) have reported more significant contributions of spliced peptides, ranging from ∼11 to 45%.
A potential factor that may have led to discrepancies in these results is incomplete or overcomplete reference databases (7). Sources from which MHC-presented peptides can emerge include proteins containing posttranscriptional modifications or single-nucleotide polymorphisms (SNPs), or proteins derived from alternative RNA splicing or RNA editing or other noncanonical processes (34). Such additional protein sources should be taken into account when annotating spliced peptides, because incomplete peptide reference databases can lead to incorrect peptide assignments. Furthermore, results from these algorithms should be experimentally validated before drawing conclusions. Not every report included less conventional source proteins in their reference databases or thoroughly validated their results, which may have led to overestimations of the proportion of spliced peptides in the immunopeptidome.
It is clear that assumptions underlying analyses of acquired peptidome data highly influence the outcome of spliced peptide identification attempts. Wrong assumptions on spliced peptide occurrence may lead to significant erroneous peptide calling in computational workflows. Thus, the current differences in workflows that lead to vast discrepancies between the reported frequencies highlight the need for a different analysis to estimate the fraction of spliced peptides in the immunopeptidome. Such analysis will contribute to the debate as a benchmark for workflow outcomes, which may support the development of more accurate immunopeptidome analyses.
The Role of PCPS in Anticancer Immunotherapy
An interesting question is which (patho)physiological role PCPS and the generated spliced peptides play, especially because the process is evolutionarily conserved dating back to yeast (37). Although this question remains unanswered, there is hope that PCPS provides new opportunities for anticancer immunotherapy.
Currently, the identification of antigen specificity of tumor-infiltrating T cells is considered key for the design of next-generation immunotherapies. Many antigens have been identified that are more or less selectively expressed by tumors, including neoantigens derived from mutated proteins. Still, a large proportion of T cells in tumor tissue have an unknown specificity. It is therefore tempting to speculate that at least some of these T cells respond to tumor-expressed spliced antigens. To determine whether (neo)antigen identification strategies need to include a focus on spliced antigen discovery, it is of utmost importance to know the fraction of T cells recognizing a spliced tumor antigen.
Until today, only a few publications have portrayed a role for spliced peptides as targets in antitumor immunotherapies. Adoptive transfer of an autologous tumor-infiltrating lymphocyte (TIL) which recognized a spliced antigen originating from Tyrosinase (Fig. 1C) led to complete tumor regression in one patient (32). Other spliced antigen-specific T cells may also exert efficient antitumor activity upon adoptive transfer or after activation by other specific immunotherapies. This is especially relevant for neoantigens, since somatic mutations may be presented by the patient’s MHC-I molecules as spliced, but not linear, antigens. For instance, mutations in codons 12 and 13 account for 90% of KRAS mutations in cancer, including the G12D and G12V mutations (38, 39). Although the G12D mutation is presented as a nonspliced epitope on HLA-C*08:02 and HLA-A*11:01, it lacks broad applicability as a linear target, because this mutation is not efficiently presented by more-prevalent MHC-I alleles in humans (40–42). A spliced peptide containing the G12V mutation is predicted to be presented by the more prevalent HLA-A*02:01 allele, suggesting that therapeutic options can be extended with PCPS antigens (43). Lastly, vaccination against PCPS products that were presented by tumor cells was efficacious in preclinical mouse models (44). Yet, the actual relevance of PCPS for immunotherapy depends on how frequently spliced peptides are generated, whether such peptides are correctly processed and presented by MHC-I molecules, whether their expression is tumor restricted, and whether they can elicit a T cell response.
Spliced Peptides Need to Be Tumor Specific, Sufficiently Expressed for T Cell Recognition, and Identifiable to Be Considered as Potential Therapeutic Antigens.
To date, there is no evidence that spliced peptides have a different expression level on tumor cells compared to regular, linear peptides. As described above, spliced peptide generation may differ between the constitutive proteasome versus immunoproteasome (23, 28), although neither of these proteasomes are tumor restricted. This means that the likelihood of spliced peptides to be genuinely tumor specific is dependent on the expression profile of the encoding gene, either with or without mutations. For example, the spliced antigen derived from Tyrosinase is presented by both healthy and malignant melanocytes (3).
So far, only a few spliced antigens have been identified that are expressed by tumors (1–6). The broad and successful clinical application of (spliced) tumor antigen-specific immunotherapy requires more than a handful of immunogenic antigens. TIL therapy, for example, is a T cell therapy that has shown complete and durable responses in advanced metastatic cancer (41, 45, 46). These TIL studies have relied on the infusion of products containing T cells expressing T cell receptors (TCRs) with various reactivities for effective elimination of tumors (45–48). In the case of TIL, the detailed composition of the product is generally unknown, and the frequency of T cells that may be effective in mounting a robust antitumor response may be limited. Therefore, T cell products with TCRs recognizing a single molecularly defined target have been evaluated. These include studies against attractive (relatively) universal targets such as NY-ESO1, WT1, GP100, and the MAGE antigens that have been identified in the past. But, while such T cell therapies utilizing a single TCR have been capable of inducing antitumor responses in a clinical setting (49, 50), recent studies have demonstrated common obstacles for broad implementation of these immunotherapies. Tumor cells can escape the selective pressure exerted by the T cells through down-regulation or mutation of the targeted antigens (51). Likewise, expression of individual HLA alleles may be lost (52–55). Additionally, solid cancers are known for their complex and heterogeneous landscape (56). Such a lack of uniform expression of the target antigen in tumor cells potentially limits the therapeutic efficacy of monospecific engineered T cell therapy (57). Lastly, the lack of broad HLA allele coverage by the TCR portfolio of individual or collaborating institutes limits broad patient inclusion in clinical studies.
