Significance
The dCache shows a ubiquitous phylogenetic distribution and is abundantly present in archaea, bacteria, and eukaryota. It’s the predominant bacterial extracytosolic sensor domain and present in all major families of transmembrane receptors. Most characterized dCache mediate responses to diversely similar ligands, which were found to bind to the membrane-distal module. Although there’s also evidence that other dCache respond to multiple ligands, the information on the corresponding molecular mechanism is currently limited. We show here that the capacity to respond to multiple ligands is due to the utilization of both dCache modules. This work provides information on the forces that have led to the evolution of dCache domains and provides features that characterize receptors with narrow and broad ligand ranges.
Keywords: signal transduction, chemotaxis, chemoreceptor, dCache sensor domain, ligand recognition
Abstract
Bacteria have evolved multiple signal transduction systems that permit an adaptation to changing environmental conditions. Chemoreceptor-based signaling cascades are very abundant in bacteria and are among the most complex signaling systems. Currently, our knowledge on the molecular features that determine signal recognition at chemoreceptors is limited. Chemoreceptor McpA of Bacillus velezensis SQR9 has been shown to mediate chemotaxis to a broad range of different ligands. Here we show that its ligand binding domain binds directly 13 chemoattractants. We provide support that organic acids and amino acids bind to the membrane-distal and membrane-proximal module of the dCache domain, respectively, whereas binding of sugars/sugar alcohols occurred at both modules. Structural biology studies combined with site-directed mutagenesis experiments have permitted to identify 10 amino acid residues that play key roles in the recognition of multiple ligands. Residues in membrane-distal and membrane-proximal regions were central for sensing organic acids and amimo acids, respectively, whereas all residues participated in sugars/sugar alcohol sensing. Most characterized chemoreceptors possess a narrow and well-defined ligand spectrum. We propose here a sensing mechanism involving both dCache modules that allows the integration of very diverse signals by a single chemoreceptor.
About half of sequenced bacterial genomes contain genes encoding chemotactic signaling cascades that confer on bacteria the capacity to explore their environment for optimal conditions (1, 2). The benefits of chemotaxis include the access to nutrients, the protection from predators, an escape from the host immune defense, an increased tolerance to antibiotics, the promotion of host colonization, the degradation of pollutant, or the localization of prey (1–7). Chemotaxis is based on chemosensory pathways that are among the most complex and studied signal transduction mechanisms in bacteria (8, 9). A chemotactic response is typically initiated by the signal recognition at the sensor or ligand binding domain (LBD) of chemoreceptors, which creates a molecular stimulus that is transduced across the membrane, where it alters the autophosphorylation rate of the histidine kinase CheA (10), and consequently transphosphorylation to the CheY response regulator. Only the phosphorylated form of CheY is able to bind to the flagellar motor causing ultimately chemotaxis (11). Since the binding of ligands at the LBD defines the specificity of the chemotactic response, the determination of the molecular features that defines ligand recognition is of central importance.
Bacteria possess a wide range of different types of transmembrane receptors (12) that are able to sense ligands at extracytosolic LBDs. A large number of different LBD types have been reported (13): for example, more than 80 in chemoreceptors, and a same LBD type is frequently employed by different families of transmembrane receptors (14, 15). The dCache type LBD is the predominant bacterial sensor domain and is found in all major families of bacterial transmembrane receptors, including chemoreceptors, sensor kinases, c-di-GMP cyclases and diesterases, serine phosphatases, adenylate- and guanylate cyclases, Ser/Thr kinases, or ion channels (16). Furthermore, dCache domains show a wide phylogenetic spread and are also found in archaea and different eukaryota (16, 17).
dCache domains are composed of a long N-terminal helix and two α/β-fold–like subdomains that are referred to as membrane-distal and membrane-proximal modules (13). dCache domains of chemoreceptors were found to recognize structurally very different ligands, such as proteinogenic amino acids, polyamines, quaternary amines, purines, organic acids (OAs), sugars, quorum sensing signals, or inorganic ions (18). However, most of the corresponding receptors showed significant ligand specificity as illustrated by chemoreceptors specific for amino acids (19), quaternary amines (20), purines (21), or polyamines (22). A significant number of dCache-ligand cocrystal structures have been solved that indicate ligands interact with the membrane distal module in most cases (13, 22–25) and there are only few examples of signal interaction with the membrane-proximal module (26–28). Tohidifar et al. (28) identified four key amino acid residues in the membrane-proximal module of a pH-responsive dCache domain, suggesting that these are involved in pH sensing. Machuca et al. (26) identified lactate as a ligand that bound to the membrane-proximal module of the TlpC chemoreceptor that was found to mediate lactate chemotaxis. Another study demonstrated that point mutations in the membrane proximal module of the chemoreceptor TlpA reduced its binding affinity for three chemoattractants (27). dCache domains were proposed to be the result of a fusion of two sCache domains (16). However, since the membrane-proximal module of most dCache domains is not involved in signal recognition, the forces that have driven this evolutionary process are not very clear. In addition, there is evidence for promiscuity in the signal recognition at dCache domains. Examples are the PctA and TlpQ chemoreceptors that recognize with high affinity at the membrane-distal module proteinogenic amino acids (29) and polyamines (25), respectively, as well as the autoinducer-2 quorum-sensing signal (30). Another example for this promiscuity is the Tlp3 chemoreceptor that bound amino acids, OAs, purines, and sugars to induce either chemoattraction or repellence responses (31). The molecular basis for the recognition of multiple ligands by dCache domains are unclear and are investigated here.
Bacillus velezensis SQR9 (former Bacillus amyloliquefaciens SQR9) is a well-studied and commercially widely used plant growth-promoting rhizobacterium (32–35). Its dCache LBD containing chemoreceptor McpA plays a critical role in chemotaxis to root exudates, biofilm formation, and rhizosphere colonization (36). We have shown previously that McpA performed chemoattraction to 23 structurally different compounds that could be classified as OAs (11), sugars (6), and proteinogenic amino acids (6). Furthermore, microcalorimetric studies revealed high-affinity binding of two of these ligands, citric and aspartic acids, to the individual McpA-LBD (37, 38).
We investigate here the molecular mechanism that permits the recognition of this diverse range of ligands by a single chemoreceptor. We were able to establish a model that involved the synergetic participation of both dCache modules in signal recognition, which provide novel insight into signal recognition mechanism by bacterial sensor domains.
Results
McpA-LBD Directly Binds Chemically Diverse Ligands.