Together, these issues suggest that targeting a single antigen is likely not sufficient to induce complete tumor regression in a broad patient population. Thus, expansion of the list of targetable tumor-specific antigens is required for broad clinical application of (multispecificity) immunotherapy (58), which will need to be personalized based on HLA allotype and antigen expression. In addition, neoantigens and minor histocompatibility (H) antigens are considered more specific and more immunogenic than tumor-associated antigens (59–61), but these are not universal targets, due to their personalized character, and suffer from the same issues for clinical translation. To select the most optimal neoantigen or minor H antigen identification strategies, it is crucial to understand which kind of peptide classes are represented in the immunopeptidome. From this perspective, the oncoimmunology community has closely followed the developments and debate on the proportion of spliced peptides among immunogenic antigens. If spliced peptides make up a large fraction of immunogenic antigens, then antigen identification strategies should also include the option to discover targetable tumor antigens, minor H antigens, and neoantigens from such a comprehensive source.
Hence, it is currently important to picture the contribution of spliced peptides to the immunopeptidome using an independent approach. We reasoned that an analysis of previously identified T cell antigens may shed new light on this matter, as it concerns the same repertoire of MHC-I-presented peptides. Of note is that the use of antigen-specific T cells, as well as the use of MS of eluted peptides, has inherent limitations in detecting a full representation of the MHC-I peptide repertoire. Furthermore, a T cell–based analysis will also reveal the physiological impact of spliced peptides. For an equitable analysis of T cell antigen data, it is essential to consider intrinsic biases of T cell antigen identification methods that may skew identification of linear over spliced antigens.
Forward Methods Allow for Relatively Unbiased Identification of Spliced Antigens.
A main group of previously identified antigens are expressed by tumors, including classical tumor (associated) antigens, neoantigens, and minor H antigens. The latter are MHC-presented peptides that are polymorphic in the population and are therefore highly immunogenic after MHC-matched transplantation from an allogeneic donor (62, 63). Their polymorphic nature is caused by natural genetic variations, including nonsynonymous SNPs, that give rise to single amino acid differences in proteins.
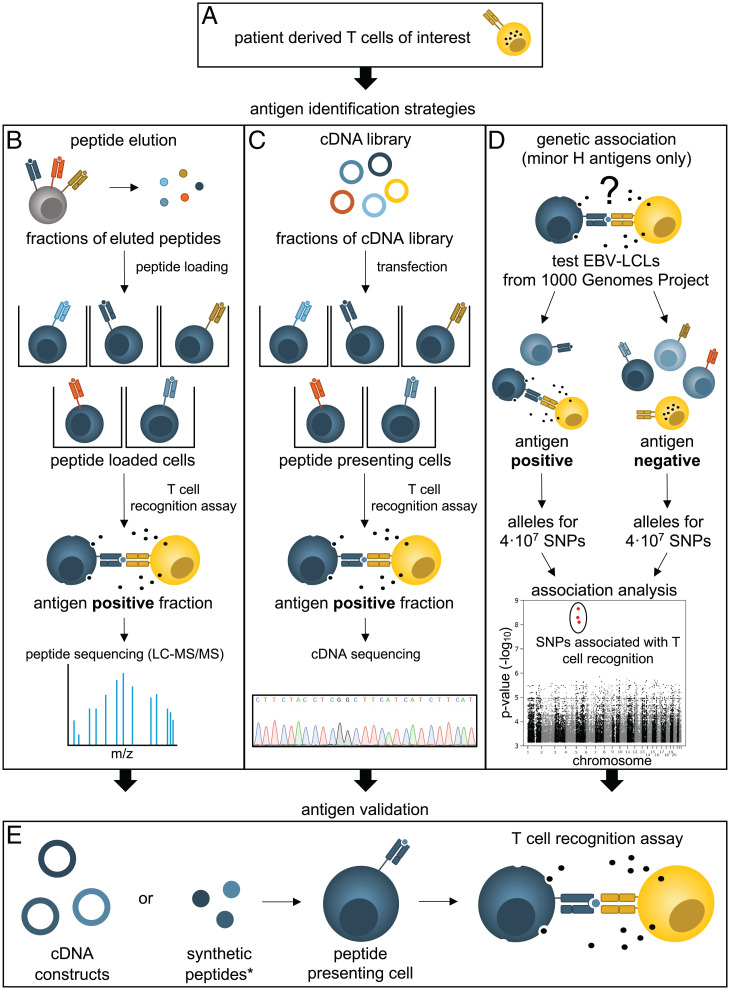
There are several approaches for the discovery of tumor and minor H antigens, each with a different power to identify spliced antigens. The approaches can be categorized into so-called forward and reverse antigen identification strategies. The forward “T cell-to-antigen” strategies aim to identify tumor antigens that are recognized by antigen-specific T cells (Fig. 2), while the reverse strategies focus on isolating an antigen-specific T cell specific for a predicted or selected antigen (64).
Fig. 2.
Schematic overview of the forward antigen identification strategies. (A) Prior to identification, T cells are isolated from a patient, and relevant tumor or minor H antigen–specific T cell clones are selected. Three main forward strategies are regularly used to identify the recognized peptide antigen (B–D). (B) Peptides are eluted from MHC molecules that are restriction elements of an antigen-specific T cell clone. Eluted peptides are fractionated high-performance LC and separately loaded on cells to measure reactivity by the antigen-specific T cell clone. The antigen-positive fraction is analyzed by MS/MS to determine the putative sequence of the recognized peptide. (C) Cells are transfected with fractions of a cDNA library and tested for their ability to activate the antigen-specific T cell clone. Antigen-positive fractions are subsequently sequenced to determine the cDNA coding for the antigen. (D) For WGAS, EBV-LCLs from the 1000 Genomes Project are phenotyped as minor H antigen positive or negative using a T cell recognition assay. The genetic variation encoding the antigen of interest can be determined by analyzing the association of the phenotypes with the publicly available genotypes of 4·107 SNPs from the same cells. Former genetic linkage analyses were based on a similar principle, but were less powerful because of the smaller number of genotyped variations. (E) Final validation of the epitope for each of those strategies (B–D) is achieved by testing specific (truncated) cDNA constructs or synthetic peptides, which are selected based on MHC-binding prediction algorithms, for recognition by the T cell clone.
Reverse antigen identification strategies.
To date, the majority of tumor antigens and a few minor H antigens were identified using a reverse approach based on in silico selection of genes of interest that are subsequently subjected to predictions for possible tumor or minor H antigens (65–69). For example, genes that are known to be polymorphic or mutated and genes that are exclusively expressed or overexpressed by tumor cells may encode clinically relevant antigens. MHC peptide binding algorithms are then applied to estimate how well the predicted antigens bind desired MHC alleles (64, 70, 71). This can be combined with prediction algorithms for proteasome processing and TAP transport to increase the specificity of the approach (72–76). Validation of candidate peptides as true antigens requires evidence of specific recognition by T cells. For example, T cells can be isolated using peptide-loaded MHC-I tetramers followed by confirmation of their antigen-specific reactivity through cytokine secretion or cytotoxicity assays (67). This reverse antigen identification approach is unquestionably biased toward conventional antigens, since nonconventional antigens—such as spliced peptides—are simply not included in current widely used T cell epitope prediction algorithms (Fig. 3A). The major challenge for inclusion of spliced peptides in reliable prediction algorithms is the high number of possible spliced products that can be generated from a region of interest. Further understanding of the ubiquity of PCPS may allow targeted inclusion of spliced peptide predictions in the future.
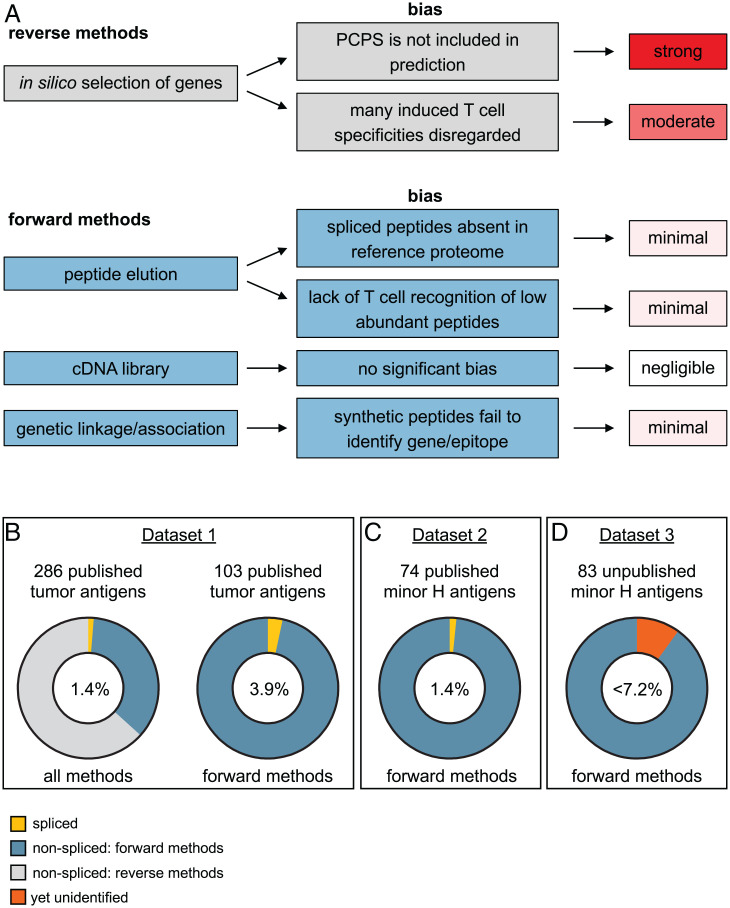
Fig. 3.