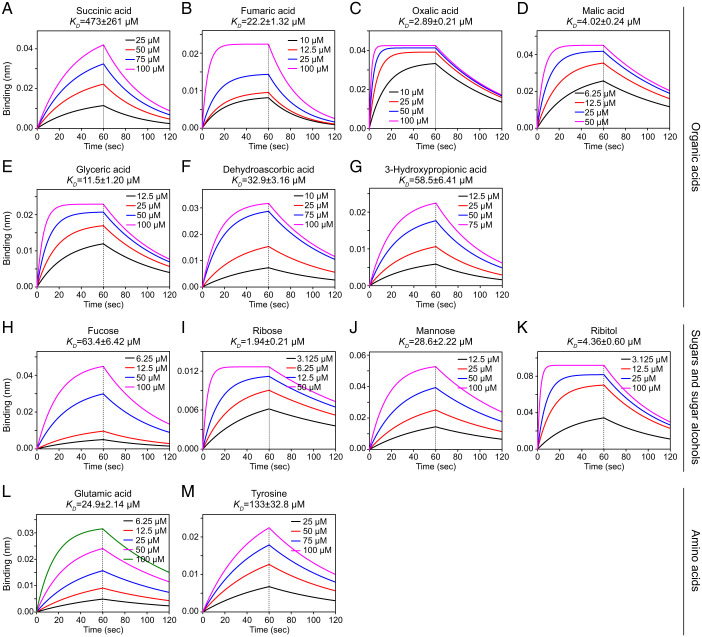
Our previous studies have shown that McpA mediates chemoattraction to 23 chemically diverse ligands (SI Appendix, Table S1), with direct binding of citric acid and aspartic acid but no direct binding of d-galactose (37, 38). For the remaining 20 attractants, herein we investigated the recognition by surface plasmon resonance (SPR) and biolayer interferometry (BLI). SPR data revealed binding for all ligands, except for serine and fucose (SI Appendix, Fig. S1). We then employed the BLI technology to verify ligand binding, and were able to observe an interaction of 13 compounds, including amino acids (glutamate and tyrosine), OAs (succinic, fumaric, oxalic, malic, glyceric, dehydroascorbic, and 3-hydroxypropionic acids), as well as sugars/sugar alcohols (SAs; fucose, ribose, mannose, and ribitol) (Fig. 1). The BLI data also permitted the calculation of the corresponding dissociation constants that ranged from 1.8 to 473 μM (Fig. 1) and that are within the range of affinities typically observed for ligand binding at chemoreceptors (18). These results indicate that the McpA-mediated chemotactic responses to structurally very different ligands (38) is due to direct ligand binding at its LBD.
Fig. 1.
Interaction between chemoreceptor the LBD of McpA and chemoattractants of different chemical categories. (A–M) Shown are BLI for the interaction between McpA-LBD (A36-L277) and members of the three ligand categories.
McpA Recognizes Ligands at Its Membrane-Proximal and Membrane-Distal Module.
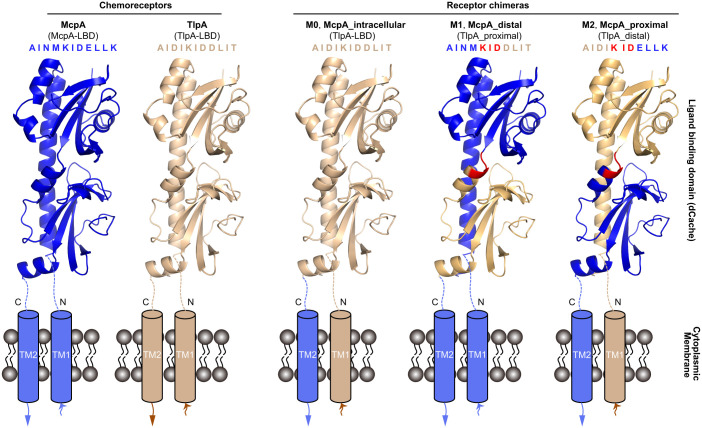
To identify whether McpA binds ligands at the membrane-distal or membrane-proximal module, we constructed a series of chimeras comprising different forms of the McpA-LBD fused to the cytosolic fragment of TlpA (Fig. 2). TlpA was selected due to its elevated sequence identity with McpA (61%), and its incapacity to sense 15 ligands recognized by McpA (38, 39). All the three receptor chimeras and C-mcpA (the SQR9Δ8mcp strain, devoid of any chemoreceptor gene, complemented with mcpA) showed similar growth (SI Appendix, Fig. S2). Based on these results, subsequent chemotaxis experiments were made with strains cultured to an OD600 of 1.0.
Fig. 2.
Schematic of the B. velezensis SQR9 McpA and TlpA chemoreceptor LBDs and the three derived receptor chimera. Shown are homology models of the LBD as generated by Phyre2 (57). The blue and beige color segments represent the corresponding region of McpA and TlpA, respectively. The amino acid sequence fragment represents the fusion site between both modules. The segment in red is conserved in McpA and TlpA and was therefore used as the fusion point between both modules. TM: transmembrane region.