Spliced antigens constitute a marginal fraction of the MHC-I-presented antigen pool. (A) Methods for antigen identification and their bias toward spliced versus nonspliced antigens. (B) We evaluated the percentage of spliced peptides within the CAP database. The CAP database contains 474 tumor antigens, of which 286 are unique MHC-I restricted. Four (1.4%) of these are spliced peptides. Literature analyses revealed that 103 of the 286 antigens were identified with largely unbiased forward methods, still including four spliced peptides (3.9%). (C) The second independent dataset contains 76 molecularly characterized MHC-I-restricted minor H antigens that were identified with forward methods. One (1.4%) of these is a spliced antigen. (D) A third independent dataset consists of recently identified but unpublished minor H antigens discovered by our largely unbiased GWAS strategy using 83 different antigen-specific T cell clones. Of the eight antigens that are currently unresolved, two are, in all likelihood, encoded by cryptic transcripts. The remaining six (7.2%) antigens are potential spliced antigens or other unconventionally encoded or processed antigens. We concluded that the maximum percentage of spliced antigens in this dataset is therefore 7.2%.
A second more targeted reverse approach selects genes of interest without further predictions, which was mainly applied for genes of the tumor-associated MAGE and LAGE protein families. Proof of antigen presentation is obtained after T cell inductions with gene-transfected or polypeptide-loaded antigen-presenting cells, such as monocyte-derived dendritic cells. Some of the T cell specificities in this culture are fine mapped using synthetic peptides, tetramers, or truncated complementary DNA (cDNA) constructs. This fine-mapping procedure is also biased against spliced antigen identification, since the process is often halted as soon as one or a few nonspliced antigens are elucidated, disregarding the specificity of other T cell clones (Fig. 3A). In addition, peptides or genes may be selected based on immunopeptidome data. The bias of this strategy is related to the peptide-calling algorithm, which may not take PCPS into account. Nevertheless, it has been successfully applied to identify a spliced and multiple linear T cell antigens (5, 77).
Forward antigen identification strategies.
Several forward strategies have been broadly used to identify the peptide specificity of relevant T cell clones isolated from naïve or immunotherapeutically treated patients.
In the first approach, peptides are eluted from MHC molecules derived from antigen-positive cells and fractionated (Fig. 2B). T cell reactivity against antigen-negative cells loaded with these fractions reveals which fraction contains the sought antigen (78–81). The sequence of the peptide(s) in the positive fraction is determined with LC-MS/MS. To validate that these sequences are the sought epitope, synthetic peptides or overexpressed genes are evaluated in T cell reactivity assays (Fig. 2E). Since efficiency of PCPS is lower than normal peptide processing, investigators may have stopped analysis of spliced antigen-specific T cell clones because of insufficient recognition of any of the peptide fractions (Fig. 3A). Another potential bias in this strategy is that sequences of spliced peptides may be harder to resolve or simply overlooked compared to linear peptides, because they are not contained in the reference proteome (Fig. 3A). It is questionable whether this leads to significant abandoning of identification attempts. This approach is therefore unlikely to impact the success rate of spliced antigen identification.
The second forward strategy utilizes cDNA library screening (Fig. 2C) (82–85). Antigen-negative cells are transfected with a cDNA library generated from antigen-positive cells. T cell reactivity assays against limiting fractions of the library reveal which cells contain the antigen-encoding cDNA. After sequencing of this cDNA, putative antigens are validated using synthetic peptides or overexpressed gene fragments (Fig. 2E). In our opinion, there is no significant bias in this strategy that will favor the identification of linear over spliced antigens (Fig. 3A). However, it depends on the research question and the perseverance of the researcher whether this genetic narrowing of the epitope is performed and whether spliced peptides are considered during this last identification step.
Finally, genome-wide association approaches are specifically applied for the identification of minor H antigens, since these antigens are encoded by inheritable genetic polymorphisms (Fig. 2D). Of the more than 70 known MHC-I-presented minor H antigens, most were identified by a genetic approach. For this methodology, EBV-LCLs derived from different individuals are phenotyped as minor H antigen positive or negative using the T cell clone with uncharacterized specificity (86, 87). The (publicly available) genetic variation of the same EBV-LCLs is associated with these phenotypes to identify a genetic locus or even specific SNPs that likely encode the minor H antigen (58, 86–96). This genetic information is used to select the most-probable candidate transcripts or MHC-binding peptides for further testing. Similar as for the other forward methods, candidate epitopes are evaluated using synthetic peptides or gene overexpression (Fig. 2D). With genetic association approaches, it is possible that the genetic locus or variation involved in generation of the antigen is discovered, but that identification of the epitope fails. This is mainly because it may be challenging to deduce the exact transcript or peptide from the identified locus or variation. Specific explanations for a lack of success include incomplete annotations in the reference genome databases (97), imprecise MHC-I binding predictions (98), or that the antigen is a posttranslationally modified peptide, including spliced variants. Since unsuccessful attempts are generally not reported, published antigens identified by genetic approaches may reflect a bias of unknown impact on the ratio of linear over spliced antigens (Fig. 3A).
Antigenic Spliced Peptides Represent Less than 7.2% of T Cell Recognized Antigens.
Knowing the capacity of different strategies to identify spliced antigens, we first assessed the proportion of spliced antigens among the well-defined set of previously discovered tumor antigens in the Cancer Antigenic Peptide (CAP) database (see SI Appendix, Table S1) (65, 99). This manually composed database (accessed on 9 December 2021) includes 286 thoroughly validated unique MHC-I-restricted tumor antigens, of which four are spliced (1.4%; Fig. 3B) (2–4, 6). Because the list is manually composed and curated following strict requirements, several antigens are not included in the database, including linear antigens and one of the GP100 spliced antigens that was identified by a reverse approach (5).
Importantly, many of the 286 tumor antigens were identified using the reverse immunology approach, which carries several biases against the discovery of spliced antigens. To make a more accurate estimation of spliced versus nonspliced antigens, we determined the antigen identification strategy for each of those 286 antigens, in an extensive literature analysis. Considering solely the tumor antigens identified with forward methods (i.e., peptide elution and cDNA library; 103 in total), spliced antigens still make up only 3.9% (4/103) of the total (Fig. 3B).
Because of the controversy in the field, we searched for a second independent set of T cell antigens to further confirm this rather low percentage. The majority of autosomal minor H antigens have been identified by forward strategies, and, although tumor expressed and rigorously validated, they are not included in the CAP database. Moreover, studies on minor H antigens represent a clearly defined research field, facilitating the unbiased inclusion of all known antigens in our analysis. After exclusion of minor H antigens identified using a reverse approach, we obtained a list of 74 nonspliced epitopes and one spliced epitope (1.4%; Fig. 3C and SI Appendix, Table S2) (1), indicating that the percentage of spliced peptides among minor H antigens is also low. Even with the potential biases kept in mind, it seems improbable that spliced peptides represent ∼30% of the MHC-I-presented peptidome, as reported by Liepe et al. (7).
We recognize that the increasing utilization of the genetic approach for high-throughput identification of minor H antigens (89–91) might lead to underreporting of spliced antigens that are more challenging to resolve and might remain unidentified (Fig. 3A). Therefore, we further analyzed our unpublished data of identified and unidentified minor H antigens using an optimized genome-wide association study (GWAS) (89). In our laboratory, we isolated a set of 83 different minor H antigen–specific T cell clones for which we found significant association in our high-throughput genome-wide association analyses (P < 10−5) (SI Appendix, SI Methods and Fig. S1). For 75 of these minor H antigens, we discovered and validated the epitopes in T cell recognition assays, all of which are nonspliced (SI Appendix, Fig. S1 and Table S3). Theoretically, the remaining eight yet unidentified antigens (9.6%) may be spliced peptides (SI Appendix, Table S4). However, the GWAS of two T cell clones yielded no associating missense SNPs, strongly suggesting that these T cells recognize other unconventional antigens such as antigens encoded by cryptic transcripts. For the remaining six T cell clones, significantly associating missense SNPs were found, but recognition of linear peptides covering these SNPs was not (yet) seen. These T cell clones are potentially specific for either spliced antigens or other unconventional antigens. Thus, the fraction of spliced antigens from this third independent dataset, which is still under investigation, is likely less than 6 out of 83 (<7.2%; Fig. 3D).
The genetic minor H antigen identification methods described above are all based on antigen presentation by EBV-LCLs. Coincidentally, the ∼30% contribution of spliced peptides to the peptidome was estimated largely from LC-MS/MS data of also EBV-LCLs. But our minor H antigen–based analyses on these EBV-LCLs reveal a much lower estimation of the spliced peptide frequency. A difference is that the T cell epitopes in this manuscript only include tumor antigens and minor H antigens. It is unlikely, however, that spliced peptides would be underrepresented or overrepresented in any of these antigen categories compared to other categories such as neoantigens, self-antigens, or autoimmune antigens. Another difference is that our estimations only include peptides that have truly elicited an immune response in cancer patients. The ability of T cells to respond to their cognate antigen relies on the affinity of their TCR for the peptide–MHC complex, but also on the abundance of the particular antigen in complex with MHC on target cells. It is conceivable that the proteasome produces lower amounts of spliced peptides compared to linear peptides, which may explain a possible discrepancy between the presentation of spliced peptides by MHC-I and their immunogenicity. On the other hand, some data in the field suggest that PCPS is as efficient as linear peptide generation by the proteasome (5). Altogether, our evaluation of the contribution of immunogenic spliced peptides to the repertoire of immunogenic tumor antigens is physiologically more relevant than analyses of solely peptides derived from MHC-I.
Concluding Remarks
To conclude, our complete and largely unbiased analyses of antigen-specific T cells from three different datasets indicate that spliced peptides represent 1.4% to maximally 7.2% of the immunogenic antigen pool. These results contribute to the efforts to develop reliable LC-MS/MS identification workflows, by serving as a rough benchmark of spliced peptides in the immunopeptidome, obtained independently of MS. Furthermore, this marginal contribution of spliced peptides to the pool of cancer antigens will therefore not serve as a comprehensive source to expand the number of targetable antigens for immunotherapies.