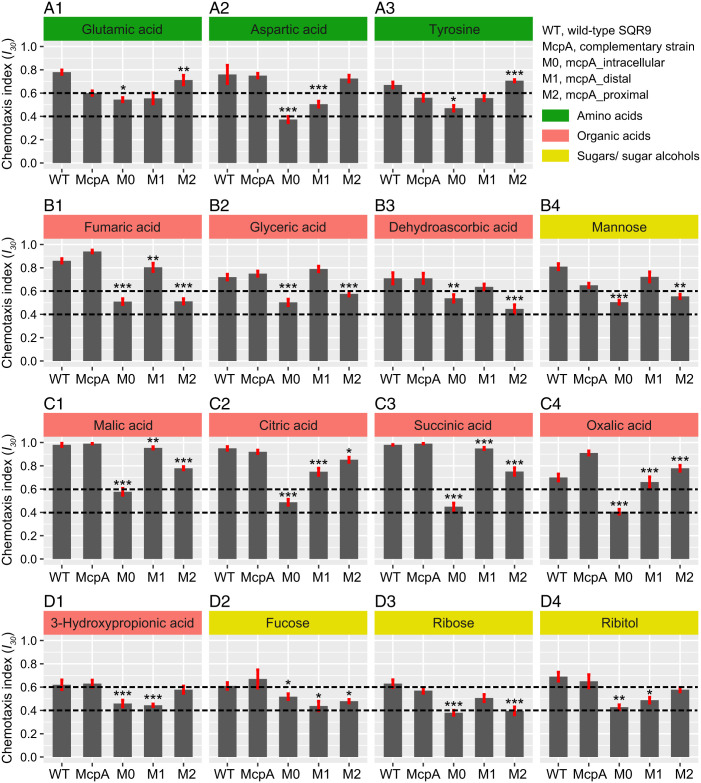
Chemotaxis assays indicated that the strain harboring TlpA-LBD had completely lost its capacity to perform chemotaxis to the 15 McpA ligands (Fig. 3 and SI Appendix, Table S2), suggesting that McpA-LBD is the domain required for signal sensing. To identify the McpA-LBD region that is necessary for signal sensing, we assessed the chemotactic responses of strains containing McpA_distal and McpA_proximal. The results from these experiments can be summarized as follows: 1) strains bearing only the membrane proximal domain of McpA retained the ability to perform chemoattraction to all three amino acids (glutamic acid, aspartic acid, and tyrosine) (Fig. 3A); 2) the strain harboring only the membrane distal domain of McpA maintained response to three OAs (fumaric, glyceric, dehydroascorbic acids) and one sugar (mannose) (Fig. 3B); 3) chimera that conserved the membrane-distal or membrane-proximal modules of McpA showed significant chemotactic response to four OAs (malic, citric, succinic, and oxalic acids) (Fig. 3C); 4) the complete LBD of McpA is necessary for the sensing of three SAs (fucose, ribose, and ribitol) and one OA (3-hydroxypropionic acid) (Fig. 3D). Subsequently, we have purified the individual LBDs of the M1 and M2 chimera that were then submitted to circular dichroism (CD) spectroscopy and SPR-based ligand binding studies. The CD spectra analysis showed only minor differences among the native McpA-LBD (WT) and the three receptor chimeras, indicating the chimera construction may not affect the secondary structure (SI Appendix, Fig. S3A). The results indicated that all three amino acids bound to M2-LBD, whereas six OAs (fumaric, glyceric, dehydroascorbic, citric, succinic, and oxalic acids) bound to M1-LBD. The four SAs failed to bind to M1-LBD and M2-LBD (SI Appendix, Fig. S4). These results indicate that McpA employs multiple modes of ligand recognition.
Fig. 3.
Chemotaxis of different McpA receptor chimeras to multiple ligands. A1–A3 refer to the function of McpA-LBD relying on the membrane-proximal module. B1–B4 refer to the function of McpA-LBD relying on the membrane-distal module. C1–C4 refer to the function of McpA-LBD relying on the membrane-distal or membrane-proximal module of McpA-LBD. D1–D4 refer to the function of McpA-LBD relying on the complete LBD. The chemotaxis data of WT SQR9 and its complementary strain mcpA have been reported previously (38). Shown are chemotaxis indices (I30) derived from microfluidics-based chemotaxis assays. I30 > 0.6 or < 0.4 indicates attractant or repellent responses, respectively, while 0.4 ≤ I30 ≤ 0.6 designates an absence of taxis. The dotted lines represent the chemotaxis cutoff used to classify the response. The asterisks indicate statistically significant differences compared with the strain containing exclusively mcpA: *P < 0.05, **P < 0.01, ***P < 0.001. Data are means and SDs from 13 independent measurements.
Identification of Amino Acids Involved in Ligand Binding Based on the McpA-sLBD Structure and In Silico Analyses.
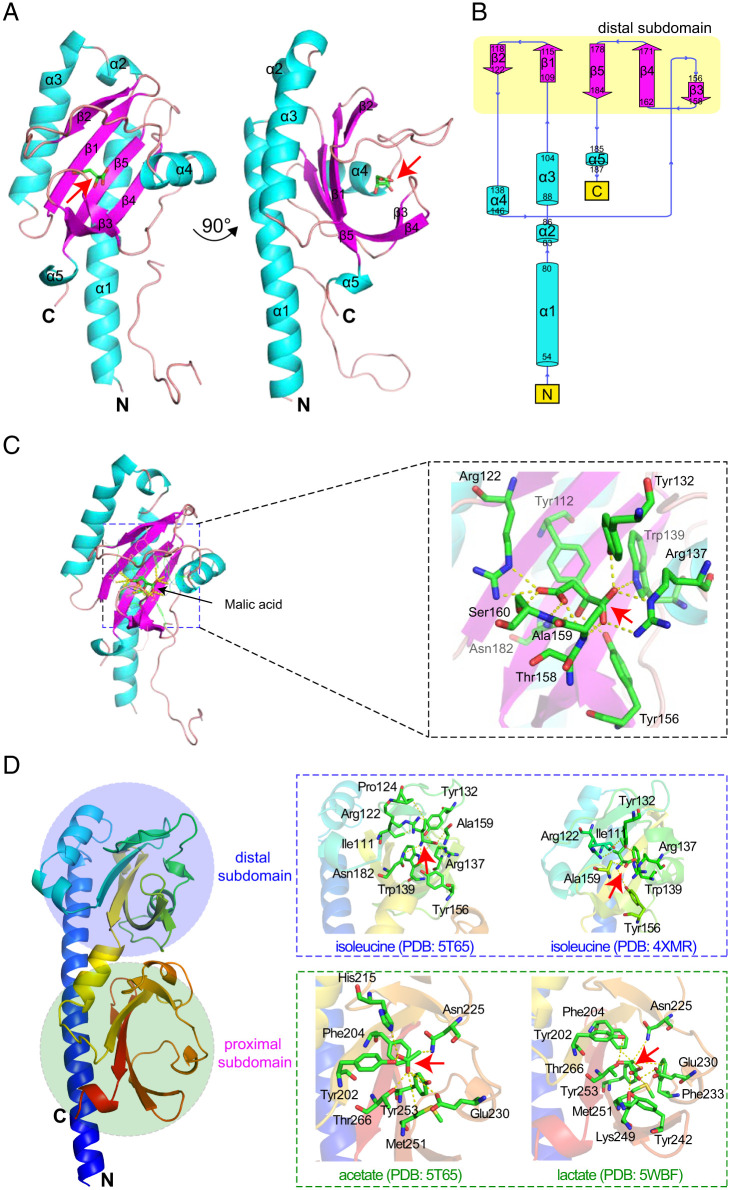
We employed a structural biology-based approach to identify amino acids involved in ligand binding. Initial crystallization trials of McpA-LBD (amino acids 36 to 277) were unsuccessful, due to the proteolytic cleavage between residues 224 and 225. Therefore, a shortened version of this LBD was generated that span from amino acids 38 to 224, referred to as McpA-sLBD. The screening of crystallization conditions (SI Appendix, Table S3) resulted in diffractable crystals of McpA-sLBD in complex with malic acid. Crystals diffracted to 2.25-Å resolution and belonged to space group P21212 (SI Appendix, Table S4). The structure was solved using molecular replacement and two protein monomers were present in the asymmetric unit (Fig. 4A). The electron density was of sufficient quality to place a molecular model comprising amino acids 54 to 187, whereas no sufficient electron density was observed for the remaining part of the structure. The final structure included the N-terminal helix, the membrane distal module, as well as the α5 connector helix (Fig. 4B). Malic acid was bound at membrane-distal subdomain revealing the amino acids that are involved in its binding (Fig. 4C).