Supplementary Material
Acknowledgments
This work was supported by grants from the Dutch Research Council (Grant NWO-VIDI 91719369 to R.M.S.), the Landsteiner Foundation for Blood Transfusion Research (LSBR Fellowship 1842F to R.M.S.), and the Dutch Cancer Society (KWF Project 10713 to M.G.).
Footnotes
Competing interest statement: R.M.S. now works as principal scientist at Neogene Therapeutics BV, which may be perceived as a competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119736119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Warren E. H., et al. , An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313, 1444–1447 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hanada K., Yewdell J. W., Yang J. C., Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427, 252–256 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Dalet A., et al. , An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. U.S.A. 108, E323–E331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigneron N., Ferrari V., Stroobant V., Habib J. A., Van Den Eynde B. J., An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Ebstein F., et al. , Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non-spliced epitopes. Sci. Rep. 6, 24032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaux A., et al. , A spliced antigenic peptide comprising a single spliced amino acid is produced in the proteasome by reverse splicing of a longer peptide fragment followed by trimming. J. Immunol. 192, 1962–1971 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Liepe J., et al. , A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Faridi P., et al. , A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, eaar3947 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Erhard F., Dölken L., Schilling B., Schlosser A., Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol. Res. 8, 1018–1026 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Rolfs Z., Müller M., Shortreed M. R., Smith L. M., Bassani-Sternberg M., Comment on “A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.” Sci. Immunol. 4, eaaw1622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faridi P., et al. , Response to Comment on “A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.” Sci. Immunol. 4, eaaw8457 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Rolfs Z., Solntsev S. K., Shortreed M. R., Frey B. L., Smith L. M., Global identification of post-translationally spliced peptides with neo-fusion. J. Proteome Res. 18, 349–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faridi P., Dorvash M., Purcell A. W., Spliced HLA-bound peptides: A Black Swan event in immunology. Clin. Exp. Immunol. 204, 179–188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mylonas R., et al. , Estimating the contribution of proteasomal spliced peptides to the HLA-I ligandome. Mol. Cell. Proteomics 17, 2347–2357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilhelm M., et al. , Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nat. Commun. 12, 3346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Admon A., Are there indeed spliced peptides in the immunopeptidome? Mol. Cell. Proteomics 20, 100099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishto M., Commentary: Are there indeed spliced peptides in the immunopeptidome? Mol. Cell. Proteomics 20, 100158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durgeau A., Virk Y., Corgnac S., Mami-Chouaib F., Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Front. Immunol. 9, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt M., Finley D., Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigneron N., Stroobant V., Ferrari V., Abi Habib J., Van den Eynde B. J., Production of spliced peptides by the proteasome. Mol. Immunol. 113, 93–102 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Kisselev A. F., Akopian T. N., Woo K. M., Goldberg A. L., The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 274, 3363–3371 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Paes W., et al. , Elucidation of the signatures of proteasome-catalyzed peptide splicing. Front. Immunol. 11, 563800 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalet A., Vigneron N., Stroobant V., Hanada K., Van den Eynde B. J., Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor-5. J. Immunol. 184, 3016–3024 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Murata S., Takahama Y., Kasahara M., Tanaka K., The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 19, 923–931 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Guillaume B., et al. , Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. U.S.A. 107, 18599–18604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesne J., et al. , Conformational maps of human 20S proteasomes reveal PA28- and immuno-dependent inter-ring crosstalks. Nat. Commun. 11, 6140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber E. M., et al. , Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 148, 727–738 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Dalet A., Stroobant V., Vigneron N., Van den Eynde B. J., Differences in the production of spliced antigenic peptides by the standard proteasome and the immunoproteasome. Eur. J. Immunol. 41, 39–46 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Vigneron N., Ferrari V., Stroobant V., Abi Habib J., Van den Eynde B. J., Peptide splicing by the proteasome. J. Biol. Chem. 292, 21170–21179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liepe J., Ovaa H., Mishto M., Why do proteases mess up with antigen presentation by re-shuffling antigen sequences? Curr. Opin. Immunol. 52, 81–86 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Bassani-Sternberg M., Pletscher-Frankild S., Jensen L. J., Mann M., Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteomics 14, 658–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robbins P. F., et al. , Recognition of tyrosinase by tumor-infiltrating lymphocytes from a patient responding to immunotherapy. Cancer Res. 54, 3124–3126 (1994). [PubMed] [Google Scholar]