Fig. 4.
Ligand recognition at the ligand binding domain of the McpA chemoreceptor. (A) Ribbon diagram of the X-ray structure of McpA-sLBD (Q38-L224). The secondary structure elements are labeled. Bound malic acid is shown in stick mode. (B) Schematic representation of the secondary structure elements of McpA-sLBD. (C) Close-up view of the malic acid-binding pocket of the membrane-distal subdomain of McpA-sLBD. (D) Homology model of McpA-LBD (A36-L277). Boxed are close-up views of the temples that were used for modeling and the amino acids involved in ligand recognition are labeled. The analogous amino acids in McpA-LBD have been mutated. A ribbon model of the McpA-LBD (A36-L277) monomer without ligand is colored in rainbow. The red arrows indicate the position of the different ligands.
In parallel, we performed in silico analyses to identify key residues in both McpA-LBD modules that may be involved in ligand binding. We initially retrieved 11 ligand complexed three-dimensional (3D) structures of McpA-LBD homologs (SI Appendix, Table S5), and extrapolated from them the analogous amino acids that may be present in both binding sites at McpA-LBD. This search resulted in the identification of 23 residues, of which 12 were in the membrane distal module and 11 in the proximal module (Fig. 4D and SI Appendix, Table S5). The results obtained by this approach coincided with the amino acids identified in the binding pocket of the 3D structure of McpA-sLBD.
Assessment of the Key Amino Acids Contributing to Ligand Binding.
To determine the role of the identified amino acids, we constructed 23 alanine replacement mutants in the mcpA gene. All mutants showed similar growth as compared to the reference strain C-mcpA (SI Appendix, Fig. S5), and then were used for chemotaxis assays. Six McpA-LBD derivatives that contain single amino acid substitutions were randomly selected, and purified for CD analysis. Comparison of the CD of the variants with that of McpA-LBD showed no significant differences, indicating that the amino acid substitutions did not alter the secondary structure (SI Appendix, Fig. S3B).
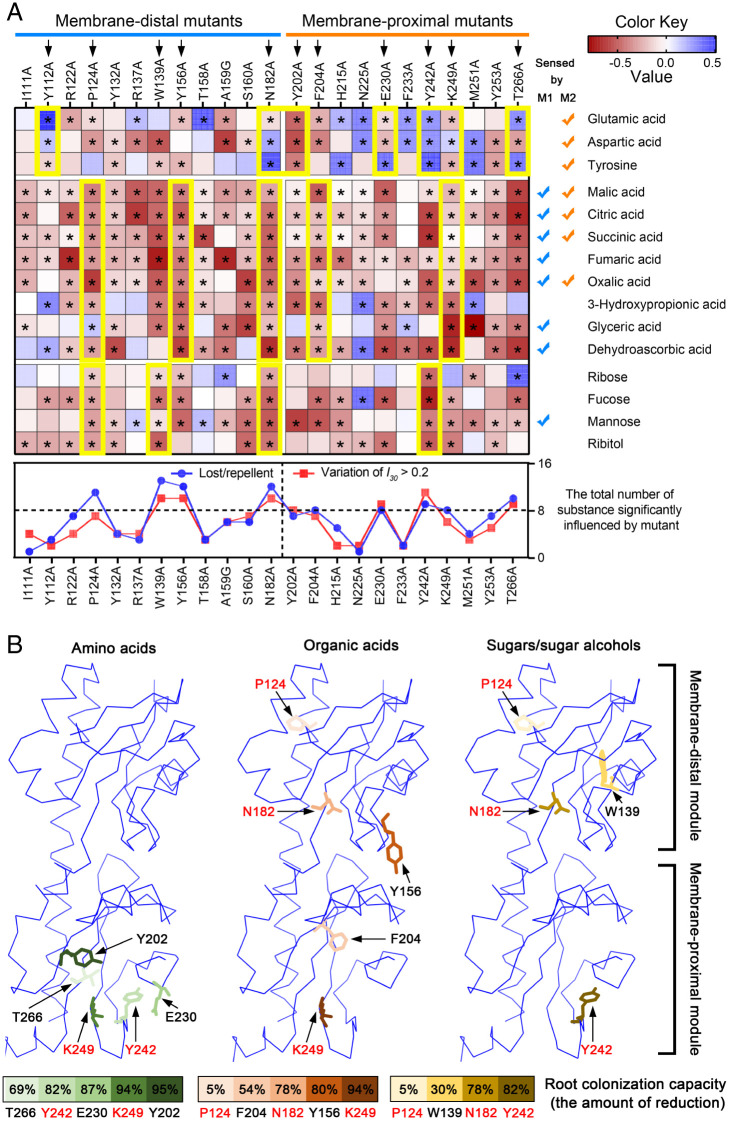
Data from chemotaxis measurements were assessed for their statistical significance (SI Appendix, Table S6), and the results can be summarized as follows. 1) Seven residues that were critical for amino acid sensing (i.e., mutant chemotaxis was significantly altered as compared to C-mcpA) (Fig. 5 and SI Appendix, Fig. S6). Five of these seven residues were in the proximal pocket (Tyr202, Glu230, Tyr242, Lys249, and Thr266), while other two were in the distal pocket (Tyr112, and Asn182) (Fig. 4). 2) Furthermore, five residues were required for chemotaxis to all eight OAs (Fig. 5 and SI Appendix, Fig. S6). Three were located to distal pocket (Pro124, Tyr156, and Asn182), in agreement with the results inform the analyses of the chimeras, while the remaining two were located in the proximal pocket (Phe204, and Lys249) (Fig. 4). 3) Finally, four residues were required for chemotaxis to the four SAs (Fig. 5 and SI Appendix, Fig. S6). Three were located in the distal pocket (Pro124, Trp139, and Asn182), whereas one (Tyr242) was located in the proximal pocket (Fig. 4).
Fig. 5.
Identification of key amino acids involved in chemotaxis to multiple ligands. (A) Heatmaps showing the magnitude of chemotaxis of different alanine replacement mutants in both modules of the McpA-LBD. The color intensity represents the percentage of mutant I30 values as compared to the reference strain C-mcpA (negative or positive values indicate weaker or stronger chemotaxis as compared to the C-mcpA reference strain, respectively). The 11 amino acids (indicated by black arrows) boxed in yellow are involved in sensing amino acids, OAs, or SAs. Asterisks indicate statistically significant differences compared to the C-mcpA reference strain (according to the Duncan’s multiple rank tests, *P < 0.05). The line chart shows the 10 key dominant amino acid residues as defined by the two criteria specified in the text. (B) The role of the 10 key dominant amino acids in ligand recognition and root colonization. McpA-LBDs and the 10 key amino acids residues are shown in ribbon and stick mode, respectively. These residues involved in sensing amino acids (green), OAs (orange), and SAs (gold) are colored according to the magnitude of the amount of reduction in root colonization (mutants harboring McpA point-mutations compared with reference strain C-mcpA). The residues in red were critical for sensing more than one ligand category.
The significant alterations of chemotactic response of these point mutants could be classified into four different phenotypes (SI Appendix, Table S7), namely repellent (I30 < 0.4), lost (0.4 < I30 < 0.6), reduced (0.6 < I30 < I30 of C-mcpA), or enhanced (I30 > the I30 of C-mcpA) chemotaxis. To get a more integral view of the role of these 23 residues in the recognition of these 15 ligands, we defined the following two different criteria: 1) chemotaxis of a point mutant toward more than half of the 15 ligands resulted in lost or repellent responses (I30 < 0.6) (SI Appendix, Fig. S7 and Table S7); 2) the variation of I30 of a point mutant toward more than half of the analyzed ligands is larger than 0.2 (ΔI30 > 0.2) (SI Appendix, Table S8). Based on these criteria (Fig. 5A), we identified the following 10 residues to be of particular importance: Pro124, Trp139, Tyr156, Asn182, Tyr202, Phe204, Glu230, Tyr242, Lys249, and Thr266. Taking into account the results from the analyses of the receptor chimeras, we also summarized the interaction intensity of these 10 key residues with ligands belonging to different chemical categories, indicating that chemotaxis to amino acids and OAs were mainly dependent on residues located in the membrane-proximal and membrane-distal pockets, respectively, while the response to SAs required both modules (Figs. 3 and 5).
Amino Acid Substitutions in both McpA-LBD Modules Alter the Root Colonization Phenotype of B. velezensis SQR9.
Finally, to assess the contribution of individual key amino acids to rhizosphere chemotaxis, we conducted root colonization experiments using these site-directed mutants. Nine mutants (except for P124A) showed a significant impairment in colonization as compared to the C-mcpA strain (Fig. 5B and SI Appendix, Fig. S8), and the amount of reduction decreased in the following order: Y202 (decreased by 94.6%; membrane-proximal module), K249 (94.1%; membrane-proximal module), E230 (86.9%; membrane-proximal module), Y242 (82.4%; membrane-proximal module), Y156 (80.1%; membrane-distal module), N182 (78.1%; membrane-distal module), T266 (69.2%; membrane-proximal module), F204 (53.7%; membrane-proximal module), and W139 (29.5%; membrane-distal module). These results demonstrate that most of the 10 key residues play important roles in rhizospheric chemotaxis and root colonization in situ. Meanwhile, we demonstrated that amino acid sensing is a key factor in bacterial root colonization, which is consistent with previous studies (36, 40).
Taking these data together, we propose a synergistic model for the capacity of McpA-LBD to recognize structurally different ligands that depends on the coordination of both membrane-distal and membrane-proximal regions (SI Appendix, Fig. S9). In this model, McpA engages the membrane-proximal subdomain (Tyr202, Glu230, Tyr242, Lys249, and Thr-266) to mediate the amino acid chemotaxis, while the membrane-distal subdomain (Pro124, Tyr156, and Asn182) mediates responses to OAs and mannose. Residues Phe204 and Lys249 in the membrane-proximal subdomain may also partially participate in the chemotaxis to four OAs (malic, citric, succinic, and oxalic acids). Both subdomains are essential for SA taxis, of which the crucial residues include Pro124, Trp139, Asn182, and Tyr242. A similar model may also be applicable for other multiligand recognizing dCache domains.
Discussion
Chemoreceptor-based signal sensing is among the most important signal transduction mechanisms and plays key roles in bacterial survival in a constantly changing environment (8). Since signal recognition defines the nature of a chemotactic response, the elucidation of the molecular mechanisms of how chemoreceptors perceive their signals is of fundamental importance. Our previous study showed the dCache chemoreceptor McpA of B. velezensis SQR9 mediates chemotaxis to multiple, structurally different compounds, and that McpA activity was essential for rhizosphere chemotaxis, biofilm formation, and root colonization (36–38). In the present study, we demonstrate that McpA employs both modules of its dCache type LBD for ligand recognition. Data indicate that the sensing of organic acids and amino acids occurs primarily at the membrane-distal and -proximal module, respectively, whereas both modules were required for the responses to SAs. Although the detection of some OAs involves either the distal or proximal module, the general synergistic working model (SI Appendix, Fig. S9) of McpA provides insight into the mode of signal recognition.
Most of the dCache chemoreceptors were reported to recognize ligands of the same type, such as the amino acid specific McpC of Bacillus subtilis (41), McpA of Pseudomonas putida (4) and CtaA in Pseudomonas fluorescens (19), the polyamine receptor McpU of P. putida, or the McpX and McpH receptors of Sinorhizobium meliloti and P. putida, that recognize quaternary amines (20) and purines (21), respectively. However, only a few receptors can sense and bind a variety of ligands of diverse chemical categories, where the representative chemoreceptors are CcmL in Campylobacter jejuni and PctA in Pseudomonas aeruginosa. In detail, CcmL was the first reported multiligand receptor in C. jejuni, which mediated chemotaxis to malic acid, fumaric acid, isoleucine, and purine as attractants, and to succinic acid, thiamine, glucosamine, arginine, and lysine as repellents (31). PctA was identified to bind 17 amino acids and the autoinducer-2 (30, 42), but also mediated chemotaxis to histamine (25, 43). The capacity of binding multiple ligands by the dCache domain-containing receptor might be due to the malleable nature of the LBD domain that can accommodate compounds of different sizes and charges (19). For the paralogous PctA, PctB, and PctC chemoreceptors of P. aeruginosa, it was proposed that the broad-range ligand chemoreceptor PctA was evolutionarily older, while receptors with a narrow ligand range arose later (29). Here we show that McpA of B. velezensis can directly bind 15 attractants, including 3 amino acids, 8 OAs, and 4 SAs, indicating that the broad response spectrum of McpA is due to a broad spectrum in the direct recognitions of ligands.