- 33.Willimsky G., et al. , In vitro proteasome processing of neo-splicetopes does not predict their presentation in vivo. eLife 10, e62019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichti C. F., Identification of spliced peptides in pancreatic islets uncovers errors leading to false assignments. Proteomics 21, e2000176 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Kuckelkorn U., et al. , Proteolytic dynamics of human 20S thymoproteasome. J. Biol. Chem. 294, 7740–7754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Specht G., et al. , Large database for the analysis and prediction of spliced and non-spliced peptide generation by proteasomes. Sci. Data 7, 146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishto M., et al. , Driving forces of proteasome-catalyzed peptide splicing in yeast and humans. Mol. Cell. Proteomics 11, 1008–1023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai B., et al. , Mutations in KRAS codon 12 predict poor survival in Chinese patients with metastatic colorectal cancer. Oncol. Lett. 15, 3161–3166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prior I. A., Hood F. E., Hartley J. L., The frequency of Ras mutations in cancer. Cancer Res. 80, 2969–2974 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S., Han M., Zhang H., Wang Y., Jiang H., Full screening and accurate subtyping of HLA-A*02 alleles through group-specific amplification and mono-allelic sequencing. Cell. Mol. Immunol. 10, 490–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran E., et al. , T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin N., et al. , Identification and validation of T-cell receptors targeting RAS hotspot mutations in human cancers for use in cell-based immunotherapy. Clin. Cancer Res. 27, 5084–5095 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishto M., et al. , An in silico-in vitro pipeline identifying an HLA-A*02:01+ KRAS G12V+ spliced epitope candidate for a broad tumor-immune response in cancer patients. Front. Immunol. 10, 2572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fidanza M., et al. , Enhancing proteasomal processing improves survival for a peptide vaccine used to treat glioblastoma. Sci. Transl. Med. 13, eaax4100 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Prickett T. D., et al. , Durable complete response from metastatic melanoma after transfer of autologous T cells recognizing 10 mutated tumor antigens. Cancer Immunol. Res. 4, 669–678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zacharakis N., et al. , Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24, 724–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Creelan B. C., et al. , Tumor-infiltrating lymphocyte treatment for anti-PD-1-resistant metastatic lung cancer: A phase 1 trial. Nat. Med. 27, 1410–1418 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Berg J. H., et al. , Tumor infiltrating lymphocytes (TIL) therapy in metastatic melanoma: Boosting of neoantigen-specific T cell reactivity and long-term follow-up. J. Immunother. Cancer 8, e000848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramachandran I., et al. , Systemic and local immunity following adoptive transfer of NY-ESO-1 SPEAR T cells in synovial sarcoma. J. Immunother. Cancer 7, 276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderson J. P., et al. , Preclinical evaluation of an affinity-enhanced MAGE-A4-specific T-cell receptor for adoptive T-cell therapy. OncoImmunology 9, 1682381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X., et al. , Mechanisms of relapse after CD19 CAR T-cell therapy for acute lymphoblastic leukemia and its prevention and treatment strategies. Front. Immunol. 10, 2664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geertsen R., et al. , Loss of single HLA class I allospecificities in melanoma cells due to selective genomic abbreviations. Int. J. Cancer 99, 82–87 (2002). [DOI] [PubMed] [Google Scholar]
- 53.McGranahan N., et al. ; TRACERx Consortium, Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fangazio M., et al. , Genetic mechanisms of HLA-I loss and immune escape in diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 118, e2104504118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doran S. L., et al. , T-cell receptor gene therapy for human papillomavirus-associated epithelial cancers: A first-in-human, phase I/II study. J. Clin. Oncol. 37, 2759–2768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dentro S. C., et al. ; PCAWG Evolution and Heterogeneity Working Group and the PCAWG Consortium, Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 184, 2239–2254.e39 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leisegang M., et al. , Eradication of large solid tumors by gene therapy with a T-cell receptor targeting a single cancer-specific point mutation. Clin. Cancer Res. 22, 2734–2743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spaapen R. M., et al. , Rapid identification of clinical relevant minor histocompatibility antigens via genome-wide zygosity-genotype correlation analysis. Clin. Cancer Res. 15, 7137–7143 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Peng M., et al. , Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 18, 128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fontaine P., et al. , Adoptive transfer of minor histocompatibility antigen-specific T lymphocytes eradicates leukemia cells without causing graft-versus-host disease. Nat. Med. 7, 789–794 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Jiang T., et al. , Tumor neoantigens: From basic research to clinical applications. J. Hematol. Oncol. 12, 93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffioen M., van Bergen C. A. M., Falkenburg J. H. F., Autosomal minor histocompatibility antigens: How genetic variants create diversity in immune targets. Front. Immunol. 7, 100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spaapen R., Mutis T., Targeting haematopoietic-specific minor histocompatibility antigens to distinguish graft-versus-tumour effects from graft-versus-host disease. Best Pract. Res. Clin. Haematol. 21, 543–557 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Mutis T., Xagara A., Spaapen R. M., The connection between minor H antigens and neoantigens and the missing link in their prediction. Front. Immunol. 11, 1162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vigneron N., Stroobant V., Van den Eynde B. J., van der Bruggen P., Database of T cell-defined human tumor antigens: The 2013 update. Cancer Immun. 13, 15 (2013). [PMC free article] [PubMed] [Google Scholar]