One of the potential mechanisms for responding to multiple ligands is engaging different modules by a chemoreceptor LBD. Three types of bimodular LBDs have been identified, which are the dCache (16), HBM (44), and DAHL (45) domains, of which the last two belong to the all-helical domain structural superfamily (13, 45). The HBM domain containing McpS of P. putida KT2440, bound malate and succinate at the membrane-proximal module, and acetate at the membrane-distal subdomain (46). The Tlp10 chemoreceptor of C. jejuni has a DAHL-type LBD that senses ligands at two distinct sites. Whereas arginine, isoleucine, and fumarate bind to one site, malate, mannose, fucose, and galactose interact with a second site. In addition, both sites were required for aspartate and thiamine binding (45). dCache domains are the predominant extracytosolic LBD-type in bacteria that are likely have arisen by a fusion of sCache domains (16). However, current information on this domain indicates that the large part of dCache domains bind ligands at the membrane-distal module (13, 22–25), whereas the function of the membrane-proximal module might be to relay the ligand-induced conformational changes to the signaling domain via the second transmembrane helix or to recognize ligand-loaded solute binding proteins (43). Until recently, only few chemoreceptors have been shown to bind ligands at the membrane-proximal module, namely the Helicobacter pylori receptors TlpC (lactate) and TlpA (fumarate) (26, 27).
Our data suggest that both regions in McpA-LBD are involved in recognizing diverse attractants in a synergistic manner. There were seven ligands that were sensed by the membrane-proximal module, namely glutamic acid, aspartic acid, tyrosine, malic acid, citric acid, succinic acid, and oxalic acid (Fig. 3 A and C and SI Appendix, Fig. S9). In contrast, current research of amino acid-sensing chemoreceptors focuses on the membrane-distal pocket, such as the highly conserved motif for amino acid recognition (Tyr121, Arg126, Trp128, Tyr129, Tyr144, and Asp173 in PctA) that were initially observed in the P. aeruginosa chemoreceptors PctA, PctB, and PctC, but were found to be present in a large number of dCache domain-containing receptors present in species throughout the tree of life (17, 29). This motif is only partially conserved in McpA (Tyr132, Arg137, Trp139, and Tyr156) since it lacks the conserved aspartate residue (Asn182 in McpA) (SI Appendix, Fig. S10A). However, this aspartate residue plays a central role in amino acid sensing, since it has been shown that its replacement with asparagine or alanine entirely abolishes the capacity of PctA to bind amino acids (17). The conserved aspartate in the sequence motif establishes a key interaction with the amino group of the bound amino acid (29). Data thus indicate that the partially conserved amino acid binding motif in the membrane-distal module of McpA-LBD is nonfunctional, a notion that agrees with amino acid recognition at the membrane-proximal module of McpA. Besides, W139 and Y156 were also considered as important residues in sensing SAs and OAs (Fig. 5B and SI Appendix, Figs. S9 and S10), respectively. Sequence alignments have shown that five key residues (Try202, Glu230, Tyr242, Lys249, and Thr266) at the proximal module in McpA are variable among the dCache chemoreceptors, including P. aeruginosa PctA, PctB, and PctC. This is consistent with the finding that the membrane proximal module of these three receptors is not involved in amino acid sensing (SI Appendix, Fig. S10A). However, previous results indicate that the McpC chemoreceptor of SQR9 is the primary amino acid receptor (38), whereas McpA appears to play a secondary role in mediating amino acid chemotaxis.
It is interesting to note that fucose, ribose, and ribitol can be directly bound to McpA, but the interaction was dependent on the entire LBD domain, since both the membrane-distal and membrane-proximal regions were required for generating the corresponding response (Fig. 3 D2–D4 and SI Appendix, Fig. S9). Currently, there are only few studies on chemoreceptors that directly bind monosaccharides. To our knowledge, Tlp11 (CcrG) of the phytopathogen C. jejuni is the only so far identified dCache containing chemoreceptor that binds a sugar, galactose, directly (47). In contrast, most chemoreceptors appear to bind sugars in an indirect way via the assistance of solute binding proteins (SBPs) (43). The response of several Escherichia coli chemoreceptors to sugars is based on the recognition of sugar-loaded SBPs (11). In addition, PctA of P. aeruginosa was shown to sense glucose through the periplasmic ligand binding protein, GltB (48). Therefore, our finding that dCache chemoreceptor McpA can directly bind sugars through a combination of both pockets may serve as a reference for analogous studies for sugar chemotaxis in bacteria.
Many bacteria use sugars as preferred carbon sources (49). Sugar chemotaxis provides a selective advantage by accessing optimal energy and carbon sources (50). The affinities of McpA-LBD for ribose (KD, 1.94 μM) and ribitol (KD, 4.36 μM) were among the highest for all ligands, suggesting ligand preference. High-affinity sugar chemotaxis may permit bacteria to better use sites with low sugar concentrations for growth.
Chemoreceptors are tightly packed in arrays and there is a solid body of evidence indicating that the interactions of different chemoreceptors within this array cause significant alterations in the magnitude of chemotactic response (51–54). Next to McpA, there are five other transmembrane chemoreceptors in B. velezensis SQR9, and McpA is very likely to interact with those receptors in the WT strain. However, in marked contrast to the WT strain, the chemoreceptor arrays in the complemented strain are exclusively composed of McpA receptors. A lacking interaction with the other receptors present in the WT chemoreceptor arrays may account for the difference in the magnitude of chemotaxis observed for several ligands between the WT and the complemented strains.
We have identified McpA-LBD homologs by BLAST searches in the National Center for Biotechnology Information database (SI Appendix, Fig. S11 and Dataset S1). Homologs are widely distributed in genus Bacillus (88.3%), and are abundantly present in the species B.cereus (20.0%), B. subtilis (7.8%), B. thuringiensis (7.1%), B. pumilus (3.5%), B. toyonensis (3.4%), B. velezensis (3.2%), and B. amyloliquefaciens (2.8%). Furthermore, McpA homologs were also found in the genera Priestia (2.4%), Paenibacillus (1.7%), Exiguobacterium (1.6%), Lysinibacillus (1.4%), Brevibacillus (1.1%), and others (3.4%) (SI Appendix, Fig. S11 and Dataset S1). We have compiled information on the ligands recognized by other dCache domain-containing chemoreceptors in SI Appendix, Table S9. These data showed that many ligands recognized by McpA have been reported previously to be recognized by homologous receptors.