- 66.Dolstra H., et al. , Bi-directional allelic recognition of the human minor histocompatibility antigen HB-1 by cytotoxic T lymphocytes. Eur. J. Immunol. 32, 2748–2758 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Mommaas B., et al. , Identification of a novel HLA-B60-restricted T cell epitope of the minor histocompatibility antigen HA-1 locus. J. Immunol. 169, 3131–3136 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Armistead P. M., et al. , Common minor histocompatibility antigen discovery based upon patient clinical outcomes and genomic data. PLoS One 6, e23217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hombrink P., et al. , Discovery of T cell epitopes implementing HLA-peptidomics into a reverse immunology approach. J. Immunol. 190, 3869–3877 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Nielsen M., et al. , NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2, e796 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bykova N. A., Malko D. B., Efimov G. A., In silico analysis of the minor histocompatibility antigen landscape based on the 1000 Genomes Project. Front. Immunol. 9, 1819 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keşmir C., Nussbaum A. K., Schild H., Detours V., Brunak S., Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 15, 287–296 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Peters B., Bulik S., Tampe R., Van Endert P. M., Holzhütter H.-G., Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol. 171, 1741–1749 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Larsen M. V., et al. , An integrative approach to CTL epitope prediction: A combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol. 35, 2295–2303 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Nielsen M., Lundegaard C., Lund O., Keşmir C., The role of the proteasome in generating cytotoxic T-cell epitopes: Insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57, 33–41 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Larsen M. V., et al. , Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8, 424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hombrink P., et al. , Identification of biological relevant minor histocompatibility antigens within the B-lymphocyte-derived HLA-ligandome using a reverse immunology approach. Clin. Cancer Res. 21, 2177–2186 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Hunt D. F., et al. , Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 (1992). [DOI] [PubMed] [Google Scholar]
- 79.Cox A. L., et al. , Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science 264, 716–719 (1994). [DOI] [PubMed] [Google Scholar]
- 80.den Haan J. M., et al. , The minor histocompatibility antigen HA-1: A diallelic gene with a single amino acid polymorphism. Science 279, 1054–1057 (1998). [DOI] [PubMed] [Google Scholar]
- 81.den Haan J. M., et al. , Identification of a graft versus host disease-associated human minor histocompatibility antigen. Science 268, 1476–1480 (1995). [DOI] [PubMed] [Google Scholar]
- 82.van der Bruggen P., et al. , A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647 (1991). [DOI] [PubMed] [Google Scholar]
- 83.Ma W., et al. , Two new tumor-specific antigenic peptides encoded by gene MAGE-C2 and presented to cytolytic T lymphocytes by HLA-A2. Int. J. Cancer 109, 698–702 (2004). [DOI] [PubMed] [Google Scholar]
- 84.Ma W., et al. , A MAGE-C2 antigenic peptide processed by the immunoproteasome is recognized by cytolytic T cells isolated from a melanoma patient after successful immunotherapy. Int. J. Cancer 129, 2427–2434 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Dolstra H., et al. , A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J. Exp. Med. 189, 301–308 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawase T., et al. , Identification of human minor histocompatibility antigens based on genetic association with highly parallel genotyping of pooled DNA. Blood 111, 3286–3294 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Spaapen R. M., et al. , Toward targeting B cell cancers with CD4+ CTLs: Identification of a CD19-encoded minor histocompatibility antigen using a novel genome-wide analysis. J. Exp. Med. 205, 2863–2872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Broen K., et al. , A polymorphism in the splice donor site of ZNF419 results in the novel renal cell carcinoma-associated minor histocompatibility antigen ZAPHIR. PLoS One 6, e21699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuchs K. J., et al. , Optimized whole genome association scanning for discovery of HLA class I-restricted minor histocompatibility antigens. Front. Immunol. 11, 659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Bergen C. A. M., et al. , High-throughput characterization of 10 new minor histocompatibility antigens by whole genome association scanning. Cancer Res. 70, 9073–9083 (2010). [DOI] [PubMed] [Google Scholar]
- 91.van Bergen C. A. M., et al. , Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J. Clin. Invest. 127, 517–529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Bergen C. A. M., et al. , Durable remission of renal cell carcinoma in conjuncture with graft versus host disease following allogeneic stem cell transplantation and donor lymphocyte infusion: Rule or exception? PLoS One 9, e85198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oostvogels R., et al. , Identification of minor histocompatibility antigens based on the 1000 Genomes Project. Haematologica 99, 1854–1859 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pont M. J., et al. , T cells specific for an unconventional natural antigen fail to recognize leukemic cells. Cancer Immunol. Res. 7, 797–804 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Kamei M., et al. , HapMap scanning of novel human minor histocompatibility antigens. Blood 113, 5041–5048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warren E. H., et al. , Therapy of relapsed leukemia after allogeneic hematopoietic cell transplantation with T cells specific for minor histocompatibility antigens. Blood 115, 3869–3878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang D., et al. , Incomplete annotation has a disproportionate impact on our understanding of Mendelian and complex neurogenetic disorders. Sci. Adv. 6, eaay8299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao W., Sher X., Systematically benchmarking peptide-MHC binding predictors: From synthetic to naturally processed epitopes. PLOS Comput. Biol. 14, e1006457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van der Bruggen P., Stroobant V., Vigneron N. J., van den Eynde B., Cancer Antigenic Peptide database. https://caped.icp.ucl.ac.be/. Accessed 9 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.