In conclusion, we demonstrate that the dCache chemoreceptor McpA directly binds 13 attractants belonging to different compound families, namely OAs, amino acids, and SAs. We show that binding was dependent on the membrane-distal, membrane-proximal, and both modules, respectively. Based on our results, we were able to propose a model for sensing multiple ligands by a specific MCP. Collectively, the synergistic working mode of McpA provides further insight into the molecular recognition mechanism between chemoreceptors and ligands.
Materials and Methods
See SI Appendix, Materials and Methods for details.
Bacterial Strains, Media, and Growth Conditions.
The strains and plasmids used in this study are shown in SI Appendix, Table S10. B. velezensis SQR9 (formerly B. amyloliquefaciens, China General Microbiology Culture Collection Center, CGMCC accession no. 5808) was isolated from the cucumber rhizosphere. The mcpA gene has been integrated into the chromosomal amyE locus of mutant SQR9Δ8mcp that is deficient in all eight chemoreceptor genes, to obtain the complemented strain SQR9Δ8mcp/mcpA (referred to as C-mcpA) (38). All stains were grown at 30 °C in low-salt Luria-Bertani (LLB) medium (10 g peptone, 5 g yeast extract, and 3 g NaCl in 1 L). E. coli BL21 (DE3) was grown at 37 °C in LB medium (10 g peptone, 5 g yeast extract, and 5 g NaCl in 1 L). If necessary, antibiotics were added to the medium at the final concentrations of 5 mg/L chloramphenicol (Cm), 20 mg/L zeocin (Zeo), and 30 mg/L kanamycin (Kan).
Chimera Construction.
Receptor chimeras were constructed combining different fragments of the McpA and TlpA chemoreceptors of B. velezensis SQR9. Chimera M0 contains the entire TlpA-LBD sequence, M1 is a fusion of the membrane-distal module of McpA and the membrane-proximal region of TlpA, and M2 comprises the membrane-distal region of TlpA fused to the membrane-proximal region of McpA. These three LBD versions were then fused the McpA fragment comprising the transmembrane regions and the cytosolic part. The resulting receptor genes were integrated into the chromosomal amyE locus of SQR9Δ8mcp (deficient in all the eight mcp genes) by homologous recombination. The primers used for chimera construction are listed in SI Appendix, Table S10 and the topology of receptor chimeras is illustrated in Fig. 2.
Site-Directed Mutagenesis of Amino Acids Involved in Ligand Binding.
Twenty-three site-directed mutants of the mcpA gene containing amino acid substitutions in the two binding pockets were integrated into the chromosomal amyE locus of the chemoreceptor-deficient SQR9Δ8mcp. The mutations in the mcpA gene were introduced by corresponding primers (SI Appendix, Table S10). The mutated mcpA gene with its native promoter, chloramphenicol resistance gene, and the upstream/downstream fragments of amyE genes were amplified from the genomic DNA of the complementary strain C-mcpA, followed by fusion through overlapping PCR. Finally, the fused regions were individually transformed into SQR9Δ8mcp and verified for the correctness of the mutation.
We randomly selected six single-point variants of McpA-LBD (W139A, A159G, E230A, Y242A, K249A, and T266A) for protein expression. These DNA sequences corresponding to A36-L277 were cloned into a pET28a expression vector to generate an N-terminal 6×His-tagged construct, with a tobacco etch virus protease site, under the control of isopropyl-β-d-thiogalactopyranoside (IPTG) (38). Proteins were purified as described above.
Chemotaxis Assay.
The chemotaxis assay was performed as described previously using a simple and reusable microfluidic SlipChip device (38, 55). The SlipChip device consists of two glass plates with reconfigurable microwells and ducts, containing 13 parallel chemotaxis measurements as duplicates. Briefly, before the assay, a 10 mg/mL bovine serum albumin (BSA) solution was injected into all channels and left for 5 min, followed by removing the BSA. Then, the compound solution, bacterial suspension, and buffer were placed into the top, middle, and bottom microwells, respectively. Subsequently, the three individual microwells of each unit were connected, which allows cells to migrate. The devices were placed into the dark for 30 min and cells present in the top and bottom microwells were counted using an inverted fluorescence microscope (Ti-Eclipse, Nikon, Japan). From these data, the chemotaxis index (It) was calculated. It is defined as Ne/(Ne + Nc), where Ne is the number of cells that have migrated to the chemoeffector, and Nc is number of cells that have migrated to the control microwells in a certain time period t (time). In this study, the chemotaxis time was kept at 30 min. I30 values (the chemotaxis index after 30 min cell migration) between 0.4 and 0.6 (0.4 ≤ I30 ≤ 0.6) indicates an absence of taxis; an I30 value above 0.6 (I30 > 0.6) indicates chemoattraction; while an I30 value below 0.4 (I30 < 0.4) indicates chemorepellence.
Statistical Analysis.
The Duncan’s multiple rang tests (P < 0.05) of the SPSS v22.0 (IBM) was used for statistical analysis. Heatmaps and growth curves were visualized using GraphPad Prism v9.2.0 for Windows (GraphPad Software, Inc.; http://www.graphpad.com).
Supplementary Material
Acknowledgments
We thank Dr. Dongwei Chen (Chinese Academy of Sciences) for the help in chemotaxis assays; Dr. Feng Wang, Dr. Zhijia Lv, and Dr. Yan Wang in Wuxi Biortus Biosciences Co. Ltd. for their technical assistance with X-ray crystallography; and the Shanghai Synchrotron Radiation Facility for X-ray data collection. This research was financially supported by the National Natural Science Foundation of China (31900080, 32072665) and China Postdoctoral Science Foundation (2019M651847); and the Spanish Ministry for Science and Innovation/Agencia Estatal de Investigación 10.13039/501100011033 (PID2020-112612GB-I00) and Junta de Andalucía (P18-FR-1621).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201747119/-/DCSupplemental.
Data Availability
Anonymized data have been deposited in the Protein Data Bank, http://www.wwpdb.org (PDB ID code 7W0W) (56). All other study data are included in the main text and supporting information.
References
- 1.Wuichet K., Zhulin I. B., Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3, ra50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchis-López C., et al. , Prevalence and specificity of chemoreceptor profiles in plant-associated bacteria. mSystems 6, e00951-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard-Massicotte R., et al. , Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. MBio 7, e01664-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corral-Lugo A., et al. , Assessment of the contribution of chemoreceptor-based signalling to biofilm formation. Environ. Microbiol. 18, 3355–3372 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Wang J., et al. , Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 176, 334–347.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., et al. ; MetaHIT Consortium, A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laventie B. J., Jenal U., Surface sensing and adaptation in bacteria. Annu. Rev. Microbiol. 74, 735–760 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Gumerov V. M., Andrianova E. P., Zhulin I. B., Diversity of bacterial chemosensory systems. Curr. Opin. Microbiol. 61, 42–50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colin R., Ni B., Laganenka L., Sourjik V., Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol. Rev. 45, fuab038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao C. V., Ordal G. W., The molecular basis of excitation and adaptation during chemotactic sensory transduction in bacteria. Contrib. Microbiol. 16, 33–64 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Parkinson J. S., Hazelbauer G. L., Falke J. J., Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin M. Y., What bacteria want. Environ. Microbiol. 20, 4221–4229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortega Á., Zhulin I. B., Krell T., Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 81, e00033-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mistry J., et al. , Pfam: The protein families database in 2021. Nucleic Acids Res. 49 (D1), D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gumerov V. M., Ortega D. R., Adebali O., Ulrich L. E., Zhulin I. B., MiST 3.0: An updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res. 48 (D1), D459–D464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upadhyay A. A., Fleetwood A. D., Adebali O., Finn R. D., Zhulin I. B., Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLOS Comput. Biol. 12, e1004862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumerov V. M., et al. , Amino acid sensor conserved from bacteria to humans. Proc. Natl. Acad. Sci. U.S.A. 119, e2110415119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matilla M. A., Velando F., Martín-Mora D., Monteagudo-Cascales E., Krell T., A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol. Rev. 46, fuab043 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Ud-Din A. I. M. S., Khan M. F., Roujeinikova A., Broad specificity of amino acid chemoreceptor CtaA of Pseudomonas fluorescens is afforded by plasticity of its amphipathic ligand-binding pocket. Mol. Plant Microbe Interact. 33, 612–623 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Webb B. A., et al. , Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol. Microbiol. 103, 333–346 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Fernández M., Morel B., Corral-Lugo A., Krell T., Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol. Microbiol. 99, 34–42 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Gavira J. A., et al. , Structural basis for polyamine binding at the dCACHE domain of the McpU chemoreceptor from Pseudomonas putida. J. Mol. Biol. 430, 1950–1963 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Liu Y. C., Machuca M. A., Beckham S. A., Gunzburg M. J., Roujeinikova A., Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr. D Biol. Crystallogr. 71, 2127–2136 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama S., et al. , Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci. Rep. 6, 20866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corral-Lugo A., et al. , High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. MBio 9, e01894-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machuca M. A., et al. , Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate. Sci. Rep. 7, 14089 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson K. S., et al. , The dCache chemoreceptor TlpA of Helicobacter pylori binds multiple attractant and antagonistic ligands via distinct sites. MBio 12, e01819-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tohidifar P., Plutz M. J., Ordal G. W., Rao C. V., The mechanism of bidirectional pH taxis in Bacillus subtilis. J. Bacteriol. 202, e00491-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavira J. A., et al. , How bacterial chemoreceptors evolve novel ligand specificities. MBio 11, e03066-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., et al. , Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11, 5371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman H., et al. , Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog. 10, e1003822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y., et al. , Bacillus subtilis SQR9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils 47, 495–506 (2011). [Google Scholar]
- 33.Chen L., et al. , Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 158, 34–44 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Shao J., Xu Z., Zhang N., Shen Q., Zhang R., Contribution of indole-3-acetic acid in the plant growth promotion by the rhizospheric strain Bacillus amyloliquefaciens SQR9. Biol. Fertil. Soils 51, 321–330 (2015). [Google Scholar]
- 35.Xu Z., et al. , Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 79, 808–815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng H., et al. , Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., et al. , Induced root-secreted D-galactose functions as a chemoattractant and enhances the biofilm formation of Bacillus velezensis SQR9 in an McpA-dependent manner. Appl. Microbiol. Biotechnol. 104, 785–797 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Feng H., et al. , Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol. Plant Microbe Interact. 31, 995–1005 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Oku S., Komatsu A., Tajima T., Nakashimada Y., Kato J., Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 27, 462–469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glekas G. D., et al. , The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J. Biol. Chem. 287, 39412–39418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rico-Jiménez M., et al. , Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol. Microbiol. 88, 1230–1243 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Matilla M. A., Ortega Á., Krell T., The role of solute binding proteins in signal transduction. Comput. Struct. Biotechnol. J. 19, 1786–1805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortega Á., Krell T., The HBM domain: Introducing bimodularity to bacterial sensing. Protein Sci. 23, 332–336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elgamoudi B. A., et al. , The Campylobacter jejuni chemoreceptor Tlp10 has a bimodal ligand-binding domain and specificity for multiple classes of chemoeffectors. Sci. Signal. 14, eabc8521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pineda-Molina E., et al. , Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc. Natl. Acad. Sci. U.S.A. 109, 18926–18931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day C. J., et al. , A direct-sensing galactose chemoreceptor recently evolved in invasive strains of Campylobacter jejuni. Nat. Commun. 7, 13206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu C., Cao Q., Lan L., Glucose-binding of periplasmic protein GltB activates GtrS-GltR two-component system in Pseudomonas aeruginosa. Microorganisms 9, 447 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Görke B., Stülke J., Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Adler J., Hazelbauer G. L., Dahl M. M., Chemotaxis toward sugars in Escherichia coli. J. Bacteriol. 115, 824–847 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sourjik V., Berg H. C., Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Ames P., Studdert C. A., Reiser R. H., Parkinson J. S., Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 7060–7065 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briegel A., et al. , Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 3766–3771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studdert C. A., Parkinson J. S., Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U.S.A. 101, 2117–2122 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen C., et al. , Bacterial chemotaxis on SlipChip. Lab Chip 14, 3074–3080 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Feng H. C., Shen Q. R., Zhang R. F., The novel membrane-proximal sensing mechanism in a broad-ligand binding chemoreceptor McpA of Bacillus velezensis. Protein Data Bank. 10.2210/pdb7W0W/pdb. Deposited 6 July 2022. [DOI]
- 57.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized data have been deposited in the Protein Data Bank, http://www.wwpdb.org (PDB ID code 7W0W) (56). All other study data are included in the main text and supporting information.